Abstract
AIM: To investigate the role of SCCA2 and other SCCA1 molecules in the process of hepatitis B virus (HBV) binding to mammalian cells.
METHODS: SCCA1 and SCCA2 were isolated from HepG2. Binding protein (BP) genes were obtained through PCR. Recombinant baculoviruses expressing SCCA1, SCCA2, BP, and different mutants were constructed and utilized to infect mammalian cells to investigate the binding ability of infected cells to HBV.
RESULTS: A SCCA1 gene (A1) was isolated from HepG2, but it appeared to lack the binding ability of infected cells to HBV. Two mutants, A1-BP and BP-A1, were constructed by interchanging the carboxyl terminal of A1 and BP. Cells expressing A1-BP showed an increased virus binding capacity, but not BP-A1. Comparison of A1 sequence with the sequence of BP indicated the presence of only three amino acid changes in the carboxyl terminal, two of them were found in the reactive site loop (RSL) of SCCA1. Primary structure assay revealed that the hydrophobicity of BP and AJ515706 in this domain was strong, but A1 was relatively weak. Changing the aa349 of A1 from low hydrophobic glutamic acid to high hydrophobic valine enhanced HBV binding. In contrast, HBV binding was reduced by changing the aa349 of BP from valine to glutamic acid.
CONCLUSION: The results suggest that the hydrophobicity of RSL of SCCA1 may play an important role in HBV binding to cells.
Keywords: Hepatitis B virus, BP, SCCA1, SCCA2
INTRODUCTION
Hepatitis B virus (HBV) is able to infect only humans or higher primates and shows a strong tropism for liver parenchymal cells. These characteristics and the lack of cell lines that support productive HBV replication in vitro have impeded the identification of HBV receptors. A number of receptor candidates for HBV have been reported, including receptors for immunoglobulin A (IgA), human interleukin-6 and annexin V. But these results are controversial, and no conclusive and convincing biological data could be obtained to fully demonstrate the functional role of these proteins in HBV binding and internalization[1]. Studies using synthetic preS1(21-47) peptides and corresponding antibodies to prevent HBV attachment to HepG2 have identified the preS1 of the L protein as one of the important sites possibly involved in HBV infection[2-4]. A recent candidate for HBV cellular receptors is HBV-binding proteins (BP), isolated from the plasma membranes of HepG2 by using a tetravalent derivative of the preS1(21-47) sequence[5]. HepG2 cells after transfection of BP cDNA showed an increased virus binding capacity, while Chinese hamster ovary cells, which normally do not bind to HBV, acquired susceptibility to HBV binding after transfection. BPs encode for a protein of 390 amino acids. Comparison of BP sequence with the sequence of SCCA1, a member of serpin superfamily cloned by Suminami et al[6], indicated the presence of only three amino acid changes (aa351, aa357 and aa389). BP is also a member of SCCA1. The serpin is a superfamily of structurally well-conserved protein presented in plants, animals, fungi, viruses, and involved in regulating of a variety of biological events[7]. When De Falcon et al[5], isolated HBV-BP from HepG2, 12 independent recombinant clones were sequenced, showing that the sequences corresponding to SCCA1, SCCA2, and HBV-BP were equally represented. SCCA2, another member of serpin, is found to be 91% close to SCCA1. Recently, Moore et al[8], isolated another SCCA1 from HepG2, presenting four amino acid substitutions to BP, and confirmed that transfection of this SCCA1 into mammalian cells resulted in increased HBV binding. But the function of SCCA2 is still not known. In order to understand the role of SCCA2 and other SCCA1 in the process of HBV binding to mammalian cells, recombinant baculoviruses expressing SCCA1, SCCA2, BP, and different mutants were constructed and the binding ability of infected cells to HBV was investigated using infected mammalian cells.
MATERIALS AND METHODS
Cloning and sequencing of SCCA1, SCCA2, and BP
Primers were synthesized according to the sequences of BPs (Table 1) by Shanghai Boya Company. Total RNA was purified from HepG2 cells using TRIzol (Invitrogen). Reverse transcription was primed using BP2. cDNA was used in a nested PCR reaction to amplified target genes (the external pair of primers were BP1 and BP2, and the internal pair of primers were BP3 and BP4). Cycling for both reactions was performed as follows: at 94 °C for 50 s, 25 cycles at 94 °C for 50 s, at 50-58 °C for 50 s, at 72 °C for 1 min, followed by 1 cycle at 72 °C for 10 min. Products of the same length to the target genes were amplified when annealing temperature ranged between 50 °C and 58 °C. PCR-amplified products under different annealing temperatures were recovered respectively using a gel extract kit (Shanghai Huashun Company) and used directly in ligation with pMD18-T (Dalian Takara Company). Sequencing was performed by Shanghai Boya Company. Sequences were compared with published sequences in GenBankTM, and submitted to GenBankTM. The product at 56 °C was one SCCA1 (named A1, accession number AY245781). The product at 52 °C was one SCCA2 (named A2, accession number AY245782). Comparison of A1 sequence with BP sequence indicated only five amino acid changes (aa185, aa202, aa349, aa351, aa389). Then A1 was mutated through PCR to obtain the same BP[5] reported. Inserted genes were confirmed by sequencing.
Table 1.
Sequences of primers.
| Primer | Sequence |
| BP1 | 5'- CAC AGG AGT TCC AGA TCA CAT CGA G -3' |
| BP2 | 5'- CTG GAA GAA AAA GTA CAT TTA TAT GTG GGC -3’ |
| BP3 | 5'- CCG CTA GCT CAC CAT GAA TTC ACT CAG -3’ |
| BP4 | 5'- CCG TCG ACT CTA CGG GGA TGA GAA TCT -3’ |
| BPf | 5'- GGA TCC ATG AAT TCA CTC AGT G -3’ |
| BPr | 5'- CTC GAG CTA CGG GGG TGA GAA TC -3’ |
| EGf1 | 5'- GAT ATC ATG GTG AGC AAG GGC G -3’ |
| EGf2 | 5'- GTC GAC TTA CTT GTA CAG CTC GTC C -3’ |
Construction of recombinant donor plasmids: pFB-A1, pFB-A2, pFB-BP, and pFB-EGFP
A1, A2, and BP were used as templates for a pair of primers (BPf and BPr, Table 1). The amplified products were inserted into the donor plasmid pFastBacl (Invitrogen) to construct pFB-A1 and pFB-A2 and pFB-BP (Figure 1). EGFP gene was amplified from pEGFP (Clontech) using primers Egf1 and Egf2. The obtained fragments were cloned into pFastBacDual (Invitrogen) to generate pFBD-EGFP (Figure 1). The sequences of the inserts were confirmed by sequencing.
Figure 1.
Map of pFB-targets (target is A1, A2 or BP respectively) and pFBD-EGFP.
Generation of recombinant baculoviruses[9]
Recombinant donor plasmids were transformed into DH10Bac competent cells (Invitrogen). Then the transformed cells were plated on LB plates containing X-gal (Sigma), IPTG (Sigma), gentamycin, kanamycin, and tetracycline for selection of white colonies. The white colonies were cultured in liquid LB containing the same three antibiotics, and the transposed bacmids were extracted by alkaline lysis. There were compl-ementary sequences of M13 primer flanking the LacZ gene of bacmids. The length of PCR-amplified products using M13 primer was used to confirm the transposition. Cycling for both reactions was performed as follows: at 94 °C for 50 s, 30 cycles at 94 °C for 50 s, at 55 °C for 50 s, at 72 °C for 3 min, followed by 1 cycle at 72 °C for 10 min.
The resultant bacmids were then used to transfect Sf9 (Invitrogen) cells by using liposome reagent (Cellfectin, Invitrogen) to produce recombinant baculoviruses BacA1, BacA2, BacBP, and BacEGFP. Supernatant of cell culture containing recombinant baculoviruses was collected after 4 d and used to reinfect Sf9 cells to obtain baculoviruses with a higher titer.
Harvest and plaque assay of baculoviruses
Sf9 cells about 80% confluence were seeded in a six-well plate, and incubated at 28 °C for 1 d. Baculoviruses were added to each well of six-well plates at serial dilution and 2 mL 0.8% low-melting agarose (Sigma) prepared using Grace’s medium (Invitrogen) was overlaid after 1 h. The plate was stored at 28 °C for 5 d, and the plaques stained with 1% neutral red were counted.
Gene transfer to mammalian cells using recombinant baculoviruses
About 5×104 mammalian cells were seeded in each well of a 24-well plate. The medium was removed after 12 h at 37 °C, and 500 µL Sf9 culture supernatant containing recombinant baculoviruses was added. New medium was used after 8 h at 37 °C and cultured for 48 h.
HBV binding assay and HBV DNA harvest
About 5×104 mammalian cells were seeded in each well of a 24-well plate. The medium was removed after 16 h at 37 °C, and washed once with D-hanks. To each well, 200 µL Dane’s particles (2×107/well, Beijing Institute of Biological Products) was added and diluted with an appropriate growth medium of mammalian cells and the cells were incubated at 37 °C in a CO2 incubator for 4 h. The entire medium was aspirated off and washed twice with PBS and the cells were collected into an Eppendorf tube. Total HBV DNA was harvested using protease K after separation of cell nuclei and plasma.
Fluorescence PCR analysis
HBV DNA was analyzed using a HBV fluorescence PCR diagnostic kit (Xiamen Xingchuang Company). Cycling was performed in a Roton-gene real-time 2000 PCR system as follows: at 94 °C for 180 s, 50 cycles at 94 °C for 1 s, at 53 °C for 20 s. The data were analyzed and exported by fluorescence PCR analysis software.
Hydrophobicity of primary structure of target genes
Kyte–Doolittle method included in protean program of DNASTAR software was used to analyze hydrophobicity. The average length of amino acid residues used was by default, nine residues. The horizontal line represented the site of amino acids, and the vertical line represented the hydrophobicity index. Positive index meant amino acids were hydrophobic, and negative meant hydrophilic. The larger the absolute value, the stronger the hydrophobicity.
RESULTS
HBV binding to HepG2 cells and WI-38 cells
The following cell lines were used to study the binding to HBV: HepG2, WI-38, BNL 1ME A.7R.1, and CHO. Cells were incubated with an appropriate medium containing Dane’s particles. Cell-associated HBV DNA was harvested after being washed with PBS and quantified using real-time fluorescence PCR (Table 2). HepG2 cells could bind to HBV in some degree, and some non-specific bindings to viruses were found in other non-tropistic cell lines. WI-38 cells were observed to have a comparatively higher efficiency of report gene transfer and expression[10]. So WI-38 cells were selected as target cells in following research.
Table 2.
Comparison of the relative binding to HBV in different cell lines.
| Cell line | Source | HBV DNA (copies/sample) |
| HepG2 | Human/hepatocellular carcinoma | 263382±16302 |
| WI-38 | Human/lung fibroblast | 186215±15236 |
| BNL 1ME A.7R.1 | Mice/liver | 169221±13489 |
| CHO | Mice/ovary | 105400±25076 |
HBV infection to WI-38 cells expressed different target proteins
Recombinant baculoviruses (BacA1, BacA2, BacBP, and BacEGFP) infected WI-38 cells, respectively. Dane’s particles were added to the cells after 48 h. HBV DNA was extracted after 4 h and quantified (Figure 2). We observed no increase in cell-associated virus DNA in cells infected with BacA1 and BacA2, a significant increase in cells infected with BacBP (P<0.05) compared with mock infected cells (infected with BacEGFP) and uninfected cells (blank control), suggesting that expression of BP could make WI-38 acquire susceptibility to HBV binding, but not A1 and A2.
Figure 2.
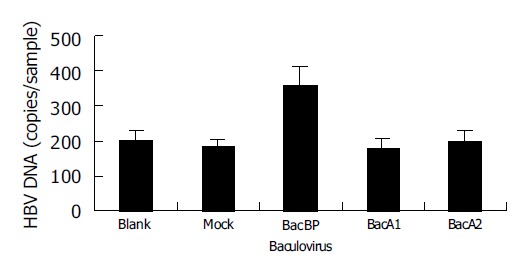
HBV binding to WI-38 cells infected with different recombinant balculoviruses; blank: uninfected cells; mock: BacEGFP infected cells.
SCCA1 protein sequence comparison
The expression of A1 could not enhance HBV binding to cells. Moore et al[8], reported that they isolated a SCCA1 gene (GenBankTM accession number AJ515706) from HepG2 cells by PCR and transfection of this SCCA1 into mammalian cells (both hepatocyte-derived and of non-hepatocyte origin) resulted in increased HBV binding. Site-directed mutagenesis in RSL was utilized to create two AJ515706 gene mutants, P3 and P14. There was no significant difference in binding between the wild type SCCA1 and either RSL mutant. Thus, except A1, the other four SCCA1 protein (BP, AJ515706, P3, P14) expression could increase cell binding to HBV. Based on the sequence comparison (Table 3), amino acid changes of A1 at aa185, aa202, aa349 and aa351 were specific.
Table 3.
Amino acid sequence differences among five SCCA1 proteins.
| Amino acid residue | 16 | 47 | 105 | 185 | 202 | 341 | 349 | 351 | 352 | 389 |
| BP | F | D | N | Q | P | A | V | A | F | P |
| AJ515706 | S | N | T | Q | P | A | V | A | F | S |
| P3 | S | N | T | Q | P | A | V | A | A | S |
| P14 | S | N | T | Q | P | R | V | A | F | S |
| A1 | F | D | N | R | S | A | E | G | F | S |
Construction of chimeric mutants of BP and A1 and their binding to HBV
The function of serpin involved exposure of the reactive site loop (RSL) to the active site of proteinases[7]. In order to understand the amino acid changes involved in HBV binding ability, two chimeric mutants, BP-A1 and A1-BP, were constructed by interchanging the fragments of BP and A1 after aa342 (Figure 3).
Figure 3.
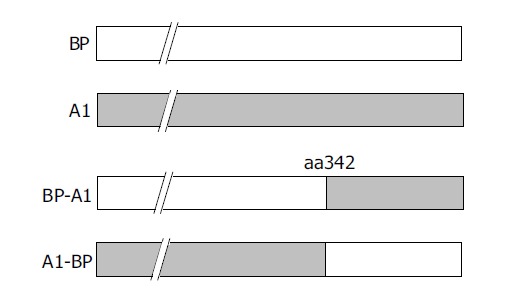
Construction of chimeric mutants of BP and A1.
Then, recombinant baculoviruses containing BP-A1 and A1-BP were generated and infected WI-38 cells were used to investigate their functions in HBV binding (Figure 4). It showed that cells expressing A1-BP could bind to HBV, but BP-A1 could not. Apparently, the susceptibility of SCCA1 to HBV was significantly affected by amino acid changes after aa342.
Figure 4.
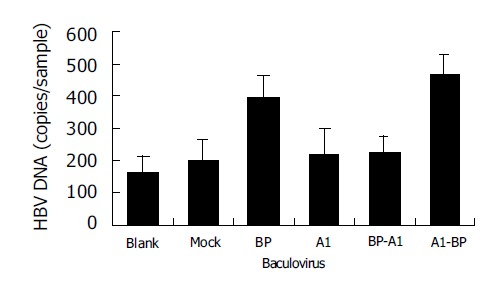
HBV binding to WI-38 cells expressing chimeric mutants of BP and A1. Blank: uninfected cells; mock: BacEGFP infected cells.
Hydrophobicity/hydrophilicity analysis of primary structure of several SCCA1 protein
Comparison of hydrophobicity/hydrophilicity of five SCCA1 proteins (BP, AJ515706, P3, P14, and A1) found that the four proteins with HBV binding ability displayed a relatively high hydrophobicity, and the peak value of hydrophobicity of these proteins was above 2.0. But the peak value of A1, the protein without HBV binding ability, was just 1.07 (Figure 5).
Figure 5.
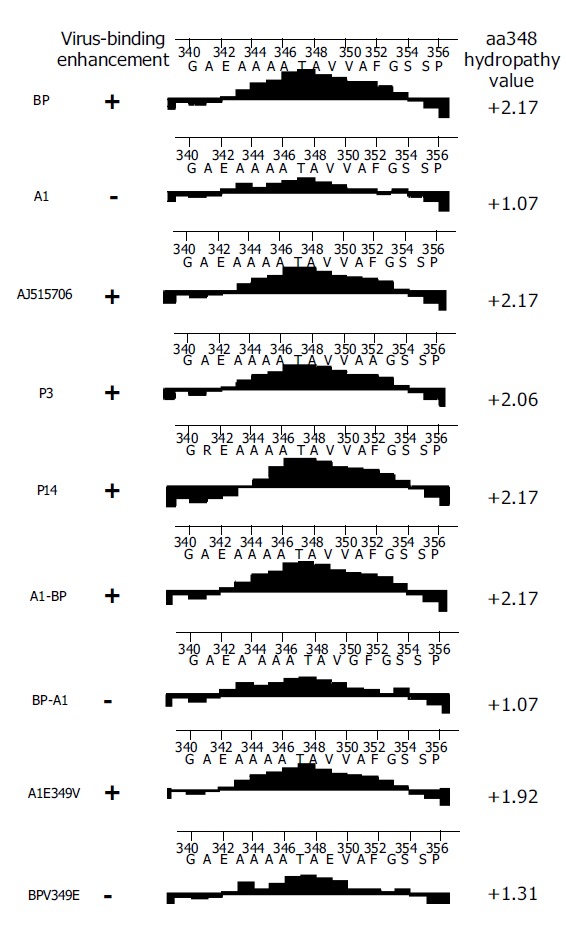
Hydrophobicity analysis of several SCCA1 proteins.
aa349 site-directed mutagenesis of BP and A1 affected HBV binding ability
A1 differed from all the other four proteins for the presence of two residue mutations, V349E and A351G between aa344 and aa354. Notably, at aa349 of A1, it was a low hydrophobic glutamine residue, but not a high hydrophobic valine residue. It was assumed that the hydrophobicity between aa344 and aa354 might have played a role in the process of SCCA1 binding to HBV. To elucidate this hypothesis, we constructed BPV349E and A1E349V utilizing the site-directed mutagenesis method at the aa349 of BP and A1, and found that a deletion mutant BPN342 deleted the fragments after BP aa342 (Figure 6). The hydrophobicity of BPV349E between aa344 and aa354 was markedly reduced, but A1E349V was reverse (Figure 5). Recombinant baculoviruses of these mutants were constructed and infected WI-38 cells were used to analyze the binding of cells to HBV. The results revealed that the expression of A1E349V could enhance cell binding to HBV, but BPV349E could not, suggesting that aa349 was of pivotal importance to the function of SCCA1 (Figure 7). Surprisingly, in the case of BPN342 expression, some virus-cell binding ability still remained, implying that the binding of BP to HBV might involve several sites, or aa344-aa354 region was not directly involved in the binding. But the altered hydrophobicity of this region might indirectly lead to conformational changes of HBV binding sites.
Figure 6.
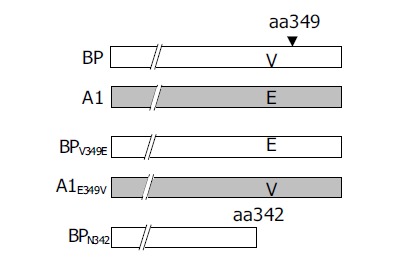
Construction of different SCCA1 mutants.
Figure 7.
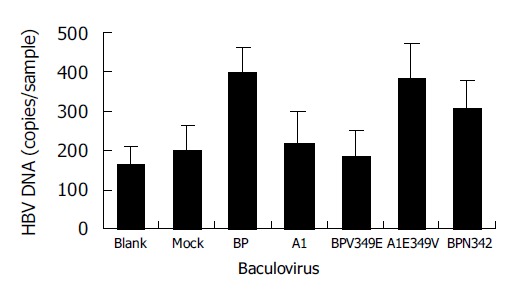
HBV binding to WI-38 cells expressing site-mutants of BP and A1. Blank: uninfected cells; mock: BacEGFP infected cells.
DISCUSSION
BP and AJ515706 are two SCCA1 genes isolated from HepG2 by De Falco et al[5], and Moore et al[8]. Mammalian cells expressing both genes showed an increased HBV binding capacity. But in this study, A1 isolated from HepG2 did not show the same capacity. A2, a SCCA2, which was found to be 91% homologous to SCCA1, also lacked this capacity.
BP, AJ515706 and A1 are of a high sequence homology, but they only have 4-7 different amino acids (Table 3). In order to understand which amino changes were related to HBV binding capacity, two mosaics, BP-A1 and A1-BP, were constructed by interchanging the fragments of BP and A1 at downstream aa342. The results showed that cells expressing A1-BP could bind to HBV, but BP-A1 could not. It seems that the changes of three amino acids between A1 and BP after aa342 were important in the binding to HBV. Two of the amino acid changes (aa349 and aa351) reside in RSL. The hydrophobicity of A1 in RSL was relatively mild, but that of BP and AJ515706 was strong. We created two mutants, A1E349V and BPV349E, by site-directed mutagenesis at the aa349 of BP and A1. The mild hydrophobic glutamic acid of A1 was changed into high hydrophobic valine. Correspondingly, valine of BP was changed into glutamic acid. Cells expressing A1E349V acquired HBV binding ability, but BPV349E lost it. The results revealed that there was an important correlation between hydrophobicity of SCCA1 RSL and HBV binding capacity. The decreased hydrophobicity in this region would lead to the reduction in levels of bound viruses.
In the hydrophobic conformation, DHBV particles were able to bind to liposome and intact cells[11]. The strong hydrophobicity of BP and AJ515706 in RSL might help them to form stable complexes with HBV or some components of cells, increase attachment to and entry in cells.
Serpin is a conformation-dependent protein. The relatio-nship between structure and function is very special. Until now, this has not been fully understood. The RSL of serpin not only plays an important role in the inhibitory activity of serpin, but also affects the whole conformation of serpin. The binding to ligands and the reciprocity between proteins can occur at any site of serpin. The conformational changes on RSL may influence other regions of serpin interacting with HBV. So, the RSL and its hydrophobicity are important in the process of cell binding to HBV, but other regions on BP may also interact with HBV. In this study, the result of BPN342 might favor this explanation. Expression of BPN342 and deletion mutants of RSL, did not result in abrogation of enhanced virus binding to infected WI-38, suggesting the complicated effect of serpin structure.
HepG2 is a cell line from hepatocellular carcinoma. The surface components of each cell are not totally identical. Some different SCCA1 genes were isolated from HepG2 using the similar methods by De Falco et al[5], and Moore et al[8]. These SCCA1 genes have a high sequence homology, but their functions in HBV binding differ markedly. In the normal hepatocytes, are there different SCCA1 genes that express simultaneously? Does the expression of different SCCA1 have a relationship with the susceptivity of cells to HBV and the cytopathological changes after HBV infection? Further research on these problems may contribute to understanding the mechanism of interaction between hepatocytes and HBV.
Footnotes
Supported by the Cross-Century Talent Training Program of Ministry of Education, China
References
- 1.Chen M, Zhang J, Chen MC, Xia NS. Progress in HBV receptor research. Bingdu Xuebao. 2002;18:185–193. [Google Scholar]
- 2.Neurath AR, Kent SB, Strick N, Parker K. Identification and chemical synthesis of a host cell receptor binding site on hepatitis B virus. Cell. 1986;46:429–436. doi: 10.1016/0092-8674(86)90663-x. [DOI] [PubMed] [Google Scholar]
- 3.Qiao M, Macnaughton TB, Gowans EJ. Adsorption and penetration of hepatitis B virus in a nonpermissive cell line. Virology. 1994;201:356–363. doi: 10.1006/viro.1994.1301. [DOI] [PubMed] [Google Scholar]
- 4.Pontisso P, Ruvoletto MG, Tiribelli C, Gerlich WH, Ruol A, Alberti A. The preS1 domain of hepatitis B virus and IgA cross-react in their binding to the hepatocyte surface. J Gen Virol. 1992;73(Pt 8):2041–2045. doi: 10.1099/0022-1317-73-8-2041. [DOI] [PubMed] [Google Scholar]
- 5.De Falco S, Ruvoletto MG, Verdoliva A, Ruvo M, Raucci A, Marino M, Senatore S, Cassani G, Alberti A, Pontisso P, et al. Cloning and expression of a novel hepatitis B virus-binding protein from HepG2 cells. J Biol Chem. 2001;276:36613–36623. doi: 10.1074/jbc.M102377200. [DOI] [PubMed] [Google Scholar]
- 6.Suminami Y, Kishi F, Sekiguchi K, Kato H. Squamous cell carcinoma antigen is a new member of the serine protease inhibitors. Biochem Biophys Res Commun. 1991;181:51–58. doi: 10.1016/s0006-291x(05)81380-4. [DOI] [PubMed] [Google Scholar]
- 7.Gettins PG. Serpin structure, mechanism, and function. Chem Rev. 2002;102:4751–4804. doi: 10.1021/cr010170+. [DOI] [PubMed] [Google Scholar]
- 8.Moore PL, Ong S, Harrison TJ. Squamous cell carcinoma antigen 1-mediated binding of hepatitis B virus to hepatocytes does not involve the hepatic serpin clearance system. J Biol Chem. 2003;278:46709–46717. doi: 10.1074/jbc.M302842200. [DOI] [PubMed] [Google Scholar]
- 9.Duisit G, Saleun S, Douthe S, Barsoum J, Chadeuf G, Moullier P. Baculovirus vector requires electrostatic interactions including heparan sulfate for efficient gene transfer in mammalian cells. J Gene Med. 1999;1:93–102. doi: 10.1002/(SICI)1521-2254(199903/04)1:2<93::AID-JGM19>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- 10.Xu CY, Cheng T, Lu WX, Chen M, Wu T, Wang YB, Zhang J, Xia NS. Mammalian gene-transfer and expression efficiencies of baculovirus bacV-CMV-EGFPA. ShengWu GongCheng XueBao. 2004;20:73–77. [PubMed] [Google Scholar]
- 11.Grgacic EV, Schaller H. A metastable form of the large envelope protein of duck hepatitis B virus: low-pH release results in a transition to a hydrophobic, potentially fusogenic conformation. J Virol. 2000;74:5116–5122. doi: 10.1128/jvi.74.11.5116-5122.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]


