Abstract
Meis2 is a homeodomain protein containing a conserved homothorax (Hth) domain that is present in all Meis and Prep family proteins and in the Drosophila homothorax protein. The Hth domain mediates interaction with Pbx homeodomain proteins, allowing for efficient DNA binding. Here we show that, like Meis1, Meis2 has a strong carboxyl-terminal transcriptional activation domain, which is required for full activation of transcription by homeodomain protein complexes comprised of Meis2 and Pbx1. We also show that the activity of the activation domain is inhibited by the Hth domain, and that this auto-inhibition can be partially relieved by the interaction of Pbx1 with the Hth domain of Meis2. Targeting the Hth domain to DNA suggests that it is not a portable trans-acting repression domain. However, the Hth domain can inhibit a linked activation domain, and this inhibition is not limited to the Meis2 activation domain. Database searching reveals that the Meis3.2 splice variant, which is found in several vertebrate species, disrupts the Hth domain by removing 17 codons from the 5’ end of exon 6. We show that the equivalent deletion in Meis2 derepresses the carboxyl-terminal activation domain and weakens interaction with Pbx1. This work suggests that the transcriptional activity of all members of the Meis/Prep homothorax protein family is subject to auto-inhibition by their Hth domains, and that the Meis3.2 splice variant encodes a protein which bypasses this auto-inhibitory effect.
Keywords: Meis, Pbx, homeodomain, transcription, repression
Homeodomain proteins were first identified in flies and are conserved across diverse species from yeasts to mammals (1,2). The characteristic DNA binding homeodomain is around 60 amino acids in length and consists of three alpha helices (3). It is the third alpha helix within the homeodomain that is the primary DNA binding region, although there are other DNA contacts outside helix three (4-7). In addition to binding DNA, the homeodomain is a protein interaction module, which mediates interactions with other DNA binding proteins and non-DNA binding transcriptional regulators. Homeodomain proteins can be recruited to DNA via direct DNA binding, and indirectly via interaction with other transcription factors (8,9). However, even when homeodomain proteins bind their cognate DNA binding site, they generally bind with other DNA-binding cofactors (10-12). Meis2 is a member of the TALE superfamily of homeodomain proteins, which are characterized by the presence of a three amino acid loop insertion between helices one and two of the homeodomain (13-15). The presence of this loop between helices one and two is unlikely to affect DNA binding directly, but plays a role in protein-protein interactions (6,7). TALE superfamily homeodomain proteins participate in both activating and repressing transcription factor complexes. For example, proteins such as Tgif1 and Tgif2, are obligate transcriptional repressors that are primarily recruited to DNA via interactions with other DNA binding proteins (16-18). In contrast, Meis-Pbx complexes appear to be primarily involved in transcriptional activation (9,19,20).
In humans and mice, there are three Meis paralogs and two Prep genes, which are closely related to the Meis group. Mammalian Meis1 (myeloid ecotropic insertion site 1) was identified initially as a common site of viral integration in mouse myeloid leukemia cells (21), and the related Meis2 and Meis3 genes were identified by sequence similarity (22,23). Meis1 plays a key role in the progression of AML and MLL leukemia, and fusion proteins generated by chromosomal rearrangements in MLL can induce increased expression of Meis1 (24-26). Prep1 plays a role in hematopoietic stem cell function, and in early T cell development (27-29). Pbx proteins, which are common partners of Meis family members, have also been implicated in tumorigenesis. Pbx1 can be fused to the transcription factor E2A as a result of the t(1;19) translocation in pre-B cell leukemia (30,31). This fusion prevents interaction with Meis proteins and converts Pbx1 to a strong transcriptional activator.
In addition to the homeodomain, Meis and Prep proteins share a second region of high sequence conservation termed the homothorax (Hth) domain (15,32,33). This domain is named for the Drosophila homothorax protein. The Hth domain interacts with Pbx proteins, thereby promoting cooperative binding of Meis-Pbx dimers to a composite DNA element (34,35). The interaction of Meis and Pbx partners also facilitates the binding of the Pbx partner to DNA (34). Interestingly, this requirement for a Meis partner is lost in oncogenic Pbx fusion proteins, such as the E2a-Pbx protein. Additionally, the interaction of Meis family proteins with a Pbx protein allows for recruitment of the Meis protein to a DNA bound Pbx-Hox complex, without the need for direct binding of the Meis protein to a consensus Meis site (8,9). A conformational change in Pbx1a and interaction with a Meis protein are required for nuclear localization of Pbx1 suggesting that both the Meis and Pbx partners are regulated by mutual interaction (36). Recent evidence has suggested that the p160 Myb-binding protein interacts with the Hth domain of Prep1 and is a negative regulator of Prep1-Pbx complexes (37). Thus the Hth region of Meis family proteins is clearly a key regulatory domain within these proteins that can mediate both positive and negative influences on transcriptional activity. Interestingly, splice variants of the mammalian Meis1 and Meis2, and Drosophila HTH have been identified, which encode proteins lacking the homeodomain (38,39). The Meis2e variant, which is truncated prior to the end of the first alpha helix of the homeodomain has been suggested to act as a dominant negative form of the Meis protein that may be able to interfere with the formation of fully functional Meis-Pbx complexes (39). The HTH protein that lacks the homeodomain can carry out many of the developmental functions of the full length HTH protein, but cannot substitute for it in all cases (38).
Here we demonstrate that the Meis2 and Prep1 Hth domains inhibit the ability of the full length proteins to activate transcription. In the case of Meis2, the carboxyl-terminus contains a strong transcriptional activation domain, the activity of which is inhibited by the Hth domain. This auto-inhibition can be relieved, in part, by interaction with Pbx1 and maps to a region of the Hth domain which also contributes to Pbx interaction. Finally, we show that the Meis3.2 splice variant generates a protein lacking 17 amino acids from the Hth domain. Removal of the equivalent region from Meis2 results in both decreased interaction with Pbx1 and weakened auto-inhibition.
Results
Meis2 contains a carboxyl-terminal activation domain
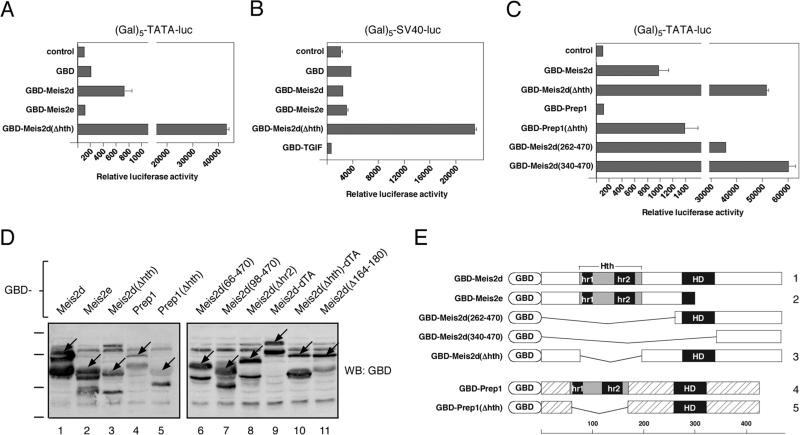
Several splice variants of Meis2 have been described, most of which affect the region carboxyl-terminal to the homeodomain, whereas Meis2e lacks most of the homeodomain and everything carboxyl-terminal to it (39). To test whether Meis2 could activate transcription we targeted both Meis2d and Meis2e to DNA by fusing them to the Gal4 DNA binding domain (GBD; see Figure 1E). When targeted to a minimal TATA element containing promoter via multiple Gal4 sites we observed several fold activation by Meis2d, but no activation by Meis2e (Figure 1A). However, this activation by Meis2d was relatively weak, particularly in light of the recent identification of a strong activation domain in the carboxyl-terminal region of the related Meis1 protein (40). Interestingly, when we deleted the Hth domain from Meis2d in the context of the GBD fusion we observed a dramatic increase in the level of transcriptional activation compared to the wild type Meis2d fusion (Figure 1A). The GBD fusion lacking the Hth domain also significantly increased transcription from the more active SV40 promoter, although the wild type Meis2d and Meis2e fusions were unable to do so (Figure 1B). No repression of SV40 promoter activity was observed by either Meis2d or Meis2e, whereas a GBD fusion to the TGIF repressor decreased activity of this reporter (Figure 1B). To test whether derepression of transcriptional activity by removal of the Hth domain might be a more general feature of Meis family proteins, we tested the activity of GBD fusions to Prep1 and a version of Prep1 lacking its Hth domain. Prep1 did not activate the TATA-containing reporter, whereas the Hth deletion mutant increased transcription at least 10-fold (Figure 1C). Importantly, the higher levels of transcriptional activation by the Hth deletion mutants did not appear to be simply a result of increased expression of these constructs compared to the wild type Meis2d or Prep1 fusions (Figure 1D). To further define the Meis2d transcriptional activation domain we tested two other GBD fusions, which contained either the Meis2 homeodomain and carboxyl-terminal region, or just the region carboxyl-terminal to the homeodomain. As shown in Figure 1C, both fusions activated gene expression to a similar degree to the Hth deletion mutant, suggesting that the approximately 150 amino acids carboxyl-terminal to the homeodomain of Meis2d contain a transcriptional activation domain.
Fig. 1.
Meis2 contains a carboxyl-terminal activation domain. HepG2 cells were transfected with the indicated GBD fusion constructs and the (Gal)5-TATA luciferase reporter (A), or the (Gal)5-SV40 reporter (B). Luciferase activity was assayed after 48 hours and is presented as the mean + s.d. of duplicate transfections (arbitrary units). C) a series of Meis2 and Prep1 deletion constructs fused to the GBD were assayed as in A. D) The relative expression of the indicated GBD-fusions was analyzed by western blot with a GBD antibody. The specific full length bands are indicated by arrows. Numbers below each lane correspond the numbered constructs in Figures 1E and 4F. The positions of molecular weight markers (95, 72, 55 and 43kD) are shown to the left. E) GBD expression constructs are shown schematically. Hth: homothorax homology domain, hr1 and hr2: homology regions 1 and 2, HD: homeodomain. Scale below shows amino acid numbers.
Both the Meis2 activation domain and the Hth domain are required for transcriptional activation by Meis/Pbx
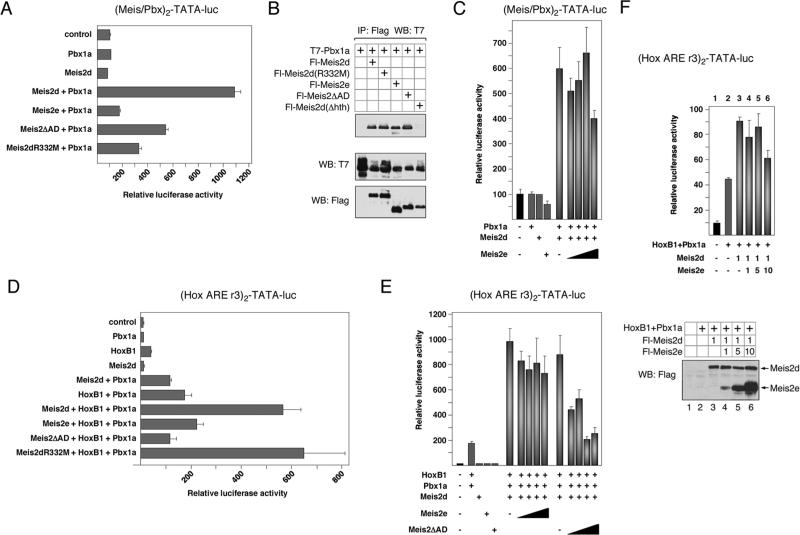
To test whether the Meis2 activation domain is required in the context of transcriptional regulation in complex with Pbx1, we tested two reporters, one in which luciferase activity is under the control of two copies of a canonical Meis/Pbx binding site and a minimal TATA element, and one with two copies of the Hoxb1 ARE r3 element (9). Coexpression of Meis2d and Pbx1 together with the Pbx/Meis reporter resulted in greater than 10-fold activation compared to the control, or to expression of either protein alone (Figure 2A). Meis2e did not activate this reporter with Pbx1, and activation was clearly impaired by deletion of the Meis2d activation domain, or by a point mutation (R332M) that decreases binding to a consensus Meis site. To confirm that these constructs were able to interact with Pbx1a, we performed co-immunoprecipitaion assays from COS1 cells transfected with T7-tagged Pbx1a and Flag-tagged Meis2d or Meis2d mutants. As shown in Figure 2B, removal of the Hth domain abolished interaction with Pbx1a. We also tested the Meis2 mutant that lacks the activation domain (Meis2d∆AD, encoding amino acids 2-345 of Meis2), and one which binds DNA poorly (R332M, contains a point mutation in helix 3 of the homeodomain, which alters a critical DNA contact residue), and both retained interaction with Pbx1a. Importantly, expression levels of both the R332M mutant and the activation domain deletion mutant were similar to wild type Meis2d.
Fig. 2.
The Meis2 activation domain is required for Pbx-dependent transcriptional activation. A) HepG2 cells were transfected with the indicated expression constructs and a luciferase reporter in which luciferase expression is driven by two copies of a Meis/Pbx consensus binding site and a minimal TATA element. Meis2d(ΔAD) encodes amino acids 2-345 of Meis2, so lacks the activation domain, and the R332M mutant has a point mutation in the homeodomain which prevents binding to a consensus Meis site. B) COS1 cells were transfected with T7-tagged Pbx1a and the indicated Flag tagged Meis2 expression constructs. Complexes were isolated on Flag agarose and analyzed for co-precipitating T7-Pbx1a. Expression in the lysates is shown below. C) Cells were transfected and analyzed as in A, with increasing amounts of co-expressed Meis2e. D) HepG2 cells were transfected with the indicated Meis2 expression constructs and HoxB1 or Pbx1 expression constructs as indicated, together with a luciferase reporter containing two copies of the Hox ARE r3 element which binds Hox and Pbx proteins. E) The effect of expressing increasing amounts of either the Meis2e splice variant, or the activation domain deletion mutant of Meis2 on Hox ARE luciferase reporter activity was assayed as in C. Triangles in C and E represent ratios of 1:1, 1:2, 1:4, 1:6 of Meis2d to Meis2e or Meis2dΔAD. F) HepG2 cells were assayed as in E, with the indicated ratios of transfected Meis2d and Meis2e. Expression of the Meis2 proteins was assayed by Flag western blot (right). Numbers 1-6 above the luciferase data correspond to lanes 1-6 of the blot.
We next tested the possibility that Meis2e might interfere with activation by Meis2d and Pbx1. However, as shown in Figure 2C, even when co-transfected at a 5-fold excess relative to Meis2d, we observed minimal inhibition by Meis2e of the Pbx/Meis reporter. Meis family proteins can also be recruited to DNA without the requirement for DNA binding, via interactions with other homeodomain proteins, such as Pbx1 and Hox proteins. To test the importance of the Meis2d activation domain for this mode of transcriptional regulation we used a reporter based on the Hoxb1 ARE, which contains a composite binding site for Pbx1 and Hoxb1, but lacks a Meis2 consensus site. Transfection of either Meis2d, Pbx1a or Hoxb1 expression constructs individually did not dramatically activate this reporter (Figure 2D). However, coexpression of either Meis2d or Hoxb1 with Pbx1a resulted in 15- to 20-fold activation, and coexpression of all three proteins together activated further. In contrast, Meis2e or the activation domain deletion mutant of Meis2d failed to increase activity over that seen with Pbx1a and Hoxb1 alone (Figure 2D). As expected, since this reporter does not contain a Meis2 binding site, the R332M mutation did not affect activity. As with the Pbx/Meis reporter, we did not observe interference by over-expression of Meis2e in the presence of Meis2d, Pbx1a and Hoxb1 (Figure 2E). However, at high levels of over-expression, the Meis2d mutant lacking the activation domain was able to inhibit activation of this reporter (Figure 2E). We next tested whether further increasing Meis2e levels, with a relatively low level of Meis2d would allow for Meis2e to interfere with Meis2d function. When Meis2e was co-transfected at a ratio of up to 10:1 with Meis2d, we did observe some interference (Figure 2F). However, it should be noted that the level of Meis2d in this experiment resulted in only modest reporter activation over that seen with HoxB1 and Pbx1a alone.
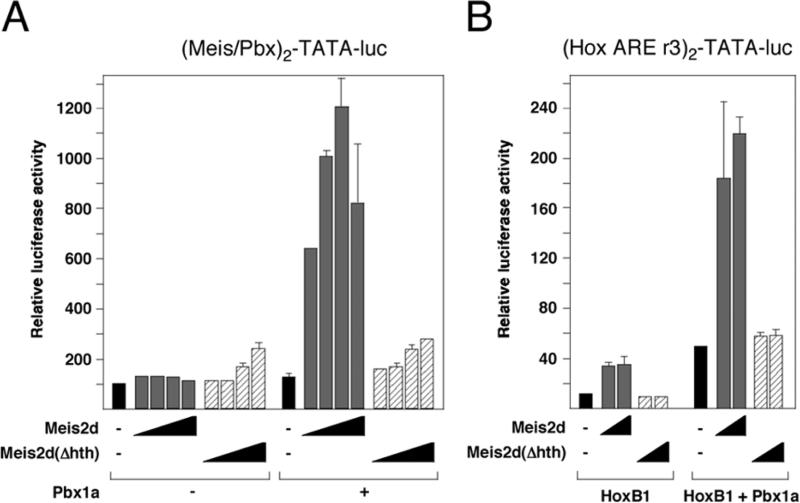
To test whether the Hth domain was required for activation of Pbx-dependent reporters by Meis2d, we expressed wild type or the Hth deletion mutant of Meis2d alone or with Pbx1a, and tested activation of the Meis/Pbx reporter and the Hoxb1 ARE. As shown in Figure 3A, we observed a small increase in activity from the Meis/Pbx reporter with the Hth deletion mutant compared to wild type Meis2d, but this mutant was unable to cooperate with Pbx1a to activate the reporter. With the Hoxb1 ARE, Meis2 lacking the Hth domain was completely non-functional, consistent with an absolute requirement for recruitment via Pbx1 (Figure 3B). Together, these results suggest that the Meis2d activation domain is required for transcriptional activation whether Meis2d binds directly to DNA or is recruited by other homeodomain proteins. Additionally, it appears that the protein encoded by the Meis2e splice variant has a limited ability to act as an effective dominant negative.
Fig. 3.
The Hth domain is required for Pbx1 dependent transcription. HepG2 cells were cotransfected with the indicated expression constructs and either the Meis/Pbx-TATA luc reporter (A) or the Hoxb1 ARE reporter (B). Luciferase activity was measured after 48 hours, and is presented as the average of duplicate transfections.
The homothorax domain inhibits the activity of a linked activation domain
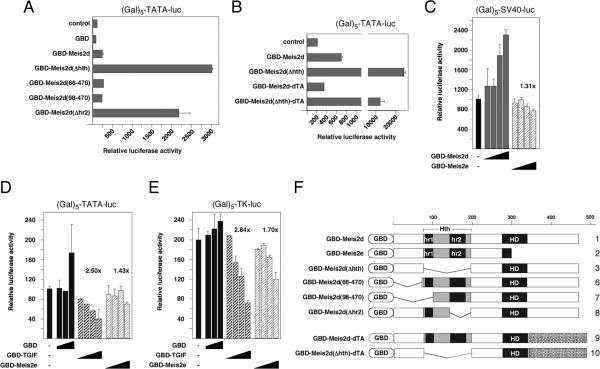
To further delineate the region required for the inhibitory effect of the Hth domain we created a series of GDB fusions (see Figure 4F). Deletion of either the amino-terminal 65 or 97 amino acids did not derepress the Meis2d activation domain, whereas, a smaller internal deletion (removing amino acids 150-193) which encompasses the hr2 region of the Hth domain derepressed to a similar degree as the full Hth deletion (Figure 4A). To test whether the inhibitory activity of the Hth domain was specific to the Meis2 activation domain, we next created an activation domain swap construct, in which the relatively proline-rich Meis2d activation domain was replaced with the acidic activation domain from the Drosophila TGIFa protein (41). As shown in Figure 4B, this chimeric construct did not activate the Gal4 reporter, but was significantly derepressed by deletion of the Hth domain, suggesting that the inhibitory effect of this domain is not specific to the Meis2d activation domain. Comparison of the relative expression levels of these GBD fusions (see Figure 1E) suggests that the increased transcriptional activation seen with Hth deletion does not correlate with expression level. To test the possibility that the Hth domain was a portable transcriptional repression domain, we targeted increasing amounts of GBD-Meis2d or GBD-Meis2e to the SV40 promoter, which has a high basal level of activity. As shown in Figure 4C, we observed a little more than 2-fold activation of this promoter by Meis2d, and little repression (1.3-fold) by Meis2e, which lacks the activation domain, but retains the Hth domain. We next compared the effects of targeting either Meis2e or TGIF to two promoters with a lower basal activity than the SV40 promoter. As shown in Figure 4D and E, the GBD-TGIF fusion resulted in a maximal repression of at least 2.5-fold for both reporters, whereas we observed much lower level repression by GBD-Meis2e. However, on the Gal-TK reporter, GBD-Meis2e was able to repress by up to 1.7-fold (a 42% reduction in activity), suggesting that it may have weak repressive activity (Figure 4E). Thus, it appears that the Hth domain is able to effectively inhibit the activity of at least two different linked activation domains, but does not act as a potent general transcriptional repression domain.
Fig. 4.
The Hth domain inhibits a linked activation domain. HepG2 cells were cotransfected with the Gal-TATA luciferase reporter (A, B), or the Gal-SV40 reporter (C) and the indicated GBD-Meis2 fusions. The effects of increasing amounts of GBD or GBD fusions to TGIF and Meis2e were tested on the Gal-TATA luciferase (D) or Gal-TK-luciferase (E) reporters. F) the GBD-Meis2 fusion constructs are shown schematically. Hth: homothorax homology domain, hr1 and hr2: homology regions 1 and 2, HD: homeodomain. The activation domain from Drosophila TGIFa is indicated as dTA.
Mutational analysis of the Hth domain
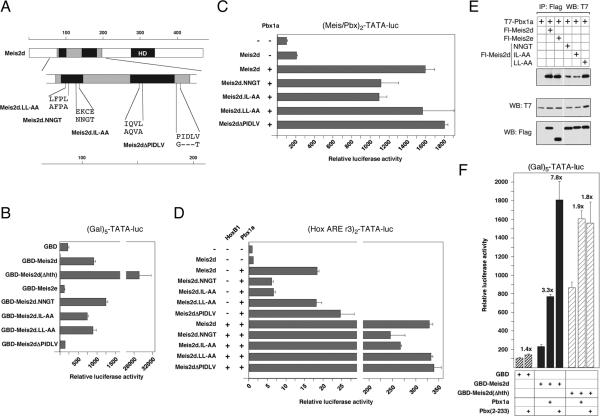
Previous work has identified point mutations within the Hth domain which weaken interaction with Pbx1 (35). An interaction between Prep1 and the transcriptional repressor, p160Mybbp1, has been mapped to the Prep1 Hth domain, and specifically to a leucine-rich motif in hr1 (37). To test whether Pbx1 or p160Mybbp1 interaction might contribute to the inhibitory effect of the Hth domain, we created three GBD-Meis2d mutants, which should affect either Pbx1 interaction (NNGT and IL-AA; see Figure 5A) or interaction with both Pbx1 and p160Mybbp1 (LL-AA). In addition, we noticed a relatively close match to the consensus interaction motif for CtBP (PxDL[R/S/T], (42); PIDLV in Meis2), which is missing from our hr2 and Hth deletion constructs. Since this sequence is conserved in most Meis relatives, except the Prep sub-family (see Figure 6A), we also created a mutant lacking the PIDLV. We first tested the effects of targeting the GBD fusions to the TATA containing luciferase reporter. As shown in Figure 5B, none of these mutations resulted in significant derepression of the GDB-Meis2d construct. When we tested the effects of the NNGT and IL-AA mutations on transcription using the Pbx/Meis and Hox ARE reporters, we observed some decrease in activity in the presence of Pbx1a relative to that seen with wild type Meis2d and Pbx1a, consistent with a weakened Pbx1 interaction (Figure 5C and D). In contrast, we did not see any effect of either the LL-AA or ΔPIDLV mutations, and none of these mutations resulted in increased Meis2d transcriptional activity as would be expected if they affected the inhibitory function of the Hth domain. To verify that the Pbx1 interaction mutants (NNGT and IL-AA) did indeed affect interaction with Pbx1, we performed co-immunoprecipitation experiments from transfected COS1 cells. As shown in Figure 4E, significantly less Pbx1a co-precipitated with the NNGT and IL-AA mutant forms of Meis2d than with the wild type, whereas the LL-AA mutant had little effect in this assay.
Fig. 5.
Pbx1 derepresses GBD-Meis2d. A) The Meis2d Hth domain is shown schematically, together with the sequence of four mutant forms of Meis2d. B) HepG2 cells were transfected with GBD-Meis2 expression constructs and the (Gal)5-TATA luciferase reporter, and luciferase activity measured after 48 hours. The indicated Meis2 expression constructs were coexpressed with Pbx1a and HoxB1, as indicated, and luciferase activity from the Meis/Pbx reporter (C) or Hox ARE reporter (D) was assayed after 48 hours. E) The indicated Flag-tagged Meis2 mutants, Meis2d or Meis2e were coexpressed with T7-tagged Pbx1a in COS1 cells. Protein complexes were isolated on Flag agarose and analyzed for coprecipitating T7-Pbx1a. Expression in the lysates is shown below. F) HepG2 cells were transfected with GBD-Meis2 expression constructs and the (Gal)5-TATA luciferase reporter, together with T7-tagged Pbx1a or a truncation mutant which encodes the amino-terminal 233 amino acids (including the Meis2 interaction domains). Luciferase activity was measured after 48 hours.
Fig. 6.
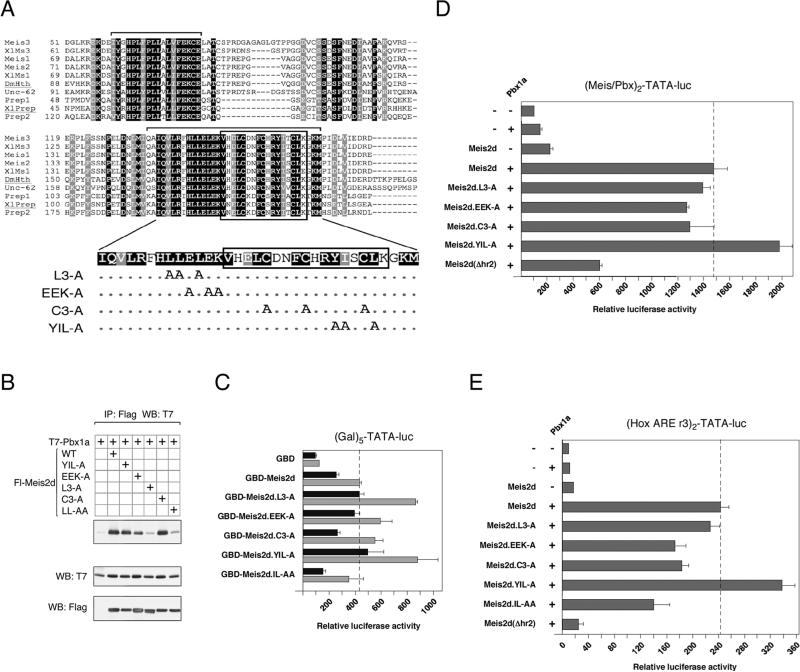
Mutational analysis of hr2. A) An alignment of the Hth domains from Meis relatives is shown. Amino acids which are identical or similar between all sequences shown are shaded black and gray respectively. The sequences shown are Meis1, Meis2, Meis3, Prep1 and Prep2 from human, Xenopus laevis Meis1, Meis3 and Prep (XlMs1, XlMs3 and XlPrep), the Drosophila melanogaster HTH protein (DmHth) and a Meis-like protein from C. elegans (Unc-62). Brackets above the sequences indicate the hr1 and hr2 regions. Mutations within the Meis2 hr2 are shown below. Dots indicate no change. B) COS1 cells were transfected with the indicated Flag-tagged Meis2 expression constructs and T7-Pbx1a. Proteins were isolated on Flag agarose and the presence of coprecipitating Pbx1a analyzed by T7 western blot. Expression in the lysates is shown below. C) Two amounts of each of the indicated GBD-Meis2d fusions were cotransfected into HepG2 cells with the (Gal)5-TATA luciferase reporter, and luciferase activity was assayed after 48 hours. The dashed line indicates the maximum activation level by Meis2d. HepG2 cells were transfected with the indicated Meis2d, Pbx1a and HoxB1 expression constructs together with the Meis/Pbx reporter (D) or Hox ARE reporter (E), and luciferase activity was determine after 48 hours. The dashed lines indicate activity with wild type Meis2d.
Since the Pbx-interaction mutants in the hr2 domain of Meis2 failed to derepress Meis2d transcriptional activity, we tested the alternative possibility, that interaction with Pbx might help alleviate the inhibitory effect of the hr2 domain. To do this, we used GBD fusions to Meis2d and the Hth deletion mutant, and coexpressed either full length Pbx1a, or the amino-terminal 233 amino acids of Pbx1a, which contains the Meis interaction domains. As shown in Figure 5F, we observed a 3.3-fold increase in the activity of GBDMeis2d with full length Pbx1a, and an almost 8-fold increase in the presence of the amino-terminal fragment of Pbx1a. In contrast, there was relatively little effect on the Hth deletion mutant of Meis2d, even when a low level of GBD-Meis2d(ΔHth) was used such that an increase in activity on this reporter would be easily detectable. This data suggests that interaction of Pbx1a with the Hth region can to some degree relieve the inhibitory effect of hr2 on transcriptional activation.
Pbx interaction is separable from auto-inhibition
The Hth domain of Meis2 contains two regions, termed hr1 and hr2, which are highly conserved from flies to mammals, and are present in multiple Meis paralogs (Figure 6A). Since the hr2 domain appeared to be most important for inhibition of transcriptional activity, we generated a series of mutant forms of Meis2d, in which we changed charged and hydrophobic residues to alanines (Figure 6A). We also noticed that the hr2 domain contains three highly conserved cysteine residues, which we also converted to alanines. We first tested whether these four Meis2d mutants were expressed at similar levels to the wild type, and whether they were able to interact with Pbx1a. As shown in Figure 6B, all four mutants were expressed at similar levels to wild type Meis2d, and all appeared to interact with Pbx1a to some degree. However, the interaction of the L3-A mutant with Pbx1a was reduced by at least as much as by the previously described LL-AA mutation. Additionally, the EEK-A mutant was somewhat impaired for Pbx1a interaction. Next we tested the effects of these mutations on transcriptional activity using the Gal4 system. Two amounts of each GBD-Meis2 fusion were transfected together with the Gal-TATA luciferase reporter. Of the four mutant forms of Meis2, we observed around two-fold derepression with two of them, the L3-A and YIL-A mutants, while the others showed a similar activity in this assay to the wild type (Figure 6C). We next tested the effect of these mutations on activation of the Pbx/Meis and Hox ARE reporters. As shown in Figure 6D and E, only the YIL-A mutant resulted in any increase in activity over that seen with the wild type Meis2d. The L3-A mutation which derepressed in the GBD fusion assay failed to do so with these reporters presumably due to its decreased interaction with Pbx1a. These data suggest that interaction with Pbx1a and the auto-inhibition activity are separable functions.
Alternate splicing of Meis3 affects the Meis auto-inhibitory domain
Several Meis2 splice variants have been identified, which primarily affect the region carboxyl-terminal to the homeodomain (39). However, we were interested to know whether alternate splicing of Meis2 or other Meis family members might affect the auto-inhibitory function of the Hth domain. Database searching revealed the presence of two isoforms of human Meis3 (termed Meis3.1 and Meis3.2), which were also found in the EST database. Although only a single mouse Meis3 isoform is listed in GenBank, two forms that are equivalent to human Meis3.1 and 3.2 can be found in the mouse EST database. Interestingly, Meis3.1 encodes a protein with the full Hth domain, whereas the Meis3.2 splice variant lacks 17 codons from the 5’ end of exon 6 (Figure 7A). The region missing in Meis3.2 encodes the equivalent of amino acids 164-180 in Meis2, which comprise about half of the hr2 domain (see Figure 6A). To confirm that the two isoforms of Meis3 were indeed expressed, we performed RT-PCR analysis on RNA from HepG2 cells using primers which span intron 5 and exon 6 of Meis2 or Meis3 and would be expected to generate two products if both isoforms were expressed. As shown in Figure 7B, we amplified PCR products of the expected size for both Meis3.1 and Meis3.2, whereas only a single longer isoform of Meis2 was detected, suggesting that the alternate splicing event is specific to Meis3. Comparison of the genomic structures of Meis1, 2 and 3 and Prep1 reveals that the three Meis genes in both mice and humans have a similar overall structure at least up to exon 6, whereas in Prep1 a single exon encompasses the equivalent of exons 5 and 6 from Meis3. Of the three Meis genes, intron 5 is considerably smaller (less than 200bp) in human and mouse Meis3 than in either of the other genes. Examination of the 5’ and 3’ splice sites surrounding intron 5 provides some clues as to why Meis3 may undergo this alternate splicing event. Position 5 of the 5’ splice site in Meis3 is a guanosine (* in Figure 7A), which is characteristic of genes that undergo alternate splicing, whereas in Meis1 and 2, this residue is an adenosine, which correlates with constitutive splicing (43). Although the 3’ splice site in Meis3 is actually a better match to the consensus than in Meis1 or 2, the region upstream of this, within intron 5 of Meis3, is almost completely devoid of adenosines (only 3 out of the first 74 bases, excluding the 3’ splice site, are adenosines). In Meis3 no good match to the branchpoint consensus is present, whereas the Meis1 and Meis2 introns have better branchpoint consensus sequences (44). Additionally, Meis1 is unlikely to undergo a similar alternate splicing event, since a match to the consensus 3’ splice site is not found at the same internal position within exon 6. To determine how widely the Meis3.2 isoform was expressed, we performed RT-PCR on RNA isolated from several human cell lines and mouse tissues, using PCR primers that span the alternate splice junction in mouse or human Meis3. The relative intensities of the bands corresponding to the Meis3.1 and Meis3.2 splice variant were then quantified. As shown in Figure 7C, the Meis3.2 variant represented around 25% of the total Meis3 message in most human cell lines tested. In the prostate cancer metastasis-derived cell line, LNCaP, the majority of the Meis3 was Meis3.2, suggesting that some variation is possible. Analysis of a panel of mouse tissues, taken form wild type C57BL/6J mice, revealed that the Meis3.2 variant represented between 20% and 50% of the total (Figure 7D). Thus it appears that this alternate splice form of Meis3 represents a significant proportion of the total Meis3 in both mouse tissues and human cell lines, at least at the mRNA level.
Fig. 7.
A Meis3 splice variant disrupts the Hth domain. A) Meis3.1 and 3.2 splice variants are shown schematically. The first few amino acids encoded at each splice junction are shown in one letter code. The sequences at the splice junctions, together with exon and intron lengths are shown below, for mouse and human Meis1, 2 and 3. The consensus splice sequences are shown below, with identical bases shaded black. The asterisk indicates the base which correlates with alternate or constitutive splicing. B) The presence of alternate splicing around the 5’ end of exon 6 of Meis2 and Meis3 was tested by RT-PCR. The positions of molecular weight markers are shown to the left, and the size in base pairs of the products to the right (the Meis2 equivalent of Meis3.2 would be expected at 149bp). C and D) RNA from a series of human cell lines (C) or mouse tissues (D) was analyzed by RT-PCR using primers that span the alternate splice site in Meis3, such that both the Meis3.1 and Meis3.2 isoforms were amplified. The relative amount of each splice form as a percentage of the total Meis3 is plotted in the upper panels. Representative RT-PCR reactions are shown below. E) The indicated Flag-tagged Meis2 constructs, were coexpressed with T7-tagged Pbx1b, or a deletion mutant lacking the homeodomain (amino acids 2-233) in HeLa cells. Protein complexes were isolated on Flag agarose and analyzed for coprecipitating T7-Pbx1b. Expression in the lysates is shown below. F) Each of the indicated GBD-Meis2d fusions, or GBD alone, was cotransfected into HepG2 cells with the (Gal)5-TATA luciferase reporter, and luciferase activity was assayed after 48 hours.
To test whether removal of the sequence encoded by the first 17 codons of exon 6 might affect Meis function, we created a version of Meis2d in which amino acids 164-180 were deleted. This generates the Meis2d equivalent of Meis3.2 to allow for comparison with our previous mutational analysis. We first tested the effects of this deletion on Pbx-dependent transcriptional reporters, and observed no increase in activity over that seen with Meis2d (data not shown). To test the possibility that the lack of effect on Pbx-dependent reporters was due to changes in the ability of the deletion mutant to interact with Pbx1, we performed coimmunoprecipitation experiments from transfected HeLa cells. As shown in Figure 7E, the mutants of Meis2d lacking either amino acids 164-180, or lacking the entire hr2 region were both dramatically reduced in their ability to interact with Pbx1. Although there was still some residual interaction of Meis2d lacking amino acids 164-180 with full length Pbx1, this was lost when we used a deletion mutant of Pbx1 [Pbx1(2-233)] which lacks the homeodomain, but not the Meis interaction domains (Figure 7E). To test effects on Pbx-independent transcriptional activation we created a GBD fusion to the Meis2d mutant lacking amino acids 164-180. As shown in Figure 7F, deletion of amino acids 164-180 from the GBD-Meis2d fusion resulted in a 3.3-fold increase in transcriptional activity over that seen with wild type Meis2d. Together, these data suggest that the Meis3.2 splice variant produces a protein which is unable to interact with Pbx1, but is also relieved of the auto-inhibitory effect of the Hth domain.
Discussion
We have shown that Meis2d, like Meis1 contains a carboxyl-terminal transcriptional activation domain. The activity of the activation domain is inhibited by the conserved Hth domain, and this auto-inhibitory activity appears to be a general feature of Meis family proteins.
Previous work has identified a transcriptional activation domain carboxyl-terminal to the homeodomain of Meis1a (40). When assayed as a GBD fusion, the carboxyl-terminal half of Meis1a (amino acids 232-390, which lacks the Hth domain) had robust transcriptional activity, as shown here for Meis2d. However, the activity of the full length Meis1a was not tested, and based on our work we expect that its activity would be inhibited by the conserved Hth domain. The Meis1a isoform, in which the carboxyl-terminal activation domain was mapped, is equivalent to the Meis2a splice variant, and these two proteins share 74% identity and 80% similarity over their carboxyl-terminal domains. Comparison of the Meis2d isoform analyzed here to the public databases reveals a predicted splice variant of Meis1 (Meis1e, gb accession: EAW99896), which shares 75% identity (86% similarity) over the 132 amino acid domain carboxyl-terminal to the homeodomain in Meis2d. We, therefore, suggest that the auto-inhibitory function of the Hth domain in Meis2d is likely to be a common feature of Meis family proteins. Switching the activation domain of Meis2d for that of an unrelated protein still allowed for auto-inhibition suggesting that this function is not dependent on a specific activation domain, and supporting the notion that it may function for all Meis paralogs. Co-expression of Pbx1a was able to partially relieve the inhibitory effect of the Hth domain on Meis2d, at least in the GBD fusion assay. However, this derepression by Pbx1 was not very robust, perhaps suggesting that another factor, or other signals are required to fully de-repress Meis2d.
The Meis2e splice variant retains the Hth domain, but lacks both the homeodomain and activation domain. It could therefore interact with Pbx, but would be unable to bind to DNA or contribute a transcriptional activation domain, if recruited to DNA. One possibility is that Meis2e represents a naturally occurring dominant negative form of Meis2, that might be able to interfere by competing with other Meis isoforms for binding to Pbx1, for example. Our attempts to test this possibility met with limited success; we observed an interfering effect of Meis2e only when expressed at very high levels relative to Meis2d. This may not be surprising when both the Pbx and Meis partners bind DNA, since the formation of a complex of Meis2d and Pbx1 on DNA would likely be more stable than one in which Meis2e is unable to contact DNA. Where Meis2 is recruited without the need for it to bind to DNA, such as via Hox/Pbx complexes, Meis2e might be expected to be better able to interfere. Even with the Hox ARE reporter we observed relatively little inhibition by even high levels of Meis2e, perhaps suggesting that it is less well incorporated into a DNA bound Pbx/Hox complex. However, it remains possible that this may represent a normal function for Meis2e and similar Meis isoforms created by alternate splicing.
Although the Hth domain was effective at limiting the activity of a linked activation domain, Meis2e had relatively little repression activity when targeted to DNA via a heterologous DNA binding domain. An alternative possibility for the function of Meis2e-like proteins is provided by work on Drosophila HTH, which has been shown to encode a full length isoform and one lacking the homeodomain (38). In Drosophila most HTH functions could be performed by both isoforms, although for antenna development only full length HTH was sufficient. It may, therefore, be that Meis2e like proteins are functional for some activities, but that some processes can only be carried out by full length Meis paralogs. The lack of a dramatic dominant negative or repressive effect of Meis2e in our assays is consistent with this interpretation, although we show that the Meis2d activation domain contributes to transcriptional activation by Meis/Pbx/Hox complexes. Since Meis2e lacks both DNA binding and activation domains, it is not clear what positive functions such a protein might have. One possibility is that if recruited to DNA via interaction with other proteins, it might act to prime specific genes for later activation by Meis2d. However, in the case of the Drosophila HTH variant which lacks the homeodomain, the full length protein was unable to substitute completely during fly development, suggesting that there may be functions specific to the homeodomain-less versions of Meis related proteins (38).
Recent work has shown an interaction between Prep1 and the repressor, p160Mybbp1, which is mediated via the hr1 domain of Prep1 (37). However, our data suggest that p160Mybbp1 recruitment is not responsible for the auto-inhibitory function of the Meis2 Hth domain. Sub-cellular localization of Meis2d might also be expected to affect its ability to activate transcription. If the Hth domain was responsible for maintaining cytoplasmic localization of Meis2d, then its deletion might be expected to derepress activity, and the auto-inhibition could be relived by binding to Pbx1, if this allowed for nuclear entry. Although the localization of Prep1 to the nucleus has been shown to be dependent on interaction with Pbx1, a deletion mutant of Prep1 lacking the Hth domain was cytoplasmic in the absence or presence of Pbx1 (45). Thus the nuclear/cytoplasmic localization of Prep1, and possibly other Meis paralogs, may play a role in regulating transcriptional activity, but it appears that the Hth domain does not maintain cytoplasmic localization of Prep1. Additionally, the greatest derepression we observed was in the context of the GBD fusions, which contain an NLS within the GBD part of the fusion. An alternative possibility for the observed auto-inhibitory activity is that the Hth domain mediates some intra-molecular interaction, or affects the conformation of Meis2d. The ability of Pbx1 to somewhat derepress the GBD-Meis2d fusion would fit with this model if interaction of Pbx1 with the Hth domain altered the conformation or intra-molecular interactions allowing access to the activation domain. Since the auto-inhibition affected an unrelated activation domain, when this was put in place of the native Meis2d activation domain, it appears that any intra-molecular interactions with the Hth domain are likely to be with regions of Meis2 other than its activation domain.
Of the three Meis and two Prep genes present in humans, alternate splicing appears to affect the Hth region of only Meis3. This alternate splicing event removes 51 nucleotides from exon 6, creating Meis3.2 in humans. The intron-exon structure of Meis1, 2 and 3 is relatively well conserved in this region of the genes – in both mouse and human the 17 codons removed in human Meis3.2 are present at the 5’ end of exon 6 of all three genes. In contrast, in the Prep1 gene, the equivalent of exons 5 and 6 in the Meis genes are present in a single exon. Database searching reveals the presence of multiple ESTs from both mouse and human Meis3, which represent the 3.2 isoform, and we show that a similar Meis3 isoform is also present in multiple mouse tissues. Semi-quantitative RT-PCR suggests that the Meis3.2 splice variant represents 20-50% of the total Meis3 mRNA expressed in most mouse tissues and human cell lines. It may, therefore, represent a significant portion of the functional Meis3 protein. However, further work will be required to determine the relative levels of the proteins encoded by these two splice variants. Some ESTs that likely encode a similar Meis3 isoform are present from pig, cow and zebra fish. Despite the overall conservation between Meis paralogs, there is no evidence for alternate splicing of Meis1 and Meis2 creating a similar isoform. It has been suggested that Pbx proteins are the major DNA binding partners for Meis proteins, consistent with the presence of an intact Hth domain in the majority of Meis isoforms (34). However, we suggest that the Meis3.2 splice variant encodes a Pbx- independent Meis protein, which will bind DNA independent of Pbx, and does not possess the auto-inhibitory function of the Hth domain.
In summary, our data suggest that one function of the conserved Hth domain is to inhibit the activity of the transcriptional activation domain of Meis family proteins. This auto-inhibition can be relieved by interaction with Pbx, suggesting that this may provide a mechanism to better control the transcriptional activity of Meis proteins.
Experimental Procedures
Plasmids
Flag and T7 epitope tagged expression constructs were generated in a modified pCMV5 by PCR. Gal4 DNA binding domain (GBD) fusions were created within pM (Clontech, Mountain View, CA). Gal4 luciferase reporters were as previously described (18). The Pbx/Meis site and Hox ARE reporters were created in pGL2 basic (Promega). Briefly, a double stranded oligonucleotide containing the Adenovirus major late TATA element was inserted into the BglII and HindIII sites, as previously described (18). Double stranded oligonucleotides containing either a consensus Meis2 and Pbx1 binding site or the Pbx/Hox binding site from the Hox B1 ARE were phosphorylated with polynucleotide kinase (NE Biolabs) and ligated into the TATA-luc vector. Oligonucleotide sequences for reporters were (upper strand only). Pbx/Meis: GATCGTTGATTGACAGA, Hox ARE: GATCGGGTGATGGATGGGCC.
Luciferase assays
HepG2 cells were transfected with firefly luciferase reporters and a phCMVRLuc control (Promega, Madison, WI), together with appropriate expression constructs, using Exgen 500 (MBI Fermentas, Hanover, MD). After 48 hours promoter activity was assayed with luciferase assay reagent (Biotium) using a Berthold LB953 luminometer. Results were standardized using Renilla luciferase activity, assayed with 0.09μM colenterazine (Biosynth, Naperville, IL).
Immunoprecipitation and western blotting
COS1 and HeLa cells were maintained in DMEM with 10% BGS (Hyclone, Logan, UT) and were transfected using LipofectAmine (Invitrogen, Carlsbad, CA). Thirty-six hours after transfection, cells were lysed by sonication in 75 mM NaCl, 50 mM HEPES, pH 7.8, 20% glycerol, 0.1% Tween 20, 0.5% NP40 with protease and phosphatase inhibitors. Immunocomplexes were precipitated with Flag M2-agarose (Sigma, St. Louis, MO). Following SDS-polyacrylamide gel electrophoresis, proteins were electroblotted to Immobilon-P (Millipore, Billerica, MA) and incubated with antisera specific for Flag (Sigma) or T7 epitope tags (Novagen). The GBD antibody was from Cell Signaling.
RT-PCR
RNA was isolated and purified using Absolutely RNA kit (Stratagene). For qRT-PCR, cDNA was generated using Superscript III (Invitrogen), and analyzed by PCR using a DNA engine cycler and Promega Taq. Intron spanning primer pairs were selected using Primer3 (http://frodo.wi.mit.edu/). Oligonucleotides for RT-PCR: Meis2-F: AGGACATCGCGGTCTTCG, Meis2-R: GAGGTCGATGGGCATTTTC, Meis3-F: GATGATCCAGGCCATCCA, Meis3-R: GGCTGGGTAGTCCTCGAAGT, mMeis3-F: GTCCAGGCCATCCAGGTACT, mMeis3R: TCCTCCCTGCAACTACCATC. The relative intensities of the Meis3.1 and Meis3.2 bands were quantified using ImageJ software, from PCR reactions that had not left the linear range.
References
- 1.McGinnis W, Garber RL, Wirz J, Kuroiwa A, Gehring WJ. A homologous protein-coding sequence in Drosophila homeotic genes and its conservation in other metazoans. Cell. 1984;37(2):403–8. doi: 10.1016/0092-8674(84)90370-2. [DOI] [PubMed] [Google Scholar]
- 2.McGinnis W, Levine MS, Hafen E, Kuroiwa A, Gehring WJ. A conserved DNA sequence in homoeotic genes of the Drosophila Antennapedia and bithorax complexes. Nature. 1984;308(5958):428–33. doi: 10.1038/308428a0. [DOI] [PubMed] [Google Scholar]
- 3.Gehring WJ, Affolter M, Burglin T. Homeodomain proteins. Annu. Rev. Biochem. 1994;63:487–526. doi: 10.1146/annurev.bi.63.070194.002415. [DOI] [PubMed] [Google Scholar]
- 4.Chang CP, Shen WF, Rozenfeld S, Lawrence HJ, Largman C, Cleary ML. Pbx proteins display hexapeptide-dependent cooperative DNA binding with a subset of Hox proteins. Genes Dev. 1995;9(6):663–74. doi: 10.1101/gad.9.6.663. [DOI] [PubMed] [Google Scholar]
- 5.Gehring WJ, Qian YQ, Billeter M, Furukubo-Tokunaga K, Schier AF, Resendez-Perez D, Affolter M, Otting G, Wuthrich K. Homeodomain- DNA recognition. Cell. 1994;78(2):211–23. doi: 10.1016/0092-8674(94)90292-5. [DOI] [PubMed] [Google Scholar]
- 6.Passner JM, Ryoo HD, Shen L, Mann RS, Aggarwal AK. Structure of a DNA-bound Ultrabithorax-Extradenticle homeodomain complex [see comments]. Nature. 1999;397(6721):714–9. doi: 10.1038/17833. [DOI] [PubMed] [Google Scholar]
- 7.Piper DE, Batchelor AH, Chang C-P, Cleary ML, Wolberger C. Structure of a HoxB1-Pbx1 heterodimer bound to DNA: Role of the hexapeptide and a fourth homeodomain helix in complex formation. Cell. 1999;96:587–597. doi: 10.1016/s0092-8674(00)80662-5. [DOI] [PubMed] [Google Scholar]
- 8.Jacobs Y, Schnabel CA, Cleary ML. Trimeric association of hox and TALE homeodomain proteins mediates hoxb2 hindbrain enhancer activity [In Process Citation]. Mol Cell Biol. 1999;19(7):5134–42. doi: 10.1128/mcb.19.7.5134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shanmugam K, Green NC, Rambaldi I, Saragovi HU, Featherstone MS. PBX and MEIS as non-DNA-binding partners in trimeric complexes with HOX proteins. Mol Cell Biol. 1999;19(11):7577–88. doi: 10.1128/mcb.19.11.7577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chang CP, Brocchieri L, Shen WF, Largman C, Cleary ML. Pbx modulation of Hox homeodomain amino-terminal arms establishes different DNA-binding specificities across the Hox locus. Mol Cell Biol. 1996;16(4):1734–45. doi: 10.1128/mcb.16.4.1734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Knoepfler PS, Kamps MP. The pentapeptide motif of Hox proteins is required for cooperative DNA binding with Pbx1, physically contacts Pbx1, and enhances DNA binding by Pbx1. Mol Cell Biol. 1995;15(10):5811–9. doi: 10.1128/mcb.15.10.5811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mann RS, Chan SK. Extra specificity from extradenticle: the partnership between HOX and PBX/EXD homeodomain proteins. Trends Genet. 1996;12(7):258–62. doi: 10.1016/0168-9525(96)10026-3. [published erratum appears in Trends Genet 1996 Aug;12(8):328].
- 13.Bertolino E, Reimund B, Wildt-Perinic D, Clerc R. A novel homeobox protein which recognizes a TGT core and functionally interferes with a retinoid-responsive motif. J. Biol. Chem. 1995;270:31178–31188. doi: 10.1074/jbc.270.52.31178. [DOI] [PubMed] [Google Scholar]
- 14.Burglin TR. Analysis of TALE superclass homeobox genes (MEIS, PBC, KNOX, Iroquois, TGIF) reveals a novel domain conserved between plants and animals. Nucl. Acids Res. 1997;25:4173–4180. doi: 10.1093/nar/25.21.4173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mukherjee K, Burglin TR. Comprehensive analysis of animal TALE homeobox genes: new conserved motifs and cases of accelerated evolution. J Mol Evol. 2007;65(2):137–53. doi: 10.1007/s00239-006-0023-0. [DOI] [PubMed] [Google Scholar]
- 16.Melhuish TA, Gallo CM, Wotton D. TGIF2 interacts with histone deacetylase 1 and represses transcription. J Biol Chem. 2001;26:26. doi: 10.1074/jbc.M103377200. [DOI] [PubMed] [Google Scholar]
- 17.Melhuish TA, Wotton D. The interaction of C-terminal binding protein with the Smad corepressor TG-interacting factor is disrupted by a holoprosencephaly mutation in TGIF. J Biol Chem. 2000 doi: 10.1074/jbc.C000416200. [DOI] [PubMed] [Google Scholar]
- 18.Wotton D, Lo RS, Swaby LA, Massague J. Multiple modes of repression by the smad transcriptional corepressor TGIF. J Biol Chem. 1999;274(52):37105–10. doi: 10.1074/jbc.274.52.37105. [DOI] [PubMed] [Google Scholar]
- 19.Berthelsen J, Zappavigna V, Ferretti E, Mavilio F, Blasi F. The novel homeoprotein Prep1 modulates Pbx-Hox protein cooperativity. EMBO J. 1998;17:1434–1445. doi: 10.1093/emboj/17.5.1434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu Y, MacDonald RJ, Swift GH. DNA binding and transcriptional activation by a PDX1.PBX1b.MEIS2b trimer and cooperation with a pancreas- specific basic helix-loop-helix complex. J Biol Chem. 2001;276(21):17985–93. doi: 10.1074/jbc.M100678200. [DOI] [PubMed] [Google Scholar]
- 21.Moskow JJ, Bullrich F, Huebner K, Daar IO, Buchberg AM. Meis1, a PBX1-related homeobox gene involved in myeloid leukemia in BXH- 2 mice. Mol Cell Biol. 1995;15(10):5434–43. doi: 10.1128/mcb.15.10.5434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nakamura T, Jenkins NA, Copeland NG. Identification of a new family of Pbx-related homeobox genes. Oncogene. 1996;13(10):2235–42. [PubMed] [Google Scholar]
- 23.Oulad-Abdelghani M, Chazaud C, Bouillet P, Sapin V, Chambon P, Dolle P. Meis2, a novel mouse Pbx-related homeobox gene induced by retinoic acid during differentiation of P19 embryonal carcinoma cells. Dev Dyn. 1997;210(2):173–83. doi: 10.1002/(SICI)1097-0177(199710)210:2<173::AID-AJA9>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- 24.Schnabel CA, Jacobs Y, Cleary ML. HoxA9-mediated immortalization of myeloid progenitors requires functional interactions with TALE cofactors Pbx and Meis. Oncogene. 2000;19(5):608–16. doi: 10.1038/sj.onc.1203371. [DOI] [PubMed] [Google Scholar]
- 25.Thorsteinsdottir U, Kroon E, Jerome L, Blasi F, Sauvageau G. Defining roles for HOX and MEIS1 genes in induction of acute myeloid leukemia. Mol Cell Biol. 2001;21(1):224–34. doi: 10.1128/MCB.21.1.224-234.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wong P, Iwasaki M, Somervaille TC, So CW, Cleary ML. Meis1 is an essential and rate-limiting regulator of MLL leukemia stem cell potential. Genes Dev. 2007;21(21):2762–74. doi: 10.1101/gad.1602107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Di Rosa P, Villaescusa JC, Longobardi E, Iotti G, Ferretti E, Diaz VM, Miccio A, Ferrari G, Blasi F. The homeodomain transcription factor Prep1 (pKnox1) is required for hematopoietic stem and progenitor cell activity. Dev Biol. 2007;311(2):324–34. doi: 10.1016/j.ydbio.2007.08.031. [DOI] [PubMed] [Google Scholar]
- 28.Penkov D, Di Rosa P, Fernandez Diaz L, Basso V, Ferretti E, Grassi F, Mondino A, Blasi F. Involvement of Prep1 in the alphabeta T-cell receptor T-lymphocytic potential of hematopoietic precursors. Mol Cell Biol. 2005;25(24):10768–81. doi: 10.1128/MCB.25.24.10768-10781.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Penkov D, Palazzolo M, Mondino A, Blasi F. Cytosolic sequestration of Prep1 influences early stages of T cell development. PLoS ONE. 2008;3(6):e2424. doi: 10.1371/journal.pone.0002424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kamps MP, Look AT, Baltimore D. The human t(1;19) translocation in pre-B ALL produces multiple nuclear E2A-Pbx1 fusion proteins with differing transforming potentials. Genes Dev. 1991;5(3):358–68. doi: 10.1101/gad.5.3.358. [DOI] [PubMed] [Google Scholar]
- 31.Kamps MP, Murre C, Sun XH, Baltimore D. A new homeobox gene contributes the DNA binding domain of the t(1;19) translocation protein in pre-B ALL. Cell. 1990;60(4):547–55. doi: 10.1016/0092-8674(90)90658-2. [DOI] [PubMed] [Google Scholar]
- 32.Moens CB, Selleri L. Hox cofactors in vertebrate development. Dev Biol. 2006;291(2):193–206. doi: 10.1016/j.ydbio.2005.10.032. [DOI] [PubMed] [Google Scholar]
- 33.Rieckhof GE, Casares F, Ryoo HD, Abu-Shaar M, Mann RS. Nuclear translocation of extradenticle requires homothorax, which encodes an extradenticle-related homeodomain protein. Cell. 1997;91(2):171–83. doi: 10.1016/s0092-8674(00)80400-6. [DOI] [PubMed] [Google Scholar]
- 34.Chang CP, Jacobs Y, Nakamura T, Jenkins NA, Copeland NG, Cleary ML. Meis proteins are major in vivo DNA binding partners for wild-type but not chimeric Pbx proteins. Mol Cell Biol. 1997;17(10):5679–87. doi: 10.1128/mcb.17.10.5679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Knoepfler PS, Calvo KR, Chen H, Antonarakis SE, Kamps MP. Meis1 and pKnox1 bind DNA cooperatively with Pbx1 utilizing an interaction surface disrupted in oncoprotein E2a-Pbx1. Proc Natl Acad Sci U S A. 1997;94(26):14553–8. doi: 10.1073/pnas.94.26.14553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Saleh M, Huang H, Green NC, Featherstone MS. A conformational change in PBX1A is necessary for its nuclear localization. Exp Cell Res. 2000;260(1):105–15. doi: 10.1006/excr.2000.5010. [DOI] [PubMed] [Google Scholar]
- 37.Diaz VM, Mori S, Longobardi E, Menendez G, Ferrai C, Keough RA, Bachi A, Blasi F. p160 Myb-binding protein interacts with Prep1 and inhibits its transcriptional activity. Mol Cell Biol. 2007;27(22):7981–90. doi: 10.1128/MCB.01290-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Noro B, Culi J, McKay DJ, Zhang W, Mann RS. Distinct functions of homeodomain-containing and homeodomain-less isoforms encoded by homothorax. Genes Dev. 2006;20(12):1636–50. doi: 10.1101/gad.1412606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yang Y, Hwang CK, D'Souza UM, Lee SH, Junn E, Mouradian MM. Tale homeodomain proteins Meis2 and TGIF differentially regulate transcription. J Biol Chem. 2000;275:20734–20741. doi: 10.1074/jbc.M908382199. [DOI] [PubMed] [Google Scholar]
- 40.Huang H, Rastegar M, Bodner C, Goh SL, Rambaldi I, Featherstone M. MEIS C termini harbor transcriptional activation domains that respond to cell signaling. J Biol Chem. 2005;280(11):10119–27. doi: 10.1074/jbc.M413963200. [DOI] [PubMed] [Google Scholar]
- 41.Hyman CA, Bartholin L, Newfeld SJ, Wotton D. Drosophila TGIF proteins are transcriptional activators. Mol Cell Biol. 2003;23(24):9262–74. doi: 10.1128/MCB.23.24.9262-9274.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chinnadurai G. CtBP, an unconventional transcriptional corepressor in development and oncogenesis. Mol Cell. 2002;9(2):213–24. doi: 10.1016/s1097-2765(02)00443-4. [DOI] [PubMed] [Google Scholar]
- 43.Ast G. How did alternative splicing evolve? Nat Rev Genet. 2004;5(10):773–82. doi: 10.1038/nrg1451. [DOI] [PubMed] [Google Scholar]
- 44.Patel AA, Steitz JA. Splicing double: insights from the second spliceosome. Nat Rev Mol Cell Biol. 2003;4(12):960–70. doi: 10.1038/nrm1259. [DOI] [PubMed] [Google Scholar]
- 45.Berthelsen J, Kilstrup-Nielsen C, Blasi F, Mavilio F, Zappavigna V. The subcellular localization of PBX1 and EXD proteins depends on nuclear import and export signals and is modulated by association with PREP1 and HTH. Genes Dev. 1999;13(8):946–53. doi: 10.1101/gad.13.8.946. [DOI] [PMC free article] [PubMed] [Google Scholar]