Abstract
AIM: To look for a rapid low-cost technique for the detection of HBV variants.
METHODS: Two patients who underwent orthotopic liver transplantation (OLT) for HBV infection were treated with lamivudine (100 mg daily) and HBV infection recurred in the grafted livers. The patients were monitored intensively for liver enzymes, hepatitis B surface antigen (HBsAg) and HBV DNA in serum. Liver biopsy was performed regularly. HBV DNA in a conserved polymerase domain (the YMDD locus) was amplified from serum of each patient by PCR and sequenced. HBV genotypes were analyzed by restriction fragment length polymorphism (RFLP) of the PCR products generated from a fragment of the polymerase gene.
RESULTS: YMDD wild-type HBV was detected in one patient by PCR-RFLP and DNA sequencing 19 mo after OLT, and YIDD mutant-type HBV in the other patient, 16 mo after OLT.
CONCLUSION: PCR-RFLP assay is an accurate and simple method for genotyping lamivudine-resistant HBV variants.
Keywords: Liver transplantation, HBV DNA, Polymerase gene, YMDD variants, PCR-RFLP
INTRODUCTION
Chronic hepatitis B is a common disease with about 350 million HBV carriers according to the figures of World Health Organization[1]. The persistence of viremia in patients often leads to progressive liver diseases including cirrhosis and hepatocellular carcinoma. For HBV-infected patients with end-stage liver disease, antiviral treatment cannot significantly alter their clinical course; orthotopic liver transplantation (OLT) is the only option. Because of the very high HBV reinfection rate of hepatic allografts and the rapid progression of HBV-induced disease in allografts[2], it is essential to prevent HBV reinfection of hepatic allografts after OLT. Interferon has been shown to be beneficial to some patients with chronic HBV infection; however, the overall response rate to interferon alpha is less than 40%[3]. Lamivudine, an enantiomer of β-L-2’, 3’-dideoxy-3’-thiacytidine (3TC), is a potent inhibitor of RNA-dependent DNA polymerase of HBV and HIV reverse transcriptase[4,5]. Treatment of HBV with oral lamivudine can suppress virus replication and reduce hepatic necro-inflammatory activity[6].
Although lamivudine can prevent HBV reinfection of hepatic allografts, long-term therapy can lead to lamivudine-resistance of HBV[7-9]. Furthermore, lamivudine-resistant HBV strains, with amino acid substitution in the YMDD (tyrosine, methionine, aspartate) motif of DNA polymerase, have been found in patients after OLT and immunosuppressive therapy[5]. The amino acid substitution from methionine to valine or isoleucine at amino acid 552 of the YMDD motif of the HBV polymerase gene is the main mutation responsible for resistance to lamivudine treatment. The reported rate of lamivudine resistance is 14% in patients with chronic HBV infection and 27% in patients with recurrent HBV infection after OLT[10-13]. The low incidence and late occurrence of YMDD mutant viruses suggest that such mutant strains originate from wild-type by point mutation during the course of therapy. Once lamivudine resistance occurs, a combination antiviral therapy (such as interferon and hepatitis B immunoglobulin) may be necessary to prevent exacerbation of hepatitis[14-17].
Occurrence of lamivudine-resistant HBV variants is commonly detected by direct sequencing of HBV DNA. However, this assay is excessively time-consuming for a large number of clinical samples. In this study, the frequency and characteristics of variants at codon position 552 of the HBV polymerase gene were examined by restriction fragment length polymorphism (RFLP) of the products generated from the HBV genome after amplification by PCR. The results of this analysis were compared with those obtained by direct HBV DNA sequencing, alanine aminotransferase levels and liver biopsy.
MATERIALS AND METHODS
Patients
Between 2000 and 2004, 72 patients underwent OLT for the end-stage liver disease secondary to HBV infection in our institute. Seven of these patients developed recurrent HBV infection in the grafted liver confirmed by serological examination and liver biopsy. All patients received standard immunosuppression treatment with cyclosporine, azathioprine, corticosteroids, anti-HBsAg immunoglobulin injection and oral lamivudine (100 mg daily). The patients were closely monitored for serum liver enzymes, hepatitis B surface antigen (HBsAg), HBeAg, and HBV DNA. Liver biopsies were performed before and after OLT. YMDD variants of HBV DNA polymerase gene were detected by direct sequencing in the YMDD motif of HBV and PCR-RFLP. YIDD mutant was found in one and wild-type YMDD in six of the seven patients. The following two patients served as their example.
Patient 1 was a 34-year-old man, his liver function was abnormal 15 years ago without any treatment. Two years ago, he developed oliguria, abdominal distension, anorexia, profound fatigue and edema and was diagnosed as cirrhosis secondary to chronic HBV infection, liver dysfunction and portal vein hypertension. He received liver protective and urinative therapy. He developed headache, furor and unconscious behavior after oral losec and was diagnosed as stage II hepatic encephalopathy 6 mo ago. His condition improved and was discharged after 20 d. Fifteen days ago, he was found to have a 1.8 cm×1.7 cm solid mass in the anterior lobe of right liver by B ultrasound, and MRI of portal vein showed occlusion and tumor thrombosis of the right liver branch of portal vein, and he was admitted on September 11, 2000. Blood test showed HBsAg (+), anti-HBs (-), HBeAg (+), anti-HBe (-), anti-HBc (+), HBV DNA (+). He underwent OLT on November 3, 2000. The resected liver showed poorly differentiated hepatocellular carcinoma with cirrhosis. Liver biopsy showed mild to moderate acute hepatic allograft rejection 1 wk and 9 mo after OLT, and acute viral hepatitis 19 mo after OLT, the immunohistological staining of HBsAg was positive. Metastatic hepatocellular carcinoma of the right adrenal gland was found 20 mo after OLT, which was confirmed by pathology. The patient stopped lamivudine therapy himself because of bad financial condition.
Patient 2 was a 56-year-old man. He suffered from viral hepatitis 22 years ago and improved after treatment. He developed cirrhosis 6 years ago. CT and B ultrasound examination showed a 10.3 cm×10.3 cm solid mass in liver and tumor thrombosis in portal vein. He was admitted on December 26, 2000. Blood test showed HBsAg (+), anti-HBs (-) HBeAg (-), anti-HBe (+), anti-HBc (+), HBV DNA (+). He underwent OLT on January 16, 2001. The resected liver showed cirrhosis secondary to viral hepatitis with massive coagulative necrosis. Liver biopsy showed no acute cellular rejection and other lesions 3 wk after OLT, and acute viral hepatitis 16 mo after OLT, the immunohistological staining of HBsAg was positive. He died of upper gastrointestinal hemorrhage and hepatic encephalopathy 25 mo after OLT.
Amplification and sequencing of HBV DNA polymerase gene fragment including YMDD motif
HBV DNA was extracted from 200 μL of serum samples after digestion with proteinase K (0.8 g/L) at 56 °C for 4 h in 20 mmol/L Tris-HCl (pH 8.0), 10 mmol/L EDTA, and 1 g/L sodium dodecyl sulfate. After a final extraction by chloroform, DNA was precipitated by absolute ethanol and dissolved in 50 µL of H2O. For amplification of the DNA polymerase gene including YMDD motif, PCR was performed using primers BF105 and BR112 (Table 1 and Figure 1)[18]. The PCR reaction mixture contained 1 µL DNA template, 0.5 µL primers, 1.5 mmol/L MgCl2, 200 µmol/L dNTP, 0.5 u pfu polymerase, 12.5 mmol/L KCl in a total volume of 25 µL. The amplification conditions included an initial denaturation for 4 min at 94 °C, 35 cycles of amplification with denaturation at 94 °C for 1 min, annealing at 58 °C for 1 min, extension at 72 °C for 3 min, followed by a final extension at 72 °C for 7 min. PCR-negative samples were further amplified by nested PCR using 1 µL of the above PCR products by primers BF111 and BR109 under the same PCR conditions. PCR-amplified DNA was purified after agarose gel electrophoresis and sequenced directly.
Table 1.
Primers used in the present study.
| Primer | Nucleotide Sequence | Nucleotide | Direction |
| BF105 | 5’-TCCTGCTGCTATGCCTCATC-3’ | 411-430 | Sense |
| BF107 | 5’-CCAGGAACATCAACCACCAG-3’ | 485-504 | Sense |
| BR112 | 5’-TTCCGTCGACATATCCCATGAAGTTAAGGGA-3’ | 895-865 | Antisense |
| BF111 | 5’-TCGGACGGAAACTGCACTTG-3’ | 581-600 | Sense |
| BF108 | 5’-ATGCCGTTTCTCCTGGCTCA-3’ | 655-674 | Sense |
| BR109 | 5’-AAGGGAGTAGCCCCAACGTT-3’ | 870-851 | Antisense |
| YNSspI | 5’-TTTCCCCCACTGTTTGGCTTTCAGTAATAT-3’ | 711-740 | Sense |
| TMPpu10I | 5’-CAGACTTGGCCCCCAATACCACATCATGCA-3’ | 769-740 | Antisense |
| TMApaLI | 5’-CAGACTTGGCCCCCAATACCACATCGTGCA-3’ | 769-740 | Antisense |
Figure 1.
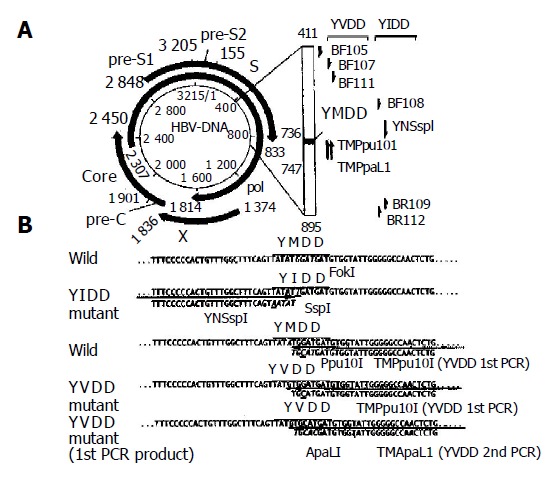
Schematic representation of analyzed partial DNA polymerase gene and primers used to detect two YMDD mutants (A) and nucleotide sequences of DNA polymerase gene including YMDD motif and primers for specific detection of both YIDD and YVDD mutants (B).
Sensitive and specific detection of YMDD mutant by PCR-RFLP-based assay
To detect two YMDD mutants, YIDD and YVDD, we designed PCR primers that could introduce a restriction enzyme recognition site into only PCR products amplified from mutant viruses. To detect YIDD mutant, 1 µL of the extracted HBV DNA solution was initially amplified using primers BF108 and BR112. Twenty-five cycles of PCR (at 94 °C for 1 min, at 58 °C for 1 min, at 72 °C for 1.5 min) were performed after initial denaturation at 94 °C for 4 min, followed by a final extension at 72 °C for 7 min. The second PCR was performed with primer YNSspI, which introduced SspI site into only mutant PCR products (Figure 1), and BR109 using 1 µL of the first PCR product and similar time-temperature conditions. Five microliters of the second PCR product was digested with 5 units of restriction enzyme Ssp I (Takara, Shuzo Co., Kusatsu, Japan) and was subjected to electrophoresis on 40 g/L agarose gel.
To detect YVDD mutant, similar amplification was performed using primers BF107 and TMPpu 10I in the first PCR, and TMApaL I and BF111 in the second PCR. Digestion by the restriction enzyme ApaLI, which digested only DNA fragment from YVDD mutant, was performed before agarose gel electrophoresis.
To enhance the detection of both YMDD mutants, we used the restriction enzyme FokI that could digest amplified DNA from wild type but not YIDD mutant, and Ppu10 I that could also digest DNA fragment amplified from the wild type using primer TMPpu 10I but not YVDD mutant. The first PCR products (1 µL) were digested by either of the two enzymes, and used in the second amplification as described above.
RESULTS
Clinicopathological data
The clinicopathological data of patient 1 before and after OLT are summarized in Figure 2A. The patient developed HBV reinfection 19 mo after OLT. Liver biopsy showed that the portal tracts were slightly enlarged because of mild to moderate chronic inflammatory cell infiltration with focal piecemeal necrosis. Disarray of single necrotic hepatocytes and chronic inflammation were found in the parenchyma (Figure 3A). Cholestasis was evident in both hepatocytes and canaliculi. Knodell score was 3 (1 score for periportal/bridging necrosis, portal inflammation, and lobular necrosis respectively; 0 score for fibrosis; the total score was 3). Immunohistochemical staining was positive for HBsAg in most hepatocytes (Figure 3B).
Figure 2.
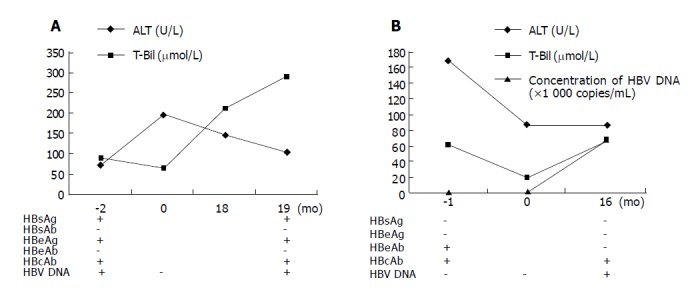
Clinicopathological data of patient 1 (A) and patient 2 (B) before and after OLT.
Figure 3.
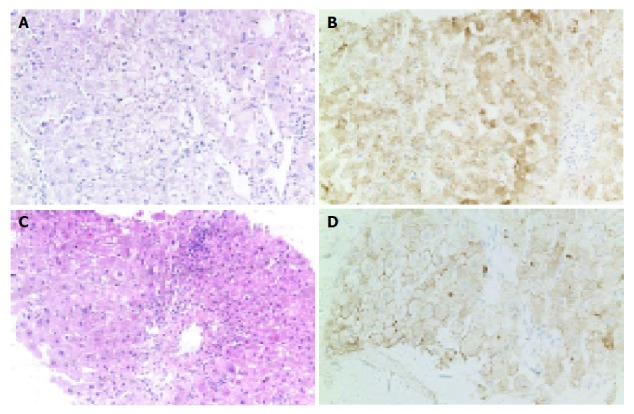
Liver biopsy of patients 1 (A and B) and 2 (C and D) 19 and 16 mo after OLT. A and C: HE, ×20; B and D: positive immunohistochemical staining for HBsAg in hepatocytes (IHC, ×20).
The clinicopathological data of patient 2 before and after OLT are summarized in Figure 2B. The patient developed HBV reinfection 16 mo after OLT. Liver biopsy showed that the portal tracts were enlarged because of moderate chronic inflammatory cell infiltration and mild fibrosis with obvious piecemeal necrosis. Disarray of many necrotic hepatocytes and chronic inflammation were found in the parenchyma (Figure 3C). Knodell score was 7 (2 scores for periportal/bridging necrosis, portal inflammation, and lobular necrosis respectively; 1 score for fibrosis; the total score was 7). Immunohistochemical staining was positive for HBsAg in hepatocytes (Figure 3D).
In short, patient 2 had more obvious hepatic lesions than patient 1, showing more evident degeneration and necrosis of hepatocytes, piecemeal necrosis and fibrosis of portal tracts. However, the immunohistochemical staining for HBsAg in patient 2 was weaker than that in patient 1.
Result of PCR-amplified HBV DNA sequencing and PCR-RFLP
The PCR-amplified products of a 289-bp fragment of the DNA polymerase gene of HBV including YMDD motif were seen in patients 1 and 2, but not in control (Figure 4). They were purified and analyzed by direct sequencing.
Figure 4.
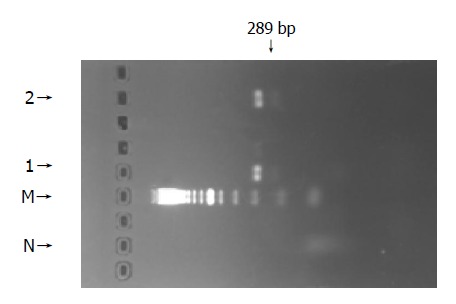
PCR-amplified products of a 289-bp fragment of DNA polymerase gene of HBV including YMDD motif in patients 1 and 2. N: normal; M: DNA marker; 1: patient 1; 2: patient 2.
Detection of YIDD mutant A weak 159-bp and a strong 159-bp products were amplified by nested PCR with and without FokI restriction enzyme digestion after first PCR. The nested PCR product was digested with SspI restriction enzyme, which did not cut the sequence, suggesting that HBV of patient 1 was YMDD wild without YIDD mutant (Figure 5A).
Figure 5.
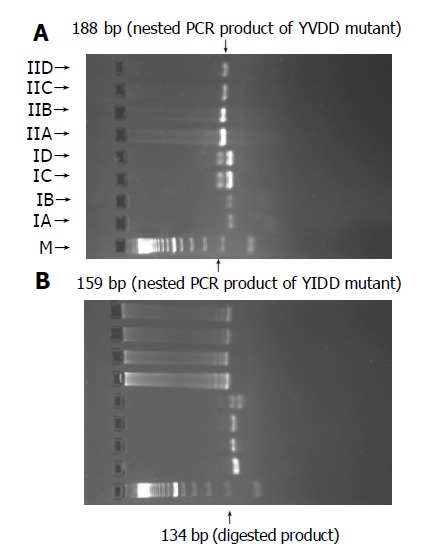
PCR-RFLP analysis pattern of patient 1 (A) and patient 2 (B).
Detection of YVDD mutant A strong 188-bp product was amplified by nested PCR with and without FokI restriction enzyme digestion after first PCR. The nested PCR product was digested with ApaLI restriction enzyme, which did not cut the sequence compared with IIC, suggesting that HBV of patient 1 was YMDD wild, without YVDD mutant (Figure 5A). The DNA sequence of patient 1 was TAT ATG GAT GAT from positions 132 to 142 of nested PCR product, its corresponding amino acid sequence was YMDD (Figure 6A).
Figure 6.
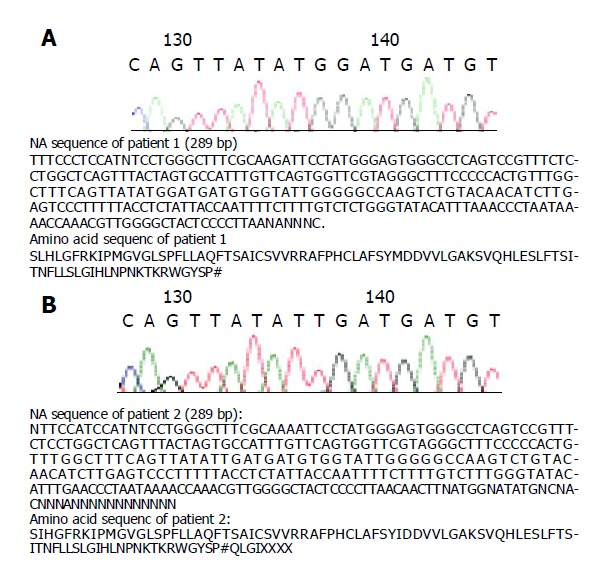
DNA sequence of patient 1 (A) and patient 2 (B).
Detection of YIDD mutant A strong 159-bp product was amplified by nested PCR with and without FokI restriction enzyme digestion after first PCR. The nested PCR product was digested with Ssp I restriction enzyme, which cut the sequence as a 134-bp fragment and a 25-bp fragment, suggesting that HBV of patient 2 was YIDD mutant (Figure 5B).
Detection of YVDD mutant A strong 188-bp product was amplified by nested PCR with and without FokI restriction enzyme digestion after first PCR. The nested PCR product was digested with ApaLI restriction enzyme, which did not cut the sequence compared with IIC, suggesting that HBV of patient 2 was not YVDD mutant (Figure 5B). The DNA sequence of patient 2 was TAT ATT GAT GAT from positions 132 to 142 of nested PCR product, its corresponding amino acid sequence was YIDD (Figure 6B).
DISCUSSION
Both patients developed HBV reinfection 19 and 16 mo after OLT, respectively. HBV was wild-type YMDD in patient 1, and YIDD mutant in patient 2. The clinicopathological data of patient 2 showed a sudden rise in HBV DNA titer during administration of lamivudine, suggesting the occurrence of YMDD mutant of HBV DNA polymerase gene and lamivudine resistance. Synthesis of HBV DNA genome occurs within the viral nucleocapsid in a mechanically ordered fashion. The nucleocapsid contains small pores that permit influx of nucleotide triphosphates and metabolites of nucleoside analogs such as lamivudine for DNA synthesis[19-22]. Lamivudine is a cytidine analog that has been shown to be effective in inhibiting HIV and HBV replication[23-26]. Lamivudine inhibits HIV and HBV replication by suppressing the activity of viral reverse transcriptase and acts as a chain terminator. HBV DNA polymerase activity in the wild-type YMDD motif is inhibited by lamivudine[27-30]. The mutation of methionine to isoleucine or valine in the YMDD motif of HBV reverse transcriptases confers resistance to lamivudine[19] because the side groups of isoleucine and valine of the YMDD mutants sterically prevent lamivudine from appropriately configuring into the nucleotide binding site of the reverse transcriptase. Moreover, in our cases, the immunohistochemical staining for HBsAg of patient 2 was weaker than that of patient 1, indicating that the replication rate of YIDD mutant HBV is lower than that of wild-type YMDD HBV, being consistent with the study of Seta et al[31]. In addition, patient 2 had more obvious hepatic lesions than patient 1, suggesting that the pathogenic mechanism of mutants may be different from that of wild-type YMDD.
Furthermore, the prevalence and characteristics of mutations identified by sequencing and PCR-RFLP are completely identical to those of Chayama et al[18]. However, our study found that the detectability of YMDD mutants was good by direct nested PCR without Fok I/Ppu10 I digestion of the first PCR product, showing that the test steps are markedly simplified.
Occurrence of lamivudine-resistant HBV variants is commonly detected by direct sequencing of HBV DNA. Although sequence analysis is considered as the gold standard for characterizing HBV DNA isolates, most laboratories do not have the necessary experience or equipments to carry this test. However, this assay is excessively time-consuming for a large number of clinical samples. Several simpler techniques have been developed to study the viral polymorphism, including type-specific PCR, PCR and single-strand conformation polymorphism (PCR-SSCP), PCR and hybridization with specific probes, but not all are suitable for identifying mutations in some fragments of the HBV sequence. PCR-SSCP assay is one of the easiest and fastest methods available for studying specific mutations and large-scale screening of polymorphisms in the general population. The advantage of PCR-SSCP over PCR-RFLP resides in the fact that it is a single-step process and does not require enzyme digestion of the PCR products. However, in a recent study, to investigate the occurrence of lamivudine-resistant HBV variants, a small fragment (only 58 bp) of a conserved sequence including position 552 of the polymerase gene is amplified and 86 mutations are found in 4 638 nucleotides, with no changes in the YMDD motif[32]. Since these mutations occur in wild-type HBV, PCR-SSCP is not suitable for the study of YMDD variants.
In summary, PCR-RFLP is a simple, rapid, accurate method for genotyping lamivudine-resistant HBV variants without requiring special equipments.
Footnotes
Supported by the National Natural Science Foundation of China, No. 30170363; the Major Science and Technology Program of Ministry of Education, No. 01003; the Major State Basic Research Development Program of China, No. 2002CB513100; the National Key Technology Research and Development Program of China during the 10th Five-Year Plan Period, No. 2001BA703B05
References
- 1.Hanazaki K. Antiviral therapy for chronic hepatitis B: a review. Curr Drug Targets Inflamm Allergy. 2004;3:63–70. doi: 10.2174/1568010043483908. [DOI] [PubMed] [Google Scholar]
- 2.Lake JR, Wright TL. Liver transplantation for patients with hepatitis B: what have we learned from our results? Hepatology. 1991;13:796–799. [PubMed] [Google Scholar]
- 3.Wong DK, Cheung AM, O'Rourke K, Naylor CD, Detsky AS, Heathcote J. Effect of alpha-interferon treatment in patients with hepatitis B e antigen-positive chronic hepatitis B. A meta-analysis. Ann Intern Med. 1993;119:312–323. doi: 10.7326/0003-4819-119-4-199308150-00011. [DOI] [PubMed] [Google Scholar]
- 4.Fu L, Liu SH, Cheng YC. Sensitivity of L-(-)2,3-dideoxythiacytidine resistant hepatitis B virus to other antiviral nucleoside analogues. Biochem Pharmacol. 1999;57:1351–1359. doi: 10.1016/s0006-2952(99)00073-8. [DOI] [PubMed] [Google Scholar]
- 5.Ling R, Mutimer D, Ahmed M, Boxall EH, Elias E, Dusheiko GM, Harrison TJ. Selection of mutations in the hepatitis B virus polymerase during therapy of transplant recipients with lamivudine. Hepatology. 1996;24:711–713. doi: 10.1002/hep.510240339. [DOI] [PubMed] [Google Scholar]
- 6.Nevens F, Main J, Honkoop P, Tyrrell DL, Barber J, Sullivan MT, Fevery J, De Man RA, Thomas HC. Lamivudine therapy for chronic hepatitis B: a six-month randomized dose-ranging study. Gastroenterology. 1997;113:1258–1263. doi: 10.1053/gast.1997.v113.pm9322520. [DOI] [PubMed] [Google Scholar]
- 7.Chan HL, Chui AK, Lau WY, Chan FK, Hui AY, Rao AR, Wong J, Lai EC, Sung JJ. Outcome of lamivudine resistant hepatitis B virus mutant post-liver transplantation on lamivudine monoprophylaxis. Clin Transplant. 2004;18:295–300. doi: 10.1111/j.1399-0012.2004.00163.x. [DOI] [PubMed] [Google Scholar]
- 8.Yuen MF, Kato T, Mizokami M, Chan AO, Yuen JC, Yuan HJ, Wong DK, Sum SM, Ng IO, Fan ST, et al. Clinical outcome and virologic profiles of severe hepatitis B exacerbation due to YMDD mutations. J Hepatol. 2003;39:850–855. doi: 10.1016/s0168-8278(03)00388-x. [DOI] [PubMed] [Google Scholar]
- 9.Beckebaum S, Cicinnati VR, Gerken G, Broelsch CE. Treatment of hepatitis B reinfection after liver transplantation. Minerva Chir. 2003;58:705–716. [PubMed] [Google Scholar]
- 10.Ben-Ari Z, Pappo O, Zemel R, Mor E, Tur-Kaspa R. Association of lamivudine resistance in recurrent hepatitis B after liver transplantation with advanced hepatic fibrosis. Transplantation. 1999;68:232–236. doi: 10.1097/00007890-199907270-00012. [DOI] [PubMed] [Google Scholar]
- 11.Perrillo R, Hann HW, Mutimer D, Willems B, Leung N, Lee WM, Moorat A, Gardner S, Woessner M, Bourne E, et al. Adefovir dipivoxil added to ongoing lamivudine in chronic hepatitis B with YMDD mutant hepatitis B virus. Gastroenterology. 2004;126:81–90. doi: 10.1053/j.gastro.2003.10.050. [DOI] [PubMed] [Google Scholar]
- 12.Lagget M, Rizzetto M. Current pharmacotherapy for the treatment of chronic hepatitis B. Expert Opin Pharmacother. 2003;4:1821–1827. doi: 10.1517/14656566.4.10.1821. [DOI] [PubMed] [Google Scholar]
- 13.Kim KH, Lee KH, Chang HY, Ahn SH, Tong S, Yoon YJ, Seong BL, Kim SI, Han KH. Evolution of hepatitis B virus sequence from a liver transplant recipient with rapid breakthrough despite hepatitis B immune globulin prophylaxis and lamivudine therapy. J Med Virol. 2003;71:367–375. doi: 10.1002/jmv.10503. [DOI] [PubMed] [Google Scholar]
- 14.Hann HW, Fontana RJ, Wright T, Everson G, Baker A, Schiff ER, Riely C, Anschuetz G, Gardner SD, Brown N, et al. A United States compassionate use study of lamivudine treatment in nontransplantation candidates with decompensated hepatitis B virus-related cirrhosis. Liver Transpl. 2003;9:49–56. doi: 10.1053/jlts.2003.50005. [DOI] [PubMed] [Google Scholar]
- 15.Ben-Ari Z, Daudi N, Klein A, Sulkes J, Papo O, Mor E, Samra Z, Gadba R, Shouval D, Tur-Kaspa R. Genotypic and phenotypic resistance: longitudinal and sequential analysis of hepatitis B virus polymerase mutations in patients with lamivudine resistance after liver transplantation. Am J Gastroenterol. 2003;98:151–159. doi: 10.1111/j.1572-0241.2003.07178.x. [DOI] [PubMed] [Google Scholar]
- 16.Perrillo RP. How will we use the new antiviral agents for hepatitis B? Curr Gastroenterol Rep. 2002;4:63–71. doi: 10.1007/s11894-002-0039-6. [DOI] [PubMed] [Google Scholar]
- 17.Bock CT, Tillmann HL, Torresi J, Klempnauer J, Locarnini S, Manns MP, Trautwein C. Selection of hepatitis B virus polymerase mutants with enhanced replication by lamivudine treatment after liver transplantation. Gastroenterology. 2002;122:264–273. doi: 10.1053/gast.2002.31015. [DOI] [PubMed] [Google Scholar]
- 18.Chayama K, Suzuki Y, Kobayashi M, Kobayashi M, Tsubota A, Hashimoto M, Miyano Y, Koike H, Kobayashi M, Koida I, et al. Emergence and takeover of YMDD motif mutant hepatitis B virus during long-term lamivudine therapy and re-takeover by wild type after cessation of therapy. Hepatology. 1998;27:1711–1716. doi: 10.1002/hep.510270634. [DOI] [PubMed] [Google Scholar]
- 19.Tipples GA, Ma MM, Fischer KP, Bain VG, Kneteman NM, Tyrrell DL. Mutation in HBV RNA-dependent DNA polymerase confers resistance to lamivudine in vivo. Hepatology. 1996;24:714–717. doi: 10.1002/hep.510240340. [DOI] [PubMed] [Google Scholar]
- 20.Sokal E. Lamivudine for the treatment of chronic hepatitis B. Expert Opin Pharmacother. 2002;3:329–339. doi: 10.1517/14656566.3.3.329. [DOI] [PubMed] [Google Scholar]
- 21.Shapira R, Daudi N, Klein A, Shouval D, Mor E, Tur-Kaspa R, Dinari G, Ben-Ari Z. Seroconversion after the addition of famciclovir therapy in a child with hepatitis B virus infection after liver transplantation who developed lamivudine resistance. Transplantation. 2002;73:820–822. doi: 10.1097/00007890-200203150-00030. [DOI] [PubMed] [Google Scholar]
- 22.Lagget M, Rizzetto M. Treatment of chronic hepatitis B. Curr Pharm Des. 2002;8:953–958. doi: 10.2174/1381612024607054. [DOI] [PubMed] [Google Scholar]
- 23.Benhamou Y, Dohin E, Lunel-Fabiani F, Poynard T, Huraux JM, Katlama C, Opolon P, Gentilini M. Efficacy of lamivudine on replication of hepatitis B virus in HIV-infected patients. Lancet. 1995;345:396–397. doi: 10.1016/s0140-6736(95)90388-7. [DOI] [PubMed] [Google Scholar]
- 24.Pramoolsinsup C. Management of viral hepatitis B. J Gastroenterol Hepatol. 2002;17 Suppl:S125–S145. doi: 10.1046/j.1440-1746.17.s1.3.x. [DOI] [PubMed] [Google Scholar]
- 25.Chan HL, Chui AK, Lau WY, Chan FK, Wong ML, Tse CH, Rao AR, Wong J, Sung JJ. Factors associated with viral breakthrough in lamivudine monoprophylaxis of hepatitis B virus recurrence after liver transplantation. J Med Virol. 2002;68:182–187. doi: 10.1002/jmv.10185. [DOI] [PubMed] [Google Scholar]
- 26.Seehofer D, Rayes N, Steinmüller T, Müller AR, Jonas S, Settmacher U, Neuhaus R, Berg T, Neuhaus P. Combination prophylaxis with Hepatitis B immunoglobulin and lamivudine after liver transplantation minimizes HBV recurrence rates unless evolution of pretransplant lamivudine resistance. Z Gastroenterol. 2002;40:795–799. doi: 10.1055/s-2002-33874. [DOI] [PubMed] [Google Scholar]
- 27.Dumortier J, Chevallier P, Scoazec JY, Berger F, Boillot O. Combined lamivudine and hepatitis B immunoglobulin for the prevention of hepatitis B recurrence after liver transplantation: long-term results. Am J Transplant. 2003;3:999–1002. doi: 10.1034/j.1600-6143.2003.00191.x. [DOI] [PubMed] [Google Scholar]
- 28.Villamil FG. Hepatitis B: progress in the last 15 years. Liver Transpl. 2002;8:S59–S66. doi: 10.1053/jlts.2002.35782. [DOI] [PubMed] [Google Scholar]
- 29.Ben-Ari Z, Mor E, Shapira Z, Tur-Kaspa R. Long-term experience with lamivudine therapy for hepatitis B virus infection after liver transplantation. Liver Transpl. 2001;7:113–117. doi: 10.1053/jlts.2001.21308. [DOI] [PubMed] [Google Scholar]
- 30.Fontana RJ, Hann HW, Wright T, Everson G, Baker A, Schiff ER, Riely C, Anschuetz G, Riker-Hopkins M, Brown N. A multicenter study of lamivudine treatment in 33 patients with hepatitis B after liver transplantation. Liver Transpl. 2001;7:504–510. doi: 10.1053/jlts.2001.24896. [DOI] [PubMed] [Google Scholar]
- 31.Seta T, Yokosuka O, Imazeki F, Tagawa M, Saisho H. Emergence of YMDD motif mutants of hepatitis B virus during lamivudine treatment of immunocompetent type B hepatitis patients. J Med Virol. 2000;60:8–16. doi: 10.1002/(sici)1096-9071(200001)60:1<8::aid-jmv2>3.0.co;2-u. [DOI] [PubMed] [Google Scholar]
- 32.Jardi R, Buti M, Rodriguez-Frias F, Cotrina M, Costa X, Pascual C, Esteban R, Guardia J. Rapid detection of lamivudine-resistant hepatitis B virus polymerase gene variants. J Virol Methods. 1999;83:181–187. doi: 10.1016/s0166-0934(99)00125-1. [DOI] [PubMed] [Google Scholar]


