Abstract
AIM: To identify the genes differentially expressed in the regenerating rat liver of 0-4-8-12 h short interval successive partial hepatectomy (SISPH) and to analyze their expression profiles.
METHODS: Five hundred and fifty-one elements screened from subtractive cDNA libraries were made into a cDNA microarray (cDNA chip). Extensive gene expression analysis following 0-4-8-12 h SISPH was conducted by microarray.
RESULTS: One hundred and eighty-three elements were selected, which were either up- or down-regulated more than 2-fold at one or more time points after SISPH. Cluster analysis and generalization analysis showed that there were five distinct temporal patterns of gene expression. Eighty-six genes were unreported, associated with liver regeneration (LR).
CONCLUSION: Microarray analysis shows that the down regulated genes are much more than the up-regulated ones in SISPH; the numbers of genes expressed consistently are fewer than that expressed immediately; the genes expressed in high abundance are much fewer than that increased 2-5-fold. The comparison of SISPH with partial hepatectomy (PH) shows that the expression trends of most genes in SISPH and in PH are similar, but the expression of 43 genes is specifically altered in SISPH.
Keywords: SISPH, Liver regeneration
INTRODUCTION
Liver has a capacity to regain original mass after partial hepatectomy (PH)[1-6]. It was confirmed that a great deal of genes participate in liver regeneration (LR), and the genes expressed in the different phases of LR were various, and that hepatocytes progressing from G0 phase to G1 phase occurred in 2-6 h after PH[7,8]. Despite numerous related papers, it is still not thoroughly elucidated how many genes participate in LR and what its molecular mechanism is[9-15]. To get sight into the mechanism of LR, we established the short interval successive partial hepatectomy (SISPH) model[16], which could offer the useful materials for studying specific gene expression at various crucial points of LR[17,18]. To seek some novel differential display genes responsible for LR, the method of subtracted suppression hybridization was used to obtain a bulk of up-regulated and down-regulated expression sequence tags (ESTs) in the regenerating rat liver. With development of cDNA microarray technology, genomewide expression of thousands and thousands of activated or suppressed genes can be simultaneously analyzed under various biological conditions[19-21]. To further explore the genes participating in LR, the present study successfully identified the gene expression profile in regenerating liver following 0-4-8-12 h SISPH by microarray, and some important information was achieved by analyzing the data using Microsoft Excel and GeneSpring.
MATERIALS AND METHODS
Rat model of SISPH
Male and female Sprague-Dawley (SD) rats, aged 10-12 wk, weighting 200-220 g, were raised in Experimental Animal Center of Henan Normal University (HNU). According to Xu et al, lobus external sinister, lobus centralis sinister, lobus centralis and lobus dexter were removed one by one at four different time points of 0, 4, 8, 12 h, which is named as 0-4-8-12 h SISPH.
Sample preparation and RNA isolation
The removed liver lobus was rinsed in cold 1×PBS and immersed and stored in -80 °C refrigerator for RNA and protein extraction. Total RNA was isolated from frozen liver lobus according to the manual of TRIzol kit of Invitrogen. In brief, 50-100 mg liver tissue was homogenized in 1 mL TRIzol reagent containing phenol and guanidinium isothiocyanate/cationic detergent, followed by phenol-chloroform extraction and isopropyl alcohol precipitation. The quantity and integrity of total RNA was examined by ultraviolet spectrometer and denaturing formaldehyde agarose electrophoresis stained by ethidium bromide.
Subtracted cDNA library construction and screening
cDNA subtracted libraries were generated from total RNA by PCR-SelectTM cDNA Subtraction kit (Clontech) following the manufacturer’s instruction. Briefly, total RNA was transcribed into double cDNA strands and digested with restriction enzymes, followed by subtracted hybridization with drivers and testers. Finally with suppression polymerase chain reaction (PCR), differential ESTs were used to construct subtracted cDNA library. The latter was cloned into T/A vector and screened by PCR with nested primer 1 and 2.
cDNA microarray construction
cDNA fragments amplified by PCR with nested PCR primers 1 and 2 and purified by NaAc/isopropyl alcohol were spotted onto glass slides (Biostar) with the help of ProSys-5510A spotting machine following designed project. Then the gene chips were ready by hydration and blockage and drying. Totally 1152 elements (double spot chip) including 50 control system (8 negative control, 12 blank control, 30 internal control) and 551 target genes to be studied comprised 8 submatrixes (12*12) occupying 9 mm×18 mm (Biostar). Then the gene chips were ready by hydration, blockage and drying.
Fluorescence-labeled cDNA preparation
RNA isolated from rat livers before SISPH was ready for a reference for all cDNA microarray analyses. Totally denatured RNA was reversely transcribed with cy3-conjugated dCTP (control group) and cy5-conjugated dCTP (test group) (Amersham-Pharmacia Biotech) using MMLV reverse transcriptase (Promega) with olig (dT) primer. After bath incubation for 2 h, labeled buffer I and II were subsequently added to the reaction. The control group and test group were mingled together symmetrically and stored in the dark until use.
Hybridization and scanning
The glass slices were prehybridized at 42 °C for 5-6 h in hybridization buffer containing freshly cooked shared salmon sperm DNA. The labeled denatured probe was hybridized against cDNA microarrays with overnight (16-18 h) incubation at 42 °C. The slides were then washed twice with 2×SSC containing 0.5% SDS for 5 min at room temperature, once with 0.2×SSC containing 0.5% SDS at 60 °C for 10 min, and finally with 0.2×SSC at 60 °C for 10 min. The slices were exposed to photographer. Hybridized images were scanned by a fluorescence laser scanning device, Gene Pix 4000A (Axon Instruments, Inc., Foster City, CA). At least two hybridizations were performed at each time point. In addition, a semiquantitative inspection of the hybridization results was performed for (1) green signal (down-regulation); (2) yellow signal (no obvious regulation); (3) red signal (up-regulation).
Data analysis
The cy3 and cy5 signal intensities were quantified by Gene Pix Pro 3.0 software (Axon Instruments, Inc., Foster City, CA). Subsequently, we normalized the obtained numerical data with classical linear regression techniques. In brief, quantified cy3 and cy5 signal intensities were obtained when foreground signal intensities were deducted by background signal intensities and cy5 signal intensities was replaced by 200 when it was <200. When Ri (Ri = cy5/cy3) was between 0.1 and 10, Ri was taken as logarithms base natural to generate Ri’[log (Ri)] and ND was taken by EXP (R) (averaged Ri’). The modified cy3* was generated by ND multiplying cy3 and was replaced by 200 when it was <200. The ratio was expressed by cy5/cy3*. Therefore, we selected genes whose ratio was more than 2 or less than 0.5 representing a 2-fold difference in expression level.
To analyze the selected gene expression data, we applied κ-means cluster analysis, and performed GeneMaths hierarchical clustering to appraise the number of groups. Whole analyses were executed with Microsoft Excel (Microsoft, Redmond, WA) and GeneSpring (Silicon Genetics, San Carlos, CA).
Chromosome location and functional prediction of the novel ESTs
All the clones of ESTs were sequenced by TaKaLa and blasted in NCBI. The unreported ESTs were searched at http://www.ncbi.nlm.nih.gov/genomeguide/rat/index.html/ for gene location in chromosome and gene corresponding whole genome shotgun. In virtue of rat genome database, electronic cloning and chromosome location of the unreported ESTs representing novel cDNA full-length were performed successfully (data not shown). By delivering the clones sequences to http://genes.mit.edu/GENSCAN.html/, we acquired corresponding coding domain sequences (CDS) supported by the ESTs. Compared with known proteins by BLASTP (http://www.ncbi.nlm.nih.gov/BLAST/), their function and accession number were achieved.
RESULTS
Category of genes identified in the 0-4-8-12 h SISPH
One hundred and eighty-three elements altered by more than 2-fold intensity at least at one time point in the 0-4-8-12 h SISPH were identified and selected, of which, 77 were up regulated and 106 were down regulated. Eighty-six of them belonged to the unreported novel genes, and the other 97 were reported, of which quite a few genes had been previously reported to be involved in LR. Following the functions of the reported genes and the time points at which they attained maximum up or down-regulation, they were respectively categorized into 21 groups: the genes involved in stress response, in glycometabolism, in stearoyl metabolism, in oxidative and reductive response, genes encoding regulation proteins, glycoproteins, lipid-proteins, nucleolar proteins, receptors, factors, hemoglobins, immunological proteises, cytoskelets, marker proteins, amino acid enzymes, proteolytic enzymes, proteinase inhibitors, phosphorylase, phosphatases, synthases and transferases (Table 1).
Table 1.
Genes related to LR altered in SISPH.
| No. | Gene description | Fold difference |
| Unreported genes | ||
| 1 | CH230-206C20 | 0.3 |
| 2 | CH230-403C20 | 0.5 |
| 3 | CH230-329D3 | 2.2 |
| 4 | CH230-372C24 | 0.1 |
| 5 | CH230-7A22 | 0.1 |
| 6 | CH230-11N5 | 2.1 |
| 7 | CH230-4L11 | 3.5 |
| 8 | CH230-211F21 | 0.4 |
| 9 | CH230-155H3 | 0.5 |
| 10 | CG31759-PA | 2.9 |
| 11 | Citb585c7 | 0.2 |
| 12 | D930042H13 | 0.3 |
| 13 | DNA segment of Chr 1 (Wsu94) | 3.8 |
| 14 | DNA segment of Chr 17 (Wsu94) | 2 |
| 15 | KIAA0205 | 0.4 |
| 16 | KIAA1376 | 0.4 |
| 17 | LOC303588 | 2 |
| 18 | LOC333273 | 2.9 |
| 19 | LRRP Cc1-27 | 0.3 |
| 20 | LRRP Aa2-028 | 3.1 |
| 21 | LRRP Aa2-111 | 0.2 |
| 22 | LRRP Aa2-174 | 0.3 |
| 23 | LRRP Aa2-258 | 0.5 |
| 24 | LRRP Ab1-021 | 9.1 |
| 25 | LRRP Ab1-022 | 0.5 |
| 26 | LRRP Ab1-114 | 3.5 |
| 27 | LRRP Ab1-216 | 3.5 |
| 28 | LRRP Ab1-334 | 4.4 |
| 29 | LRRP Ab2-001 | 0.3 |
| 30 | LRRP Ab2-034 | 0.3 |
| 31 | LRRP Ab2-037 | 0.5 |
| 32 | LRRP Ab2-051 | 2.1 |
| 33 | LRRP Ab2-131 | 0.3 |
| 34 | LRRP Ab2-132 | 0.4 |
| 35 | LRRP Ab2-225 | 0.3 |
| 36 | LRRP Ab2-232 | 0.2 |
| 37 | LRRP Ab2-255 | 0.2 |
| 38 | LRRP Ab2-371 | 0.1 |
| 39 | LRRP Ab2-389 | 0.5 |
| 40 | LRRP Ab2-402 | 0.2 |
| 41 | LRRP Ab2-416 | 0.3 |
| 42 | LRRP Ab2-417 | 0.2 |
| 43 | LRRP Ab2-427 | 0.5 |
| 44 | LRRP Ab2-440 | 0.3 |
| 45 | LRRP Ac1-060 | 0.3 |
| 46 | LRRP Ac1-158 | 2.1 |
| 47 | LRRP Ac1-177 | 7.3 |
| 48 | LRRP Ac1-233 | 5.6 |
| 49 | LRRP Ac1-873 | 4.6 |
| 50 | LRRP Ac2-061 | 6.4 |
| 51 | LRRP Ac2-067 | 2.6 |
| 52 | LRRP Ac2-143 | 2 |
| 53 | LRRP Ac2-210 | 2.3 |
| 54 | LRRP Ba1-647 | 3.3 |
| 55 | LRRP Bm403207 | 7.7 |
| 56 | LRRP Cb1-727 | 0.4 |
| 57 | LRRP Cb1-739 | 2.2 |
| 58 | LRRP Cc1-27 | 0.3 |
| 59 | LRRP Cc1-38 | 0.1 |
| 60 | LRRP Cc1-9 | 3.5 |
| 61 | LRRP zbs559 | 3.5 |
| 62 | LRRPAb2-132 | 0.2 |
| 63 | MGC10946 | 2.8 |
| 64 | MGC38937 | 2.4 |
| 65 | MGC5178 | 0.4 |
| 66 | mKIAA0665 | 2.4 |
| 67 | RIKEN 1300002A08 | 0.2 |
| 68 | RIKEN 1600027G01 | 0.3 |
| 69 | RP11-281N10 | 0.3 |
| 70 | RP11-586K2 | 2.6 |
| 71 | RP23-100C5 | 2 |
| 72 | RP23-165H7 | 0.5 |
| 73 | RP23-235O1 | 0.4 |
| 74 | RP23-28G130 | 2.3 |
| 75 | RP23-32O9 | 2.3 |
| 76 | RP23-417P22 | 0.3 |
| 77 | RP23-476D16 | 1.6 |
| 78 | RP23-480P21 | 2.9 |
| 79 | RP23-92K11 | 2.9 |
| 80 | RP24-347B22 | 0.3 |
| 81 | Rp32-28p17 | 0.3 |
| 82 | U48828 | 2.4 |
| 83 | Adult male liver cDNA | 0.1 |
| 84 | Mouse mRNA | 0.4 |
| 85 | Open reading frame 31 | 0.5 |
| 86 | 13 d embryo liver cDNA | 3.6 |
| Stress response | ||
| 87 | Acute-phase protein alpha-1-inhibitor 3 | 0.3 |
| 88 | alpha-1 major acute phase protein prepeptide | 6.8 |
| 89 | Angiotensinogen (Agt) | 11.5 |
| 90 | Kininogen | 2.5 |
| 91 | T-kininogen (Kng) | 5 |
| Glycometabolism | ||
| 92 | alpha enolase (Enolase 1) | 2.7 |
| 93 | Isocitrate dehydrogenase 1 (Idh1) | 0.1 |
| 94 | 3-Phosphoglycerate dehydrogenase | 2.2 |
| Fatty and stearoyl metabolism | ||
| 95 | Bile acid CoA ligase | 2 |
| 96 | Malonyl-CoA decarboxylase | 0.4 |
| 97 | Methylmalonate semialdehyde dehydrogenase | 0.4 |
| 98 | P450 cholesterol 7-alpha-hydroxylase (P450 VII) | 0.1 |
| 99 | NAD (P) dependent steroid dehydrogenase | 0.1 |
| 100 | 3-alpha-hydroxysteroid dehydrogenase | 0.1 |
| Oxidation and reduction | ||
| 101 | Acyl-coA oxidase | 0.5 |
| 102 | Alcohol dehydrogenase (ADH) | 0.1 |
| 103 | Cytochrome b5 (Cyb5) | 0.3 |
| 104 | Cytochrome P450 | 0.3 |
| 105 | Cytochrome P450 15-beta gene (Cyp2c12) | 0.4 |
| 106 | Cytochrome P450 2E1 | 0.2 |
| 107 | Cytochrome P450, 2c39 (Cyp2c39) | 0.3 |
| 108 | Cytochrome P450 (PNCN inducible, Cyp3A1) | 0.4 |
| 109 | Flavin-containing monooxygenase 1 (Fmo1) | 0.1 |
| 110 | Peroxisomal sarcosine oxidase (PSO) | 0.1 |
| 111 | Plasma selenoprotein P1 (Sepp1) | 0.4 |
| 112 | Small subunit precursor of RuBPCase | 0.1 |
| Regulation proteins | ||
| 113 | G-protein beta polypeptide 2-like 1 (Gnb2l1) | 2.1 |
| 114 | II-Protein with tetratricopeptide repeats 3 | 0.2 |
| 115 | RAKb | 0.2 |
| Glycoproteins | ||
| 116 | apha-1-B glycoprotein (A1bg) | 0.3 |
| 117 | Fibronectin 1 (Fn1) | 3 |
| 118 | Fibrinogen, gamma polypeptide (Fgg) | 8 |
| 119 | Myelin-associated glycoprotein (L-MAG) | 8.4 |
| 120 | UDP-glucuronosyltransferase 2B3 (Udpgt) | 0.4 |
| 121 | TRAM1 | 0.3 |
| Lipid-proteins | ||
| 122 | Adipose differentiation-related protein | 4.6 |
| 123 | Solute carrier family 20, mem 1 (Slc20a1) | 0.4 |
| Nucleolar proteins | ||
| 124 | Damage-specific DNA binding protein 1 (Ddb1) | 0.5 |
| 125 | Nuclear protein 1 (Nupr1) | 2.9 |
| 126 | Nucleolar protein family A, mem 2 | 2.9 |
| 127 | ORF2 endonuclease and Rnase H | 0.5 |
| 128 | RNase A family 4 | 0.3 |
| Receptors | ||
| 129 | ATP-binding cassette, sub-family B | 0.5 |
| 130 | ATP-binding cassette, sub-family C | 0.1 |
| 131 | Interleukin 1 receptor, type I (Il1r1) | 7.3 |
| 132 | Nuclear receptor subfamily 0, member 2 (Nr0b2) | 0.4 |
| Factors | ||
| 133 | Amphoterin | 0.5 |
| 134 | Angiogenin | 0.4 |
| 135 | Eukaryotic translation initiation factor 4A1 | 3.5 |
| 136 | Early growth response factor 1 (Egr1) | 4.3 |
| 137 | Neuropeptide Y (Npy) | 11.3 |
| 138 | NF-E2-related factor 2 (Nfe2l2) | 0.3 |
| 139 | Pre-B-cell colony-enhancing factor (Pbef) | 5.3 |
| 140 | ADP-ribosylation factor 3 (Arf3) | 0.8 |
| 141 | Angiopoietin-like 3 | 0.3 |
| Hemoglobins | ||
| 142 | Hemoglobin, alpha 1 (Hba1) | 2.4 |
| 143 | Hemoglobin beta chain (Hbb) | 2.5 |
| Immunological proteises | ||
| 144 | Achaete-scute complex homolog-like 1 (Ascl1) | 0.4 |
| 145 | Complement component 5 (C5) | 19.4 |
| 146 | Complement component 6 (C6) | 0.4 |
| 147 | Immunoglobulin C kappa gene | 0.3 |
| 148 | JE/MCP-1 | 2.5 |
| Cytoskelets | ||
| 149 | Actin -beta | 2.3 |
| 150 | Actin gamma | 5.1 |
| 151 | Keratin 8 (Krt8) | 2.8 |
| Marker proteins | ||
| 152 | alpha globin | 2.4 |
| 153 | Amyloid a-5 protein | 53.9 |
| 154 | Subchromosomal transferable fragment | 0.4 |
| Amino acid enzymes | ||
| 155 | Arginase 1 (Arg1) | 6.4 |
| 156 | Argininosuccinate lyase (Asl) | 5.6 |
| 157 | Cytosolic aspartate aminotransferase | 6.8 |
| 158 | 2-Hydroxyphytanoyl-CoA lyase (Hpcl2) | 0.3 |
| Proteolytic enzymes | ||
| 159 | Cathepsin C (Ctsc) | 0.4 |
| 160 | Cathepsin D (Ctsd) | 0.5 |
| 161 | Coagulation factor 2 (F2) | 0.5 |
| 162 | Neutrophil collagenase (Mmp8) | 0.2 |
| Proteinase inhibitors | ||
| 163 | alpha-2-macroglobulin (A2m) | 5.1 |
| 164 | alpha-trypsin inhibitor heavy chain | 0.4 |
| 165 | Contrapsin-like protease inhibitor related protein | 6.1 |
| 166 | Leuserpin-2 (Serpind1) | 0.3 |
| 167 | Serine protease inhibitor 1 | 5.6 |
| Phosphorylase | ||
| 168 | CDK103 | 1.2 |
| 169 | Purine-nucleoside phosphorylase | 0.3 |
| Phosphatases | ||
| 170 | Phosphatidylserine-specific phospholipase A1 | 4.9 |
| 171 | Pyrophosphatase/phosphodiesterase 1(Enpp1) | 3.1 |
| 172 | Protein phosphatase 1 (GL-subunit) | 0.2 |
| Synthase | ||
| 173 | Carbamyl phosphate synthetase I | 2.5 |
| 174 | Fatty acid elongase 1 (rELO1) | 0.1 |
| 175 | Glutamyl-prolyl-tRNA synthetase (Eprs) | 2.2 |
| 176 | RNA cyclase | 2 |
| Transferases | ||
| 177 | Glutathione S-transferase, alpha 1 (Gsta1) | 0.3 |
| 178 | Glutathione S-transferase, type 3 (Yb3) (Gstm3) | 0.5 |
| 179 | Glutathione S-transferase Y(b) subunit | 0.5 |
| 180 | Serine hydroxymethyl transferase 1 | 0.4 |
| 181 | Sialyltransferase 1 (Siat1) | 0.2 |
| 182 | Sulfotransferase K2 | 0.2 |
| 183 | UDP-glucuronosyltransferase 2, mem 5 (Ugt2b5) | 0.5 |
Gene expression differences at the various time points of the 0-4-8-12 h SISPH
The gene expression differences induced or suppressed at 4, 4-8, 4-12, 8, 8-12 h of SISPH are shown in Figure 1. At 4 h of SISPH, 5 genes expressed up over 2-fold (Figure 1A); at 4-8 h of SISPH, 2 genes expressed up over 2-fold (Figure 1A); at 4-12 h of SISPH, 29 genes expressed up (Figures 1B-1E), of which 10 genes up over 5-fold (Figure 1B), and 7 genes up over 10-fold (Figure 1C), and 10 genes expressed down (Figures 1D and Figures 1E); at 8 h of SISPH, 2 genes expressed up (Figure 1G) and 7 genes down (Figure 1F); at 8-12 h of SISPH, 25 genes expressed down (Figures 1H and Figures 1I).
Figure 1.
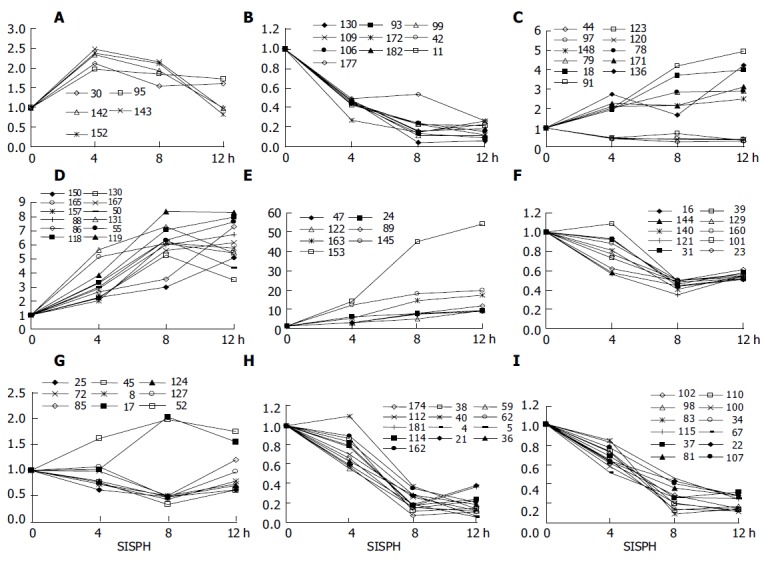
Gene expression differences in 0-4-8-12 SISPH. A: 4 h, 4-8 h; B-E: 4-12 h; F and G: 8 h; H and I: 8-12 h.
Gene expression level in the 0-4-8-12 h SISPH
According to the up- and down-regulated density of genes in the 0-4-8-12 h SISPH, we categorized them into three groups: (1) 106 genes were down-regulated by less than 50% (Figure 2A); (2) 54 genes were up-regulated by 2-5-fold (Figure 2B); (3) 23 genes were strongly up-regulated by more than 5-fold (Figure 2C).
Figure 2.
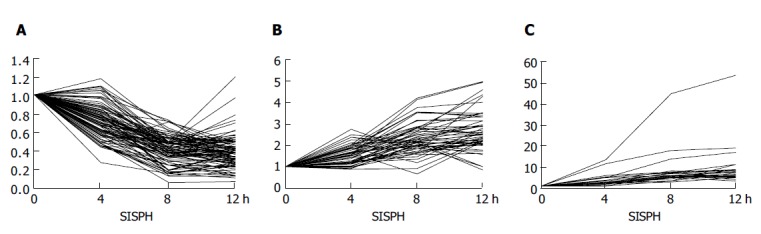
Expression level of genes in the 0-4-8-12 h SISPH. A: Down-regulated genes; B: up-regulated genes; C: strong up-regulated genes.
Hierarchical cluster analysis of genes expressed in the 0-4-8-12 h SISPH
The expression profile of the 183 genes altered by more than a 2-fold intensity at least at one time point in the 0-4-8-12 h SISPH was emanative to the last time point, which indicated that LR at 12 h has not been completed yet, quite active on the contrary (Figure 3A). We undertook hierarchical clustering of four time points 0, 4, 8 and 12 h of SISPH using GeneSpring software and discovered that gene expression profiles had no similarity between the four time points (Figure 3B). To facilitate the visualization and interpretation of the gene expression program represented in this very large body of data, we used the method of κ-means to order genes on the basis of similarities in their expression patterns and displayed the results in a compact graphical format, generating seven kinds of ramose gene expression clusters (Figure 3C). We then categorized the 183 elements into five distinct temporal induction or suppression patterns following immediate induction, late induction, consistent induction, late suppression, consistent suppression (Figure 3D).
Figure 3.
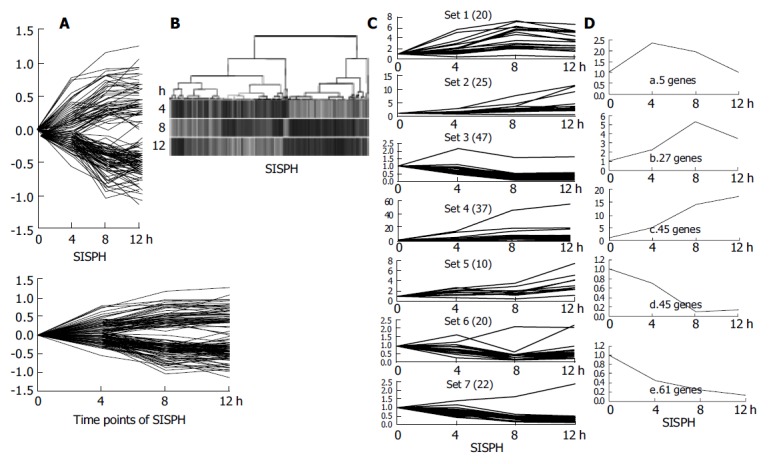
Cluster analysis of 185 elements. A: The difference of their intensity was identified more than 2-fold at least at one time point; B: A hierarchical clustering of five time points indicated that the genes in these time points hardly had a common expression profile; C: The k-means method was used and these genes were classified into seven clusters; D: Five distinct temporal patterns were designated. a. Immediate induction; b. Late induction; c. Consistent induction; d. Late suppression; e. Consistent suppression.
Comparison of gene expression in SISPH with that in PH
Comparing gene expression profile in SISPH with that in PH, we discovered that 43 genes were specially induced by SISPH, and the expression of 140 genes were altered simultaneously in both SISPH and PH, but the time points of their expression and extent of up- and down-regulation were different (Table 2).
Table 2.
Comparison of difference of gene expression in SISPH with that in PH.
| Gene description |
Change |
|
| SISPH | PH | |
| Unreported genes | ||
| CH230-206C20 | 0.3 | 0.3 |
| CH230-403C20 | 0.5 | 0.2 |
| CH230-372C24 | 0.1 | 0.1 |
| CH230-7A22 | 0.1 | 0.1 |
| CH230-11N5 | 2.1 | 4.5 |
| CH230-4L11 | 3.5 | 4.5 |
| CG31759-PA | 2.9 | 0.5 |
| D930042H13 | 0.3 | 0.3 |
| LRRP Cc1-27 | 0.3 | 0.3, 2.1 |
| LRRP Aa2-028 | 3.1 | 3 |
| LRRP Aa2-174 | 0.3 | 0.1 |
| LRRP Ab1-021 | 9.1 | 10 |
| LRRP Ab1-022 | 0.5 | 8.1 |
| LRRP Ab1-114 | 3.5 | 4.2 |
| LRRP Ab1-216 | 3.5 | 6.8 |
| LRRP Ab1-334 | 4.4 | 2.7 |
| LRRP Ab2-001 | 0.3 | 0.2 |
| LRRP Ab2-132 | 0.4 | 0.1 |
| LRRP Ab2-225 | 0.3 | 0.3 |
| LRRP Ab2-232 | 0.2 | 0.2 |
| LRRP Ab2-371 | 0.1 | 0.4 |
| LRRP Ab2-402 | 0.2 | 0.1 |
| LRRP Ab2-417 | 0.2 | 0.2 |
| LRRP Ab2-427 | 0.5 | 0.4 |
| LRRP Ab2-440 | 0.3 | 0.3 |
| LRRP Ac1-060 | 0.3 | 0.4, 2.3 |
| LRRP Ac1-177 | 7.3 | 0.5, 4.9 |
| LRRP Ac1-233 | 5.6 | 4.2 |
| LRRP Ac1-873 | 4.6 | 0.3 |
| LRRP Ac2-061 | 6.4 | 7.6 |
| LRRP Ac2-067 | 2.6 | 2.5 |
| LRRP Ac2-143 | 2 | 3.3 |
| LRRP Ac2-210 | 2.3 | 2.4 |
| LRRP Ba1-647 | 3.3 | 3.2 |
| LRRP Bm403207 | 7.7 | 2.9 |
| LRRP Cb1-739 | 2.2 | 2.3 |
| LRRP Cc1-27 | 0.3 | 0.1, 2.2 |
| LRRP Cc1-38 | 0.1 | 0.2 |
| LRRP Cc1-9 | 3.5 | 3.6 |
| LRRP zbs559 | 3.5 | 3.1 |
| LRRPAb2-132 | 0.2 | 0.1 |
| MGC10946 | 2.8 | 2.8 |
| MGC5178 | 0.4 | 0.4 |
| RIKEN 1300002A08 | 0.2 | 0.3, 2.4 |
| RIKEN 1600027G01 | 0.3 | 0.1 |
| RP11-281N10 | 0.3 | 0.2 |
| RP23-100C5 | 2 | 3 |
| RP23-235O1 | 0.4 | 0.2 |
| RP23-32O9 | 2.3 | 2.8 |
| RP23-417P22 | 0.3 | 0.1 |
| Rp32-28p17 | 0.3 | 0.3 |
| U48828 | 2.4 | 0.2 |
| Open reading frame 31 | 0.5 | 0.3, 2.8 |
| 13 d embryo liver cDNA | 3.6 | 5.9 |
| Stress response | ||
| alpha-1 major acute phase protein prepeptide | 6.8 | 66.4 |
| Angiotensinogen (Agt) | 11.5 | 8.4 |
| Kininogen | 2.5 | 5.9 |
| T-kininogen (Kng) | 5 | 0.2 |
| Glycometabolism | ||
| Alpha enolase (Enolase 1) | 2.7 | 3.9 |
| Isocitrate dehydrogenase 1 (Idh1) | 0.1 | 0.2 |
| Fatty and stearoyl metabolism | ||
| Bile acid CoA ligase | 2 | 2.3 |
| Malonyl-CoA decarboxylase | 0.4 | 0.3 |
| Methylmalonate semialdehyde dehydrogenase | 0.4 | 0.2 |
| 3-alpha-hydroxysteroid dehydrogenase | 0.1 | 0.2 |
| Oxidation and reduction | ||
| Alcohol dehydrogenase (ADH) | 0.1 | 0.1, 2.4 |
| Cytochrome b5 (Cyb5) | 0.3 | 0.2 |
| Cytochrome P450 | 0.3 | 0.2 |
| Cytochrome P450 15-beta gene (Cyp2c12) | 0.4 | 0.2 |
| Cytochrome P450 2E1 | 0.2 | 0.1 |
| Cytochrome P450, 2c39 (Cyp2c39) | 0.3 | 0.1 |
| Cytochrome P450 (PNCN inducible, Cyp3A1) | 0.4 | 0.2 |
| Flavin-containing monooxygenase 1 (Fmo1) | 0.1 | 0.1 |
| Peroxisomal sarcosine oxidase (PSO) | 0.1 | 0.1 |
| Plasma selenoprotein P1 (Sepp1) | 0.4 | 0.3 |
| Small subunit precursor of RuBPCase | 0.1 | 0.1, 3.3 |
| Regulation proteins | ||
| G-protein beta polypeptide 2-like 1 (Gnb2l1) | 2.1 | 2.4 |
| II-Protein with tetratricopeptide repeats 3 | 0.2 | 0.2 |
| RAKb | 0.2 | 0.2 |
| Glycoproteins | ||
| apha-1-B glycoprotein (A1bg), | 0.3 | 0.3 |
| Fibronectin 1 (Fn1) | 3 | 7.2 |
| Fibrinogen, gamma polypeptide (Fgg) | 8 | 5.1 |
| Myelin-associated glycoprotein (L-MAG) | 8.4 | 0.3 |
| UDP-glucuronosyltransferase 2B3 (Udpgt) | 0.4 | 0.3 |
| TRAM1 | 0.3 | 0.4, 3.3 |
| Lipid-proteins | ||
| Adipose differentiation-related protein | 4.6 | 7.3 |
| Solute carrier family 20, mem1 (Slc20a1) | 0.4 | 0.4 |
| Nucleolar proteins | ||
| Damage-specific DNA binding protein 1 (Ddb1) | 0.5 | 0.3 |
| Nuclear protein 1 (Nupr1) | 2.9 | 0.5, 5.1 |
| Nucleolar protein family A, mem 2 | 2.9 | 3.9 |
| ORF2 endonuclease and Rnase H | 0.5 | 0.4 |
| RNase A family 4 | 0.3 | 0.2 |
| Receptors | ||
| ATP-binding cassette, sub-family C | 0.1 | 0.2 |
| Interleukin 1 receptor, type I (Il1r1) | 7.3 | 7.9 |
| Nuclear receptor subfamily 0, member 2 (Nr0b2) | 0.4 | 0.2 |
| Factors | ||
| Amphoterin | 0.5 | 0.3 |
| Angiogenin | 0.4 | 0.2 |
| Eukaryotic translation initiation factor 4A1 | 3.5 | 3.8 |
| Early growth response factor 1 (Egr1) | 4.3 | 3.5 |
| Neuropeptide Y (Npy) | 11.3 | 18.1 |
| NF-E2-related factor 2 (Nfe2l2) | 0.3 | 0.4 |
| Pre-B-cell colony-enhancing factor (Pbef) | 5.3 | 3.3 |
| Angiopoietin-like 3 | 0.3 | 0.2 |
| Hemoglobins | ||
| Hemoglobin, alpha 1 (Hba1) | 2.4 | 0.3 |
| Hemoglobin beta chain (Hbb) | 2.5 | 0.3 |
| Immunological proteises | ||
| Complement component 5 (C5) | 19.4 | 16.5 |
| Immunoglobulin C kappa gene | 0.3 | 0.3 |
| JE/MCP-1 | 2.5 | 4 |
| Cytoskelets | ||
| Actin gamma | 5.1 | 4.8 |
| Keratin 8 (Krt8) | 2.8 | 3.8 |
| Marker proteins | ||
| Amyloid a-5 protein | 53.9 | 90.4 |
| Subchromosomaltransferable fragment | 0.4 | 0.3, 2.4 |
| Amino acid enzymes | ||
| Arginase 1 (Arg1) | 6.4 | 3.5 |
| Argininosuccinate lyase (Asl) | 5.6 | 4.9 |
| Cytosolic aspartate aminotransferase | 6.8 | 7.4 |
| 2-Hydroxyphytanoyl-CoA lyase (Hpcl2) | 0.3 | 0.4 |
| Proteolytic enzymes | ||
| Cathepsin C (Ctsc) | 0.4 | 0.4 |
| Cathepsin D (Ctsd) | 0.5 | 0.3 |
| Coagulation factor 2 (F2) | 0.5 | 0.2 |
| Neutrophil collagenase (Mmp8) | 0.2 | 0.2 |
| Proteinase inhibitors | ||
| alpha-2-macroglobulin (A2m) | 5.1 | 21.3 |
| alpha-trypsin inhibitor heavy chain | 0.4 | 0.2 |
| Contrapsin-like protease inhibitor related protein | 6.1 | 6.6 |
| Leuserpin-2 (Serpind1) | 0.3 | 0.2 |
| Serine protease inhibitor 1 | 5.6 | 5 |
| Phosphatases | ||
| Phosphatidylserine-specific phospholipase A1 | 4.9 | 2.8 |
| Pyrophosphatase/phosphodiesterase 1(Enpp1) | 3.1 | 5 |
| Protein phosphatase 1 (GL-subunit) | 0.2 | 0.2 |
| Synthase | ||
| Carbamyl phosphate synthetase I | 2.5 | 2.9 |
| Glutamyl-prolyl-tRNA synthetase (Eprs) | 2.2 | 4.5 |
| RNA cyclase | 2 | 0.2 |
| Transferases | ||
| Glutathione S-transferase, alpha 1 (Gsta1) | 0.3 | 0.1 |
| Glutathione S-transferase, type 3 (Yb3) (Gstm3) | 0.5 | 0.3, 2.2 |
| Glutathione S-transferase Y(b) subunit | 0.5 | 0.3 |
| UDP-glucuronosyltransferase 2 , mem 5 (Ugt2b5) | 0.5 | 0.3 |
DISCUSSION
This study found that hemoglobin alpha and beta were immediately induced to synthesize in the residual liver at 4 h in SISPH and after PH and returned to normal level there after, and that the activity peak of bile acid CoA ligase appeared at the same time, indicating a role of hemoglobin and amino acids transport in early phase of LR[22,23].
Thirty-six genes were up-regulated and reached a maximum level at 8 h in SISPH, and then were down regulated, in which 18 were unreported new genes and 18 were reported genes, with little knowledge about their relationship with LR. These results showed that their expression products could be necessary for the initiation of S phase. Myelin-associated glycoprotein-binding activity of novel sulfated GM1b, high-affinity ligands for neural siglecs is reported important to nerve system regeneration[24]. It may be available to repair liver or hepatocytes damage caused by SISPH. A CPi-21 encoded by contrapsin-like protease inhibitor is found to inhibit the proteolysis of Edman to glycosylation proteins[25]. Abrupt increase of CPi-21 is presumed to be beneficial to cell adherence in SISPH, in which it was severely destroyed. Aspartate amino transformation reaction catalyzed by aspartate aminotransferase is necessary for accommodated purine, indicating liver cells need a great deal of purine for DNA synthesis in SISPH and after PH. Pre-B-cell colony-enhancing factor is known up regulated in the infected fetal membranes[26], which was continuously induced in the early phase of SISPH and PH, and lately suppressed, suggesting that it is associated with recovery of damage in LR. Translation initiation factor eIF-4A1 is reportedly over expressed in human melanoma cells[27], whose abundant expression in SISPH shows that it may participate in protein synthesis in LR.
Forty-five genes were consistently up regulated and reached a maximum level at 12 h in SISPH, in which 21 were unreported new genes, and the others were reported genes, but whether they have relationship with LR is unknown yet. JE/MCP-1 is known as a CC chemokine attracting monocytes, basophils and T-lymphocytes responsible for IL-1β and TNF-α[28,29]. Its consistent induction is assumed to be involved in relinquishing inflammation caused by SISPH. It was confirmed that keratin 8 could form characteristically intermediate filaments of liver and internal epithelia, including derivative cancers, whose consistent induction may help maintain hepatocyte structural and functional integrity following trauma after SISPH. T-kininogen is important in the acute phase response to trauma as a cysteine proteinase inhibitor[30]. IL-6 is confirmed to act as a direct hepatic mitogen and potent liver growth factor with potential clinical utility for increasing liver mass following injury[31]. The augment of T-kininogen and kininogen level in SISPH and PH may reveal that there is an increasing response of kininogen promoter to IL-6 as an LR signal. Serum amyloid α-5 protein, precursor of serum amyloid A, was successively and dramatically up-regulated for more than 50-fold in SISPH and PH, probably on account of IL-6 simulation in LR[32], indicating that they have some positive role in LR, which is worth studying. Angiotensinogen is already known to be induced by TNF-NFκB and the IL-6-STAT3 signaling pathways in PHx[33], whose level was consistently increased by 11-fold in SISPH, implying a role in avoiding excessive loss of hepatic blood in LR. The successive increase of G-protein in SISPH and PH suggests that DNA synthesis by cAMP in hepatocytes are largely activated for LR[34]. The ensuing variations of cAMP levels are supposed to modulate both growth and differentiated functions during LR of SISPH. The advanced transcription level of enolase 1 (α-enolase) is supposed to provide sufficient VEGF for LR of SISPH. Activity and marked content increase of arginase in the previous period of regenerating liver co-ordinated with other urea cycles’ enzymes[35,36]. The consistent induction of glutamyl-prolyl-tRNA synthetase (Eprs) may indicate that there is a great demand for glutamyl to induce anti-toxin in LR after SISPH. NPY localized exclusively to hepatic vascular walls is conceived and involved in more complex physiological response[37]. The ascending mRNA level in SISPH was suggested to be correlated to recover damaged vascular walls after hepatectomy. The continuous elevation of nucleolar protein family A and nuclear protein 1 (Nupr1) may ascribe to the need of DNA synthesis for nuclear reconstruction of hepatocytes. The successive induction of alpha 2-macroglobulin, as homotetrimeric protease inhibitor, binding TGF-β family, is premised to participate in restraining protein degradation in SISPH for accumulation of protein and prevention of TGF-β termination for longer LR[38]. Hemoglobin secreted by hepatocytes was replacer of hemachrome in liver without hemachrome[22]. Hemoglobin alpha and beta were always induced by SISPH according to the change in PHx, exerting the same function of hepatocytes nourishment in LR. Fibronectin and fibrinogen gamma polypeptide (Fgg) were all induced abundantly through the process of SISPH for the sake of cell adherence in LR. Compared with normal beta actin which has no obvious change in PHx, mutant beta actin was continuously increased, indicating that house keeping gene is additionally expressed for LR of SISPH.
It was revealed that 45 genes were included in consistent suppression and late induction at 8 h time point, of which 22 are unreported genes and others are reported genes hardly known to be related to LR previously. Trypsin and its substrate are supposed to accelerate hepatocytes regeneration under growth factor, but excessive inhibition by inter alpha-trypsin inhibitor may lead to imbalance of glucose concentration in blood, therefore, there was a late induction at 8 h of SISPH to keep the balance[39]. Commonly, acyl-coA plays many important roles in numerous biochemical reactions, such as tricarboxylic acid cycle, glyoxylate bypass, fatty acid synthesis. As a result, the mRNA level of acyl-coA oxidase is assumed to have dropped to retain the level of fat and glycos vital to LR of SISPH and later ascended to eliminate over expressed acyl-coA. Sialyl is a vital constituent of ganglioside, whose sialyltransferase down-regulation may be intervened to prevent liver from successive injury by SISPH and immigrate to other functional substance in late phase. Mutant glutathione S-transferase (GST) mRNA was not always hampered in SISPH, which indicates that its overfull accumulation may lead to opposite reaction in LR. Fatty binding protein is well known to transfer fat from cytoplasm to nuclear or membrane, and fatty acid elongase 1 (rELO1) catalyzes short chain fat transition to long chain fat whose repression of mRNA in SISPH manifested that long chain fat was not badly needed until 12 h in LR. The decrease of purine-nucleoside phosphorylase and phosphatase 1 (GL-subunit) in SISPH may suggest that purine-nucleoside and eser are in demand until 8 h in LR. Nrf2 was reported to lead to antioxidant reaction element to drive gene expression[29], which was suppressed and induced at 8 h, implying that antioxidant reaction was mild in LR of SISPH. Cathepsin D (aspirin endopeptidase) was discovered to participate in basal membrane dissolving, promoting tumor cell migration[40]. The continuous suppression but late induction suggests that basal membrane dissolving was not necessary in early phase of LR of SISPH. Coagulation factor 2 protease inhibitor was supposed to decrease to facilitate blood circulation for LR but at 12-h time point to increase to hamper great loss of blood by SISPH[23]. Damage-specific DNA binding protein was reported to recognize many types of DNA lesions and inducible by DNA damage agent. Its down regulation shows that hepatocytes suffer no severe DNA damage in SISPH, different from other injuries.
Sixty-one genes were consistently suppressed and reached minimum level at 12-h time point of SISPH, of which 22 were novel genes and the others were reported genes that may or may not be related to LR. The genes in the group are presumed to promote LR by various ways. A large number of them are the genes related to energy metabolism, suggesting that the demand for energy in LR of SISPH is not as important as for other demands, differing from partial hepatectomy. The mRNA level of cytochrome P450 2c39 (Cyp2c39), cytochrome P450 2E1, cytochrome b5 (Cyb5), cytochrome P450 15-beta, cytochrome P450, flavin-containing monooxygenase 1 (Fmo1), peroxisomal sarcosine oxidase, malonyl-CoA decarboxylase, methylmalonate semialdehyde dehydrogenase (Mmsdh), alcohol dehydrogenase, microsomal UDP-glucuronosyltransferase 2B3 (Udpgt) are all consistently suppressed from 0 to 12 h of SISPH. The substrate or product of these enzymes related to oxidation and reduction were in urgent need for or toxic to LR, for instance, imine is a cell toxic substance by isocitrate dehydrogenase reaction holding back LR[41]. Angiopoietin-like protein 3 (Angptl3) is reported to activate lipolysis in adipocytes as a vascular endothelial growth factor by response to the liver X receptor[42]. The sustaining suppression of angiopoietin-like protein 3 mRNA in SISPH suggested that the activity of lipolysis of hepatocytes is very low in LR. Neutrophil collagenase specially degrading type I collagen is generated by osteoblastic progenitors, differentiated osteoblasts, osteocytes, and chondrocytes in the growth plate[43]. The decrease of neutrophil collagenase showed that collagen is in great demand for cell skeletal conformation or osteoblasts, osteocytes and chondrocytes were not distinctly harmed in LR after SISPH. Cathepsin C (dipeptidyl aminopeptidase I) in rat liver was discovered playing significant role in protein degradation and the activation of proenzyme[44]. The decreased mRNA level of cathepsin C was supposed to satisfy requisite of peptide for protein construction in SISPH. 2-Hydroxyphytanoyl-CoA lyase, a peroxisomal pyrophosphate-dependent enzyme, was involved in the carbon-carbon bond cleavage during α-oxidation of 3-methyl-branched fatty acids[45]. 2-Hydroxyphytanoyl-CoA lyase was always repressed through SISPH, implying that degradation of long and branch chain fat by α-oxidation was not important in LR. GST was reported to catalyze nucleophilic addition of tripeptide glutathione to xenobiotics carcinogens and endogenous lipophilic compounds[46]. Different kinds of GST were all reduced in SISPH, implying that xenobiotics carcinogens and endogenous lipophilic compounds may bring some uncertain toxic effect on LR. Leuserpin-2 (Sperpind1) is confirmed to participate in fibrinolysis complement activation and inflammatory response[47], which was continuously repressed in SISPH, suggesting that it can regulate inflammatory response to resume severe injured hepatocytes in LR. In previous research, selenoprotein P can defense against peroxynitrite. Its down-regulation may be the result from activation of vast peroxynitrite in SISPH. Amphoterin is believed to enhance process of extension and migration of embryonic neurons and tumor cells by binding to advanced glycation end products (RAGE)[48], whose down-regulation in SISPH makes clear that LR has not much to do with hepatocytes or other cells’ migration.
ACKNOWLEDGMENTS
The authors are very grateful to Dr. JR. Schrock for his correction of and valuable suggestion to the paper. We also thank BioStar for microarray.
Footnotes
Supported by the National Natural Science Foundation of China, No. 30270673 and National Key Laboratory Funds, China
References
- 1.Fausto N. Liver regeneration and repair: hepatocytes, progenitor cells, and stem cells. Hepatology. 2004;39:1477–1487. doi: 10.1002/hep.20214. [DOI] [PubMed] [Google Scholar]
- 2.Taub R. Liver regeneration 4: transcriptional control of liver regeneration. FASEB J. 1996;10:413–427. [PubMed] [Google Scholar]
- 3.Fausto N. Liver regeneration. J Hepatol. 2000;32:19–31. doi: 10.1016/s0168-8278(00)80412-2. [DOI] [PubMed] [Google Scholar]
- 4.Zimmermann A. Liver regeneration: the emergence of new pathways. Med Sci Monit. 2002;8:RA53–RA63. [PubMed] [Google Scholar]
- 5.Nagy P, Bisgaard HC, Schnur J, Thorgeirsson SS. Studies on hepatic gene expression in different liver regenerative models. Biochem Biophys Res Commun. 2000;272:591–595. doi: 10.1006/bbrc.2000.2811. [DOI] [PubMed] [Google Scholar]
- 6.Michalopoulos GK, DeFrances MC. Liver regeneration. Science. 1997;276:60–66. doi: 10.1126/science.276.5309.60. [DOI] [PubMed] [Google Scholar]
- 7.Fukuhara Y, Hirasawa A, Li XK, Kawasaki M, Fujino M, Funeshima N, Katsuma S, Shiojima S, Yamada M, Okuyama T, et al. Gene expression profile in the regenerating rat liver after partial hepatectomy. J Hepatol. 2003;38:784–792. doi: 10.1016/s0168-8278(03)00077-1. [DOI] [PubMed] [Google Scholar]
- 8.Gressner AM. Cytokines and cellular crosstalk involved in the activation of fat-storing cells. J Hepatol. 1995;22:28–36. [PubMed] [Google Scholar]
- 9.Cressman DE, Diamond RH, Taub R. Rapid activation of the Stat3 transcription complex in liver regeneration. Hepatology. 1995;21:1443–1449. [PubMed] [Google Scholar]
- 10.FitzGerald MJ, Webber EM, Donovan JR, Fausto N. Rapid DNA binding by nuclear factor kappa B in hepatocytes at the start of liver regeneration. Cell Growth Differ. 1995;6:417–427. [PubMed] [Google Scholar]
- 11.Sato Y, Igarashi Y, Hakamata Y, Murakami T, Kaneko T, Takahashi M, Seo N, Kobayashi E. Establishment of Alb-DsRed2 transgenic rat for liver regeneration research. Biochem Biophys Res Commun. 2003;311:478–481. doi: 10.1016/j.bbrc.2003.09.230. [DOI] [PubMed] [Google Scholar]
- 12.Mars WM, Kim TH, Stolz DB, Liu ML, Michalopoulos GK. Presence of urokinase in serum-free primary rat hepatocyte cultures and its role in activating hepatocyte growth factor. Cancer Res. 1996;56:2837–2843. [PubMed] [Google Scholar]
- 13.Jensen SA. Liver gene regulation in rats following both 70 or 90% hepatectomy and endotoxin treatment. J Gastroenterol Hepatol. 2001;16:525–530. doi: 10.1046/j.1440-1746.2001.02475.x. [DOI] [PubMed] [Google Scholar]
- 14.Enami Y, Kato H, Murakami M, Fujioka T, Aoki T, Niiya T, Murai N, Ohtsuka K, Kusano M. Anti-transforming growth factor-beta1 antibody transiently enhances DNA synthesis during liver regeneration after partial hepatectomy in rats. J Hepatobiliary Pancreat Surg. 2001;8:250–258. doi: 10.1007/s005340170025. [DOI] [PubMed] [Google Scholar]
- 15.Tang W, Liang K, Wang J, Du L, Zhang W. Effects of pHGF on hepatocyte DNA synthesis after partial hepatectomy in rats. J Tongji Med Univ. 1998;18:25–27. doi: 10.1007/BF02888274. [DOI] [PubMed] [Google Scholar]
- 16.Xu CS, Li YH, Duan RF, Lu AL, Xia M, Ji AL. Effects of the short interval successive partial hepatectomy on rat survival and liver tissue structure. Dongwu Xuebao. 2001;47:659–665. [Google Scholar]
- 17.Li YC, Lin JT, Li WQ, Zhang HY, Wei MX, Xu CS. Cloning and functional analysis of up-regulated expressed genes in rat liver regeneration following short interval successive partial hepatectomy. Developmental Reproductive Biol. 2002;11:151–160. [Google Scholar]
- 18.Li YC, Ma ZQ, Xu CS. Change of TNF-α, c-myc, p53, p21, PCNA, Bcl-2, TGF-β related with the cell prolification in rat liver regeneration following short interval successive partial hepatectomy. Developmental Reproductive Biol. 2002;11:253–260. [Google Scholar]
- 19.Yang GP, Ross DT, Kuang WW, Brown PO, Weigel RJ. Combining SSH and cDNA microarrays for rapid identification of differentially expressed genes. Nucleic Acids Res. 1999;27:1517–1523. doi: 10.1093/nar/27.6.1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Callow MJ, Dudoit S, Gong EL, Speed TP, Rubin EM. Microarray expression profiling identifies genes with altered expression in HDL-deficient mice. Genome Res. 2000;10:2022–2029. doi: 10.1101/gr.10.12.2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Arai M, Yokosuka O, Chiba T, Imazeki F, Kato M, Hashida J, Ueda Y, Sugano S, Hashimoto K, Saisho H, et al. Gene expression profiling reveals the mechanism and pathophysiology of mouse liver regeneration. J Biol Chem. 2003;278:29813–29818. doi: 10.1074/jbc.M212648200. [DOI] [PubMed] [Google Scholar]
- 22.Atha DH, Riggs A. Tetramer-dimer dissociation in homoglobin and the Bohr effect. J Biol Chem. 1976;251:5537–5543. [PubMed] [Google Scholar]
- 23.Dent SB, Peterson CT, Brace LD, Swain JH, Reddy MB, Hanson KB, Robinson JG, Alekel DL. Soy protein intake by perimenopausal women does not affect circulating lipids and lipoproteins or coagulation and fibrinolytic factors. J Nutr. 2001;131:2280–2287. doi: 10.1093/jn/131.9.2280. [DOI] [PubMed] [Google Scholar]
- 24.Ito H, Ishida H, Collins BE, Fromholt SE, Schnaar RL, Kiso M. Systematic synthesis and MAG-binding activity of novel sulfated GM1b analogues as mimics of Chol-1 (alpha-series) gangliosides: highly active ligands for neural siglecs. Carbohydr Res. 2003;338:1621–1639. doi: 10.1016/s0008-6215(03)00245-3. [DOI] [PubMed] [Google Scholar]
- 25.Ohkubo K, Ogata S, Misumi Y, Takami N, Ikehara Y. Molecular cloning and characterization of rat contrapsin-like protease inhibitor and related proteins. J Biochem. 1991;109:243–250. [PubMed] [Google Scholar]
- 26.Ognjanovic S, Bao S, Yamamoto SY, Garibay-Tupas J, Samal B, Bryant-Greenwood GD. Genomic organization of the gene coding for human pre-B-cell colony enhancing factor and expression in human fetal membranes. J Mol Endocrinol. 2001;26:107–117. doi: 10.1677/jme.0.0260107. [DOI] [PubMed] [Google Scholar]
- 27.Eberle J, Fecker LF, Bittner JU, Orfanos CE, Geilen CC. Decreased proliferation of human melanoma cell lines caused by antisense RNA against translation factor eIF-4A1. Br J Cancer. 2002;86:1957–1962. doi: 10.1038/sj.bjc.6600351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ueno M, Sonoda Y, Funakoshi M, Mukaida N, Nose K, Kasahara T. Differential induction of JE/MCP-1 in subclones from a murine macrophage cell line, RAW 264.7: role of kappaB-3 binding protein. Cytokine. 2000;12:207–219. doi: 10.1006/cyto.1999.0544. [DOI] [PubMed] [Google Scholar]
- 29.Caulin C, Ware CF, Magin TM, Oshima RG. Keratin-dependent, epithelial resistance to tumor necrosis factor-induced apoptosis. J Cell Biol. 2000;149:17–22. doi: 10.1083/jcb.149.1.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cole TJ, Schreiber G. The structure and expression of the genes for T-kininogen in the rat. Agents Actions Suppl. 1992;38(Pt 1):292–299. doi: 10.1007/978-3-0348-7321-5_37. [DOI] [PubMed] [Google Scholar]
- 31.Zimmers TA, Pierce RH, McKillop IH, Koniaris LG. Resolving the role of IL-6 in liver regeneration. Hepatology. 2003;38:1590–1591; author reply 1591. doi: 10.1016/j.hep.2003.09.021. [DOI] [PubMed] [Google Scholar]
- 32.Ray A, Schatten H, Ray BK. Activation of Sp1 and its functional co-operation with serum amyloid A-activating sequence binding factor in synoviocyte cells trigger synergistic action of interleukin-1 and interleukin-6 in serum amyloid A gene expression. J Biol Chem. 1999;274:4300–4308. doi: 10.1074/jbc.274.7.4300. [DOI] [PubMed] [Google Scholar]
- 33.Sherman CT, Brasier AR. Role of signal transducers and activators of transcription 1 and -3 in inducible regulation of the human angiotensinogen gene by interleukin-6. Mol Endocrinol. 2001;15:441–457. doi: 10.1210/mend.15.3.0609. [DOI] [PubMed] [Google Scholar]
- 34.Diehl AM, Yang SQ, Wolfgang D, Wand G. Differential expression of guanine nucleotide-binding proteins enhances cAMP synthesis in regenerating rat liver. J Clin Invest. 1992;89:1706–1712. doi: 10.1172/JCI115771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sonoki T, Nagasaki A, Gotoh T, Takiguchi M, Takeya M, Matsuzaki H, Mori M. Coinduction of nitric-oxide synthase and arginase I in cultured rat peritoneal macrophages and rat tissues in vivo by lipopolysaccharide. J Biol Chem. 1997;272:3689–3693. doi: 10.1074/jbc.272.6.3689. [DOI] [PubMed] [Google Scholar]
- 36.Garrard LJ, Bui QT, Nygaard R, Raushel FM. Acid-base catalysis in the argininosuccinate lyase reaction. J Biol Chem. 1985;260:5548–5553. [PubMed] [Google Scholar]
- 37.Ding WG, Tooyama I, Kitasato H, Fujimura M, Kimura H. Phylogenetic and ontogenetic study of neuropeptide Y-containing nerves in the liver. Histochem J. 1994;26:453–459. doi: 10.1007/BF00160059. [DOI] [PubMed] [Google Scholar]
- 38.Niemuller CA, Randall KJ, Webb DJ, Gonias SL, LaMarre J. Alpha 2-macroglobulin conformation determines binding affinity for activin A and plasma clearance of activin A/alpha 2-macroglobulin complex. Endocrinology. 1995;136:5343–5349. doi: 10.1210/endo.136.12.7588280. [DOI] [PubMed] [Google Scholar]
- 39.Starzl TE, Francavilla A, Porter KA, Benichou J, Jones AF. The effect of splanchnic viscera removal upon canine liver regeneration. Surg Gynecol Obstet. 1978;147:193–207. [PMC free article] [PubMed] [Google Scholar]
- 40.Bond JS, Aronson NN. Endocytosis and degradation of native, cathepsin D-degraded, and glutathione-inactivated aldolase by perfused rat liver. Arch Biochem Biophys. 1983;227:367–372. doi: 10.1016/0003-9861(83)90465-4. [DOI] [PubMed] [Google Scholar]
- 41.Zalkin H, Sprinson DB. An investigation of imine formation in the isocitrate dehydrogenase reaction. J Biol Chem. 1966;241:1067–1071. [PubMed] [Google Scholar]
- 42.Shimamura M, Matsuda M, Kobayashi S, Ando Y, Ono M, Koishi R, Furukawa H, Makishima M, Shimomura I. Angiopoietin-like protein 3, a hepatic secretory factor, activates lipolysis in adipocytes. Biochem Biophys Res Commun. 2003;301:604–609. doi: 10.1016/s0006-291x(02)03058-9. [DOI] [PubMed] [Google Scholar]
- 43.Herman MP, Sukhova GK, Libby P, Gerdes N, Tang N, Horton DB, Kilbride M, Breitbart RE, Chun M, Schönbeck U. Expression of neutrophil collagenase (matrix metalloproteinase-8) in human atheroma: a novel collagenolytic pathway suggested by transcriptional profiling. Circulation. 2001;104:1899–1904. doi: 10.1161/hc4101.097419. [DOI] [PubMed] [Google Scholar]
- 44.Cigic B, Pain RH. Location of the binding site for chloride ion activation of cathepsin C. Eur J Biochem. 1999;264:944–951. doi: 10.1046/j.1432-1327.1999.00697.x. [DOI] [PubMed] [Google Scholar]
- 45.Foulon V, Antonenkov VD, Croes K, Waelkens E, Mannaerts GP, Van Veldhoven PP, Casteels M. Purification, molecular cloning, and expression of 2-hydroxyphytanoyl-CoA lyase, a peroxisomal thiamine pyrophosphate-dependent enzyme that catalyzes the carbon-carbon bond cleavage during alpha-oxidation of 3-methyl-branched fatty acids. Proc Natl Acad Sci USA. 1999;96:10039–10044. doi: 10.1073/pnas.96.18.10039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Atkins WM, Wang RW, Bird AW, Newton DJ, Lu AY. The catalytic mechanism of glutathione S-transferase (GST). Spectroscopic determination of the pKa of Tyr-9 in rat alpha 1-1 GST. J Biol Chem. 1993;268:19188–19191. [PubMed] [Google Scholar]
- 47.Ragg H, Ulshöfer T, Gerewitz J. On the activation of human leuserpin-2, a thrombin inhibitor, by glycosaminoglycans. J Biol Chem. 1990;265:5211–5218. [PubMed] [Google Scholar]
- 48.Huttunen HJ, Kuja-Panula J, Sorci G, Agneletti AL, Donato R, Rauvala H. Coregulation of neurite outgrowth and cell survival by amphoterin and S100 proteins through receptor for advanced glycation end products (RAGE) activation. J Biol Chem. 2000;275:40096–40105. doi: 10.1074/jbc.M006993200. [DOI] [PubMed] [Google Scholar]


