Abstract
AIM: To express the complete PreS region of HBV in E.coli with good solubility and stability, and to establish an effective method for purification of the recombinant PreS protein.
METHODS: The complete PreS region (PreS1 and PreS2) was fused into a series of tags including glutathione S-transferase (GST), dihydrofolate reductase (DHFR), maltose binding protein (MBP), 6×histidine, chitin binding domain (CBD), and thioredoxin, respectively. Expression of recombinant PreS fusion proteins was examined by SDS-PAGE analysis and confirmed by Western blot. Two fusion proteins, thio-PreS, and PreS-CBD, with desirable solubility and stability, were subjected to affinity purification and further characterization.
RESULTS: Recombinant PreS fusion proteins could be synthesized with good yields in E.coli. However, most of these proteins except for thio-PreS and PreS-CBD were vulnerable to degradation or insoluble as revealed by SDS-PAGE and Western blot. Thio-PreS could be purified by affinity chromatography with nickel-chelating sepharose as the matrix. However, some impurities were also co-purified. A simple freeze-thaw treatment yielded most of the thio-PreS proteins in solution while the impurities were in the precipitate. Purified thio-PreS protein was capable of inhibiting the binding of HBV virion to a specific monoclonal antibody against an epitope within the PreS1 domain.
CONCLUSION: Increased solubility and stability of the complete PreS region synthesized in E.coli can be achieved by fusion with the thioredoxin or the CBD tag. A simple yet highly effective method has been established for the purification of the thio-PreS protein. Purified thio-PreS protein likely assumes a native conformation, which makes it an ideal candidate for studying the structure of the PreS region as well as for screening antivirals.
Keywords: Hepatitis B virus, PreS, Expression, Purification
INTRODUCTION
Virally encoded small (HBs), middle (MHBs), and large (LHBs) surface proteins together with cellular phospholipids form the envelope of HBV virion. These proteins are translated from distinct initiation codons, but share a common reading frame and stop codon. The HBs protein contains 226 amino acids and is the major component of the viral envelope. The MHBs protein has 55 extra amino acids (PreS2) located to the N-terminal of HBs and the LHBs protein carries an additional 119 amino acid (or 109 amino acids depending on the viral subtype, PreS1) N-terminal extension with respect to the MHBs protein[1]. All envelope proteins are co-translationally inserted into the endoplasmic reticulum (ER) membrane directed by the topogenic elements in HBs[2,3].
LHBs protein plays pivotal roles in infection and budding processes during the HBV life cycle. The N-terminal PreS region of the LHBs protein can adopt one of two topological conformations depending on whether it undergoes a posttranslational translocation. Thus, LHBs proteins in virions exhibit a mixed population with their PreS region (PreS1 and PreS2) located either inside or outside of viral envelopes[2,3]. Encapsidation of viral nucleocapsids and secretion of mature viral particles require LHBs proteins with cytoplasmic PreS region[4-8]. A stretch of amino acids across the PreS1 and the PreS2 is thought to be involved in the interaction with the cytosolic nucleocapsid before the budding event[7,8]. During the infection process, externally exposed PreS region may mediate the binding of virion to a putative cellular receptor [9,10]. Amino acids 21-47 of the PreS1 domain likely bear the major epitope for cell attachment[9-11].
Given its important functional role in the HBV life cycle and being a potential target for the development of novel antivirals, efforts have been made to express the PreS region for structural analysis, however, with little success. To date, the PreS region synthesized in E.coli is either insoluble or quickly degraded[12-14]. Nunez et al[12], produced the PreS region fused with a 6×histidine (6×His) tag that is largely insoluble. Purification of the His-tagged PreS protein under non-denaturing condition failed owing to a severe proteolysis. Structure analysis is difficult due to the fragile nature of the PreS region synthesized in E.coli after denature-renaturing cycles. Study with UV-CD spectra indicates that almost half of the proteins display non-ordered conformations[12], which may reflect an inherent instability of the PreS region or the improper folding of the recombinant PreS region. Alternative ways have been adopted to synthesize partial regions of PreS[15,16]. However, it is not clear whether these partial regions retain their original structures.
In this study, we fused the PreS region with different tags respectively and studied the expression of these PreS fusion proteins in E.coli, in the hope of stabilizing the PreS region by a structurally linked tag. We observed increased solubility and stability of the PreS fused with the thioredoxin or the CBD tag. We have further established a simple yet highly effective method for purification of thio-PreS. The thio-PreS protein purified with this method is capable of inhibiting the binding of viral particle to a PreS1-specific monoclonal antibody.
MATERIALS AND METHODS
Plasmid construction
Primers (PreS-forward: 5’-ATGGGAGGTTGGTCTTC-CAAAC-3’; PreS-reverse: 5’-GTTCGGTGCAGGGTC-CCCAGTC-3’) with adaptors harboring appropriate restriction endonuclease sites were used to amplify the fragment encoding the PreS region from the p3.6II plasmid[17,18] that contains a terminally redundant HBV genome (subtype adr-1). After digestion with appropriate enzymes, the PCR product was inserted into several prokaryotic expression vectors, respectively. Expression vectors used in this study were pET28a+ (Novagen), pQE40 (Qiagen), pMalC2x (NEB), pGEX2T (Pharmacia), pThioHisA (Invitrogen), and pTXB1 (NEB) (Figure 1). Clones containing correct inserts were verified by sequencing (Bioasia, Shanghai).
Figure 1.
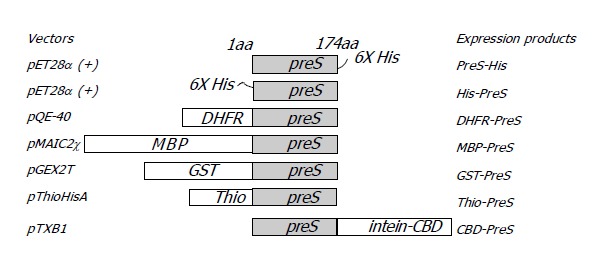
Diagram of various PreS fusion proteins.
Expression of fusion proteins
E.coli Top10 (F’) strain was used for the expression of DHFR-PreS and thio-PreS while the BL21 (DE3) strain was used for the expression of other fusion proteins. Expression was induced by IPTG for 4 h at room temperature. Optimized IPTG concentrations were 0.1 mmol/L for DHFR-PreS, 1 mmol/L for thio-PreS, and 0.3 mmol/L for other fusion proteins. Bacteria were harvested and resuspended in PBS with 1 mmol/L EDTA and 100 mmol/L PMSF. Lysates were prepared by sonication on ice and centrifuged before further SDS-PAGE analysis.
Purification of fusion proteins
Purification of PreS-CBD was performed with chitin resin (NEB) according to the manufacturer’s instructions. PreS-CBD coupled on the resin was subjected to an inter-mediated self-cleavage, induced by incubation overnight with 50 mmol/L DTT in PBS at 4 °C. Purification of thio-PreS was performed with ProBondTM nickel-chelating sepharose resin (Invitrogen), taking advantage of a conformational His-Patch motif within the thioredoxin tag. Supernatant of the bacteria lysate was mixed with the resin and rocked gently. The resin was washed thoroughly with PBS, followed by a stringent wash with five column volumes of wash buffer (500 mmol/L NaCl, 5 mmol/L imidazole, 20 mmol/L phosphate, pH 6.0) at a flow rate of 1 mL/min. Recombinant proteins were collected in 2.5 column volumes of elution buffer (500 mmol/L NaCl, 250 mmol/L imidazole, 20 mmol/L phosphate, pH 6.0). Impurities were removed by centrifugation of the thawed sample having undergone an overnight freeze at -70 °C. Purified proteins were subjected to desalting with Sephadex G25 (Pharmacia). Protein concentration was determined by the Bradford assay.
Virus capture assay
125E11[19,20] is a monoclonal antibody against PreS1. For virus capturing, the antibody was immobilized on a microplate (Nunc) in carbonate buffer. PBS diluted sera of HBV patients (provided by Ruijin Hospital, Shanghai) were added at 106 infectious virions per well and incubated for 1 h at room temperature. HBV virions were then detected with HRP conjugated anti-HBs antibody (Sino-America Biotech) and subjected to TMB developed color reaction. The optical density values were measured at 450 nm (A450) with an automatic photometer (Bio-rad). For competitive binding assays, different amount of purified thio-PreS, or thioredoxin as the control, was added to the diluted sera prior to virus capturing.
Western blot
Western blot analyses of PreS fusion proteins were performed according to a standard method[21]. Supernatants of the expression lysates were separated on SDS-PAGE, and transferred to nitrocellulose filters. 125E11 (1:1000) was used as the primary antibody for PreS detection. Blots were developed using the ECL method (PerfectBio) with HRP-labeled rabbit anti-mouse Ig (1:2000, Dako).
RESULTS
Construction and expression of PreS fusion proteins
A series of prokaryotic expression vectors (Figure 1) were employed to construct PreS expression plasmids. The DNA fragment encoding the PreS region of HBV was amplified by PCR and inserted into these vectors respectively. 6×His tag was fused to either the N-terminal or the C-terminal of PreS. The CBD tag was joined to the C-terminal of PreS by an intein (Figure 1). Other tags (GST, DHFR, MBP, and thioredoxin) were located at N-terminal respectively.
SDS-PAGE was performed to examine the synthesis of fusion proteins. His-tagged PreS fusion proteins were insoluble, though fairly high yields were achieved (Figure 2A). GST-PreS was found in soluble fraction of the bacteria lysate. However, severe degradation was obvious. Even worse degradation was observed with DHFR-PreS (data not shown). Previous studies by Cho et al[14], suggested a more stable recombinant product of MBP fused PreS. In our study, however, moderate proteolysis still occurred. Besides the full-length 60 ku protein, other bands with a smaller molecular weight were observed as indicated on SDS-PAGE, probably represented degraded proteins (Figure 2B). On the contrary, thio-PreS showed an excellent stability in solution (Figure 2C) with a high yield reaching about 0.2-0.5 mg per ml of the bacteria culture. Similarly, PreS-CBD was found predominantly in the supernatant of the bacteria lysate, and importantly, without any obvious degradation by SDS-PAGE analysis (Figure 2D).
Figure 2.
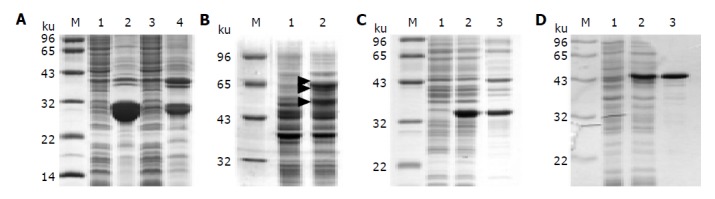
SDS-PAGE of the PreS fusion proteins. A: His-tagged PreS. Lanes 1 and 2: His-PreS; lanes 3 and 4: PreS-His; lanes 1 and 3: supernatants of the E.coli lysate; lanes 2 and 4: insoluble pellets; B: MBP-PreS. Lane 1: bacteria lysate without IPTG induction; lane 2: supernatant of the bacteria lysate after IPTG induction. The 60 kD intact protein and degradation products of MBP-PreS are indicated with triangles; C: thio-PreS. Lane 1: bacteria lysate without IPTG induction; lane 2: whole lysate after IPTG induction; lane 3: supernatant of the E.coli lysate after IPTG induction; D: PreS-CBD. Lane 1: bacteria lysate without IPTG induction; lane 2: PreS-CBD in the supernatant of the bacteria lysate after IPTG induction; lane 3: Purified PreS-CBD coupled to the chitin resin. Molecular weight marker is denoted as M in all figures.
Purification of PreS fusion proteins
Given their desirable solubility and stability, PreS-CBD and thio-PreS were subjected to further purification. The chitin matrix was used to purify the PreS-CBD protein. As shown in Figure 2D, PreS-CBD could be successfully separated from the lysate mixture and coupled to the chitin resin. Binding of PreS-CBD to the chitin resin was highly specific, and few contaminants were detected by SDS-PAGE analysis (lane 3, Figure 2D). However, the purification process was handicapped in the elution step. The intact fusion product was difficult to be eluted off the chitin resin, and inter-based cleavage was unsuccessful due to the fragile nature of the free PreS region (data not shown).
Thioredoxin tag as in thio-PreS harbored a His-Patch motif that allowed purification of recombinant proteins on nickel chelating sepharose. The His-Patch motif differed from other linear His-tags in conformation. Thus, recombinant proteins could be purified under a non-denaturing condition. Figure 3A shows the affinity purification of thio-PreS with nickel chelating sepharose. Proteins were eluted with a buffer containing imidazole at different concentrations. An elution buffer containing 250 mmol/L imidazole, 500 mmol/L NaCl and 20 mmol/L sodium phosphate, pH 6.0, gave an optimized elution of thio-PreS. However, some impurities were apparently present even under a highly stringent wash condition. We also tried the resin of thio-bond (Invitrogen) that is specific for the thioredoxin tag rather than the His-Patch motif, but with little success (data not shown).
Figure 3.
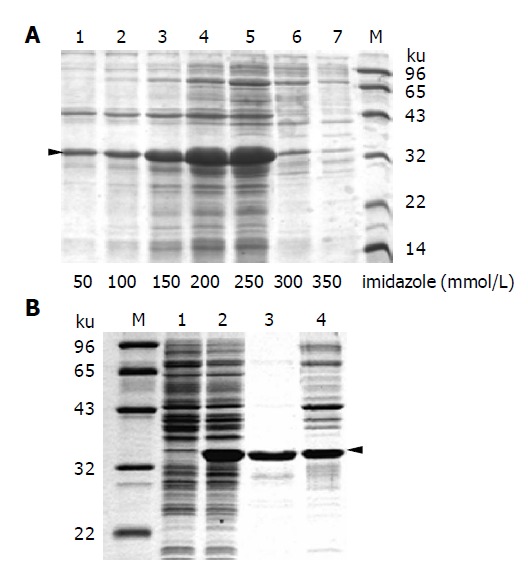
Purification of thio-PreS. A: Fractions of thio-PreS eluted with increasing imidazole concentrations listed at the bottom of each lane. The triangle indicates the position of thio-PreS; B: Further purification with a freeze-thaw treatment. Lanes 1 and 2: supernatants of the bacteria lysate before or after IPTG induction, respectively; lanes 3 and 4: supernatant and precipitate of the affinity purified thio-PreS sample after a freeze-thaw treatment. Molecular weight maker is denoted as M.
Since thio-PreS showed a great solubility, freezing the affinity purified sample followed by a quick thaw might improve the purity of thio-PreS by eliminating the impurities that were slow in redissolving. As shown in Figure 3B, with this approach, most impurities were completely fractioned into the precipitate while the majority of thio-PreS remained in the supernatant without any proteolysis. Densitometry scan (Bio-Rad) of the sample on SDS-PAGE suggested a high purity of above 95%. The purified thio-PreS protein was very stable. A procedure of desalting chromatography with Sephadex G25 did little harm to the integrality of this fusion protein even at room temperature (data not shown).
Characterization of the PreS fusion protein
To verify the correctness of synthesized fusion proteins, Western blot was performed with the monoclonal antibody 125E11. As shown in Figure 4, MBP-PreS, PreS-CBD, and thio-PreS with a correct molecular weight could be detected, respectively. Proteolysis of MBP-PreS was also apparent, while only tiny traces of degradation of thio-PreS and PreS-CBD were found. 125E11 could recognize a PreS1 epitope exposed on the surface of the virion. As shown in Figure 5, when immobilized on the solid surface of microplate wells, the antibody could specifically capture viral particles from HBV patient sera. Addition of purified thio-PreS protein to the sera could inhibit the capturing process efficiently and in a dose-dependent manner, suggesting that the PreS region in the thio-PreS protein likely adopts a native conformation that mimics that of the HBV virion.
Figure 4.
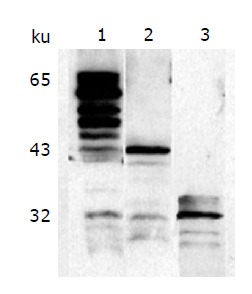
Characterization of the fusion proteins with Western blot.
Figure 5.
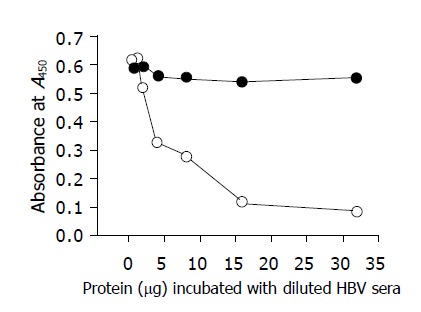
Virus capture assay.
DISCUSSION
The PreS region of LHBs has been implicated in the attachment of HBV virion to the putative receptor on host cells, while its inward conformation is thought to be involved in viral morphogenesis. Other important roles have also been assigned to PreS, including regulation of viral replication and transactivation of a variety of promoter elements[22-24]. Thus, multiple virological functions of the PreS region provide a useful target for anti-HBV drug intervention. Inhibition on viral infection by E.coli expressed PreS occurs through a direct interference with the binding of HBV to the putative cell surface receptor[25]. Recombinant PreS product could also be developed as a protein vaccine that elicits B and T cell immune responses on a broader range of MHC haplotypes[26-29]. Nevertheless, detailed structural studies of PreS are complicated by the difficulties in isolating large quantities of LHBs from viral particles and in expressing soluble and stable PreS proteins.
The complete PreS on the surface of HBV virions has been shown as an independent domain with a native structure[2,3,9,10]. Thus, recombinant PreS proteins expressed in the E.coli system may accurately reflect the proper structure. However, according to previous works, the PreS region is susceptible to heavy proteolysis during expression and purification. In this paper, attempts have been made to express the PreS fused with various tags in E.coli, in the hope of stabilizing the recombinant products by structurally linked tags[30]. Although most PreS fusion proteins expressed in this study are still vulnerable to degradation, or exist as insoluble inclusion bodies, a stable and soluble PreS fusion protein containing the thioredoxin or the CBD tag has been synthesized with a good yield. Thioredoxin is a highly hydrophilic protein tag, designed for achieving increased solubility of fusion proteins expressed in E.coli, and usually accumulates at sites on the cytoplasmic inner membrane known as adhesion zones[31]. We proposed that the environment of adhesion zones as well as the great solubility of thioredoxin could contribute to the better folding of the recombinant PreS protein. The impact of intein-CBD (Figure 1) on a better stability of PreS is also obvious. The underlying mechanism is currently unknown. It is possible that the intein-CBD tag forms a structure that may hinder the access of proteases to the proteolytic sites in the PreS region.
Affinity purification based on the His-Patch motif under a non-denaturing condition is not always efficient. In purification of thio-PreS, the contamination of impurities is obvious. Interestingly, after a freeze-thaw treatment, the affinity purified thio-PreS sample is dramatically purified. The majority of thio-PreS remains in the supernatant while the impurities are largely precipitated. This process may take advantage of the differential solubility between thio-PreS and the impurities or act as a salting-out step. The purified thio-PreS might adopt a native structure, since the soluble expression product in a stable fashion usually suggests a proper folding, and His-Patch based affinity purification would imply a correct conformation of the thioredoxin tag. Importantly, thio-PreS can efficiently inhibit the binding of viral particle to the PreS1 specific monoclonal antibody, suggesting exposed eptitopes in the recombinant protein as those on the surface of the native virion. PreS-CBD is also soluble and shows a high stability. Purified PreS-CBD coupled to the chitin resin could be obtained, but the following intein-based cleavage by DTT was unsuccessful which may be due to the fragile nature of the PreS region. Similarly, thio-PreS was subjected to an enterokinase-cleavage that is engineered in the thio-fusion expression system to separate the desired PreS region from the thioredoxin tag. However, severe proteolysis was observed during this treatment (data not shown), suggesting an important role of the thioredoxin tag in stabilizing the PreS region.
In conclusion, the complete PreS region fused with the thioredoxin or CBD tag has been successfully synthesized. A simple yet efficient method has been established for purification of the thio-PreS protein. The thio-PreS protein is highly stable and soluble. The purified thio-PreS protein may be a valuable candidate for studying the structure of the PreS region as well as for screening antivirals.
ACKNOWLEDGMENTS
125E11 is a kind gift from Professor Zhu-Chuan Zhang. The authors thank Mr. Jie-Hong Jiang, and Mr. Dong-Hua Zhang, Ruijin Hospital, for providing serum samples of HBV patients.
Footnotes
Supported by the Basic Research Program from Ministry of Science and Technology of China, No. G1999054105, and special funds for Sino-France Center for Life Science and Genome Research from Chinese Academy of Sciences and Pasteur Institute in France
Co-correspondents: Yuan Wang and You-Hua Xie
Science Editor Wang XL Language Editor Elsevier HK
References
- 1.Neurath AR, Kent SB. The pre-S region of hepadnavirus envelope proteins. Adv Virus Res. 1988;34:65–142. doi: 10.1016/s0065-3527(08)60516-3. [DOI] [PubMed] [Google Scholar]
- 2.Ostapchuk P, Hearing P, Ganem D. A dramatic shift in the transmembrane topology of a viral envelope glycoprotein accompanies hepatitis B viral morphogenesis. EMBO J. 1994;13:1048–1057. doi: 10.1002/j.1460-2075.1994.tb06353.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Prange R, Streeck RE. Novel transmembrane topology of the hepatitis B virus envelope proteins. EMBO J. 1995;14:247–256. doi: 10.1002/j.1460-2075.1995.tb06998.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Poisson F, Severac A, Hourioux C, Goudeau A, Roingeard P. Both pre-S1 and S domains of hepatitis B virus envelope proteins interact with the core particle. Virology. 1997;228:115–120. doi: 10.1006/viro.1996.8367. [DOI] [PubMed] [Google Scholar]
- 5.Bruss V, Vieluf K. Functions of the internal pre-S domain of the large surface protein in hepatitis B virus particle morphogenesis. J Virol. 1995;69:6652–6657. doi: 10.1128/jvi.69.11.6652-6657.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bruss V, Ganem D. The role of envelope proteins in hepatitis B virus assembly. Proc Natl Acad Sci USA. 1991;88:1059–1063. doi: 10.1073/pnas.88.3.1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bruss V. A short linear sequence in the pre-S domain of the large hepatitis B virus envelope protein required for virion formation. J Virol. 1997;71:9350–9357. doi: 10.1128/jvi.71.12.9350-9357.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ponsel D, Bruss V. Mapping of amino acid side chains on the surface of hepatitis B virus capsids required for envelopment and virion formation. J Virol. 2003;77:416–422. doi: 10.1128/JVI.77.1.416-422.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.De Meyer S, Gong ZJ, Suwandhi W, van Pelt J, Soumillion A, Yap SH. Organ and species specificity of hepatitis B virus (HBV) infection: a review of literature with a special reference to preferential attachment of HBV to human hepatocytes. J Viral Hepat. 1997;4:145–153. doi: 10.1046/j.1365-2893.1997.00126.x. [DOI] [PubMed] [Google Scholar]
- 10.Cooper A, Paran N, Shaul Y. The earliest steps in hepatitis B virus infection. Biochim Biophys Acta. 2003;1614:89–96. doi: 10.1016/s0005-2736(03)00166-4. [DOI] [PubMed] [Google Scholar]
- 11.Neurath AR, Kent SB, Strick N, Parker K. Identification and chemical synthesis of a host cell receptor binding site on hepatitis B virus. Cell. 1986;46:429–436. doi: 10.1016/0092-8674(86)90663-x. [DOI] [PubMed] [Google Scholar]
- 12.Núñez E, Wei X, Delgado C, Rodríguez-Crespo I, Yélamos B, Gómez-Gutiérrez J, Peterson DL, Gavilanes F. Cloning, expression, and purification of histidine-tagged preS domains of hepatitis B virus. Protein Expr Purif. 2001;21:183–191. doi: 10.1006/prep.2000.1368. [DOI] [PubMed] [Google Scholar]
- 13.Delos S, Villar MT, Hu P, Peterson DL. Cloning, expression, isolation and characterization of the pre-S domains of hepatitis B surface antigen, devoid of the S protein. Biochem J. 1991;276(Pt 2):411–416. doi: 10.1042/bj2760411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cho EW, Park JH, Yoo OJ, Kim KL. Translocation and accumulation of exogeneous hepatitis B virus preS surface proteins in the cell nucleus. J Cell Sci. 2001;114:1115–1123. doi: 10.1242/jcs.114.6.1115. [DOI] [PubMed] [Google Scholar]
- 15.Lin Y, Liu YX, Cislo T, Mason BL, Yu MY. Expression and characterization of the preS1 peptide of hepatitis B surface antigen in Escherichia coli. J Med Virol. 1991;33:181–187. doi: 10.1002/jmv.1890330308. [DOI] [PubMed] [Google Scholar]
- 16.Maeng CY, Oh MS, Park IH, Hong HJ. Purification and structural analysis of the hepatitis B virus preS1 expressed from Escherichia coli. Biochem Biophys Res Commun. 2001;282:787–792. doi: 10.1006/bbrc.2001.4641. [DOI] [PubMed] [Google Scholar]
- 17.Feng Y, Kong YY, Wang Y, Qi GR. Inhibition of hepatitis B virus by hammerhead ribozyme targeted to the poly(A) signal sequence in cultured cells. Biol Chem. 2001;382:655–660. doi: 10.1515/BC.2001.077. [DOI] [PubMed] [Google Scholar]
- 18.Fu L, Wu X, Kong YY, Wang Y. Regulation of HBV gene expression by core promoter and its upstream sequence. Zhongguo Bingduxue. 1997;13:215–223. [Google Scholar]
- 19.Yang HL, Jin Y, Cao HT, Xu X, Li GD, Wang Y, Zhang ZC. Affinity Purification of Hepatitis B Virus Surface Antigen Containing PreS1 Region. Sheng Wu Hua Xue Yu Sheng Wu Wu Li Xue Bao (Shanghai) 1996;28:412–417. [PubMed] [Google Scholar]
- 20.Hui J, Li G, Kong Y, Wang Y. Expression and characterization of chimeric hepatitis B surface antigen particles carrying preS epitopes. J Biotechnol. 1999;72:49–59. doi: 10.1016/s0168-1656(99)00049-8. [DOI] [PubMed] [Google Scholar]
- 21.Sambrook J, Fritsch EF, Maniatis T. Molecular cloning: A laboratory manual.2nd ed. Cold Spring Harbor Laboratory Press, 1989 [Google Scholar]
- 22.Lenhoff RJ, Summers J. Coordinate regulation of replication and virus assembly by the large envelope protein of an avian hepadnavirus. J Virol. 1994;68:4565–4571. doi: 10.1128/jvi.68.7.4565-4571.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Caselmann WH. Transactivation of cellular gene expression by hepatitis B viral proteins: a possible molecular mechanism of hepatocarcinogenesis. J Hepatol. 1995;22:34–37. [PubMed] [Google Scholar]
- 24.Natoli G, Avantaggiati ML, Balsano C, De Marzio E, Collepardo D, Elfassi E, Levrero M. Characterization of the hepatitis B virus preS/S region encoded transcriptional transactivator. Virology. 1992;187:663–670. doi: 10.1016/0042-6822(92)90469-6. [DOI] [PubMed] [Google Scholar]
- 25.Urban S, Gripon P. Inhibition of duck hepatitis B virus infection by a myristoylated pre-S peptide of the large viral surface protein. J Virol. 2002;76:1986–1990. doi: 10.1128/JVI.76.4.1986-1990.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shouval D. Hepatitis B vaccines. J Hepatol. 2003;39 Suppl 1:S70–S76. doi: 10.1016/s0168-8278(03)00152-1. [DOI] [PubMed] [Google Scholar]
- 27.Pride MW, Bailey CR, Muchmore E, Thanavala Y. Evaluation of B and T-cell responses in chimpanzees immunized with Hepagene, a hepatitis B vaccine containing pre-S1, pre-S2 gene products. Vaccine. 1998;16:543–550. doi: 10.1016/s0264-410x(97)00242-9. [DOI] [PubMed] [Google Scholar]
- 28.Milich DR, Jones JE, McLachlan A, Bitter G, Moriarty A, Hughes JL. Importance of subtype in the immune response to the pre-S(2) region of the hepatitis B surface antigen. II. Synthetic Pre-S(2) immunogen. J Immunol. 1990;144:3544–3551. [PubMed] [Google Scholar]
- 29.Hui J, Mancini M, Li G, Wang Y, Tiollais P, Michel ML. Immunization with a plasmid encoding a modified hepatitis B surface antigen carrying the receptor binding site for hepatocytes. Vaccine. 1999;17:1711–1718. doi: 10.1016/s0264-410x(98)00430-7. [DOI] [PubMed] [Google Scholar]
- 30.Terpe K. Overview of tag protein fusions: from molecular and biochemical fundamentals to commercial systems. Appl Microbiol Biotechnol. 2003;60:523–533. doi: 10.1007/s00253-002-1158-6. [DOI] [PubMed] [Google Scholar]
- 31.LaVallie ER, DiBlasio EA, Kovacic S, Grant KL, Schendel PF, McCoy JM. A thioredoxin gene fusion expression system that circumvents inclusion body formation in the E. coli cytoplasm. Biotechnology (N Y) 1993;11:187–193. doi: 10.1038/nbt0293-187. [DOI] [PubMed] [Google Scholar]


