SUMMARY
Hepatitis C virus (HCV) is a leading cause of liver disease, but insight into virus-host interactions remains limited. We systematically used affinity purification/mass spectrometry to define the host interactions of all 10 HCV proteins in hepatoma cells. We combined these studies with RNAi knockdown of corresponding genes using a two-step scoring approach to generate a map of 139 high-confidence HCV-host protein-protein interactions. We found mitochondrial proteins highly involved in HCV infection and characterized a new interaction between the viral core protein and host protein within bgcn homolog (WIBG). Expression of core prevents WIBG from binding its regular interaction partners Y14 and Magoh, two known mediators of the nonsense-mediated mRNA decay pathway. We discovered that this surveillance pathway is disrupted in HCV-infected cells, causing potentially harmful transcripts to accumulate. Our study provides the first comprehensive view of HCV-host interactions and uncovers new mechanisms for how HCV perturbs host functions during infection.
INTRODUCTION
Hepatitis C virus (HCV) has infected ~200 million people and newly infects 3–4 million people each year. Nearly 3% of the world's population is infected, but rates are higher in developing nations. Of those affected, 85% develop a chronic infection that can lead to liver cirrhosis, liver failure, and hepatocellular carcinoma (Moradpour et al., 2007). No vaccine exists, and traditional interferon-based therapies have limited efficacy. While new antivirals promise to dramatically lessen the disease burden in developed countries, they are likely too expensive for most (Lawitz and Gane, 2013).
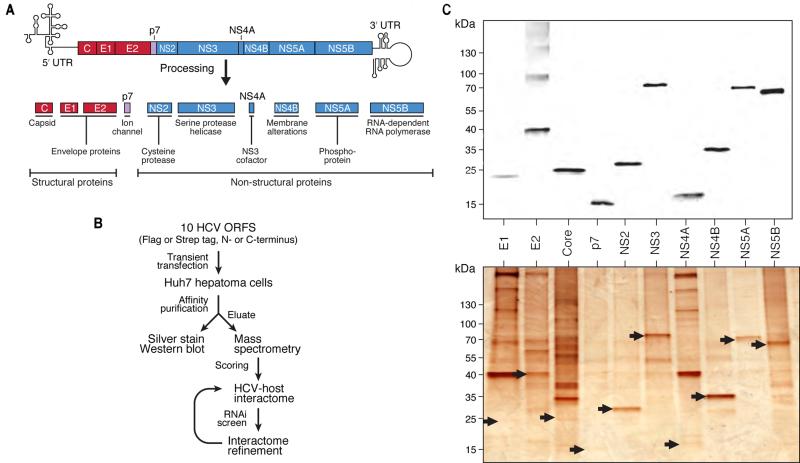
HCV is a positive-strand, RNA virus of the Flaviviridae family that replicates primarily in hepatocytes. HCV enters via receptor-mediated endocytosis, releases its genome into the cytoplasm and HCV RNA is translated into a polyprotein at the rough endoplasmic reticulum (ER) that is processed into 10 functional HCV proteins (Figure 1A) (Kim and Chang, 2013). The structural proteins core, E1 and E2 aid in viral assembly and virion release. The nonstructural proteins NS2, NS3, NS4A, NS4B, NS5A, and NS5B process the polyprotein and facilitate viral RNA replication. When expressed without structural proteins, nonstructural proteins form an HCV RNA replication unit called the replicon that autonomously propagates HCV RNA (Murray and Rice, 2011). The p7 protein is a cation channel or “viroporin” and its function is not yet fully understood (Carrere-Kremer et al., 2002; OuYang et al., 2013).
Figure 1. Affinity-purification of HCV proteins and interactome strategy.
(A) Schematic of HCV genome and 10 proteins derived from the viral polyprotein.
(B) Flowchart of the strategy to generate the HCV-host protein-protein interactome.
(C) Anti-flag western blot (upper) and silver staining (lower) of 10 flag-tagged HCV proteins after transient transfection and affinity purification from Huh7 cells. Silver-stained bands corresponding to HCV bait proteins are indicated by arrows. See also Figure S1 and Tables S1, S2, S3.
Additionally, HCV relies on host factors to complete its infection cycle. Several factors interact with HCV proteins and critically influence either viral RNA replication or particle formation. These include the ER-resident membrane trafficking and protein sorting factor phosphatidylinositol 4-kinase III alpha (PI4KA), the molecular chaperone cyclophilin A (CYPA), the RNA helicase DDX3X, the triglyceride-synthesizing protein diacylglycerol O-acyltransferase 1 (DGAT1), and the 47-kDa endosome-to-Golgi transport factor tail-interacting protein (TIP47) (Borawski et al., 2009; Herker et al., 2010; Ploen et al., 2013; Vogt et al., 2013; Yang et al., 2008). Also, several HCV proteins (i.e. NS3/NS4A protease) benefit the virus by interfering with host factors in the interferon response pathway, dampening the immune response (Horner, 2014). Two vesicle-associated membrane proteins, VAPA and VAPB, interact with HCV proteins NS5A and NS5B and are required for RNA replication (Evans et al., 2004; Hamamoto et al., 2005; Tu et al., 1999). Others (i.e., BIN1, binds transcription factor Myc; USP19, ER-resident deubiquitinase) interact with HCV proteins, but the relevance is unclear (de Chassey et al., 2008; Pichlmair et al., 2012).
Systematic approaches have identified host proteins key to HCV infection, including RNAi screens (Li et al., 2009; Suratanee et al., 2010; Tai et al., 2009) and a two-hybrid method (de Chassey et al., 2008; Dolan et al., 2013). A systematic affinity tag purification/mass spectrometry (AP-MS) approach was first used to construct a comprehensive host-pathogen protein-protein interaction (PPI) map for HIV-1 (Jager et al., 2011). This method is also used by Davis et al. to decipher host-pathogen interactions of Kaposi's sarcoma-associated herpesvirus reported in the current issue of Molecular Cell. Similar AP-MS approaches have partially been used in HCV research (Germain et al., 2014; Pichlmair et al., 2012). However, these studies were performed in HEK293T cells and lacked the full set of HCV proteins. Here, we describe a new approach we used to test for global host-pathogen interactions involving all 10 HCV proteins in Huh7 hepatoma cells, a more physiologically relevant cell type for HCV infection than HEK293T cells.
Protein-protein interaction (PPI) studies produce large amounts of data that must be filtered to identify robust interactions by scoring systems specifically for AP-MS (Jager et al., 2011; Sowa et al., 2009). With a two-step scoring system that incorporates results from an RNAi screen into a second round of scoring, we generated a high-confidence interaction map. We found mitochondria were more involved in HCV infection than previously thought. We also explored known virus-host protein interactions that we find are more complex than reported. Importantly, we uncovered a new interaction of HCV core and the host protein within bgcn homolog (WIBG) involved in the nonsense-mediated mRNA decay (NMD) surveillance pathway that, when functioning properly, senses and degrades damaged mRNA transcripts (Bono et al., 2004; Gehring et al., 2009). Thus, a combined proteomic/genomic approach is a powerful tool for identifying unexpected new host pathways relevant to HCV and possibly other viral infections.
RESULTS
Constructing an HCV-Host Protein-Protein Interactome
To generate a HCV-host PPI map, we cloned all 10 HCV proteins in vectors encoding N-terminal 2X-Strep or 3X-Flag affinity tags (Figure 1A). To minimize bias toward tag-specific effects, we also generated C-terminally tagged versions, except for core protein, which is cleaved at its C terminus in processing (McLauchlan et al., 2002). Robustly expressed, affinity-tagged HCV protein-encoding plasmids were transiently transfected into Huh7 hepatoma cells, and the proteins were affinity-purified and eluted (Figure 1B). Eluates were examined by western blotting and silver staining (Figure 1C) and in-solution mass spectrometry. Studies in HEK293T cells were used for comparison with other studies (Figure S1A, B, Tables S1 and S2). We identified 5085 unique HCV-host PPIs in hepatoma cells from 141 AP-MS samples (Tables S1 and S3).
To address concerns that the affinity tags may alter host factor interactions with HCV bait proteins, we used two analyses: (1) we analyzed the overlap of host interactors with HCV bait proteins for which data from N- and C-terminally tagged fusion proteins were available (Figure S1C). A high degree of overlap (in most cases between 80–100%) indicated the location of the affinity tag does not significantly alter binding; (2) We used immunoprecipitations of untagged NS5A protein in Huh7.5 cells constitutively replicating HCV RNA (replicon cells) and showed by MS that most validated host interactors identified with tagged NS5A were in the immunoprecipitated material (Figure S1D).
RNAi Screening Defines a Set of New HCV Cofactors across All Viral Proteins
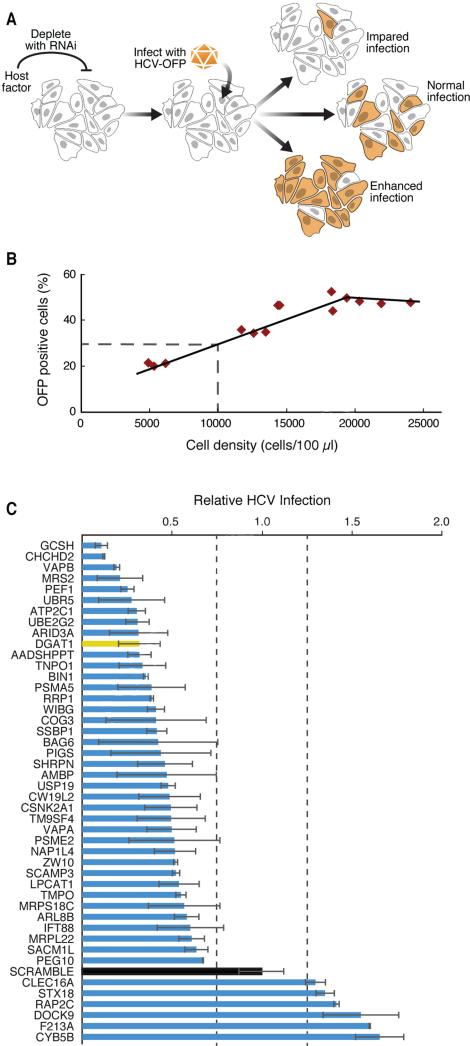
We used RNAi to knock down HCV-interacting host factors and assessed how it affected HCV infection (Figure 2A). The initial protein-protein interaction data set was analyzed with two APMS scoring algorithms, MIST (Jager et al., 2011) and COMPPASS (Sowa et al., 2009) (Table S3). For the RNAi screen, we selected 110 host proteins with a range of interaction scores representing binding partners for all 10 HCV proteins (Table S4). Although we focused on proteins with robust reproducibility or specificity scores, we also selected proteins with lesser scores to reduce bias from the default parameters in our first scoring round. We included the known HCV cofactors diacylglycerol O-acyltransferase 1 (DGAT1), BIN1, USP19, DDX3X, VAPA, and VAPB, but most of the factors we tested had not been reported.
Figure 2. A lentiviral shRNA screen identifies new HCV cofactors.
(A) Schematic of the HCV infection assay with an HCV-OFP (orange fluorescent protein mKO2) reporter virus.
(B) For each infection, we generated a standard curve of expected infection rates in scrambled shRNA-treated cells by plating the cells at indicated densities and obtaining the percentage of OFP-positive cells.
(C) Results of the HCV infection screen. Interacting host proteins that caused changes in infection levels are indicated on the y-axis. Relative infection levels (normalized to cell density) are indicated on the x-axis. The positive control, DGAT1, is shown in yellow, and the negative control, scrambled, in black. Results are shown as average (±SD) of a minimum of six experiments with two independent shRNAs (Experimental Procedures, S5). See also Figure S2 and Table S4.
Huh7.5 cells, a subclone of Huh7 cells and highly permissive for HCV infection (Blight et al., 2002), were transduced with several lentiviral shRNAs per target or scrambled shRNAs as controls (Table S4). After selecting cells with puromycin and confirming knockdown by quantitative RT-PCR, we inoculated cells with a monocistronic infectious clone of HCVJc1 encoding orange fluorescent protein (OFP) (Figures S2A, B) (Webster et al., 2013). After 6 days, we assessed viral spread by flow cytometry to quantify the OFP-expressing cells, giving the observed infection rate. The rate was normalized to the expected infection rate, based on the cell density in the infected cultures, a factor that affects viral spread (Figure 2B; Experimental Procedures) (Liu and He, 2013). In this system, cells expressing scrambled shRNAs consistently had infection scores of 1 (observed infection rate = expected infection rate), while shRNAs against DGAT1 markedly decreased the infection score (observed < expected infection rate), as anticipated (Figure 2C).
To determine if a candidate affected the infection rate, we used three criteria: (1) target transcript expression had to decrease by ≥25%, (2) infection rates had to increase or decrease by ≥25%, and (3) two or more shRNAs against the same target had to induce a similar change in HCV infection. We found that 75 of 110 host factors (68%) influenced HCV infection (Figure 2C, Table S4). Most factors supported infection, as infection rates decreased when they were knocked down. This was most prominent for mitochondrial proteins—glycine cleavage system protein H (GCSH), coiled-coil-helix-coiled-coil-helix domain containing 2 (CHCHD2), and mitochondrial matrix magnesium transporter (MRS2)—and an E3 ubiquitin-protein ligase (UBR5). No toxicity was observed in cells after knockdown of mitochondrial proteins (Figure S2C, D). We identified new restriction factors of HCV infection, because their depletion caused HCV infection rates to increase. These included the outer mitochondrial membrane-localized hemoprotein cytochrome B5 type B (CYB5B), the cytoplasmic proteins dedicator of cytokinesis 9 (DOCK9) and Ras-related GTPase protein (RAP2C), and the Golgi-derived retrograde transport associated protein syntaxin 18 (STX18). Overall, we identified 69 novel host factors regulating HCV infection.
Identifying 133 High-Confidence HCV-Interacting Proteins
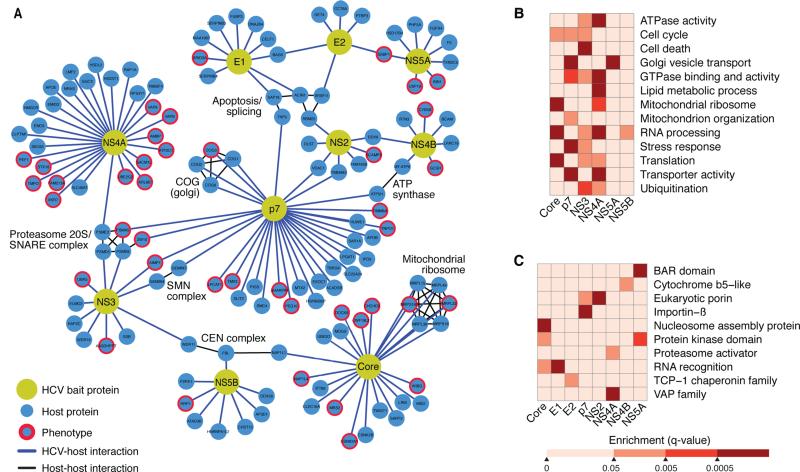
The accuracy of scoring systems (e.g., MIST and COMPPASS) is routinely benchmarked and optimized by “gold standard” interactors, which are not available in sufficient numbers for the HCV proteome. However, with our new set of 75 HCV-specific cofactors that both interacted with HCV proteins and regulated HCV infection, we exhaustively trained all possible combinations of the MIST and COMPPASS parameters for our interactome on two-thirds of the RNAi hits, and we validated results on the remaining one-third. The simulations allowed us to assemble a smaller, high-confidence interaction network of 139 HCV-host interactions, containing all 10 HCV proteins and 133 host factors (Figures 3A).
Figure 3. A complete HCV-host protein-protein interaction network in hepatoma cells.
(A) Network representation of the HCV-host interactome in Huh7 cells generated with Cytoscape. There are 10 HCV bait proteins (yellow nodes) and 134 interacting host proteins (blue nodes). Interactions of HCV and host proteins are indicated by blue lines. Curated host-host protein interactions from the CORUM database are indicated by black lines.
(B–C) Heat maps representing manually curated enriched biological processes, molecular functions, and pathways (B) and protein domains (C) of the host factors that interact with HCV bait proteins. Colors represent statistical significance (q value) as indicated by the accompanying scale. See also Table S6.
Comparing to previous AP-MS studies (Germain et al., 2014; Pichlmair et al., 2012), we found an overlap of 18 proteins (p = 2.92E−10) (Figure S1A, Table S5), including HCV cofactors USP19 and BIN1. We found greater overlap of previous AP-MS data (from HEK293T cells) and our HEK293T cell-derived data, indicating that some host-virus PPIs are cell-type specific (25 proteins, p = 4.16E−17; Figure S1A, Table S5). We found highly significant overlap (p = 3.92E−18; Figure S1B; Table S5) in the combined Huh7- and HEK293T-derived data and previous PPI results, including data from two-hybrid methods and co-immunoprecipitations. We saw the same trend when our data were compared to previous RNAi studies, but the overlap was only significant with data from Huh7 cells (p = 0.018), indicating that PPIs in Huh7 cells are functionally relevant (Table S5).
Next, we used a database of known human-human protein interactions to overlay additional connections between host proteins to identify protein complexes (Figure 3A) (Ruepp et al., 2010). We identified several complexes that interact with HCV (e.g., mitochondrial ribosome complex binds viral core). This bolsters reports that core associates with mitochondria (Okuda et al., 2002; Schwer et al., 2004). Interestingly, viral p7 and NS4B proteins interacted independently with components of the mitochondrial ATP synthase complex, ATP5H and MTATP6. This establishes new links between HCV and mitochondria and with a critical cellular ATP-producing enzyme. HCV p7 also interacted with four components of the conserved oligomeric Golgi (COG) complex, a multi-subunit tethering complex that maintains intra-Golgi vesicular trafficking and Golgi glycosylation (Willett et al., 2013). This interaction highlights the ways in which p7 may contribute to HCV particle assembly and release (Steinmann and Pietschmann, 2010). Many host complexes we identified interacted with multiple HCV proteins: the 20S proteasome is bound by p7, NS3, and NS4A. This complex forms the catalytic core of the 26S proteasome, which interacts with HIV Vif, Vpr, gp120 and gp160 (Jager et al., 2011).
A customized term enrichment test was performed for each viral protein using GO, KEGG, PFAM, and Uniprot ontologies (for details see Experimental Procedures). These tests confirmed that HCV has strong ties with biological processes, such as the Golgi vesicle transport (via p7 and NS4A) and mitochondrial ribosome functions (via core, p7, and NS4A) (Figure 3B, Table S6). Furthermore, this analysis reinforces that the viral NS3 protein is involved in apoptosis (Aweya and Tan, 2011) and that the core protein influences translation (Mamiya and Worman, 1999; Shimoike et al., 1999) and nuclear processes (Falcon et al., 2005) (Figure 3C). As expected, VAP family members were enriched in the interactome, but the strongest enrichment was found for NS4A, not NS5A and NS5B, as previously reported (Evans et al., 2004; Hamamoto et al., 2005; Tu et al., 1999) (Figure 3C).
Among the host proteins that bind to viral p7, we observed enrichment of domains associated with transporter activity and eukaryotic porins, further supporting that p7 is a viroporin targeting vesicular transport (Figures 3B, 3C, Table S6). We also identified an unexpectedly strong new link between cellular RNA processing and the viral core, NS3, NS4A, and NS5B proteins, which we discuss later on.
HCV NS4B Protein Localizes to Mitochondria
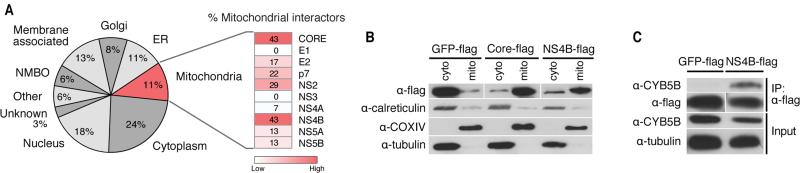
Most interacting proteins (24%) localized to the cytoplasm, which was expected as HCV replicates in the cytoplasm (Figure 4A). We also detected many nuclear proteins (18%)—mostly nuclear-cytoplasmic proteins (11%), but some that are considered exclusively nuclear (7%)—suggesting that HCV could be more involved in nuclear processes than originally thought. Many HCV-interacting host proteins were membrane-bound (18%), consistent with the membrane anchoring of most HCV proteins (Dubuisson et al., 2002), and localized to the ER (11%) and the Golgi apparatus (8%), both of which are implicated in HCV RNA replication and virion assembly and release (Suzuki et al., 2005; Tai et al., 2009). Interestingly, many host interactors (11%) localized to the mitochondria, underscoring that mitochondrial processes were enriched in the interactome and that mitochondria play an important role in HCV infection.
Figure 4. Cellular localization of HCV-interacting host proteins.
(A) Localization of HCV-interacting host proteins as a pie chart. Each category contains the percentage of interactors of the total refined interactome that localize to a cell compartment. The heat map indicates the percentage of mitochondrial proteins within each individual bait protein's interactome.
(B) Differential centrifugation of HEK293T cells expressing GFP-flag, flag-core, or NS4B-flag to examine protein localization to mitochondria or cytosolic fractions. Control antibodies recognize calreticulin (endoplasmic reticulum), tubulin (cytosol) and COXIV (mitochondria).
(C) Immunoprecipitations with Huh7 cell lysates expressing GFP-flag or NS4B-flag were performed with α-flag affinity resin and blotted with the indicated antibodies to demonstrate that NS4B specifically interacts with endogenous CYB5B, a mitochondrial outer membrane protein. See also Figure S3.
Individual analysis of proteins co-purifying with HCV proteins indicated that the highest percentage of cellular proteins interacting with core and NS4B localize to mitochondria (Figure S3). Core localizes to mitochondria (Okuda et al., 2002; Schwer et al., 2004), and this has also been suggested for NS4B (Liefhebber et al., 2009). To determine if NS4B is enriched at mitochondria, we used membrane fractionation experiments in HEK293T cells transiently expressing NS4B-flag protein and used the core-flag construct as a positive control. Both proteins were enriched in mitochondrial fractions, marked by COXIV protein, and the GFP-flag control protein was not (Figure 4B). As expected, tubulin was not found in mitochondrial fractions, but a small signal for the ER protein calreticulin was detected, indicating the presence of ER membranes associated with mitochondria, mitochondrial-associated membranes (MAMs). Co-immunoprecipitation experiments in Huh7 cells overexpressing NS4B-flag and endogenous CYB5B, a protein localized to the outer mitochondrial membrane that associates with NS4B in the interactome (Figure 4C), further support that NS4B can localize to the mitochondria.
HCV NS4A Protein Binds Cellular VAPA and VAPB Proteins
Because E1 and E2, as well as NS3 and NS4A, interact in infected cells (Moradpour et al., 2007), we performed AP-MS in cells co-expressing both interaction partners, scored interactions with host proteins as above, and determined the overlap in the interactome of co-expressed and singly expressed viral proteins (Figures S4A and S4B) (Tables S2 and S3). The overlap between proteins interacting with at least one singly expressed protein and the viral complex was greater for NS3-4A than E1-2 and, in the former group, involved mostly NS4A, showing NS4A is an important PPI interface of the NS3-4A protease (Bartenschlager et al., 1995; Kim et al., 1996). Proteins that interacted with NS4A alone and the NS3-4A complex included VAPA and VAPB, previously shown to interact with viral NS5A and NS5B (Evans et al., 2004; Hamamoto et al., 2005; Tu et al., 1999) (Figure 5A). We uncovered a weak interaction of NS5A with VAPA and VAPB, and another with NS5B and VAPB, but these scored below our cut-off (Figure 5A). In co-immunoprecipitation experiments in Huh7 cells with flag-tagged NS3, NS4A, and NS5A and V5-tagged VAP proteins, VAPA and VAPB interacted robustly with NS4A, markedly less with NS5A, and almost undetectably with NS3, reflecting our individual AP-MS results (Figures 5B–5D). When NS3-flag was co-expressed with NS4A-strep, VAPA coimmunoprecipitated with NS3, indicating VAPA binds the NS3-4A complex (Figures 5B–5D). No interaction was observed with GFP-flag in control experiments. These results were confirmed when endogenous VAPA was immunoprecipitated from HCV replicon-expressing hepatoma cells, and viral proteins were analyzed by MS (Figure S5C). Similar results were obtained with NS5B-flag, which co-immunoprecipitated with VAPB but not with VAPA (Figures 5A, S5A). NS5B immunoprecipitated with endogenous VAPA in replicon-expressing cells, indicating the interaction is supported by the other viral proteins with a functional replicase complex (Figure S5C).
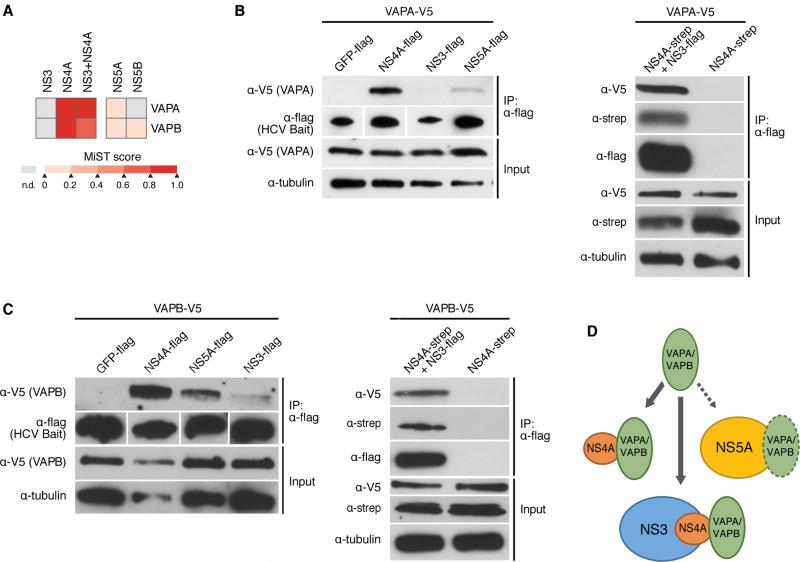
Figure 5. HCV NS4A interacts with host proteins VAPA and VAPB.
(A) Heat map indicating the interaction scores of VAPA or VAPB with NS3, NS4A, NS3+NS4A, NS5A, and NS5B, as identified by AP-MS. MIST scores in the color scale represent the confidence in the observed interaction.
(B) Immunoprecipitations with α-flag resin of Huh7 cell lysates expressing the indicated HCV bait proteins, or GFP as a control, and VAPA. VAPA-V5 interacts strongly with NS4A-flag, weakly with NS5A-flag, and very little with NS3-flag (left). V5-VAPA interacts with NS3-flag when NS4A-strep is coexpressed (right). Results are representative of three independent experiments.
(C) Same experiment as in (B) only with VAPB-V5. Results are representative of three independent experiments.
(D) Model of the interaction of VAPA and VAPB with NS4A alone, or while NS4A is bound to NS3. See also Figures S4, S5.
HCV Infection Disrupts the Host NMD Surveillance Pathway
RNA processing factors were significantly enriched in the hepatoma PPI data set (Figure 3B), so we next focused on cellular WIBG, a protein implicated in mRNA translation and surveillance (Bono et al., 2004; Diem et al., 2007; Gehring et al., 2009). WIBG was found to interact with core (Figure 3A), and HCV infection decreased after knocking WIBG down with five shRNAs (Figures 2C and 6A). We validated the core-WIBG interaction by co-immunoprecipitation in HEK293T cells with WIBG-HA or WIBG-V5 and flag-core proteins. WIBG readily interacted with flag-core proteins from HCV genotypes 1b and 2a, but not with the control (Figures 6B and S6A). Similar results were obtained in hepatoma cells with overexpressed WIBG-V5 (Figure S6B) and with endogenous WIBG in the context of HCV infection (Figure 6E). Interactions of WIBG were also identified with capsid proteins from dengue and West Nile virus, but not HIV-1, indicating the WIBG interaction is conserved among members of the Flaviviridae family (Figure 6C).
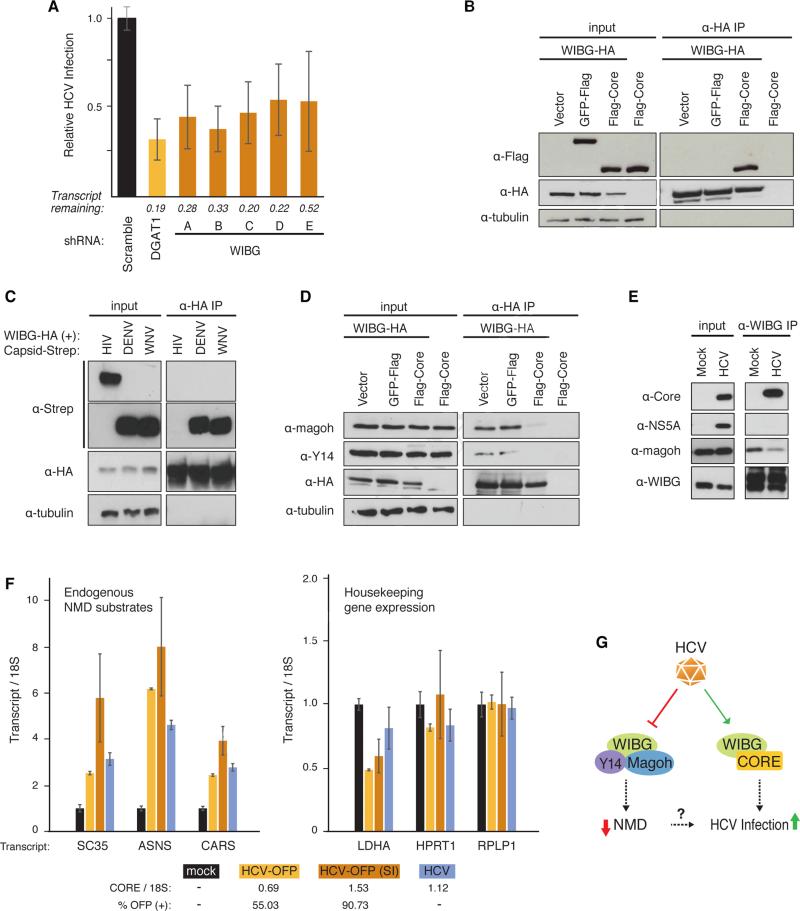
Figure 6. HCV infection requires WIBG and disrupts the host nonsense-mediated decay surveillance pathway.
(A) Relative infection rates (Figure 2A, B) after knock-down of WIBG with five shRNAs or shRNA targeting DGAT1 or scrambled shRNA as positive and negative controls, respectively. Shown is the average (±SD) of at least 12 replicates in two independent experiments.
(B) and (D) Immunoprecipitations in HEK293T cells expressing the indicated constructs along with the flag-tagged HCV core from genotype 2a. Results are representative of three independent experiments.
(C) Immunoprecipitations in HEK293T cells expressing WIBG-HA along with the indicated viral capsids from dengue virus, West Nile virus or HIV-1. Results are representative of three independent experiments.
(E) Immunoprecipitations of endogenous WIBG in mock- and HCV-OFP SI-infected Huh7.5 cells. Results are representative of two independent experiments.
(F) Huh7.5 cells were infected with the indicated HCV constructs. Three endogenous NMD substrate transcripts y (left) and transcripts of three housekeeping genes (right) were analyzed by quantitative RT-PCR. Shown is a representative of three independent experiments. Infection rates based on OFP expression or on quantitative RT-PCR are indicated below.
(G) Model of the WIBG-HCV interaction. See text for details. See also Figure S6.
WIBG was originally identified as a cytoplasmic factor that co-purifies with Y14 and Magoh proteins and is also known as PYM (partner of Y14 and Magoh) (Forler et al., 2003). Y14 and Magoh are components of the exon junction complex (EJC), which facilitates mRNA splicing, export, translation, and surveillance (Tange et al., 2004). To assess how HCV core affects WIBG-EJC interactions, we transiently transfected WIBG-HA into HEK293T cells with and without flag-core or GFP-flag as a control. After immunoprecipitations with HA antibodies, WIBG-HA interacted with endogenous Y14 and Magoh proteins in vector- and GFP-expressing cells (Figure 6D). However, with flag-core, these interactions were markedly reduced (Figure 6D). Similar results were obtained with core from HCV genotype 1b (Figure S6C). This dissociation was also seen in hepatoma cells when WIBG and core were overexpressed (Figure S6D) and with endogenous WIBG in HCV infection (Figure 6E), underscoring its physiological relevance.
The EJC is important in the nonsense-mediated decay (NMD) pathway, an mRNA surveillance process that degrades transcripts with premature termination codons (PTC), which may produce toxic, C-terminally truncated proteins (Nicholson et al., 2010; Tange et al., 2004). Because WIBG influences NMD (Bono et al., 2004; Gehring et al., 2009), we hypothesized that the NMD pathway may be perturbed in HCV infection. To investigate this hypothesis, we monitored transcript levels of three known NMD substrates—SR protein SC35, asparagine synthetase (ASNS), and cysteinyl-tRNA synthetase (CARS) (Linde et al., 2007; Mendell et al., 2004; Singh et al., 2013; Sureau et al., 2001). SC35 is a splicing factor that auto-regulates its expression by inducing alternative splicing of its mRNA and generating transcripts that are degraded via NMD (Sureau et al., 2001). Transcripts for ASNS and CARS were found upregulated in cells that lack the critical NMD factor hUpf1, identifying them as NMD targets (Mendell et al., 2004).
Huh7.5 cells were infected with three different infectious clones of HCV: untagged wild-type HCV Jc1; HCV-OFP (as above); or HCV-OFP (SI), a “super-infectious” HCV Jc1 clone with several adaptive mutations that enhance infectivity (Webster et al., 2013). HCV infection was monitored by flow cytometry (for OFP-expressing virus) and quantitative RT-PCR analysis of the HCV RNA with a core amplicon (for all viruses). Transcript levels of SC35, ASNS, and CARS accumulated significantly upon infection with all three HCV clones, as measured by quantitative RT-PCR and correlated with infection rates of HCV-OFP and HCV-OFP (SI) viruses (55% and 91%, respectively) (Figure 6F). Knockdown of WIBG in HCV-infected cells reduced the accumulation of ASNS transcripts suggesting WIBG expression is required for HCV-induced accumulation of NMD substrates (Figure S6E).
We also analyzed transcript levels of three housekeeping genes—lactate dehydrogenase A (LDHA), hypoxanthine phosphoribosyltransferase 1 (HPRT1), and ribosomal protein large, P1 (RPLP1)—to ensure that HCV-induced accumulation of transcripts was specific to NMD substrates. These remained unchanged or, as with LDHA, were slightly downregulated during HCV infection (Figure 6F). No change in RPLP1 transcript levels was observed when WIBG was downregulated in infected cells (Figure S6E). Collectively, these results suggest HCV infection disrupts NMD mRNA surveillance through manipulation of WIBG's function.
DISCUSSION
Here we report the first complete HCV-host PPI map in hepatoma cells. We used established scoring systems, COMPPASS and MIST, but also examined ~30% of the interactors in a functional RNAi screen with infectious HCV to find 69 new cofactors of HCV infection. We developed a second iteration of scoring, in which we utilized these cofactors as standards to train the MIST scoring system and optimize the identification of relevant interactors in our dataset. We found 133 high-confidence HCV-interacting host factors, 99 of which are cellular proteins that have never been associated with HCV infection. Biological functions enriched with HCV-binding PPIs include expected ones, such as cell-cycle control and apoptosis (Deng et al., 2008; Walters et al., 2009). In addition, through interactions with p7, we confirmed Golgi vesicle transport as an important biological process associated with HCV infection (Li et al., 2009; Tai et al., 2009) and find new biological functions such as RNA processing enriched with HCV interactors. By refining interactions of VAPA and VAPB with viral proteins, we found the strongest interactions involve viral NS3/4A protease via NS4A.
We compared our results to other virus-host interactomes to find pathways targeted by multiple viruses and those unique to HCV. Comparison with the HIV-host PPI study (Jager et al., 2011), identifies 42 proteins targeted by both HIV and HCV. These include the 26S proteasome, the mitochondrial ribosome, and ubiquitination-related enzymes, including a shared interaction with the ubiquitin-conjugating enzyme UBE2O. HIV Rev interacts with MRS2, a mitochondrial magnesium transporter that interacts with HCV core and is required for HCV infection. Also HIV gp41 protein binds VAPA, which interacts with NS3/4A protease and other non-structural HCV proteins.
A proposed mechanism for HCV's engagement with VAPA/VAPB is that the interaction aids viral non-structural proteins to associate with intracellular membranes for efficient RNA replication (Tu et al., 1999). We speculate that the interaction of NS4A and VAPA/VAPB is important to the recruitment, but other cellular functions recently linked to the VAPB protein may be relevant to HCV infection. VAPB is required for membrane bridges to form between the ER and mitochondria (Stoica et al., 2014). VAPB is linked to the unfolded protein response, a cellular protein rescue/degradation pathway induced when unfolded proteins accumulate at the ER and altered by HCV infection (Ke and Chen, 2011; Tardif et al., 2004). The recruitment of VAPB along with several ubiquitin-associated host proteins bound by HCV proteins, including USP19 that rescues misfolded proteins from ER-associated degradation (Hassink et al., 2009; Vembar and Brodsky, 2008), may prevent host cells from detecting and degrading viral proteins at the ER.
Surprisingly, many mitochondrial proteins interact with HCV proteins. While this is expected for viral proteins that localize to mitochondria, such as core and p7, we highlight a new interaction between viral NS4B and host CYB5B, a predicted electron carrier localized at the mitochondrial outer membrane. We confirm that NS4B localizes to mitochondria or the interface between ER and mitochondria, the MAMs. NS4B induces ER membrane reorganization to facilitate HCV replication (membranous web formation) (Egger et al., 2002), and the interaction of NS4B with mitochondria implicates recruitment of mitochondrial membranes or MAMs for this purpose. As HCV infection induces apoptosis (Deng et al., 2008), the NS4B/CYB5B interaction may affect this process, but this requires further study.
The high number of host proteins that interact with p7 was unexpected. P7 is a very small (7 kDa) integral membrane protein that can self-assemble into a large cation channel or viroporin (OuYang et al., 2013). Other viroporins (e.g. influenza A virus M2, HIV-1 Vpu) affect the cell vesicle system, glycoprotein trafficking, and membrane permeability (Nieva et al., 2012). We found p7 interactors are enriched for proteins with porin domains and those involved in Golgi vesicle transport, supporting the function of p7 as a bona fide viroporin. A new finding is that p7 associates with the COG complex, an intra-Golgi vesicle-tethering complex composed of eight subunits, four of which interact with p7. Knocking down the COG3 component impaired HCV infection (Table S4), consistent with the notion that p7 engages the COG complex and Golgi vesicular transport to facilitate virus production or release (Steinmann et al., 2007).
Our work further uncovers a new connection between HCV infection and the NMD RNA surveillance pathway. The NMD pathway identifies and degrades premature termination codon (PTC)-containing transcripts, which can otherwise encode for potentially toxic C-terminally truncated proteins (Nicholson et al., 2010). We show that HCV core interacts with cellular WIBG, and core expression disrupts interaction of WIBG with Magoh/Y14. Magoh and Y14 are central components of the EJC, a dynamic structure deposited on pre-mRNA during splicing in the nucleus, and remains associated with the mRNA until the first round of translation in the cytoplasm (Tange et al., 2004). The EJC is central in NMD: when located downstream of a PTC, it recruits NMD factors that degrade the transcript (Tange et al., 2004). We hypothesize that disrupting Magoh and Y14 from WIBG impairs NMD, given that NMD substrates accumulated in HCV-infected cells. A potential mechanism is that WIBG normally facilitates EJC disassembly from cytoplasmic mRNAs, thus allowing for EJC to recycle back to the nucleus (Gehring et al., 2009), a process that is likely disrupted in HCV-infected cells. We find additional interactions between HCV and the EJC and NMD pathway through binding of E1 to two mRNA splicing factors peripherally associated with the EJC complex –SAP18 (sin3-associated protein 18 kDa) and ACIN1 (apoptotic chromatin condensation inducer 1) and binding of several NMD factors including Magoh, Y14, UPF1 (Up-Frameshift Mutation 1), and UPF3B to different HCV proteins with MIST and COMPASS scores below cut-off (Figure S6G).
How does disrupting the NMD pathway benefit infection? A similar phenomenon was reported for human T-cell leukemia virus type-1 (HTLV-1), a retrovirus, which encodes an RNA-binding protein Rex that globally suppresses NMD (Nakano et al., 2013). This Rex-mediated block of NMD is instrumental for HTLV-1 gene expression as alternative splicing and programmed ribosomal frameshifting cause accumulation of viral transcripts with multiple stop codons that likely trigger NMD. In contrast, HCV replicates in the cytoplasm and does not undergo nuclear RNA processing. Thus, reasons why HCV (and maybe other Flaviviridae) blocks the NMD pathway are likely more complex.
During submission of this paper, three studies were published describing the NMD pathway as a natural restriction mechanism for certain RNA viruses in plants and animal cells (Balistreri et al., 2014; Garcia et al., 2014; Gloggnitzer et al., 2014). The model is that the viral RNA itself may function as an NMD substrate due to the presence of internal termination codons and long 3′UTRs (Garcia et al., 2014). Our finding that NMD is inactivated in HCV-infected cells supports the model that NMD restricts RNA virus infections. But important differences exist. While previous studies show knockdown of NMD factors increases viral replication, we find knockdown of WIBG decreases HCV infection, pointing to an important role of WIBG in the viral lifecycle. We knocked down other NMD factors (e.g., Magoh, UPF1 and UPF2) and saw no effect on viral infection. We hypothesize that, as NMD is already blocked in HCV-infected cells, any additional effects caused by the knockdown of these NMD factors are masked. HCV may evade NMD by many strategies, including polyprotein processing, lack of internal termination codons, a short 3’UTR, disrupting the WIBG-Magoh/Y14 interaction and subverting host WIBG into a critical cofactor for infection.
Disruption of the NMD pathway in HCV infection could also be a byproduct of engagement of WIBG into the viral lifecycle, possibly causing liver damage in chronically infected individuals. Notably, NMD was previously implicated in the pathogenesis of diseases such as Duchenne muscular dystrophy, Marfan syndrome, cystic fibrosis, and various cancers, where PTC-containing transcripts result in altered protein production or mRNA abundance (Nicholson et al., 2010). Why WIBG is necessary for HCV infection is unclear, but its interaction with capsid proteins from dengue and West Nile virus underscores a key role of this host factor in Flavivirus biology. Because WIBG targets viral capsid proteins, it could directly support virion assembly. Alternatively, it may facilitate viral translation by associating with ribosomal proteins (Diem et al., 2007). Indeed, WIBG recruits the cellular translation machinery to unspliced transcripts of the Kaposi's sarcoma-associated herpesvirus, where ORF57 protein facilitates a WIBG-RNA interaction (Boyne et al., 2010).
Notably, NMD is not only an RNA quality control mechanism but in eukaryotic cells appears to play a more global role by controlling the stability of 3–10% of the transcriptome (Nicholson et al., 2010). The WIBG-core interaction may thus cause alterations of a wider range of host mRNAs beyond PTC-containing mRNAs, possibly altering the cellular transcriptome in favor of HCV infection. A recent study found 197 transcripts to be upregulated in HeLa cells with silenced expression of hUpf1, a critical NMD factor (Mendell et al., 2004). By mining published RNA-seq data (Papic et al., 2012), we find that 43 of these transcripts are upregulated in Huh7.5 cells infected with HCV, supporting our model that the NMD pathway is perturbed during HCV infection. Future studies will determine how WIBG, the EJC, and the NMD pathway regulate HCV infection and how they reprogram the host cell to foster viral spread while possibly damaging infected liver cells.
EXPERIMENTAL PROCEDURES
For a detailed description of Experimental Procedures, refer to the Supplemental Information.
Cell Culture and Transfections
HEK293T and Huh7 cell lines were obtained from the American Type Culture Collections. Huh7.5 cells were provided by Charles M. Rice. All cultures were grown under standard conditions. Huh7 cells were transfected with X-treme GENE 9 (Roche Applied Science) and HEK293T using calcium phosphate.
Affinity Purification, Mass Spectrometry, and Interactome Scoring
Affinity purifications were performed as described (Jager et al., 2011). The first iteration of the HCV interactome was compiled by selecting bait-prey pairs with a MIST score >.70, computed with the published feature weights for reproducibility (.31), abundance (.01) and specificity (.68) (Jager et al., 2011), or a top 5% COMPPASS WD score per bait (Sowa et al., 2009). The new weights for the MIST score, MIST threshold and COMPASS threshold were derived by testing the prediction performance of the top 10 optimal weight combinations, trained on two thirds of a benchmark set of 5200 interactions (52 positive interactions and 5148 negative interactions) randomly sampled from the mass-spectrometry results. The refined interactome was compiled through selecting bait-prey pairs with a retrained MIST score >0.68, with new weights for reproducibility (.36), abundance (.09) and specificity (.55), or a top 1% COMPASS WD score per bait.
Functional and Domain Ontology Enrichments
All scored bait-prey pairs were joined with the GO biological process (BP), molecular function (MF), and KEGG ontologies for functional annotation and the PFAM and Uniprot domain for domain annotations with their Uniprot accession codes and represented as bait-term-protein entries. Terms were manually curated and analyzed for significance of enrichment with a resampling procedure on the MIST and COMPPASS WD scores.
HCV Infections
Huh7.5 cells were infected with 4–6 independent lentiviruses expressing shRNAs against the indicated factor and a puromycin resistance gene for 24 h. After a 2-day puromycin selection, knockdown of the host factor was determined by quantitative RT-PCR, and cells were infected with monocistronic Jc1 HCV encoding orange fluorescent protein (OFP) and a blasticidin resistance cassette (Bsd): Jc1/NS5AB-OFP-Bsd at an MOI of 0.06 or 0.03 (Figure S1). The number of live cells and the percentage of OFP-positive cells were determined by flow cytometry on the MACSQuant VYB instrument.
RNA Isolation and Quantitative RT-PCR
Total cellular RNA was isolated with RNAstat60 (Tel-test), and cleaned and DNase-treated with RNA Clean & Concentrator (Zymo). Complementary DNA (cDNA) was synthesized with an oligo(dT)18 primer (Thermo) and/or random hexamer primers (Life Technologies) using the AMV reverse transcriptase (Promega) and analyzed by qPCR using a Bio-Rad ABI 7900. Relative values for each transcript were normalized to GAPDH (gene/GAPDH; HCV infection screen) or 18S rRNA (gene/18S; all other qPCR analyses) and compared to the scramble or mock controls, respectively.
Mitochondrial Enrichment
Mitochondria were isolated from HEK293T cells with the Mitochondrial Isolation Kit for Cultured Cells (Pierce) with minor modifications (Supplemental Experimental Procedures). The mitochondrial fraction and cytoplasmic fractions were normalized for protein concentration and analyzed by SDS-PAGE and western blotting.
Supplementary Material
ACKNOWLEDGEMENTS
We thank all members of the Ott Lab, particularly Gregory Camus, Eva Herker, Dorothee A. Vogt and Andrew Kondratowicz for helpful discussions, advice and reagents. We thank members of the Krogan lab, especially Stefanie Jäger for technical advice. We also thank Brian R. Webster for sharing reagents and providing technical advice, Marielle Cavrois and the Gladstone Flow Cytometry Core for assistance with FACS (NIH-funded CFAR grant P30 AI027763). We thank Ralf Bartenschlager (University of Heidelberg) for the Jc1 construct, Charles Rice (Rockefeller University) for Huh7.5 cells, Takaji Wakita (National Institute of Infectious Disease, Japan) for the original JFH1 construct. We thank Mike Shales for graphical support and Celeste Brennecke for editorial assistance. This work was supported by funds from the UCSF Liver Center, Gladstone, and NIH (M.O.: RO56 AI069090, R01 AI097552 and P30 DK026743; N.J.K.: P50 GM082250, P01 AI090935, P50 GM081879, P01 AI091575 and U19 AI106754). N.J.K. is a Keck Young Investigator. We gratefully acknowledge support from the A.P. Giannini Foundation and the UCSF Program for Breakthrough Biomedical Research (H.R.R.) as well as a Ruth L. Kirschstein NRSA Hepatology Training Grant (G.R.K.; T32 DK060414).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
AUTHOR CONTRIBUTIONS
H.R.R. and G.R.K. designed, conducted and analyzed experiments. E.V. and J.V.D. performed all bioinformatics analyses and provided graphical support. J.R.J., B.N., J.H., T.J. and P.S. were involved in mass spectrometry analysis. M.O. and N.J.K. supervised this project. The manuscript was written by H.R.R. and G.R.K. with input from E.V., N.J.K. and M.O.
REFERENCES
- Aweya JJ, Tan YJ. Modulation of programmed cell death pathways by the hepatitis C virus. Front Biosci (Landmark Ed) 2011;16:608–618. doi: 10.2741/3709. [DOI] [PubMed] [Google Scholar]
- Balistreri G, Horvath P, Schweingruber C, Zund D, McInerney G, Merits A, Muhlemann O, Azzalin C, Helenius A. The host nonsense-mediated mRNA decay pathway restricts Mammalian RNA virus replication. Cell Host Microbe. 2014;16:403–411. doi: 10.1016/j.chom.2014.08.007. [DOI] [PubMed] [Google Scholar]
- Bartenschlager R, Lohmann V, Wilkinson T, Koch JO. Complex formation between the NS3 serine-type proteinase of the hepatitis C virus and NS4A and its importance for polyprotein maturation. J Virol. 1995;69:7519–7528. doi: 10.1128/jvi.69.12.7519-7528.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blight KJ, McKeating JA, Rice CM. Highly permissive cell lines for subgenomic and genomic hepatitis C virus RNA replication. J Virol. 2002;76:13001–13014. doi: 10.1128/JVI.76.24.13001-13014.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bono F, Ebert J, Unterholzner L, Guttler T, Izaurralde E, Conti E. Molecular insights into the interaction of PYM with the Mago-Y14 core of the exon junction complex. EMBO Rep. 2004;5:304–310. doi: 10.1038/sj.embor.7400091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borawski J, Troke P, Puyang X, Gibaja V, Zhao S, Mickanin C, Leighton-Davies J, Wilson CJ, Myer V, Cornellataracido I, et al. Class III phosphatidylinositol 4-kinase alpha and beta are novel host factor regulators of hepatitis C virus replication. J Virol. 2009;83:10058–10074. doi: 10.1128/JVI.02418-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyne JR, Jackson BR, Taylor A, Macnab SA, Whitehouse A. Kaposi's sarcoma-associated herpesvirus ORF57 protein interacts with PYM to enhance translation of viral intronless mRNAs. EMBO J. 2010;29:1851–1864. doi: 10.1038/emboj.2010.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrere-Kremer S, Montpellier-Pala C, Cocquerel L, Wychowski C, Penin F, Dubuisson J. Subcellular localization and topology of the p7 polypeptide of hepatitis C virus. J Virol. 2002;76:3720–3730. doi: 10.1128/JVI.76.8.3720-3730.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Chassey B, Navratil V, Tafforeau L, Hiet MS, Aublin-Gex A, Agaugue S, Meiffren G, Pradezynski F, Faria BF, Chantier T, et al. Hepatitis C virus infection protein network. Mol Syst Biol. 2008;4:230. doi: 10.1038/msb.2008.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng L, Adachi T, Kitayama K, Bungyoku Y, Kitazawa S, Ishido S, Shoji I, Hotta H. Hepatitis C virus infection induces apoptosis through a Bax-triggered, mitochondrion-mediated, caspase 3-dependent pathway. J Virol. 2008;82:10375–10385. doi: 10.1128/JVI.00395-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diem MD, Chan CC, Younis I, Dreyfuss G. PYM binds the cytoplasmic exon-junction complex and ribosomes to enhance translation of spliced mRNAs. Nat Struct Mol Biol. 2007;14:1173–1179. doi: 10.1038/nsmb1321. [DOI] [PubMed] [Google Scholar]
- Dolan PT, Zhang C, Khadka S, Arumugaswami V, Vangeloff AD, Heaton NS, Sahasrabudhe S, Randall G, Sun R, LaCount DJ. Identification and comparative analysis of hepatitis C virus-host cell protein interactions. Mol Biosyst. 2013;9:3199–3209. doi: 10.1039/c3mb70343f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubuisson J, Penin F, Moradpour D. Interaction of hepatitis C virus proteins with host cell membranes and lipids. Trends Cell Biol. 2002;12:517–523. doi: 10.1016/s0962-8924(02)02383-8. [DOI] [PubMed] [Google Scholar]
- Egger D, Wolk B, Gosert R, Bianchi L, Blum HE, Moradpour D, Bienz K. Expression of hepatitis C virus proteins induces distinct membrane alterations including a candidate viral replication complex. J Virol. 2002;76:5974–5984. doi: 10.1128/JVI.76.12.5974-5984.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans MJ, Rice CM, Goff SP. Phosphorylation of hepatitis C virus nonstructural protein 5A modulates its protein interactions and viral RNA replication. Proc Natl Acad Sci U S A. 2004;101:13038–13043. doi: 10.1073/pnas.0405152101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falcon V, Acosta-Rivero N, Shibayama M, Chinea G, Gavilondo JV, de la Rosa MC, Menendez I, Gra B, Duenas-Carrera S, Vina A, et al. HCV core protein localizes in the nuclei of nonparenchymal liver cells from chronically HCV-infected patients. Biochem Biophys Res Commun. 2005;329:1320–1328. doi: 10.1016/j.bbrc.2005.02.107. [DOI] [PubMed] [Google Scholar]
- Forler D, Kocher T, Rode M, Gentzel M, Izaurralde E, Wilm M. An efficient protein complex purification method for functional proteomics in higher eukaryotes. Nat Biotechnol. 2003;21:89–92. doi: 10.1038/nbt773. [DOI] [PubMed] [Google Scholar]
- Garcia D, Garcia S, Voinnet O. Nonsense-mediated decay serves as a general viral restriction mechanism in plants. Cell Host Microbe. 2014;16:391–402. doi: 10.1016/j.chom.2014.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gehring NH, Lamprinaki S, Kulozik AE, Hentze MW. Disassembly of exon junction complexes by PYM. Cell. 2009;137:536–548. doi: 10.1016/j.cell.2009.02.042. [DOI] [PubMed] [Google Scholar]
- Germain MA, Chatel-Chaix L, Gagne B, Bonneil E, Thibault P, Pradezynski F, de Chassey B, Meyniel-Schicklin L, Lotteau V, Baril M, et al. Elucidating novel hepatitis C virus-host interactions using combined mass spectrometry and functional genomics approaches. Mol Cell Proteomics. 2014;13:184–203. doi: 10.1074/mcp.M113.030155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gloggnitzer J, Akimcheva S, Srinivasan A, Kusenda B, Riehs N, Stampfl H, Bautor J, Dekrout B, Jonak C, Jimenez-Gomez JM, et al. Nonsense-mediated mRNA decay modulates immune receptor levels to regulate plant antibacterial defense. Cell Host Microbe. 2014;16:376–390. doi: 10.1016/j.chom.2014.08.010. [DOI] [PubMed] [Google Scholar]
- Hamamoto I, Nishimura Y, Okamoto T, Aizaki H, Liu M, Mori Y, Abe T, Suzuki T, Lai MM, Miyamura T, et al. Human VAP-B is involved in hepatitis C virus replication through interaction with NS5A and NS5B. J Virol. 2005;79:13473–13482. doi: 10.1128/JVI.79.21.13473-13482.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassink GC, Zhao B, Sompallae R, Altun M, Gastaldello S, Zinin NV, Masucci MG, Lindsten K. The ER-resident ubiquitin-specific protease 19 participates in the UPR and rescues ERAD substrates. EMBO Rep. 2009;10:755–761. doi: 10.1038/embor.2009.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herker E, Harris C, Hernandez C, Carpentier A, Kaehlcke K, Rosenberg AR, Farese RV, Jr., Ott M. Efficient hepatitis C virus particle formation requires diacylglycerol acyltransferase-1. Nat Med. 2010;16:1295–1298. doi: 10.1038/nm.2238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horner SM. Activation and evasion of antiviral innate immunity by hepatitis C virus. J Mol Biol. 2014;426:1198–1209. doi: 10.1016/j.jmb.2013.10.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jager S, Cimermancic P, Gulbahce N, Johnson JR, McGovern KE, Clarke SC, Shales M, Mercenne G, Pache L, Li K, et al. Global landscape of HIV-human protein complexes. Nature. 2011;481:365–370. doi: 10.1038/nature10719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ke PY, Chen SS. Activation of the unfolded protein response and autophagy after hepatitis C virus infection suppresses innate antiviral immunity in vitro. J Clin Invest. 2011;121:37–56. doi: 10.1172/JCI41474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim CW, Chang KM. Hepatitis C virus: virology and life cycle. Clin Mol Hepatol. 2013;19:17–25. doi: 10.3350/cmh.2013.19.1.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JL, Morgenstern KA, Lin C, Fox T, Dwyer MD, Landro JA, Chambers SP, Markland W, Lepre CA, O'Malley ET, et al. Crystal structure of the hepatitis C virus NS3 protease domain complexed with a synthetic NS4A cofactor peptide. Cell. 1996;87:343–355. doi: 10.1016/s0092-8674(00)81351-3. [DOI] [PubMed] [Google Scholar]
- Lawitz E, Gane EJ. Sofosbuvir for previously untreated chronic hepatitis C infection. N Engl J Med. 2013;369:678–679. doi: 10.1056/NEJMc1307641. [DOI] [PubMed] [Google Scholar]
- Li Q, Brass AL, Ng A, Hu Z, Xavier RJ, Liang TJ, Elledge SJ. A genome-wide genetic screen for host factors required for hepatitis C virus propagation. Proc Natl Acad Sci U S A. 2009;106:16410–16415. doi: 10.1073/pnas.0907439106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liefhebber JM, Brandt BW, Broer R, Spaan WJ, van Leeuwen HC. Hepatitis C virus NS4B carboxy terminal domain is a membrane binding domain. Virol J. 2009;6:62. doi: 10.1186/1743-422X-6-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linde L, Boelz S, Nissim-Rafinia M, Oren YS, Wilschanski M, Yaacov Y, Virgilis D, Neu-Yilik G, Kulozik AE, Kerem E, et al. Nonsense-mediated mRNA decay affects nonsense transcript levels and governs response of cystic fibrosis patients to gentamicin. J Clin Invest. 2007;117:683–692. doi: 10.1172/JCI28523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, He JJ. Cell-cell contact-mediated hepatitis C virus (HCV) transfer, productive infection, and replication and their requirement for HCV receptors. J Virol. 2013;87:8545–8558. doi: 10.1128/JVI.01062-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mamiya N, Worman HJ. Hepatitis C virus core protein binds to a DEAD box RNA helicase. J Biol Chem. 1999;274:15751–15756. doi: 10.1074/jbc.274.22.15751. [DOI] [PubMed] [Google Scholar]
- McLauchlan J, Lemberg MK, Hope G, Martoglio B. Intramembrane proteolysis promotes trafficking of hepatitis C virus core protein to lipid droplets. EMBO J. 2002;21:3980–3988. doi: 10.1093/emboj/cdf414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendell JT, Sharifi NA, Meyers JL, Martinez-Murillo F, Dietz HC. Nonsense surveillance regulates expression of diverse classes of mammalian transcripts and mutes genomic noise. Nat Genet. 2004;36:1073–1078. doi: 10.1038/ng1429. [DOI] [PubMed] [Google Scholar]
- Moradpour D, Penin F, Rice CM. Replication of hepatitis C virus. Nat Rev Microbiol. 2007;5:453–463. doi: 10.1038/nrmicro1645. [DOI] [PubMed] [Google Scholar]
- Murray CL, Rice CM. Turning hepatitis C into a real virus. Annu Rev Microbiol. 2011;65:307–327. doi: 10.1146/annurev-micro-090110-102954. [DOI] [PubMed] [Google Scholar]
- Nakano K, Ando T, Yamagishi M, Yokoyama K, Ishida T, Ohsugi T, Tanaka Y, Brighty DW, Watanabe T. Viral interference with host mRNA surveillance, the nonsense-mediated mRNA decay (NMD) pathway, through a new function of HTLV-1 Rex: implications for retroviral replication. Microbes Infect. 2013;15:491–505. doi: 10.1016/j.micinf.2013.03.006. [DOI] [PubMed] [Google Scholar]
- Nicholson P, Yepiskoposyan H, Metze S, Zamudio Orozco R, Kleinschmidt N, Muhlemann O. Nonsense-mediated mRNA decay in human cells: mechanistic insights, functions beyond quality control and the double-life of NMD factors. Cell Mol Life Sci. 2010;67:677–700. doi: 10.1007/s00018-009-0177-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieva JL, Madan V, Carrasco L. Viroporins: structure and biological functions. Nat Rev Microbiol. 2012;10:563–574. doi: 10.1038/nrmicro2820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okuda M, Li K, Beard MR, Showalter LA, Scholle F, Lemon SM, Weinman SA. Mitochondrial injury, oxidative stress, and antioxidant gene expression are induced by hepatitis C virus core protein. Gastroenterology. 2002;122:366–375. doi: 10.1053/gast.2002.30983. [DOI] [PubMed] [Google Scholar]
- OuYang B, Xie S, Berardi MJ, Zhao X, Dev J, Yu W, Sun B, Chou JJ. Unusual architecture of the p7 channel from hepatitis C virus. Nature. 2013;498:521–525. doi: 10.1038/nature12283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papic N, Maxwell CI, Delker DA, Liu S, Heale BS, Hagedorn CH. RNA-sequencing analysis of 5' capped RNAs identifies many new differentially expressed genes in acute hepatitis C virus infection. Viruses. 2012;4:581–612. doi: 10.3390/v4040581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pichlmair A, Kandasamy K, Alvisi G, Mulhern O, Sacco R, Habjan M, Binder M, Stefanovic A, Eberle CA, Goncalves A, et al. Viral immune modulators perturb the human molecular network by common and unique strategies. Nature. 2012;487:486–490. doi: 10.1038/nature11289. [DOI] [PubMed] [Google Scholar]
- Ploen D, Hafirassou ML, Himmelsbach K, Sauter D, Biniossek ML, Weiss TS, Baumert TF, Schuster C, Hildt E. TIP47 plays a crucial role in the life cycle of hepatitis C virus. J Hepatol. 2013;58:1081–1088. doi: 10.1016/j.jhep.2013.01.022. [DOI] [PubMed] [Google Scholar]
- Ruepp A, Waegele B, Lechner M, Brauner B, Dunger-Kaltenbach I, Fobo G, Frishman G, Montrone C, Mewes HW. CORUM: the comprehensive resource of mammalian protein complexes--2009. Nucleic Acids Res. 2010;38:D497–501. doi: 10.1093/nar/gkp914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwer B, Ren S, Pietschmann T, Kartenbeck J, Kaehlcke K, Bartenschlager R, Yen TS, Ott M. Targeting of hepatitis C virus core protein to mitochondria through a novel C-terminal localization motif. J Virol. 2004;78:7958–7968. doi: 10.1128/JVI.78.15.7958-7968.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimoike T, Mimori S, Tani H, Matsuura Y, Miyamura T. Interaction of hepatitis C virus core protein with viral sense RNA and suppression of its translation. J Virol. 1999;73:9718–9725. doi: 10.1128/jvi.73.12.9718-9725.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh KK, Wachsmuth L, Kulozik AE, Gehring NH. Two mammalian MAGOH genes contribute to exon junction complex composition and nonsense-mediated decay. RNA Biol. 2013;10:1291–1298. doi: 10.4161/rna.25827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sowa ME, Bennett EJ, Gygi SP, Harper JW. Defining the human deubiquitinating enzyme interaction landscape. Cell. 2009;138:389–403. doi: 10.1016/j.cell.2009.04.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinmann E, Penin F, Kallis S, Patel AH, Bartenschlager R, Pietschmann T. Hepatitis C virus p7 protein is crucial for assembly and release of infectious virions. PLoS Pathog. 2007;3:e103. doi: 10.1371/journal.ppat.0030103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinmann E, Pietschmann T. Hepatitis C virus p7-a viroporin crucial for virus assembly and an emerging target for antiviral therapy. Viruses. 2010;2:2078–2095. doi: 10.3390/v2092078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoica R, De Vos KJ, Paillusson S, Mueller S, Sancho RM, Lau KF, Vizcay-Barrena G, Lin WL, Xu YF, Lewis J, et al. ER-mitochondria associations are regulated by the VAPB-PTPIP51 interaction and are disrupted by ALS/FTD-associated TDP-43. Nat Commun. 2014;5:3996. doi: 10.1038/ncomms4996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suratanee A, Rebhan I, Matula P, Kumar A, Kaderali L, Rohr K, Bartenschlager R, Eils R, Konig R. Detecting host factors involved in virus infection by observing the clustering of infected cells in siRNA screening images. Bioinformatics. 2010;26:i653–658. doi: 10.1093/bioinformatics/btq398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sureau A, Gattoni R, Dooghe Y, Stevenin J, Soret J. SC35 autoregulates its expression by promoting splicing events that destabilize its mRNAs. EMBO J. 2001;20:1785–1796. doi: 10.1093/emboj/20.7.1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki R, Sakamoto S, Tsutsumi T, Rikimaru A, Tanaka K, Shimoike T, Moriishi K, Iwasaki T, Mizumoto K, Matsuura Y, et al. Molecular determinants for subcellular localization of hepatitis C virus core protein. J Virol. 2005;79:1271–1281. doi: 10.1128/JVI.79.2.1271-1281.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tai AW, Benita Y, Peng LF, Kim SS, Sakamoto N, Xavier RJ, Chung RT. A functional genomic screen identifies cellular cofactors of hepatitis C virus replication. Cell Host Microbe. 2009;5:298–307. doi: 10.1016/j.chom.2009.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tange TO, Nott A, Moore MJ. The ever-increasing complexities of the exon junction complex. Curr Opin Cell Biol. 2004;16:279–284. doi: 10.1016/j.ceb.2004.03.012. [DOI] [PubMed] [Google Scholar]
- Tardif KD, Mori K, Kaufman RJ, Siddiqui A. Hepatitis C virus suppresses the IRE1-XBP1 pathway of the unfolded protein response. J Biol Chem. 2004;279:17158–17164. doi: 10.1074/jbc.M312144200. [DOI] [PubMed] [Google Scholar]
- Tu H, Gao L, Shi ST, Taylor DR, Yang T, Mircheff AK, Wen Y, Gorbalenya AE, Hwang SB, Lai MM. Hepatitis C virus RNA polymerase and NS5A complex with a SNARE-like protein. Virology. 1999;263:30–41. doi: 10.1006/viro.1999.9893. [DOI] [PubMed] [Google Scholar]
- Vembar SS, Brodsky JL. One step at a time: endoplasmic reticulum-associated degradation. Nat Rev Mol Cell Biol. 2008;9:944–957. doi: 10.1038/nrm2546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogt DA, Camus G, Herker E, Webster BR, Tsou CL, Greene WC, Yen TS, Ott M. Lipid droplet-binding protein TIP47 regulates hepatitis C Virus RNA replication through interaction with the viral NS5A protein. PLoS Pathog. 2013;9:e1003302. doi: 10.1371/journal.ppat.1003302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walters KA, Syder AJ, Lederer SL, Diamond DL, Paeper B, Rice CM, Katze MG. Genomic analysis reveals a potential role for cell cycle perturbation in HCV-mediated apoptosis of cultured hepatocytes. PLoS Pathog. 2009;5:e1000269. doi: 10.1371/journal.ppat.1000269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webster B, Ott M, Greene WC. Evasion of superinfection exclusion and elimination of primary viral RNA by an adapted strain of hepatitis C virus. J Virol. 2013;87:13354–13369. doi: 10.1128/JVI.02465-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willett R, Ungar D, Lupashin V. The Golgi puppet master: COG complex at center stage of membrane trafficking interactions. Histochem Cell Biol. 2013;140:271–283. doi: 10.1007/s00418-013-1117-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang F, Robotham JM, Nelson HB, Irsigler A, Kenworthy R, Tang H. Cyclophilin A is an essential cofactor for hepatitis C virus infection and the principal mediator of cyclosporine resistance in vitro. J Virol. 2008;82:5269–5278. doi: 10.1128/JVI.02614-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.