Abstract
C4.4A and Haldisin belong to the Ly6/uPAR/α-neurotoxin protein domain family. They exhibit highly regulated expression profiles in normal epidermis, where they are confined to early (C4.4A) and late (Haldisin) squamous differentiation. We have now explored if dysregulated expressions occur in non-invasive and invasive skin lesions. In non-invasive lesions, their expression signatures were largely maintained as defined by that of normal epidermis. The scenario was, however, markedly different in the progression towards invasive squamous cell carcinomas. In its non-invasive stage (carcinoma in situ), a pronounced attenuation of C4.4A expression was observed, but upon transition to malignant invasive squamous cell carcinomas, the invasive fronts regained high expression of C4.4A. A similar progression was observed for the early stages of benign infiltrating keratoacanthomas. Interestingly, this transition was accompanied by a shift in the predominant association of C4.4A expression with CK1/10 in the normal epidermis to CK5/14 in the invasive lesions. In contrast, Haldisin expression maintained its confinement to the most-differentiated cells and was hardly expressed in the invasive lesions. Because this altered expression of C4.4A was seen in the invasive front of benign (keratoacanthomas) and malignant (squamous cell carcinomas) neoplasms, we propose that this transition of expression is primarily related to the invasive process.
Keywords: C4.4A, Haldisin, stratum spinosum, stratum granulosum, skin lesions, invasive front, cytokeratins, LYPD3, LYPD5, PRO4356
Introduction
The epidermis provides a vital, first-line barrier for protection against environmental assaults such as microbial infections and transepidermal loss of fluids. Homeostasis of this exquisitely ordered organ is fuelled by different resident stem cells that enable constitutive renewal of the stratified squamous epithelium as well as its various epidermal appendages (Blanpain and Fuchs 2009). The integrity of this physical barrier function is, however, occasionally compromised by congenital dysregulation of, for example, cytokeratins (Coulombe et al. 2009), proteases (de Veer et al. 2014; Sales et al. 2010) and ion pumps (Sakuntabhai et al. 1999). In addition, certain autoimmune diseases targeting such antigens also perturb skin homeostasis (Nishie 2014; Stanley and Amagai 2006) and the subsequent pathogenesis (e.g., severity of blister formation) may be further exacerbated by local plasminogen activation (Liu et al. 2005). Being a highly exposed and proliferative tissue, the epidermis is furthermore prone to developing a number of neoplastic lesions that may be invasive, such as keratoacanthoma, basal cell carcinoma, squamous cell carcinoma, benign nevi and malignant melanoma (Madan et al. 2010; Schwartz 1994; Thompson et al. 2005).
In the present study, we have investigated various congenital and acquired skin lesions for the expression patterns of two glycolipid-anchored membrane biomarkers, C4.4A and Haldisin, which are associated with early and late squamous differentiation, respectively. These two proteins are predicted structural homologs belonging to the Ly6/uPAR/α-neurotoxin (LU) protein domain family. Along with the urokinase-type plasminogen activator receptor, uPAR (Ploug 2013), TEX101 (Fujihara et al. 2013) and CD177 (Hu et al. 2014), they constitute the few multidomain members of this family, all of which are encoded by a small gene cluster located on human chromosome region 19q13 (Jacobsen and Ploug 2008). Immunohistochemical surveys of resected organs from mice demonstrate that C4.4A is predominantly expressed in stratum spinosum of the squamous epithelium (Kriegbaum et al. 2011), whereas Haldisin expression primarily is confined to stratum granulosum (Gårdsvoll et al. 2013). The biochemical and cellular functions of these proteins in the squamous epithelium remain speculative, but circumstantial experimental evidence has led to the proposition that C4.4A may be involved in cell adhesion and cell migration (Paret et al. 2005; Rösel et al. 1998; Smith et al. 2001). In line with this, C4.4A was originally identified in two independent screens for metastasis-associated proteins (Claas et al. 1996; Matzku et al. 1989) and proteins involved in wound healing of the urothelium (Smith et al. 2001). In pathological conditions, high C4.4A expression levels have been correlated with poor survival of patients with non-small cell lung adenocarcinoma, but not for patients with squamous cell carcinoma (Hansen et al. 2007; Jacobsen et al. 2013), which most likely pertains to the robust induction of C4.4A in the bronchial epithelium already during the early non-malignant stages of hyperplasia and metaplasia (Jacobsen et al. 2012). High C4.4A levels are furthermore reported to be correlated to poor prognosis in patients with colorectal cancer (Konishi et al. 2010; Oshiro et al. 2012), gastric cancer (Cheng et al. 2014) and esophageal cancer (Ohtsuka et al. 2013a). The expression pattern of C4.4A has additionally been investigated in a number of other human cancer lesions and their corresponding metastases, including urothelial transitional cell carcinoma (Smith et al. 2001), esophageal squamous cell carcinoma (Hansen et al. 2008; Ohtsuka et al. 2013b), mammary adenocarcinoma (Miyake et al. 2013) and malignant melanoma (Seiter et al. 2001).
Due to the potential role of C4.4A in cell adhesion and the strict regulation of both C4.4A and Haldisin expression during squamous differentiation, we have focused our present study on a selection of blistering diseases, including subtypes of pemphigus, bullous pemphigoid and Darier disease. They are all characterized by the pathological detachment of keratinocytes but differ by the locations of the lesions in the stratified squamous epithelium. We also included a heterogeneous group of skin lesions with dysregulated differentiation programs such as psoriasis, lichen ruber planus and ichthyosis. Because previous studies have correlated C4.4A expression to survival in various human carcinoma patients (Cheng et al. 2014; Hansen et al. 2007; Jacobsen et al. 2013; Konishi et al. 2010; Ohtsuka et al. 2013a), we have also expanded our study on C4.4A and Haldisin expression to include various skin cancer precursor lesions, such as actinic keratosis, carcinoma in situ, and the benign, but often infiltrating, lesion of keratoacanthoma as well as invasive malignant lesions including basal cell carcinoma, squamous cell carcinoma and malignant melanoma.
Materials & Methods
Proteins and Antibodies
Recombinant human C4.4A and Haldisin as well as rabbit anti-C4.4A and anti-Haldisin polyclonal antibodies (pAb) were generated and purified as previously described (Gårdsvoll et al. 2013; Hansen et al. 2004). Rabbit anti-cytokeratin (CK) 5 pAb (ab24647), mouse anti-CK14 monoclonal antibody (mAb) (ab7800) and rabbit anti-loricrin pAb (ab24722) were purchased from Abcam (Cambridge, UK). Mouse anti-CK1 mAb (MMS-194I, clone 34βB4, ready-to-use lot. 14813601) and anti-profilaggrin pAb (51-2100) were from Covance (Princeton, NJ) and Zymed (San Francisco, CA), respectively. Mouse anti-CK10 mAb (M7002), rabbit IgG of irrelevant specificity (X903) as well as horseradish peroxidase–labeled EnVision rabbit (K4003) and mouse (K4001) reagent were purchased from Dako (Glostrup, Denmark).
Patient Samples
Ninety-five skin specimens from a total of 86 patients were included in the study. The resected tissues were collected with diagnostic intent in the period from 2006 to 2013 at Oslo University Hospital (Oslo, Norway) and the histopathological diagnoses are listed in Tables 1 and 2. Normal human skin collected during resection of mammary cancer was received from Rigshospitalet (Copenhagen, Denmark). All specimens were paraformaldehyde fixed and paraffin embedded. The experiments were approved by the Regional Scientific Ethics Committee (H-1-2012-141).
Table 1.
Overview of C4.4A and Haldisin Expression in Non-invasive Skin Lesions.
| Type | Lesion | No. of cases | Lesion Description | C4.4A | Haldisin |
|---|---|---|---|---|---|
| Blistering disorders | Bullous pemphigoid | 4 | Detachment of basal layer from the basement membrane due to IgG autoantibodies targeting type XVII collagen of hemidesmosomes | 4/4: No alteration | 4/4: No alteration |
| Epidermolysis bullosa dystrophica | 2 | Detachment of basal layer from the basement membrane due to mutations in collagen VII | 2/2: No alteration | 2/2: No alteration | |
| Pemphigus vulgaris | 2 | Acantholysis and detachment of keratinocytes primarily in the basal and lower spinous layer due to IgG autoantibodies against desmoglein 3 | 2/2: No alteration | 2/2: No alteration | |
| Pemphigus foliaceus | 7 | Acantholysis and detachment of keratinocytes primarily in the granular layer due to IgG autoantibodies against desmoglein 1 | 7/7: No alteration | 6/7: Loss of expression due to loss of granular layer | |
| Subcorneal pustular dermatosis | 3 | Cytolysis of keratinocytes of the granular layer and formation of subcorneal pustules | 3/3: No alteration | 1/3: Loss of expression under pustule | |
| Darier disease | 5 | Acantholysis of suprabasal keratinocytes and dyskeratinization due to a mutation in the Ca2+ ATPase SERCA2 causing breakdown of desmosome-keratin intermediate filaments and desmosome loss. | 1/5: Membrane associated expression in basal cells 4/5) No alteration |
2/5: Thickening of Haldisin expressing cell layer 5/5) Loss of expression with acantholysis |
|
| Lesions harboring dysregulated differentiation | Ichthyosis | 4 | Heterogeneous group of scaling disorders caused by known gene mutations Examined types 1 ichthyosis congenital. 1 nonbullous congenital ichthyosiform erythroderma (NBCiE) 1 autosomal dominant lamellar ichthyosis1 ichthyosis congenital type IV |
4/4: No alteration | 1/4: Thickening of Haldisin expressing cell layer in the case of NBCiE |
| Lichen ruber planus | 7 | Inflammatory disorder where an unknown antigen leads to lymphocyte activation and subsequent keratinocyte apoptosis, basal membrane disruption, subepidermal cleft formation and thickening of the granular layer. | 7/7: No alteration | 5/7: Thickening of Haldisin expressing cell layer which largely recapitulates the expression of profilaggrin/loricrin | |
| Psoriasis | 4 | Inflammatory disease with excessive hyperproliferation of keratinocytes both in the basal- and suprabasal layer. Loss of the granular layer and formation of para- and hyperkeratosis. | 4/4: No alteration | 3/4: Thick Haldisin expressing cell layer despite loss of profilaggrin/ loricrin expression |
Sections of blistering lesions as well as lesions harboring a dysregulated differentiation were stained for C4.4A and Haldisin. The table lists the lesion types, the number of cases examined, a short description of the main characteristics of the diseases and, finally, an evaluation of the C4.4A and Haldisin expression patterns. The numbers of cases with the described expression signatures are noted.
Table 2.
Overview of C4.4A, Haldisin, CK1, CK10, CK5 and CK14 expression patterns in precursor and invasive lesions.
| Lesion | No. of cases | C4.4A | Haldisin | CK1/10 | CK5/14 |
|---|---|---|---|---|---|
|
Actinic keratosis
Precursor lesion of squamous cell carcinoma mostly through carcinoma in situ |
5 | 5/5: No alteration | 4/5: Sites with loss of or attenuated expression | 5/5: Complete loss or attenuated expression | 5/5: Expression confined to basal and suprabasal layers. |
|
Keratoacanthoma
In early stage infiltrating, despite being a benign lesion |
7 | 4/7: Attenuated expression in core and high expression in invasive front | 7/7: Sustained expression in differentiated layers | 7/7: Sustained expression in most differentiated layers and complete loss or attenuated expression in core | 6/7: Attenuated (CK5: partial loss) expression in core and high expression in invasive front |
|
Carcinoma in situ
Pre-cancerous |
5 | 3/5: Attenuated expression in lower suprabasal layers | 4/5: Complete or partial loss of expression | 3/5: Attenuated or partial loss of expression | 5/5: Expression primarily in the basal layer |
|
Basal cell carcinoma
Malignant tumor Low metastatic risk 1 keratotic, 1 nodular, 1 nodular/cystic, 3 superficial |
6 | 5/6: Tumor islands with expression. Four of the cases have overlapping CK1 expression | 6/6: Negative tumor 1/6: Weak expression in a few cells around keratin pearls |
5/6: CK1 expression in tumor islands 1/6: CK10 expression in tumor islands |
6/6: Expression in tumor cells |
|
Squamous cell carcinoma
Malignant tumor Low metastatic risk |
6 | 4/6: Attenuated expression in tumor core and recovery of high expression in the invasive front | 6/6: Loss of expression and only expression in differentiated cells often surrounding keratin pearls | 6/6: Loss of expression or expression in a few single cancer cells or tumor cell islands often surrounding keratin pearls | 5/6: Attenuated (CK5: partial loss) expression in tumor core and recovery of high expression in the invasive front |
| Normal nevi | 7 | 7/7: Negative (3 cases were examined by ISH) | 7/7: Negative | ND | ND |
|
Malignant melanoma
High metastatic risk 5 superficial spreading 4 lentigo maligna 4 nodular 3 desmoplastic |
16 | 12/16: Negative tumor 4/16: Very weak expression in minor parts of the tumor of 2 superficial spreading and 2 nodular malignant melanomas (5 superficial and 1 nodular malignant melanoma were examined by ISH) |
16/16: Negative tumor | ND | ND |
| Metastases from melanomas | 5 | 5/5: Negative (4 cases were examined by ISH) | 5/5: Negative | ND | ND |
Tissue sections of precursor and invasive lesions were stained for C4.4A, Haldisin, CK1, CK10, CK5 and CK14. The lesion type, the number of examined cases as well as a description of the main findings are listed in the table. The numbers of cases with the described staining patterns are indicated. ISH, in situ hybridization.
Immunohistochemistry
Immunohistochemical staining was performed as previously described (Kriegbaum et al. 2011). In brief, 3-µm sections were deparaffinized and pre-treated with Proteinase K for 15 min at 37C for antigen retrieval of C4.4A and CK1 (10 μg/ml Proteinase K) and Haldisin (5 μg/ml Proteinase K). For CK10, CK5 and CK14 staining, the sections were treated in a T/T Micromed microwave processor (Milestone, Sorisol, Italy) at 98C for 10 min in 10 mM Tris, 0.5 mM EGTA (TEG) at pH 9.0. Subsequently, sections were incubated with 1% (v/v) hydrogen peroxide for 15 min followed by an overnight incubation at 4C with primary antibodies diluted in Antibody Diluent (S0809, Dako, Glostrup, Denmark) using the following concentrations: 2 μg/ml anti-C4.4A pAb, 1 μg/ml anti-Haldisin pAb, 1:1.5 dilution of anti-CK1 mAb, 0.3 μg/ml anti-CK10 mAb, 0.3 μg/ml anti-CK5 pAb and 1.25 μg/ml anti-CK14 mAb. Envision horseradish peroxidase-labeled anti-rabbit or anti-mouse IgG were used for detection (45 min at room temperature) followed by color development with NovaRED (SK-4800, Vector Laboratories, Burlingame, CA) for 9 min, as specified by the manufacturer. Counterstaining was performed with Mayer’s hematoxylin. Staining specificities were validated by 1-hr pre-absorptions of the antibodies with a 10-fold molar excess of the corresponding purified recombinant C4.4A and Haldisin before addition to the sections. Staining with matching concentrations of a rabbit IgG of irrelevant specificity served as additional controls and was performed in parallel.
In Vitro Transcription
Antisense and sense probes were generated from a pBlueScript KS+ plasmid containing a 677-base pair nucleotide sequence covering position 313–989 of human C4.4A cDNA (accession number AJ223603), as previously described (Hansen et al. 2004). In vitro transcription was performed using 35S-labelled UTP (NEG039H, Perkin Elmer, Skovlunde, Denmark) as well as T3 (11 031 171 001) and T7 (10 881 775 001) RNA polymerases from Roche Applied Science (Indianapolis, IN). The DNA template was digested with DNase (10 776 785 001; Roche Applied Science) and excess 35S-labelled UTP and DNA were removed using MinElute RNeasy spin columns (74204, Qiagen, Copenhagen, Denmark) according to the manufacturer’s descriptions. The 35S-radioactivities of the probes were normalized to approximately 500,000 cpm/µl by dilution.
In Situ Hybridization
Under RNase-free conditions, paraffin sections (3 µm) were deparaffinized in xylene, hydrated through a series of ethanol/water solutions, and washed in phosphate-buffered saline (PBS) prior to heat treatment in TEG at 98C for 10 min, as described previously for the immunoperoxidase staining. Sections were subsequently dehydrated before incubation with the 35S-labeled RNA sense and anti-sense probes (2×106 cpm/slide) in 16 µl hybridization mixture (50% deionized formamide, 0.02% (w/v) Ficoll 400, 10% dextran sulfate, 1 µg/µl t-RNA, 0.02% (w/v) polyvinylpyrrolidone, 0.02% (w/v) bovine serum albumin (BSA) Fraction V, 10 mM DTT, 300 mM NaCl, 0.5 mM EDTA, 10 mM Tris-HCl and 10 mM NaH2PO4, pH 6.8) overnight at 55C in a humidified chamber. The slides were then extensively washed in sodium citrate buffers (1×SCC: 0.015 M sodium citrate, 0.15 M sodium chloride, pH 7.0) containing 0.1% (w/v) sodium dodecyl sulfate (SDS) and 10 mM dithiothreitol (DTT) as follows: 10 min in 2×SCC, 10 min in 1×SCC, and 10 min in 0.2×SCC. This was followed by a 5-min wash in NTE buffer (10 mM Tris-HCl, 0.5 M sodium chloride, 1 mM EDTA, 10 mM DTT, pH 7.2) before any nonspecifically bound probe was removed enzymatically through an incubation for 10 min at 44C with RNase-A (20 µg/ml) in the above buffer without DTT. Finally, sections were washed twice for 5 min at 44C in the NTE buffer and twice for 10 min at 55C in 0.2×SCC with 10 mM DTT. All washes/incubations in SCC and NTE buffer were performed at 100 RPM in a Bühler incubation shaker (Johanna Otto, Hechingen, Germany). Finally, the sections were dehydrated in graded ethanol solutions containing 0.3 M ammonium acetate, soaked in an autoradiographic emulsion (Ilford, Imaging UK Limited, Mobberley, England), and exposed for 14 days at 4C before being developed. Slides were counterstained with Mayer’s hematoxylin and eosin.
Results
C4.4A and Haldisin Expression in Normal Skin and Its Appendages
To establish a reference expression profile in normal homeostatic skin, we initially performed a comparative expression analysis of C4.4A and Haldisin by immunohistochemistry in normal healthy skin from human breast. As shown in Figure 1A and 1B, C4.4A and Haldisin were confined to two distinct compartments in the suprabasal layer of the epidermis; i.e. C4.4A being predominantly expressed in stratum spinosum and Haldisin in stratum granulosum, with only a marginal overlap in their expression occurring at the interface of these layers. This differential expression pattern was recapitulated in skin appendages including hair follicles, sebaceous and sweat glands. In hair follicles, C4.4A was found in the suprabasal layers of the outer root sheath as well as in the inner root sheath, whereas Haldisin expression was confined to the inner root sheath only (Fig. 1C, 1D). Squamous epithelium of the infundibulum of sebaceous glands also cohered to this differential expression (Fig. 1E, 1F). Finally, in the eccrine sweat glands, we found C4.4A to be expressed by the cuboidal epithelium of the distal ducts, whereas a patchy Haldisin expression appeared in the secretory compartment giving rise to a complementary expression pattern along the secretory pathway of the gland (Fig. 1G, 1H).
Figure 1.
C4.4A and Haldisin are expressed in normal skin and appendages. Neighboring sections of normal human skin were stained using our polyclonal rabbit antibodies against C4.4A (A, C, E, G) and Haldisin (B, D, F, H). In the epidermis, C4.4A is primarily expressed in stratum spinosum (A) whereas the most prominent Haldisin expression is observed in stratum granulosum (B). In accordance with this, C4.4A is expressed in the suprabasal layer of the outer and inner root sheaths of the hair follicle (C), and in the epidermis of the sebaceous gland (E). Haldisin expression is observed in the inner root sheath of the hair follicles (D) and the granular layer of the sebaceous glands (F). In the sweat glands, C4.4A is expressed in ducts (G) and Haldisin in the gland structures (H). Insets show higher power micrographs of C4.4A and Haldisin expression in the hair follicle (panels C and D) and in the ducts and secretory structures of the sweat gland (panels G and H). d: dermis; sb: stratum basale; ss: stratum spinosum; sg: stratum granulosum; sc: stratum corneum; closed arrows: outer root sheath; open arrows: inner root sheath. Scale (A, B), 15 µm; (C–F), 100 µm; (G, H), 80 µm.
The specificity of the immunohistochemical detection of C4.4A and Haldisin in human skin was validated by preabsorption controls (Supplemental Fig. S1, compare panels A and D with panels B and E), using an irrelevant rabbit IgG (Supplemental Fig. S1C and S1F), and by in situ hybridization (Supplemental Fig. S1G–S1I).
Non-invasive Skin Lesions
Due to the highly regulated expression of C4.4A and Haldisin in different compartments of squamous epithelia in general (Gårdsvoll et al. 2013; Kriegbaum et al. 2011) and in normal skin in particular, we conducted an expression survey of these proteins by immunohistochemistry in a large selection of human skin disorders. Initially, we focused on non-invasive human skin lesions, which we divided into blistering lesions with known underlying genetic aberrations and lesions harboring dysregulated differentiation (summarized in Table 1).
Blistering Skin Lesions
The heterogeneous group of blistering skin disorders comprised lesions with selective disruptions of cell–matrix or cell–cell adhesion in distinct epidermal layers spanning the stratum basale to the stratum granulosum. Based on circumstantial evidence, a role for C4.4A in cell adhesion has been proposed (Paret et al. 2005; Rösel et al. 1998; Smith et al. 2001) and we therefore speculated that the stringent expression in normal squamous epithelia could be affected by the attenuated adhesive force defining blistering diseases. Nonetheless, all skin lesions examined (bullous pemphigoid, epidermolysis bullosa dystrophica, pemphigus vulgaris, pemphigus foliaceus, subcorneal pustular dermatosis and Darier disease) revealed essentially unaltered expression profiles of C4.4A and Haldisin, even at the site of blister formation, when analyzed by immunohistochemistry. Only minor deviations from the normal expression patterns were observed, and these are noted in Table 1. Examples of C4.4A expression in pemphigus vulgaris and Haldisin expression in pemphigus foliaceus are shown in Figure 2A and 2B, respectively, where the blistering foci are located just adjacent to the predominant expression sites for these proteins. Their expressions in Darier disease, where all suprabasal layers are acantholytic, are shown in Figure 2C and 2D, respectively.
Figure 2.
Expression of C4.4A and Haldisin in blistering skin lesions. C4.4A and Haldisin expression in blistering skin diseases are exemplified by staining of lesions affected at the sites of C4.4A (stratum spinosum) and Haldisin (stratum granulosum) expression. Panel (A) therefore shows C4.4A expression in pemphigus vulgaris and panel (B) shows Haldisin expression in pemphigus foliaceus. C4.4A (C) and Haldisin (D) expression patterns in Darier disease are also shown, where some overlap in localization is seen. Scale, (A, B), 25 µm; (C, D), 50 µm.
Lesions with Dysregulated Differentiation
We next focused on skin lesions exhibiting dysregulated differentiation programs with manifestations in stratum spinosum and stratum granulosum, where C4.4A and Haldisin are expressed. Lichen ruber planus and psoriasis are inflammatory diseases where the dysregulated differentiation programs cause a thickening or loss of stratum granulosum, respectively. Ichthyosis, on the other hand, is a heterogeneous group of diseases characterized by a generalized scaling of the skin.
In accordance with the morphological manifestation and etiology of these diseases, we found that the molecular markers of late differentiation (profilaggrin and loricrin) were expressed in the thickened stratum granulosum and in stratum corneum of lichen ruber planus, but were attenuated or completely lost in psoriasis (Supplemental Fig. S2). In the normal epidermis as well as that of lichen ruber planus patients, Haldisin expression recapitulated the expression pattern of profilaggrin/loricrin (Supplemental Fig. S2A–2F), except for its absence in stratum corneum. Somewhat unexpected, Haldisin expression was sustained in the psoriatic skin despite the absence of profilaggrin/loricrin (Supplemental Fig. S2G–2I), suggesting that Haldisin defines a slightly earlier granular differentiation state than profilaggrin and loricrin. No obvious dysregulation of C4.4A and Haldisin were observed in the heterogeneous lesions of ichthyosis patients (Table 1).
Precursor and Invasive Skin Lesions
Circumstantial evidence tends to favor a role for C4.4A in invasive and remodeling processes, as its expression is reported to be up-regulated in bladder wound healing in vitro (Smith et al. 2001) as well as in invasive areas of esophageal squamous cell carcinoma (Hansen et al. 2008). As a consequence, we next focused our attention on neoplastic skin diseases with and without invasion, including cancer precursor lesions as well as infiltrating lesions (Table 2). Special focus was given to C4.4A expression, as Haldisin expression is confined to a later and perhaps more terminal keratinocyte differentiation stage.
Precursor Lesions, Squamous Cell Carcinoma, Keratoacanthoma and Basal Cell Carcinoma
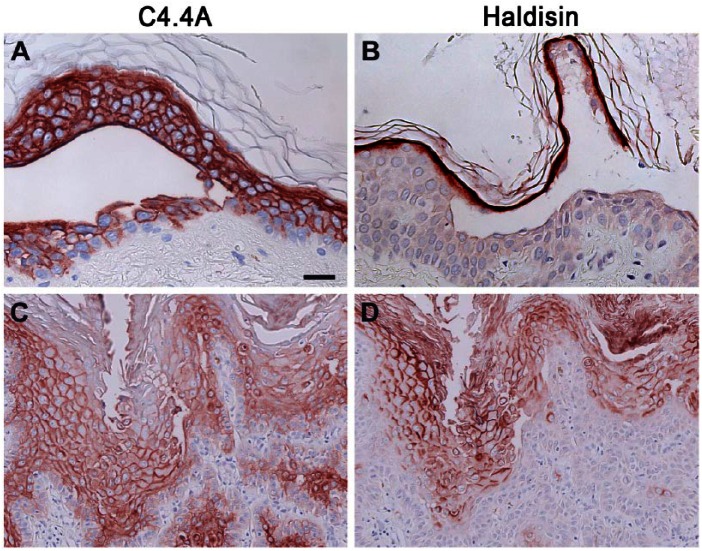
In the normal, well-stratified, and homeostatic epidermis CK1 and CK10 are exclusively confined to the suprabasal layers (CK1; Fig. 3E), whereas CK5 and CK14 are highly expressed in the basal layer and are gradually lost upon acquisition of the spinous differentiation (CK14; Fig. 3H). In the normal homeostatic skin C4.4A expression thus co-localizes with CK1 and CK10 (Fig. 3A). To simplify and improve clarity of the following figures, only staining for CK1 and CK14 is shown.
Figure 3.
Localization of C4.4A, CK1 and CK14 in normal skin and carcinoma in situ. Serial sections of either normal skin from human breast (A, E, H) or carcinoma in situ (B–D, F–G, I–J) were analyzed for C4.4A (A–D), CK1 (E–G) or CK14 (H–J) expression by immunohistochemistry. The C4.4A expression pattern primarily follows CK1 expression in both normal epidermis and carcinoma in situ. Scale, (A, C–E, G, H, J), 25 µm; (B, F, I), 100 µm.
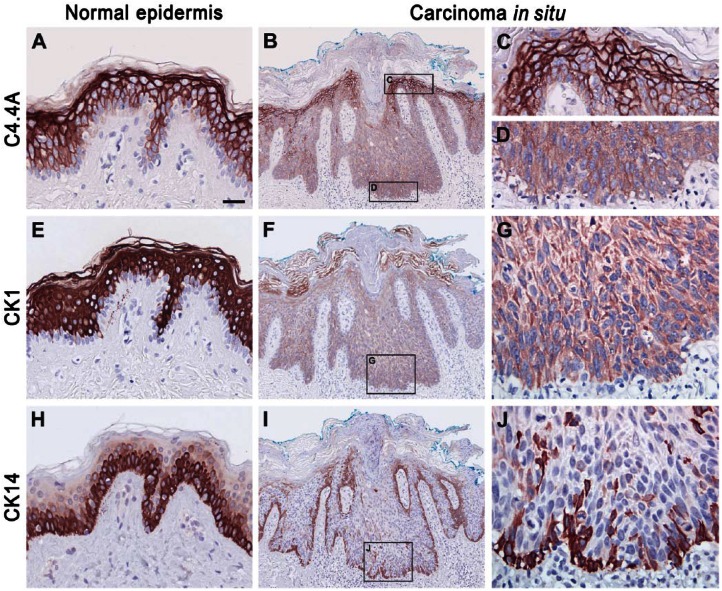
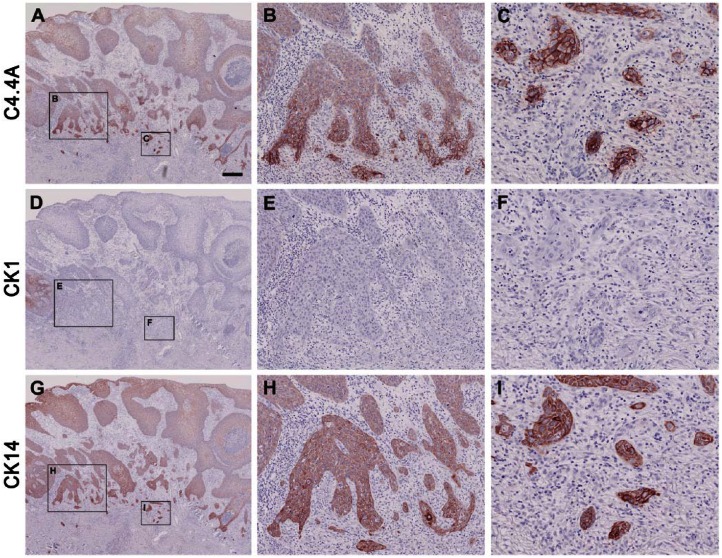
A high C4.4A expression level was maintained in all five cases of actinic keratosis (Table 2), which is considered as a precursor lesion of squamous cell carcinoma through the development of carcinoma in situ (Ko 2010). In carcinoma in situ, 3 out of 5 cases revealed an attenuated C4.4A expression in the more basally localized keratinocytes of the spinous layer (Fig. 3B–3D). This was recapitulated by the expression pattern for CK1 and CK10 (Fig. 3F–3G) and complementary to that of CK5 and CK14 (Fig. 3I–3J). With the progression to manifest highly invasive squamous cell carcinoma, the C4.4A expression of the tumor core remained consistently weak but, interestingly, high C4.4A expression level was regained in the deeply invading cancer cells of the invasive zones in 4 of 6 cases (Fig. 4A–4C). Surprisingly, this was no longer recapitulated by the CK1 and CK10 expression, which was entirely lost or confined to only a few tumor cell islands of high differentiation (Fig. 4D–4F). Instead, the C4.4A expression profile in these deeply invasive lesions appeared to have shifted towards a differentiation state correlating with CK5 and CK14 expression levels (Fig. 4G–4I). Substantiating this intriguing shift in the cytokeratin association of C4.4A during invasion is the parallel data recorded for the benign but distinctly infiltrating cases of the early proliferative stage of keratoacanthomas (Fig. 5). In this skin lesion, C4.4A expression also became attenuated in the center of the hyperproliferative core of the keratoacanthomas but a high expression was recovered in the deeply infiltrating cells in 4 of the 7 cases examined (Fig. 5A, 5B). This is closely associated with the patterns of CK5 and CK14 expression (Fig. 5G, 5H) rather than those for CK1 and CK10 (Fig. 5D, 5E); thus, recapitulating the previous association found in squamous cell carcinoma. A particularly conspicuous illustration of this unique shift in cytokeratin association was observed during perineural invasion (Fig. 5, compare panels C, F and I).
Figure 4.
Comparison of C4.4A, CK1 and CK14 expression profiles in squamous cell carcinoma. The expression profiles of C4.4A (A–C), CK1 (D–F) and CK14 (G–I) in squamous cell carcinoma were compared after immunohistochemical staining of serial sections of paraffin-embedded tissue. Panels A, D, and G show low magnification overviews of the C4.4A, CK1 and CK14 staining, respectively. Note the weak C4.4A and CK14 staining (A, G) in the tumor core and recovery of high expression in the invasive front (B, H), and in the deeply infiltrating cancer cells (C, I), as well as the complete loss of CK1 expression (D–F). Scale (A, D, G), 400 µm; (B, E, H), 100 µm; (C, F, I), 50 µm.
Figure 5.
C4.4A, CK1 and CK14 expression signatures in proliferating invasive keratoacanthoma. Paraffin-embedded serial tissue sections were incubated with primary antibodies against C4.4A (A–C), CK1 (D–F) and CK14 (G–I) and stained using immunoperoxidase and NovaRED. The low magnification pictures in panels A, D, and G show an example of a whole keratoacanthoma, where weakened expression in the lesion center (A, C4.4A; G, CK14) or complete loss of expression (D, CK1) can be appreciated. Panels B, E, and F show high magnifications of the invasive fronts, with high C4.4A and CK14 expression and a lack of CK1 expression. C4.4A- and CK14-positive and CK1-negative perineural infiltrations are shown in (C), (F) and (I), respectively. Note the similarity in expression pattern of the three proteins in invasive squamous cell carcinoma and the invasive front of proliferating keratoacanthoma. Scale (A–C), 400 µm; (D–F), 100 µm; (G–I), 50 µm.
In contrast to the C4.4A expression patterns in squamous cell carcinomas and keratoacanthomas, C4.4A expression in basal cell carcinomas was only observed in scattered islands of cancer cells often in the vicinity of keratin pearls in keratotic subtypes, which revealed a confinement to more differentiated cells (as CK1; see Table 2) and C4.4A expression was consequently absent in the invasive front (data not shown).
Haldisin expression was partially or completely lost in all cases of carcinoma in situ, actinic keratosis, squamous cell carcinoma and basal cell carcinoma reflecting the general loss of highly differentiated cells in these lesions (Table 2). Haldisin expression was consequently retained in differentiated layers of keratoacanthomas and correlated with the expression signatures of CK1 and CK10.
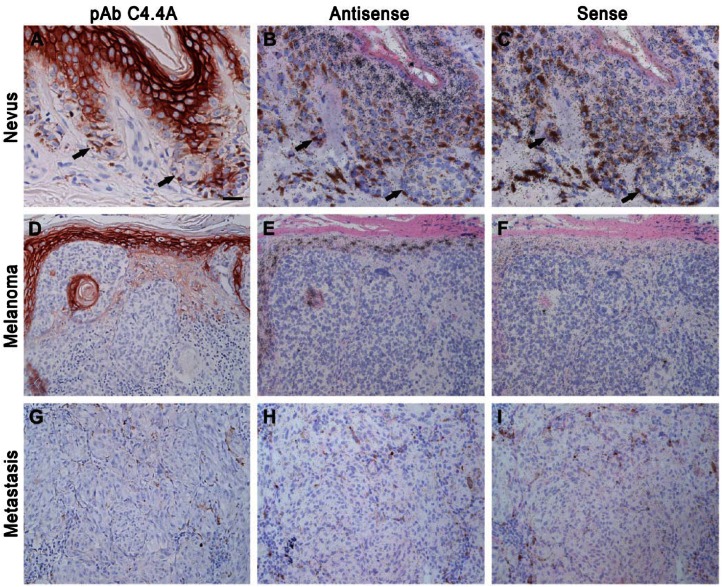
Nevi, Malignant Melanomas and Metastases
A previous study showed that C4.4A mRNA levels were upregulated in malignant melanomas as well as in skin and lymph node metastases as compared to normal nevi when evaluated by RT-PCR and in situ hybridization (Seiter et al. 2001). To further validate this, we performed both immunohistochemistry and in situ hybridization on nevi, malignant melanomas and metastases. In agreement with the original results reported by Seiter and coworkers, we did not observe neither protein nor mRNA expression of C4.4A in nevi (Fig. 6A–6C). In contrast to the previous report, we nevertheless found that C4.4A expression remained absent in 12 of the examined 16 cases of malignant melanomas encompassing the most common subtypes, such as superficial spreading malignant melanoma, nodular melanoma, lentigo maligna melanoma, as well as in the less frequently occurring desmoplastic melanoma (Fig. 6D–6F). A very weak expression was only observed in minor parts of 2 superficial spreading and 2 nodular malignant melanomas, whereas the majority of the tumor cells were C4.4A negative. C4.4A was also completely absent in all five examined cases of metastases (Fig. 6G–6I).
Figure 6.
C4.4A protein and mRNA expression in nevi, malignant melanoma and metastases. Immunohistochemistry and in situ hybridization was performed for the detection of C4.4A protein and mRNA, respectively, in normal nevi (A–C), superficial spreading melanoma (D–F) and metastases (G–I). Neighboring sections were either processed for immunohistochemical staining using our polyclonal anti-C4.4A antibody (A–C) or in situ hybridization using 35S-labelled 677 base pair nucleotide antisense (D–F) or sense (G–I) probes. Arrows point to the C4.4A-negative melanocytes. Scale (A–C), 25 µm; (D–I), 50 µm.
Discussion
In the present study, we have examined the expression patterns of C4.4A and Haldisin, two newly discovered squamous differentiation markers (Gårdsvoll et al. 2013; Kriegbaum et al. 2011), in a collection of skin diseases. Whereas we found no dramatic alterations in their expression signatures in the various blistering lesions covering most of the strata in the squamous epithelium, or in the lesions with dysregulated differentiation, we did observe attenuation in C4.4A expression in the lesion core of invasive squamous cell carcinomas and keratoacanthomas and a recovery of high expression in the deeply invasive fronts. Importantly, this was accompanied by an unexpected shift in the coupling of C4.4A expression to the traditional cytokeratin markers of differentiation. A similar shift in cytokeratin association of C4.4A was not observed in basal cell carcinomas originating from the C4.4A-negative stratum basale or in malignant melanomas, which hardly gained any C4.4A expression.
During normal homeostatic conditions, we observed a pronounced membrane-associated expression of C4.4A in those keratinocytes that were confined to the stratum spinosum of human skin and were positive for CK1 and CK10 (Fig. 3A, 3E). Although this expression pattern is almost complementary to that of CK5 and CK14, which are prototypic markers of dividing keratinocytes in the stratum basale (Alam et al. 2011), we, nevertheless, also observed an overlap in the expression of C4.4A with CK5/CK14 in the immediate suprabasal cell layer (Fig. 3A, 3H). Bearing this in mind, it is noteworthy that the invasive squamous cell carcinomas and keratoacanthomas underwent a distinct shift in their association of membrane-bound C4.4A expression from predominantly CK1/10 to CK5/14 in the deeply infiltrating sheets of neoplastic keratinocytes (Figs. 4, 5). Based on these observations and given the suggested role of C4.4A in cell adhesion, it is tempting to speculate that such invading keratinocytes recapitulate a differentiation state, where the balance between cell proliferation and cell adhesion is favored for collective cell migration and invasion. This furthermore seems independent of the chosen strategy for cell escape; i.e., invasion through the generation of de novo space through proteolytic degradation of the extracellular matrix in the deeper dermal layers as seen in both squamous cell carcinomas and keratoacanthomas (Figs. 4, 5) or cell movement along pre-existing structures, as seen in the perineural infiltrating keratoacanthomas (Fig. 5C, 5I) (Alexander et al. 2013; Friedl and Alexander 2011). Perineural invasion is a feature that is also observed in malignant squamous cell carcinomas of, for example, the esophagus (Chen et al. 2014), skin (Adams et al. 2014), tongue (Shen et al. 2014) and oral cavity (Chatzistefanou et al. 2014).
The presented CK5 and CK10 expression profiles mimic that reported in skin wound healing, which is often used as a surrogate model of cancer invasion (Johnsen et al. 1998). During re-epithelialization of human wounds, CK5 is consistently expressed in the basal cells, but it also appears in the leading suprabasal keratinocytes closing the wound; CK10, on the other hand, is absent in these “activated” keratinocytes and is only expressed in the original intact epidermis (Patel et al. 2006). C4.4A expression is sustained in these leading suprabasal keratinocytes of cutaneous wounds (Hansen et al. 2004) and is therefore also, in this setting, coupled to CK5 expression, which, as mentioned, may define a preferred differentiation state for collective cell migration.
Despite the invasive potential, keratoacanthomas are benign lesions and they regress spontaneously with time. The fact that high C4.4A expression is observed in the invasive fronts of both benign (keratoacanthoma) and malignant (squamous cell carcinoma) skin lesions points to an involvement of C4.4A in the invasive process independent of malignant potential. It should, however, be emphasized that invasion is a part of the malignant profile and that C4.4A positivity could be one step on the multistep pathway to obtaining a malignant phenotype. Along these lines of considerations, it is relevant that high C4.4A expression is observed in the invasive fronts of distinct carcinoma lesions, and high C4.4A levels are correlated with poor survival of patients suffering from different cancer types (Cheng et al. 2014; Hansen et al. 2007; Jacobsen et al. 2013; Konishi et al. 2010; Ohtsuka et al. 2013a). Accordingly, the present C4.4A expression patterns in carcinoma in situ, squamous cell carcinoma and keratoacanthoma of the skin recapitulate the expression pattern previously observed in squamous cell carcinomas of the esophagus (Hansen et al. 2008), wherein the expression is attenuated during carcinoma in situ and dysplasia, and high C4.4A expression is regained in the invasive areas and corresponding lymph node metastases. High C4.4A expression is furthermore correlated with poor survival of patients suffering from distinct carcinoma lesions including adenocarcinomas of the lung (Hansen et al. 2007; Jacobsen et al. 2013). Whether the presence of C4.4A on the membranes of cancer cells can be further translated to a direct involvement in the ability of a tumor cell to metastasize, as initially proposed by Claas and coworkers (Claas et al. 1996), remains nonetheless to be verified experimentally in vivo. Pertaining to the translational aspects of these findings, it is going to be interesting to follow the progress of a recently announced dose escalation Phase 1 study using a monoclonal anti-C4.4A antibody conjugated to auristatin E (BAY1129980) with intended use in patients with advanced solid tumors (ClinicalTrials.gov; NCT02134197).
Supplementary Material
Acknowledgments
We would like to acknowledge Lotte Frederiksen (The Finsen Laboratory, Copenhagen, Denmark) for her technical assistance in the laboratory and John Post (The Finsen Laboratory, Copenhagen, Denmark) for assisting the photographic artwork.
Footnotes
Supplementary material for this article is available on the Journal of Histochemistry & Cytochemistry Web site at http://jhc.sagepub.com/supplemental.
Declaration of Conflicting Interests: The authors declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The authors disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This work was supported financially by The Danish Cancer Society, Denmark [A1765]; The Danish National Research Foundation (Danish-Chinese Centre for Proteases and Cancer) [26-331-6] and Arvid Nilssons Fond, Denmark.
References
- Adams CC, Thomas B, Bingham JL. (2014). Cutaneous squamous cell carcinoma with perineural invasion: a case report and review of the literature. Cutis 93:141-144. [PubMed] [Google Scholar]
- Alam H, Sehgal L, Kundu ST, Dalal SN, Vaidya MM. (2011). Novel function of keratins 5 and 14 in proliferation and differentiation of stratified epithelial cells. Mol Biol Cell 22:4068-4078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander S, Weigelin B, Winkler F, Friedl P. (2013). Preclinical intravital microscopy of the tumour-stroma interface: invasion, metastasis, and therapy response. Curr Opin Cell Biol 25:659-671. [DOI] [PubMed] [Google Scholar]
- Blanpain C, Fuchs E. (2009). Epidermal homeostasis: a balancing act of stem cells in the skin. Nat Rev Mol Cell Biol 10:207-217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatzistefanou I, Lubek J, Markou K, Ord RA. (2014). The role of neck dissection and postoperative adjuvant radiotherapy in cN0 patients with PNI-positive squamous cell carcinoma of the oral cavity. Oral Oncol 50:753-758. [DOI] [PubMed] [Google Scholar]
- Chen JW, Xie JD, Ling YH, Li P, Yan SM, Xi SY, Luo RZ, Yun JP, Xie D, Cai MY. (2014). The prognostic effect of perineural invasion in esophageal squamous cell carcinoma. BMC Cancer 14:313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng DQ, Gu XD, Li ZY, Xiang JB, Chen ZY. (2014). Expression of C4.4A is a Potential Independent Prognostic Factor for Patients with Gastric Cancer. Asian Pac J Cancer Prev 15:3895-3899. [DOI] [PubMed] [Google Scholar]
- Claas C, Herrmann K, Matzku S, Möller P, Zöller M. (1996). Developmentally regulated expression of metastasis-associated antigens in the rat. Cell Growth Differ 7:663-678. [PubMed] [Google Scholar]
- Coulombe PA, Kerns ML, Fuchs E. (2009). Epidermolysis bullosa simplex: a paradigm for disorders of tissue fragility. J Clin Invest 119:1784-1793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Veer SJ, Furio L, Harris JM, Hovnanian A. (2014). Proteases and proteomics: Cutting to the core of human skin pathologies. Proteomics Clin Appl 8:389-402. [DOI] [PubMed] [Google Scholar]
- Friedl P, Alexander S. (2011). Cancer invasion and the microenvironment: plasticity and reciprocity. Cell 147:992-1009. [DOI] [PubMed] [Google Scholar]
- Fujihara Y, Tokuhiro K, Muro Y, Kondoh G, Araki Y, Ikawa M, Okabe M. (2013). Expression of TEX101, regulated by ACE, is essential for the production of fertile mouse spermatozoa. Proc Natl Acad Sci U S A 110:8111-8116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gårdsvoll H, Kriegbaum MC, Hertz EP, Alpizar-Alpizar W, Ploug M. (2013). The urokinase receptor homolog Haldisin is a novel differentiation marker of stratum granulosum in squamous epithelia. J Histochem Cytochem 61:802-813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen LV, Gårdsvoll H, Nielsen BS, Lund LR, Danø K, Jensen ON, Ploug M. (2004). Structural analysis and tissue localization of human C4.4A: a protein homologue of the urokinase receptor. Biochem J 380:845-857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen LV, Lærum OD, Illemann M, Nielsen BS, Ploug M. (2008). Altered expression of the urokinase receptor homologue, C4.4A, in invasive areas of human esophageal squamous cell carcinoma. Int J Cancer 122:734-741. [DOI] [PubMed] [Google Scholar]
- Hansen LV, Skov BG, Ploug M, Pappot H. (2007). Tumour cell expression of C4.4A, a structural homologue of the urokinase receptor, correlates with poor prognosis in non-small cell lung cancer. Lung cancer 58:260-266. [DOI] [PubMed] [Google Scholar]
- Hu N, Mora-Jensen H, Theilgaard-Monch K, Doornbos-van der Meer B, Huitema MG, Stegeman CA, Heeringa P, Kallenberg CG, Westra J. (2014). Differential Expression of Granulopoiesis Related Genes in Neutrophil Subsets Distinguished by Membrane Expression of CD177. PloS One 9:e99671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobsen B, Muley T, Meister M, Dienemann H, Christensen IJ, Santoni-Rugiu E, Lærum OD, Ploug M. (2013). Ly6/uPAR-related protein C4.4A as a marker of solid growth pattern and poor prognosis in lung adenocarcinoma. J Thorac Oncol 8:152-160. [DOI] [PubMed] [Google Scholar]
- Jacobsen B, Ploug M. (2008). The urokinase receptor and its structural homologue C4.4A in human cancer: expression, prognosis and pharmacological inhibition. Curr Med Chem 15:2559-2573. [DOI] [PubMed] [Google Scholar]
- Jacobsen B, Santoni-Rugiu E, Illemann M, Kriegbaum MC, Laerum OD, Ploug M. (2012). Expression of C4.4A in precursor lesions of pulmonary adenocarcinoma and squamous cell carcinoma. Int J Cancer 130:2734-2739. [DOI] [PubMed] [Google Scholar]
- Johnsen M, Lund LR, Rømer J, Almholt K, Danø K. (1998). Cancer invasion and tissue remodeling: common themes in proteolytic matrix degradation. Curr Opin Cell Biol 10:667-671. [DOI] [PubMed] [Google Scholar]
- Ko CJ. (2010). Actinic keratosis: facts and controversies. Clin Dermatol 28, 249-253. [DOI] [PubMed] [Google Scholar]
- Konishi K, Yamamoto H, Mimori K, Takemasa I, Mizushima T, Ikeda M, Sekimoto M, Matsuura N, Takao T, Doki Y, Mori M. (2010). Expression of C4.4A at the invasive front is a novel prognostic marker for disease recurrence of colorectal cancer. Cancer Sci 101:2269-2277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kriegbaum MC, Jacobsen B, Hald A, Ploug M. (2011). Expression of C4.4A, a structural uPAR homolog, reflects squamous epithelial differentiation in the adult mouse and during embryogenesis. J Histochem Cytochem 59:188-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Li N, Diaz LA, Shipley M, Senior RM, Werb Z. (2005). Synergy between a plasminogen cascade and MMP-9 in autoimmune disease. J Clin Invest 115:879-887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madan V, Lear JT, Szeimies RM. (2010). Non-melanoma skin cancer. Lancet 375:673-685. [DOI] [PubMed] [Google Scholar]
- Matzku S, Wenzel A, Liu S, Zöller M. (1989). Antigenic differences between metastatic and nonmetastatic BSp73 rat tumor variants characterized by monoclonal antibodies. Cancer Res 49:1294-1299. [PubMed] [Google Scholar]
- Miyake T, Ito T, Yanai A, Inoue N, Miyagawa Y, Murase K, Imamura M, Ichii S, Takatsuka Y, Nishizaki T, Hirota S, Ohtsuka M, Yamamoto H, Noguchi S, Miyoshi Y. (2013). C4.4A highly expressed in HER2-positive human breast cancers may indicate a good prognosis. Breast Cancer. August 6 [Epub ahead of print]. [DOI] [PubMed] [Google Scholar]
- Nishie W. (2014). Update on the pathogenesis of bullous pemphigoid: an autoantibody-mediated blistering disease targeting collagen XVII. J Dermatol Sci 73:179-186. [DOI] [PubMed] [Google Scholar]
- Ohtsuka M, Yamamoto H, Masuzawa T, Takahashi H, Uemura M, Haraguchi N, Nishimura J, Hata T, Yamasaki M, Miyata H, Takemasa I, Mizushima T, Takiguchi S, Doki Y, Mori M. (2013a). C4.4A expression is associated with a poor prognosis of esophageal squamous cell carcinoma. Ann Surg Oncol 20:2699-2705. [DOI] [PubMed] [Google Scholar]
- Ohtsuka M, Yamamoto H, Oshiro R, Takahashi H, Masuzawa T, Uemura M, Haraguchi N, Nishimura J, Hata T, Yamasaki M, Takemasa I, Miyata H, Mizushima T, Takiguchi S, Doki Y, Mori M. (2013b). Concurrent expression of C4.4A and Tenascin-C in tumor cells relates to poor prognosis of esophageal squamous cell carcinoma. Int J Oncol 43:439-446. [DOI] [PubMed] [Google Scholar]
- Oshiro R, Yamamoto H, Takahashi H, Ohtsuka M, Wu X, Nishimura J, Takemasa I, Mizushima T, Ikeda M, Sekimoto M, Matsuura N, Doki Y, Mori M. (2012). C4.4A is associated with tumor budding and epithelial-mesenchymal transition of colorectal cancer. Cancer Sci 103:1155-1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paret C, Bourouba M, Beer A, Miyazaki K, Schnölzer M, Fiedler S, Zöller M. (2005). Ly6 family member C4.4A binds laminins 1 and 5, associates with galectin-3 and supports cell migration. Int J Cancer 115:724-733. [DOI] [PubMed] [Google Scholar]
- Patel GK, Wilson CH, Harding KG, Finlay AY, Bowden PE. (2006). Numerous keratinocyte subtypes involved in wound re-epithelialization. J Invest Dermatol 126:497-502. [DOI] [PubMed] [Google Scholar]
- Ploug M. (2013). Structure-driven design of radionuclide tracers for non-invasive imaging of uPAR and targeted radiotherapy. The tale of a synthetic peptide antagonist. Theranostics 3:467-476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rösel M, Claas C, Seiter S, Herlevsen M, Zöller M. (1998). Cloning and functional characterization of a new phosphatidyl-inositol anchored molecule of a metastasizing rat pancreatic tumor. Oncogene 17:1989-2002. [DOI] [PubMed] [Google Scholar]
- Sakuntabhai A, Ruiz-Perez V, Carter S, Jacobsen N, Burge S, Monk S, Smith M, Munro CS, O”Donovan M, Craddock N, Kucherlapati R, Rees JL, Owen M, Lathrop GM, Monaco AP, Strachan T, Hovnanian A. (1999). Mutations in ATP2A2, encoding a Ca2+ pump, cause Darier disease. Nat Genet 21:271-277. [DOI] [PubMed] [Google Scholar]
- Sales KU, Masedunskas A, Bey AL, Rasmussen AL, Weigert R, List K, Szabo R, Overbeek PA, Bugge TH. (2010). Matriptase initiates activation of epidermal pro-kallikrein and disease onset in a mouse model of Netherton syndrome. Nat Genet 42:676-683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz RA. (1994). Keratoacanthoma. J Am Acad Dermatol 30:1-19. [DOI] [PubMed] [Google Scholar]
- Seiter S, Stassar M, Rappl G, Reinhold U, Tilgen W, Zöller M. (2001). Upregulation of C4.4A expression during progression of melanoma. J Invest Dermatol 116:344-347. [DOI] [PubMed] [Google Scholar]
- Shen WR, Wang YP, Chang JY, Yu SY, Chen HM, Chiang CP. (2014). Perineural invasion and expression of nerve growth factor can predict the progression and prognosis of oral tongue squamous cell carcinoma. J Oral Pathol 43:258-264. [DOI] [PubMed] [Google Scholar]
- Smith BA, Kennedy WJ, Harnden P, Selby PJ, Trejdosiewicz LK, Southgate J. (2001). Identification of genes involved in human urothelial cell-matrix interactions: implications for the progression pathways of malignant urothelium. Cancer Res 61:1678-1685. [PubMed] [Google Scholar]
- Stanley JR, Amagai M. (2006). Pemphigus, bullous impetigo, and the staphylococcal scalded-skin syndrome. N Engl J Med 355:1800-1810. [DOI] [PubMed] [Google Scholar]
- Thompson JF, Scolyer RA, Kefford RF. (2005). Cutaneous melanoma. Lancet 365:687-701. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.