Abstract
The 1,000 plants (1KP) project is an international multi-disciplinary consortium that has generated transcriptome data from over 1,000 plant species, with exemplars for all of the major lineages across the Viridiplantae (green plants) clade. Here, we describe how to access the data used in a phylogenomics analysis of the first 85 species, and how to visualize our gene and species trees. Users can develop computational pipelines to analyse these data, in conjunction with data of their own that they can upload. Computationally estimated protein-protein interactions and biochemical pathways can be visualized at another site. Finally, we comment on our future plans and how they fit within this scalable system for the dissemination, visualization, and analysis of large multi-species data sets.
Keywords: Viridiplantae, Biodiversity, Transcriptomes, Phylogenomics, Interactions, Pathways
Introduction
The 1,000 plants (1KP) project is an international multi-disciplinary consortium that has now generated transcriptome data from over 1,000 plant species. One of the goals of our species selection process was to provide exemplars for all of the major lineages across the Viridiplantae (green plants), representing approximately one billion years of evolution, including flowering plants, conifers, ferns, mosses and streptophyte green algae. Whereas genomics has long strived for completeness within species (e.g., every gene in the species), we were focused on completeness across an evolutionary clade – obviously not every species, but one representative species for everything at some phylogenetic level (e.g., one species per family, and perhaps more than one species when the family is especially large). Because many of our species had never been subjected to large-scale sequencing, 2 gigabases (Gb) of data per sample was sufficient to increase the number of plant genes by approximately 100-fold in comparison to the totality of the public databases.
The 1KP project began as a public-private partnership, with 75% of the funding provided by the Government of Alberta and 25% by Musea Ventures. Significant in-kind contributions were provided by BGI-Shenzhen in the form of reduced sequencing costs and by the NSF-funded iPlant collaborative [1] in the form of computational informatics support. Many plant scientists from around the world were involved in the collection of live tissue samples and in the extraction of RNA. Additional computing resources were provided by Compute Canada and by the China National GeneBank. Despite the constraints of this funding model, we released our data (on a collaborative basis) to scientists who approached us with goals that did not compete with ours. For the general community, access was provided through a BLAST portal [2].
We believed that there would be intrinsic value in data of this nature that is beyond our imagination. But for the initial publication, we agreed on two objectives. Firstly, by adopting a phylogenomics approach we hoped to resolve many of the lingering uncertainties in species relationships, especially in the early lineages of streptophyte green algae and land plants, where previous analyses were based on comparatively sparse taxonomic densities. And secondly, despite the limitations of these data, we hoped to identify some of the gene changes associated with the major innovations in Viridiplantae evolution, such as multicellularity, transitions from marine to freshwater or terrestrial environments, maternal retention of zygotes and embryos, complex life history involving haploid and diploid phases, vascular systems, seeds and flowers.
Our RNA extraction protocols [3] and our RNA-Seq transcriptome assembly algorithms [4] have already been published. Here, we are publishing the second of two linked papers. The first is a review of the state-of-knowledge for Viridiplantae species relationships and our initial foray into the phylogenomics on a subset of 1KP [5]. The other is a description of the websites that we created in order to provide access to the data (from raw reads to computed results), visualize the results, and perform custom analyses in conjunction with external data that the users can upload. An initial gene annotation is also provided, which focuses on the functional relationships between proteins and their associated metabolites.
Review
Access to raw and processed data
Our initial phylogenomics effort used sequences from multiple sources. They include transcriptomes from 1KP representing 85 species, transcriptomes from other sources representing 7 species, and genomes representing an additional 11 species. A summary of these data sources is given in Table 1. We submitted all of the unassembled reads from the 1KP transcriptomes to the Short Reads Archive (SRA) under project accession PRJEB4921 “1000 Plant (1KP) Transcriptome: The Pilot Study.” Note that, with the exception of Eschscholzia californica, we sequenced only one sample per species.
Table 1.
Data sources for phylogenomics analyses
| Species | Type | Accession | iPlant ID |
|---|---|---|---|
|
Arabidopsis thaliana |
genome |
n/a |
n/a |
|
Brachypodium distachyon |
genome |
n/a |
n/a |
|
Carica papaya |
genome |
n/a |
n/a |
|
Medicago truncatula |
genome |
n/a |
n/a |
|
Oryza sativa |
genome |
n/a |
n/a |
|
Physcomitrella patens |
genome |
n/a |
n/a |
|
Populus trichocarpa |
genome |
n/a |
n/a |
|
Selaginella moellendorffii |
genome |
n/a |
n/a |
|
Sorghum bicolor |
genome |
n/a |
n/a |
|
Vitis vinifera |
genome |
n/a |
n/a |
|
Zea mays |
genome |
n/a |
n/a |
|
Aquilegia formosa |
meta-assembly |
PlantGDB |
AQUI |
|
Cycas rumphii |
meta-assembly |
SRX022306, SRX022215 |
CYCA |
|
Liriodendron tulipifera |
meta-assembly |
PRJNA46857 |
LIRI |
|
Persea americana |
meta-assembly |
PRJNA46857 |
PERS |
|
Pinus taeda |
meta-assembly |
PRJNA79733 |
PINU |
|
Pteridium aquilinum |
meta-assembly |
PRJNA48473 |
PTER |
|
Zamia vazquezii |
meta-assembly |
PRJNA46857 |
ZAMI |
|
Acorus americanus |
OneKP meta-assembly |
ERR364395, PRJNA46857 |
ACOR |
|
Amborella trichopoda |
OneKP meta-assembly |
ERR364329, PRJNA46857 |
AMBO |
|
Catharanthus roseus |
OneKP meta-assembly |
ERR364390, PRJNA79951, PRJNA236160 |
CATH |
|
Eschscholzia californica |
OneKP meta-assembly |
ERR364338, ERR364335, ERR364336, ERR364337, ERR364334, SRX002988, SRX002987, PlantGDB |
ESCH |
|
Ginkgo biloba |
OneKP meta-assembly |
ERR364401, PlantGDB |
GINK |
|
Nuphar advena |
OneKP meta-assembly |
ERR364330, PRJNA46857 |
NUPH |
|
Ophioglossum petiolatum |
OneKP meta-assembly |
ERR364410, SRX666586 |
OPHI |
|
Saruma henryi |
OneKP meta-assembly |
ERR364383, PRJNA46857 |
SARU |
|
Welwitschia mirabilis |
OneKP meta-assembly |
ERR364404, PRJNA46857 |
WELW |
|
Allamanda cathartica |
OneKP |
ERR364389 |
MGVU |
|
Angiopteris evecta |
OneKP |
ERR364409 |
NHCM |
|
Anomodon attenuatus |
OneKP |
ERR364349 |
QMWB |
|
Bazzania trilobata |
OneKP |
ERR364415 |
WZYK |
|
Boehmeria nivea |
OneKP |
ERR364387 |
ACFP |
|
Bryum argenteum |
OneKP |
ERR364348 |
JMXW |
|
Cedrus libani |
OneKP |
ERR364342 |
GGEA |
|
Ceratodon purpureus |
OneKP |
ERR364350 |
FFPD |
|
Chaetosphaeridium globosum |
OneKP |
ERR364369 |
DRGY |
|
Chara vulgaris |
OneKP |
ERR364366 |
CHAR |
|
Chlorokybus atmophyticus |
OneKP |
ERR364371 |
AZZW |
|
Colchicum autumnale |
OneKP |
ERR364397 |
NHIX |
|
Coleochaete irregularis |
OneKP |
ERR364367 |
QPDY |
|
Coleochaete scutata |
OneKP |
ERR364368 |
VQBJ |
|
Cosmarium ochthodes |
OneKP |
ERR364376 |
STKJ |
|
Cunninghamia lanceolata |
OneKP |
ERR364340 |
OUOI |
|
Cyathea (Alsophila) spinulosa |
OneKP |
ERR364412 |
GANB |
|
Cycas micholitzii |
OneKP |
ERR364405 |
XZUY |
|
Cylindrocystis brebissonii |
OneKP |
ERR364378 |
YOXI |
|
Cylindrocystis cushleckae |
OneKP |
ERR364373 |
JOJQ |
|
Dendrolycopodium obscurum |
OneKP |
ERR364346 |
XNXF |
|
Dioscorea villosa |
OneKP |
ERR364396 |
OCWZ |
|
Diospyros malabarica |
OneKP |
ERR364339 |
KVFU |
|
Entransia fimbriata |
OneKP |
ERR364372 |
BFIK |
|
Ephedra sinica |
OneKP |
ERR364402 |
VDAO |
|
Equisetum diffusum |
OneKP |
ERR364408 |
CAPN |
|
Gnetum montanum |
OneKP |
ERR364403 |
GTHK |
|
Hedwigia ciliata |
OneKP |
ERR364352 |
YWNF |
|
Hibiscus cannabinus |
OneKP |
ERR364388 |
OLXF |
|
Houttuynia cordata |
OneKP |
ERR364332 |
CSSK |
|
Huperzia squarrosa |
OneKP |
ERR364407 |
GAON |
|
Inula helenium |
OneKP |
ERR364393 |
AFQQ |
|
Ipomoea purpurea |
OneKP |
ERR364392 |
VXKB |
|
Juniperus scopulorum |
OneKP |
ERR364341 |
XMGP |
|
Kadsura heteroclita |
OneKP |
ERR364331 |
NWMY |
|
Klebsormidium subtile |
OneKP |
ERR364370 |
FQLP |
|
Kochia scoparia |
OneKP |
ERR364385 |
WGET |
|
Larrea tridentata |
OneKP |
ERR364386 |
UDUT |
|
Leucodon brachypus |
OneKP |
ERR364353 |
ZACW |
|
Marchantia emarginata |
OneKP |
ERR364417 |
TFYI |
|
Marchantia polymorpha |
OneKP |
ERR364416 |
JPYU |
|
Mesostigma viride |
OneKP |
ERR364365 |
KYIO |
|
Mesotaenium endlicherianum |
OneKP |
ERR364377 |
WDCW |
|
Metzgeria crassipilis |
OneKP |
ERR364359 |
NRWZ |
|
Monomastix opisthostigma |
OneKP |
ERR364362 |
BTFM |
|
Mougeotia sp. |
OneKP |
ERR364374 |
ZRMT |
|
Nephroselmis pyriformis |
OneKP |
ERR364363 |
ISIM |
|
Netrium digitus |
OneKP |
ERR364379 |
FFGR |
|
Nothoceros aenigmaticus |
OneKP |
ERR364356 |
DXOU |
|
Nothoceros vincentianus |
OneKP |
ERR364357 |
TCBC |
|
Penium margaritaceum |
OneKP |
ERR364382 |
AEKF |
|
Podophyllum peltatum |
OneKP |
ERR364384 |
WFBF |
|
Polytrichum commune |
OneKP |
ERR364413 |
SZYG |
|
Prumnopitys andina |
OneKP |
ERR364343 |
EGLZ |
|
Pseudolycopodiella caroliniana |
OneKP |
ERR364345 |
UPMJ |
|
Psilotum nudum |
OneKP |
ERR364411 |
QVMR |
|
Pyramimonas parkeae |
OneKP |
ERR364361 |
TNAW |
|
Rhynchostegium serrulatum |
OneKP |
ERR364355 |
JADL |
|
Ricciocarpos natans |
OneKP |
ERR364358 |
WJLO |
|
Rosmarinus officinalis |
OneKP |
ERR364391 |
FDMM |
|
Rosulabryum cf. capillare |
OneKP |
ERR364351 |
XWHK |
|
Roya obtusa |
OneKP |
ERR364380 |
XRTZ |
|
Sabal bermudana |
OneKP |
ERR364400 |
HWUP |
|
Sarcandra glabra |
OneKP |
ERR364333 |
OSHQ |
|
Sciadopitys verticillata |
OneKP |
ERR364344 |
YFZK |
|
Selaginella stauntoniana |
OneKP |
ERR364347 |
ZZOL |
|
Smilax bona-nox |
OneKP |
ERR364398 |
MWYQ |
|
Sphaerocarpos texanus |
OneKP |
ERR364360 |
HERT |
|
Sphagnum lescurii |
OneKP |
ERR364414 |
GOWD |
|
Spirogyra sp. |
OneKP |
ERR364375 |
HAOX |
|
Spirotaenia minuta |
OneKP |
ERR364381 |
NNHQ |
|
Tanacetum parthenium |
OneKP |
ERR364394 |
DUQG |
|
Taxus baccata |
OneKP |
ERR364406 |
WWSS |
|
Thuidium delicatulum |
OneKP |
ERR364354 |
EEMJ |
|
Uronema sp. |
OneKP |
ERR364364 |
ISGT |
| Yucca filamentosa | OneKP | ERR364399 | ICNN |
Meta-assembly refers to a transcriptome assembled from more than one sequenced sample. Some of these were a combination of 1KP and other data; some were entirely non-1KP. Accession numbers (SRA or otherwise) are given for all of the transcriptomes that we used.
To make it easier for others to reproduce our phylogenomics analyses, we are releasing our intermediate computations, not just the final results. Everything is hosted at the iPlant Data Store, a high performance, large capacity, distributed storage system. The contents include transcriptome assemblies, putative coding sequences, orthogroups (i.e., from the 11 reference genomes), as well as gene and species trees with related sequence alignments. There are quite a lot of files and their total sizes are not negligible; so before users begin to download these files, we suggest that they consult Table 2 for a description of what to expect.
Table 2.
Number and size of data files on websites
| File count | Median size (Mb) | Average size (Mb) | Largest size (Mb) | Total size (Mb) | Similar directories | iPlant directory name |
|---|---|---|---|---|---|---|
| 68,253 |
0.0 |
0.3 |
481.1 |
23,116.6 |
|
onekp_pilot |
| 48,053 |
0.0 |
0.3 |
481.1 |
14,956.7 |
|
onekp_pilot/orthogroups |
| 19,220 |
0.1 |
0.7 |
243.8 |
13,276.5 |
|
onekp_pilot/orthogroups/alignments |
| 9,610 |
0.1 |
0.3 |
79.8 |
3,289.6 |
|
onekp_pilot/orthogroups/alignments/FAA |
| 9,610 |
0.2 |
1.0 |
243.8 |
9,986.9 |
|
onekp_pilot/orthogroups/alignments/FNA |
| 28,833 |
0.0 |
0.1 |
481.1 |
1,680.2 |
|
onekp_pilot/orthogroups/gene_trees |
| 9,611 |
0.0 |
0.1 |
481.1 |
583.3 |
|
onekp_pilot/orthogroups/gene_trees/FAA |
| 9,610 |
0.0 |
0.0 |
0.5 |
102.2 |
|
onekp_pilot/orthogroups/gene_trees/FAA/trees |
| 19,222 |
0.0 |
0.1 |
458.0 |
1,096.8 |
|
onekp_pilot/orthogroups/gene_trees/FNA |
| 9,611 |
0.0 |
0.1 |
458.0 |
556.6 |
|
onekp_pilot/orthogroups/gene_trees/FNA/12_codon |
| 9,610 |
0.0 |
0.0 |
0.5 |
98.5 |
|
onekp_pilot/orthogroups/gene_trees/FNA/12_codon/trees |
| 9,611 |
0.0 |
0.1 |
439.1 |
540.3 |
|
onekp_pilot/orthogroups/gene_trees/FNA/all_codon |
| 9,610 |
0.0 |
0.0 |
0.5 |
101.2 |
|
onekp_pilot/orthogroups/gene_trees/FNA/all_codon/dna_tree |
| 19,919 |
0.0 |
0.2 |
175.2 |
3,468.8 |
|
onekp_pilot/phylogenetic_analysis |
| 2,556 |
0.1 |
0.1 |
1.0 |
292.7 |
|
onekp_pilot/phylogenetic_analysis/alignments |
| 852 |
0.0 |
0.0 |
0.3 |
41.8 |
|
onekp_pilot/phylogenetic_analysis/alignments/FAA |
| 852 |
0.1 |
0.1 |
1.0 |
125.5 |
|
onekp_pilot/phylogenetic_analysis/alignments/FNA |
| 852 |
0.1 |
0.1 |
0.9 |
125.4 |
|
onekp_pilot/phylogenetic_analysis/alignments/FNA2AA |
| 17,197 |
0.0 |
0.1 |
0.4 |
1,827.3 |
|
onekp_pilot/phylogenetic_analysis/gene_trees |
| 1,704 |
0.0 |
0.1 |
0.4 |
238.3 |
|
onekp_pilot/phylogenetic_analysis/gene_trees/FAA |
| 2 |
0.3 |
0.1 |
0.4 |
0.3 |
852 |
onekp_pilot/phylogenetic_analysis/gene_trees/FAA/raxmlboot.#### |
| 1,704 |
0.0 |
0.1 |
0.4 |
238.3 |
|
onekp_pilot/phylogenetic_analysis/gene_trees/FNA |
| 2 |
0.3 |
0.1 |
0.4 |
0.3 |
852 |
onekp_pilot/phylogenetic_analysis/gene_trees/FNA/raxmlboot.#### |
| 3,408 |
0.0 |
0.1 |
0.4 |
476.7 |
|
onekp_pilot/phylogenetic_analysis/gene_trees/FNA2AA |
| 2 |
0.3 |
0.1 |
0.4 |
0.3 |
852 |
onekp_pilot/phylogenetic_analysis/gene_trees/FNA2AA/raxmlboot.#### |
| 2 |
0.3 |
0.1 |
0.4 |
0.3 |
852 |
onekp_pilot/phylogenetic_analysis/gene_trees/FNA2AA/raxmlboot.####.c1c2 |
| 10,381 |
0.0 |
0.1 |
0.4 |
874.0 |
|
onekp_pilot/phylogenetic_analysis/gene_trees/filtered |
| 2,548 |
0.0 |
0.1 |
0.4 |
169.3 |
|
onekp_pilot/phylogenetic_analysis/gene_trees/filtered/FAA |
| 1 |
0.0 |
0.0 |
0.0 |
0.0 |
852 |
onekp_pilot/phylogenetic_analysis/gene_trees/filtered/FAA/raxmlboot.####.f25 |
| 1 |
0.2 |
0.1 |
0.4 |
0.2 |
852 |
onekp_pilot/phylogenetic_analysis/gene_trees/filtered/FAA/raxmlboot.####.filterlen33 |
| 852 |
0.0 |
0.0 |
0.0 |
3.8 |
|
onekp_pilot/phylogenetic_analysis/gene_trees/filtered/FNA |
| 1 |
0.0 |
0.0 |
0.0 |
0.0 |
852 |
onekp_pilot/phylogenetic_analysis/gene_trees/filtered/FNA/raxmlboot.####.f25 |
| 6,980 |
0.0 |
0.1 |
0.4 |
700.9 |
|
onekp_pilot/phylogenetic_analysis/gene_trees/filtered/FNA2AA |
| 2 |
0.3 |
0.1 |
0.4 |
0.3 |
852 |
onekp_pilot/phylogenetic_analysis/gene_trees/filtered/FNA2AA/raxmlboot.####.GAMMA.2 |
| 2 |
0.3 |
0.1 |
0.4 |
0.3 |
852 |
onekp_pilot/phylogenetic_analysis/gene_trees/filtered/FNA2AA/raxmlboot.####.c1c2.GAMMA.2 |
| 1 |
0.0 |
0.0 |
0.0 |
0.0 |
852 |
onekp_pilot/phylogenetic_analysis/gene_trees/filtered/FNA2AA/raxmlboot.####.c1c2.f25 |
| 1 |
0.0 |
0.0 |
0.0 |
0.0 |
852 |
onekp_pilot/phylogenetic_analysis/gene_trees/filtered/FNA2AA/raxmlboot.####.f25 |
| 2 |
0.2 |
0.1 |
0.4 |
0.2 |
844 |
onekp_pilot/phylogenetic_analysis/gene_trees/filtered/FNA2AA/raxmlboot.####.filterlen33 |
| 1 |
0.3 |
0.3 |
0.4 |
0.3 |
180 |
onekp_pilot/phylogenetic_analysis/gene_trees/filtered/FNA2AA/raxmlboot.####.filtered25.GAMMA.2 |
| 166 |
0.0 |
8.1 |
175.2 |
1,348.8 |
|
onekp_pilot/phylogenetic_analysis/species_level |
| 50 |
15.0 |
27.0 |
175.2 |
1,348.1 |
|
onekp_pilot/phylogenetic_analysis/species_level/alignments |
| 15 |
14.7 |
14.3 |
58.3 |
214.2 |
|
onekp_pilot/phylogenetic_analysis/species_level/alignments/FAA |
| 35 |
29.4 |
32.4 |
175.2 |
1,133.9 |
|
onekp_pilot/phylogenetic_analysis/species_level/alignments/FNA |
| 116 |
0.0 |
0.0 |
0.0 |
0.6 |
|
onekp_pilot/phylogenetic_analysis/species_level/trees |
| 276 |
10.0 |
17.0 |
157.4 |
4,691.1 |
|
onekp_pilot/taxa |
| 3 |
9.7 |
17.0 |
157.4 |
51.0 |
92 |
onekp_pilot/taxa/####-############ |
| 1 |
30.8 |
17.0 |
157.4 |
36.0 |
92 |
onekp_pilot/taxa/####-############/assemblies |
| 2 |
9.7 |
7.5 |
45.2 |
15.0 |
92 |
onekp_pilot/taxa/####-############/translations |
| 5 |
0.0 |
0.0 |
0.0 |
0.1 |
|
onekp_pilot/tools |
|
File count |
Median size (Mb) |
Average size (Mb) |
Largest size (Mb) |
Total size (Mb) |
Similar directories |
Contents at SRA (PRJEB4921) |
| 178 |
1,915.0 |
2,045.5 |
3,371.0 |
364,100.0 |
|
total of all short reads -- uncompressed, but downloads are compressed to a quarter of these sizes |
| 2 | 1,915.0 | 2,045.5 | 3,371.0 | 4,091.0 | 89 | expecting per sample -- uncompressed, but downloads are compressed to a quarter of these sizes |
In some instances, users will find many directories with similar names, as indicated in this table by hash (#) marks. The total number of directories is given in the preceding column.
At the simplest level, anonymous downloads are permitted from a designated area of the iPlant Data Store [6]. However, much greater functionality is available through the iPlant resources that we describe in the following sections.
Visualization and custom analyses
To take full advantage of the iPlant computational infrastructure, it is necessary to first register at [7]. Accounts are free, and in addition to 1KP data, users will find high performance computing and cloud-based services. Multiple access modalities are supported: anonymous and secure web interfaces, desktop clients and high-speed command lines. However, we feel that for most users the best option is the iPlant discovery environment (DE), a web-based interface that provides users with high-performance computing resources and data storage. Most contemporary web browsers are supported, including Safari v. 6.1, Firefox v. 24, and Chrome v. 34. The caveat is that some of these functionalities (see below) require Java 1.6.
To guide users through its resources, iPlant is constantly producing new tutorials and teaching materials, including live and recorded webinars. The full catalog can be found at [8]. Here, we describe the new resources specifically created for 1KP.
Discovery environment (DE)
For access to the 1KP files, users should visit [9] and search for a folder called Community Data/onekp_pilot Figure 1.
Figure 1.
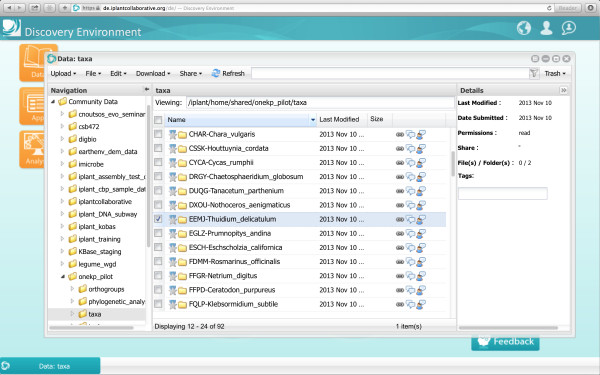
iPlant DE data window.
From the data window it is possible to download individual files or perform bulk downloads of multiple files and directories through a Java plugin. Note that for security reasons, some operating systems will not allow users to run Java applets. In this instance, a window will pop up to tell the user that there is a problem, and the user should follow the instructions that are given to configure an iDrop desktop [10] Figure 2.
Figure 2.
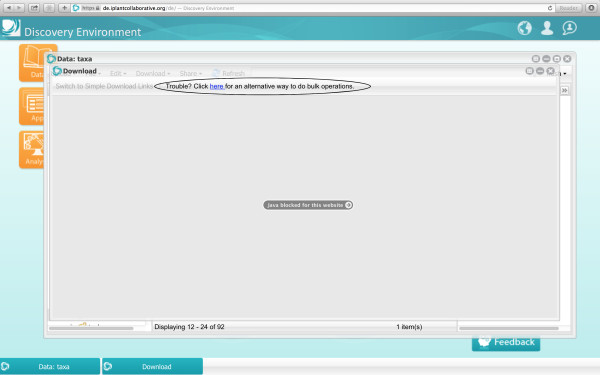
Bulk download window if Java is disabled. Click on the circled link to access the instructions to install and configure an iDrop desktop.
It is possible to perform analyses directly in the DE using any of the 1KP files as input; for example, users can re-compute the sequence alignments and gene trees using different algorithms and parameters [11] Figure 3. More generally, users can select from a variety of applications in the Apps catalogue, which is constantly growing, and includes many popular bioinformatics tools for large-scale phylogenetics, genome-wide associations and next generation sequence analyses.
Figure 3.
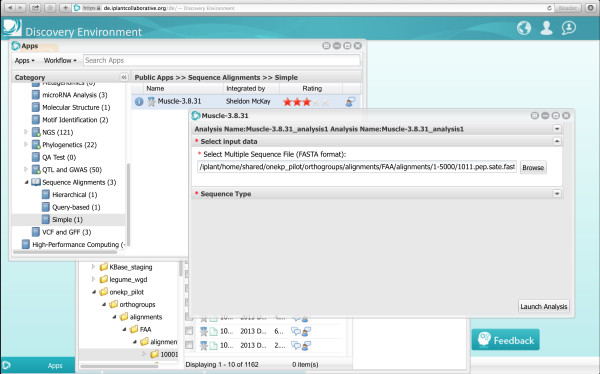
Realigning a group of sequences using Muscle.
Species and gene trees can be explored with the iPlant tree viewer, Phylozoom, a newly developed web-based phylogenetic tree viewer that supports trees with hundreds of thousand leaves and allows for semantic zooming Figure 4. To access the tree viewer, users need only click on a tree file. This will open a preview window with two tabs: one for the tree’s newick string (a format for graph-theoretical trees as defined at [12]) and another for the web link that opens a window to the tree display. Notice that pop-ups must be enabled on the user’s browser.
Figure 4.
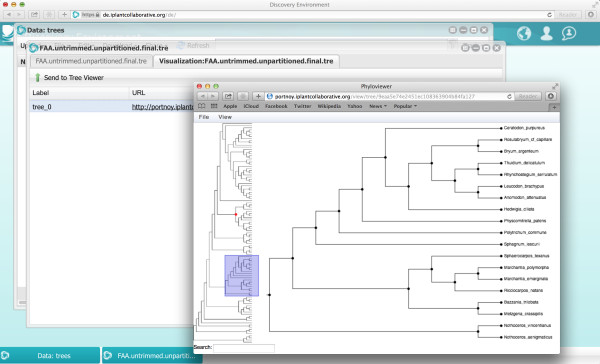
Phylozoom display of 1KP species phylogeny.
To zoom in and expand the collapsed clades, click on the node of interest. To zoom out, click and drag the tree figure to the left. To zoom out completely, click the space bar. The web address is a unique identifier that can be shared with others to let them to visualize the tree.
For more advanced users wanting to perform more complicated procedures, iPlant capabilities are available from a command line. It is based on the integrated rule-oriented data system (iRODS) [13]. All the user has to do is install a command line utility, icommands, which mimics UNIX and enables high-speed parallel data transfers. Instructions are available at [14].
Interactions and pathways
In addition to the tree-based species and gene relationships at the iPlant site, functional relationships between proteins and their associated metabolites are available from the Computational Biology Group at the University of Washington, developers of CANDO [15]. Sequence similarity-based methods are used to map 1KP proteins to curated repositories of protein-protein interactions (i.e., BioGRID [16]) and biochemical pathways (i.e., Kyoto Encylopedia of Genes and Genomes [KEGG] [17]). The user can select any metabolic pathway defined by KEGG and, within this context, see all the 1KP proteins from their chosen species, with functional annotations inferred from KEGG. This website is at [18] Figure 5.
Figure 5.
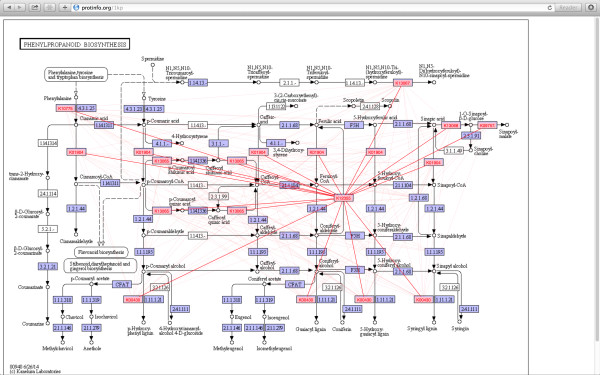
Phenylpropanoid synthesis pathway for Colchicum autumnale. Labelled rectangles are proteins. Small circles are metabolites. Black lines show the KEGG pathway. Red lines show the BioGRID interactions emanating from protein (K12355), which was interactively selected. A right-click on the protein will display the inferred function and a link to the sequence(s).
Note that, over the course of this project, there have been many improvements in the transcriptome assemblies. The phylogenomics work (now being published) was done with the SOAPdenovo algorithm. A second assembly was subsequently done with the newer SOAPdenovo-trans algorithm, which we incorporated into the newer interactions and pathways work. However, both sets of assemblies are available through the iPlant data store.
Conclusions
The rest of the 1KP data will be released, on much the same platform, along with our analyses of all one thousand species. Our scientific objectives are given at [19]. We have always been open about our intentions, because we wanted to avoid conflict among the scientists who were already working with 1KP and offer early pre-publication access to other non-competing scientists. As soon as we see a draft of a paper, we track its progress through the review process at [20]. Some of these papers have already been published, and more than a few required years of follow-up experiments, resulting for example in fundamental discoveries for molecular evolution [21] and (surprisingly) new tools for mammalian neurosciences [22].
Many of these studies were not anticipated when 1KP was conceived. We only knew that, just as there was value in sequencing every gene in a genome, despite not knowing a priori what many of the genes might do, there would be value in sequencing across an ancient and ecologically dominant clade, even when many of the species have no obvious economic or scientific value that would justify a genome sequencing effort. Transcriptomes were a less expensive way to explore plant diversity, and demonstrate value beyond the obvious species.
Abbreviations
1KP: 1,000 Plants project; DE: Discovery Environment; KEEG: Kyoto Encyclopedia of Genes and Genomes; NSF: National Science Foundation; SRA: Short Reads Archive.
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
CWD, BRR, NWM, SWG, S Ma, BS, MM, DES, PSS, CR, LP, JAS, LD, DWS, JCV, TC, TMK, MR, RSB, MKD, and JLM collected the plant samples. NM, NJW, S Mi, NN, TW, SA, MB, JGB, MAG, EW, JPD, CWD, BR, HP, BRR, and JLM performed the phylogenomic analyses. NM, LHH, ZY, and EJC setup and maintained web-resources used to communicate data. LHH and RS performed the protein and KEGG pathway analyses. EJC, ZT, XW, XS, YZ, JW, and GKW generated the sequence data. GKW and JLM designed and oversaw the research. All authors read and approved the final manuscript.
Contributor Information
Naim Matasci, Email: nmatasci@iplantcollaborative.org.
Ling-Hong Hung, Email: lhhunghimself@gmail.com.
Zhixiang Yan, Email: yanzhixiang@genomics.cn.
Eric J Carpenter, Email: ejc@ualberta.ca.
Norman J Wickett, Email: nwickett@chicagobotanic.org.
Siavash Mirarab, Email: smirarab@gmail.com.
Nam Nguyen, Email: namphuon@cs.utexas.edu.
Tandy Warnow, Email: tandy@cs.utexas.edu.
Saravanaraj Ayyampalayam, Email: raj@plantbio.uga.edu.
Michael Barker, Email: msbarker@email.arizona.edu.
J Gordon Burleigh, Email: gburleigh@ufl.edu.
Matthew A Gitzendanner, Email: magitz@ufl.edu.
Eric Wafula, Email: ekw10@psu.edu.
Joshua P Der, Email: jpd18@psu.edu.
Claude W dePamphilis, Email: cwd3@psu.edu.
Béatrice Roure, Email: beatrice.roure@umontreal.ca.
Hervé Philippe, Email: herve.philippe@ecoex-moulis.cnrs.fr.
Brad R Ruhfel, Email: ruhfel@ufl.edu.
Nicholas W Miles, Email: nicmiles@ufl.edu.
Sean W Graham, Email: swgraham@mail.ubc.ca.
Sarah Mathews, Email: smathews@oeb.harvard.edu.
Barbara Surek, Email: barbara.melkonian@uni-koeln.de.
Michael Melkonian, Email: michael.melkonian@uni-koeln.de.
Douglas E Soltis, Email: dsoltis@ufl.edu.
Pamela S Soltis, Email: psoltis@flmnh.ufl.edu.
Carl Rothfels, Email: crothfels@yahoo.ca.
Lisa Pokorny, Email: pokorny@duke.edu.
Jonathan A Shaw, Email: shaw@duke.edu.
Lisa DeGironimo, Email: ldegironimo@nybg.org.
Dennis W Stevenson, Email: dws@nybg.org.
Juan Carlos Villarreal, Email: jcarlos.villarreal@gmail.com.
Tao Chen, Email: taochen.mobg@gmail.com.
Toni M Kutchan, Email: tmkutchan@danforthcenter.org.
Megan Rolf, Email: mrolf@danforthcenter.org.
Regina S Baucom, Email: gina.baucom@gmail.com.
Michael K Deyholos, Email: deyholos@ualberta.ca.
Ram Samudrala, Email: ram@compbio.washington.edu.
Zhijian Tian, Email: tianzj@genomics.cn.
Xiaolei Wu, Email: wuxiaolei@genomics.cn.
Xiao Sun, Email: sunx@genomics.cn.
Yong Zhang, Email: zhangy@genomics.cn.
Jun Wang, Email: wangj@genomics.org.cn.
Jim Leebens-Mack, Email: jleebensmack@plantbio.uga.edu.
Gane Ka-Shu Wong, Email: gane@ualberta.ca.
Acknowledgments
The 1000 Plants (1KP) initiative, led by GKW, is funded by the Alberta Ministry of Innovation and Advanced Education, Alberta Innovates Technology Futures (AITF), Innovates Centre of Research Excellence (iCORE), Musea Ventures, BGI-Shenzhen and China National GeneBank (CNGB). We thank the many people responsible for sample collection on 1KP and the staff at BGI-Shenzhen for doing our sequencing. Phylogenomic analyses were supported by the US National Science Foundation through the iPlant collaborative. CANDO was funded by an NIH Director’s Pioneer Award 1DP1OD006779-01.
References
- Goff SA, Vaughn M, McKay S, Lyons E, Stapleton AE, Gessler D, Matasci N, Wang L, Hanlon M, Lenards A, Muir A, Merchant N, Lowry S, Mock S, Helmke M, Kubach A, Narro M, Hopkins N, Micklos D, Hilgert U, Gonzales M, Jordan C, Skidmore E, Dooley R, Cazes J, McLay R, Lu Z, Pasternak S, Koesterke L, Piel WH. et al. The iPlant Collaborative: Cyberinfrastructure for Plant Biology. Front Plant Sci. 2011;2:34. doi: 10.3389/fpls.2011.00034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1KP BLAST Search Portal. [ http://www.bioinfodata.org/app/Blast4OneKP/home]
- Johnson MT, Carpenter EJ, Tian Z, Bruskiewich R, Burris JN, Carrigan CT, Chase MW, Clarke ND, Covshoff S, dePamphilis CW, Edger PP, Goh F, Graham S, Greiner S, Hibberd JM, Jordon-Thaden I, Kutchan TM, Leebens-Mack J, Melkonian M, Miles N, Myburg H, Patterson J, Pires JC, Ralph P, Rolf M, Sage RF, Soltis D, Soltis P, Stevenson D, Stewart CN Jr. et al. Evaluating methods for isolating total RNA and predicting the success of sequencing phylogenetically diverse plant transcriptomes. PLoS One. 2012;7:e50226. doi: 10.1371/journal.pone.0050226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Y, Wu G, Tang J, Luo R, Patterson J, Liu S, Huang W, He G, Gu S, Li S, Zhou X, Lam TW, Li Y, Xu X, Wong GK, Wang J. SOAPdenovo-Trans: De novo transcriptome assembly with short RNA-Seq reads. Bioinformatics. 2014;30:1660–1666. doi: 10.1093/bioinformatics/btu077. [DOI] [PubMed] [Google Scholar]
- Wickett NJ, Mirarab S, Nguyen N, Warnow T, Carpenter E, Matasci N, Ayyampalayam S, Barker M, Burleigh JG, Gitzendanner MA, Ruhfel B, Wafula E, Der JP, Graham SW, Mathews S, Melkonian M, Soltis DE, Soltis PS, Miles NW, Rothfels C, Pokorny L, Shaw AJ, deGironimo L, Stevenson DW, Surek B, Villarreal JC, Roure B, Philippe H, dePamphilis CW, Chen T, A phylotranscriptomics analysis of the origin and early diversification of land plants. Proc Natl Acad Sci U S A. IN PRESS. [DOI] [PMC free article] [PubMed]
- iPlant Data Store for 1KP Pilot. [ http://mirrors.iplantcollaborative.org/browse/iplant/home/shared/onekp_pilot]
- iPlant User Registration. [ http://user.iplantcollaborative.org]
- iPlant Learning Center. [ http://www.iplantcollaborative.org/learning-center/all-tutorials]
- iPlant Discovery Environment. [ http://de.iplantcollaborative.org]
- Using the iDrop Desktop. [ https://pods.iplantcollaborative.org/wiki/display/DS/Using+iDrop+Desktop]
- Matasci N, McKay SJ. Phylogenetic analysis with the iPlant discovery environment. Curr Protoc Bioinformatics. 2013;6:Unit 6.13. doi: 10.1002/0471250953.bi0613s42. [DOI] [PubMed] [Google Scholar]
- Newick Trees Format. [ http://evolution.genetics.washington.edu/phylip/newicktree.html]
- iRODS Data Management Software. [ http://irods.org]
- Using iCommands (Unix) [ https://pods.iplantcollaborative.org/wiki/display/DS/Using+iCommands]
- Minie M, Chopra G, Sethi G, Horst J, White G, Roy A, Hatti K, Samudrala R. CANDO and the infinite drug discovery frontier. Drug Discov Today. 2014;19:1353–1363. doi: 10.1016/j.drudis.2014.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BioGRID Interactions. [ http://thebiogrid.org]
- Kyoto Encylopedia of Genes and Genomes (KEGG) [ http://www.genome.jp/kegg]
- 1KP Protein-Protein Interactions Mapped to Metabolic Pathways. [ http://protinfo.org/1kp]
- 1KP Capstone Objective. [ https://pods.iplantcollaborative.org/wiki/display/iptol/OneKP+Capstone+Wiki]
- 1KP Papers in Progress. [ https://pods.iplantcollaborative.org/wiki/display/iptol/OneKP+companion+papers]
- Sayou C, Monniaux M, Nanao MH, Moyroud E, Brockington SF, Thévenon E, Chahtane H, Warthmann N, Melkonian M, Zhang Y, Wong GK, Weigel D, Parcy F, Dumas R. A promiscuous intermediate underlies the evolution of LEAFY DNA binding specificity. Science. 2014;343:645–648. doi: 10.1126/science.1248229. [DOI] [PubMed] [Google Scholar]
- Klapoetke NC, Murata Y, Kim SS, Pulver SR, Birdsey-Benson A, Cho YK, Morimoto TK, Chuong AS, Carpenter EJ, Tian Z, Wang J, Xie Y, Yan Z, Zhang Y, Chow BY, Surek B, Melkonian M, Jayaraman V, Constantine-Paton M, Wong GK, Boyden ES. Independent optical excitation of distinct neural populations. Nat Methods. 2014;11:338–346. doi: 10.1038/nmeth.2836. [DOI] [PMC free article] [PubMed] [Google Scholar]


