Abstract
Background
The objective of this study was to perform a systematic review of correlations between the single-nucleotide polymorphism at nucleotide 309 (single-nucleotide polymorphism, SNP309) in the murine double-minute 2 (MDM2) gene promoter and susceptibility to leukemia.
Material/Methods
We performed a computer search of relevant case-control studies published from January 1990 to Jan 2014 in databases such as Ovid, EBSCO, PubMed, CNKI, CBMDISC, VIP, and WanFang Data. The literature was screened based on inclusion and exclusion criteria. The data were retrieved, and the quality of the methodology used in the studies was evaluated. A meta-analysis was performed by calculating the combined odds ratios (OR) and 95% confidence intervals (CI) using RevMan 5.0 and Stata 10.0 software. Sensitivity was analyzed and publication bias was assessed.
Results
A total of ten case-control studies from nine research papers were selected in this study, which included 1889 cases and 5707 controls. Meta-analysis showed that people who carried the G allele had increased susceptibility to leukemia compared to people who carried the T allele [OR=1.24, 95% CI (1.06, 1.45), P=0.007]. In a recessive model, the GG homozygotic population had a higher risk of leukemia than the heterozygotic GT+TT population [OR=1.47, 95% CI (1.11, 1.96), P=0.008]. We did not find significant difference in a dominant model [GG+GT vs. TT: OR=1.22, 95% CI (0.98, 1.52), P=0.07]. Publication bias was not significant.
Conclusions
SNP309 polymorphism in the MDM2 gene is associated with susceptibility to leukemia. The G allele may be a risk factor for leukemia.
MeSH Keywords: Core Binding Factor Alpha 2 Subunit; Meta-Analysis as Topic; Polymorphism, Single Nucleotide
Background
Leukemia is a group of malignant clonal diseases with a high degree of heterogeneity. The pathological basis of leukemia is changes in pathways regulating cell proliferation, differentiation, and apoptosis due to gene mutation [1]. However, the cause of leukemia is not yet fully understood. During the development of leukemia, the tumor suppressor gene p53 plays an important role in regulating cellular functions. The TP53 tumor suppressor pathway helps to maintain genomic integrity, thereby preventing tumor formation [2]. The murine double-minute gene 2 (MDM2) plays a key role in this pathway. MDM2 directly binds to the p53 protein and inhibits p53 activity [3]. In addition, MDM2 regulates the TP53 pathway through the ubiquitination and degradation of p53. In contrast, p53 upregulates MDM2 expression, thus forming a negative feedback loop [4,5]. In the MDM2 gene promoter region, the single-nucleotide polymorphism at nucleotide 309 (SNP309, 309T>G) can enhance the binding affinity of transcription factor Spl, thereby increasing MDM2 mRNA and protein expression levels and decreasing the tumor suppression function of p53 [6]. Currently, there are several case-control studies on the relationship between SNP309T>G and susceptibility to leukemia. However, these studies have certain limitations, including variations in research quality, small sample size, and different regional and ethnic backgrounds of the subjects. Thus, the conclusions drawn from these studies are not very firm and have limited credibility. Our study aimed to perform a meta-analysis of the relationships among MDM2 SNP309 polymorphism and susceptibility to leukemia by using data from case-control studies in China and other countries. The results from this study will provide more reliable evidence for basic research and clinical treatment.
Material and Methods
Inclusion and exclusion criteria
Inclusion criteria
1) The study design is case-control; 2) the association between MDM2 SNP309 polymorphism and susceptibility to leukemia was assessed; 3) the disease group consisted of clinically and pathologically diagnosed leukemia patients, while the control group consisted of healthy individuals; 4) among studies published by the same authors, the studies with the highest quality and largest sample size were selected; 5) studies were selected that reported proper statistical methods and highly reliable data, definitive results, various genotype data, OR, and 95% CI (or where such values could be calculated from the original data).
Exclusion criteria
Studies that were not on human subjects were excluded from the study.
Search strategy
A Medical Subject Headings (MeSH) method was used to retrieve studies from databases. “SNP309”or “MDM2 Polymorphism(s)” or “MDM2 variants” or “MDM2 genotype” and “leukemia” were used as keywords. Studies published between January 1990 and January 2014 were searched.
Quality assessment and data retrieval
The research quality of the selected case-control studies was assessed using the Oxford Critical Appraisal Skill program (Oxford-CASP, 2004). The criteria include the following: 1) whether the diagnostic criteria were clearly indicated; 2) the methods of randomization and matching; 3) whether the controls were comparable with the cases; 4) whether the gene detection method was appropriate; 5) whether the sample size was adequate; and 6) whether the data were adequate. Two investigators assessed the quality of the studies and retrieved data from the literature based on the same quality standards and then performed a cross-check. Any disagreement was resolved by discussion or by a third investigator [7].
Statistical methods
Meta-analysis was performed using the RevMan 5.0 and Stata 10.0 software. Cochran’s Q was used for the analysis of heterogeneity between the results of each study (test level=0.10). When there was no heterogeneity between studies, a fixed-effects model was used for the meta-analysis. When there was heterogeneity, a random-effects model was used for the meta-analysis. The OR and 95% CI of each allele and genotype frequency were calculated for each study. The Hardy-Weinberg equilibrium of the control group was calculated. P<0.05 was considered statistically significant. Sensitivity analysis was conducted using the individual exclusion method. The overall effects were re-assessed and compared with the overall effects prior to exclusion. Begg’s test and Egger’s test were applied to determine whether there was publication bias in the studies.
Results
Literature search and quality evaluation
Eighty-nine papers were initially retrieved. After screening based on the criteria, ten case-control studies from nine papers were included [8–16]. This study included 1889 cases and 5707 controls. The basic characteristics of the cases included are shown in Table 1. The quality assessment results showed that the studies included had clear diagnostic criteria; comparable patient and control groups; appropriate genetic testing methods; and clearly documented data and results. Six studies recruited Caucasian subjects, and another four studies recruited Asian subjects. All ten studies contained allelic data. All studies were consistent with the Hardy-Weinberg equilibrium.
Table 1.
The characteristics of included studies.
| Included studies | Country | Ethnicities | Methods for genotyping | Number (case/control) | Genotypes (case) | Genotypes (control) | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| GG | GT | TT | GG | GT | TT | |||||
| Ellis 2008a | United States | Caucasian | TaqMan | 78/2271 | 13 | 34 | 31 | 286 | 1027 | 958 |
| Ellis 2008b | Britain | Caucasian | TaqMan | 89/721 | 14 | 40 | 35 | 88 | 303 | 330 |
| Zenz 2008 | Germany | Caucasian | DHPLC | 617/1065 | 79 | 299 | 239 | 150 | 470 | 445 |
| Phang 2008 | Singapore | Asian | TaqMan | 44/160 | 14 | 13 | 17 | 50 | 80 | 30 |
| Chen 2009 | China Taiwan | Asian | SSCP-CE | 43/138 | 17 | 20 | 6 | 26 | 83 | 29 |
| Do 2009 | China | Asian | AS-PCR | 231/128 | 76 | 123 | 32 | 25 | 68 | 35 |
| Xiong 2009 | Canada | Caucasian | TaqMan | 114/414 | 13 | 55 | 46 | 60 | 167 | 187 |
| Phillips 2010 | United States | Caucasian | TaqMan | 432/485 | 78 | 178 | 176 | 62 | 229 | 194 |
| Dong 2012 | China | Asian | PCR-RFLP | 173/260 | 50 | 84 | 39 | 37 | 141 | 82 |
| Ebid 2012 | Egypt | Caucasian | PCR-RFLP | 77/77 | 14 | 33 | 21 | 6 | 29 | 30 |
Meta-analysis
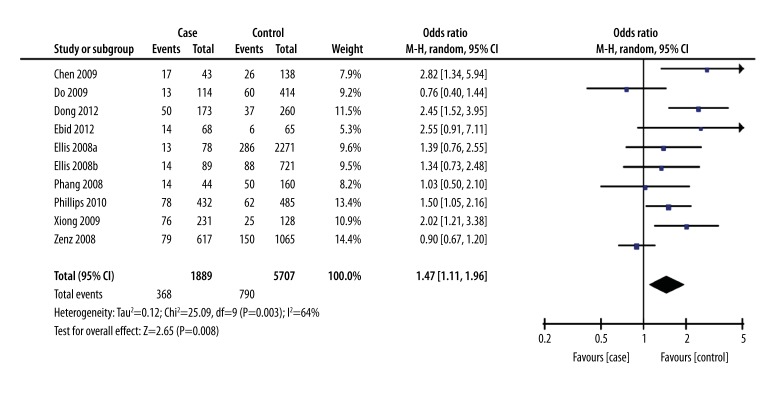
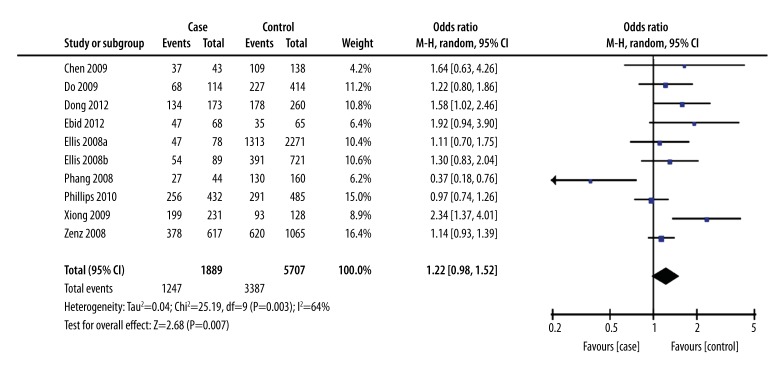
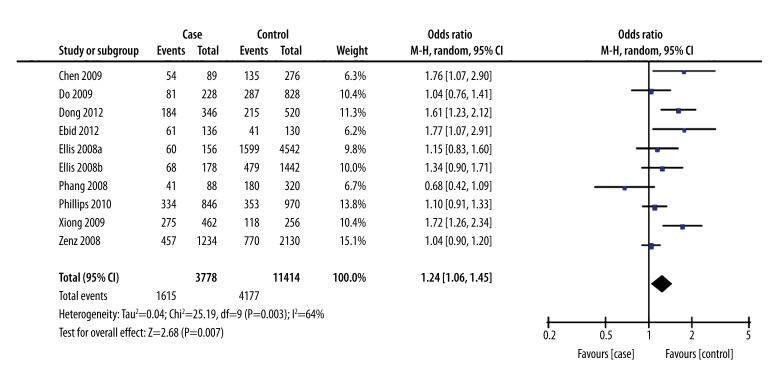
Heterogeneity analysis showed significant heterogeneity among the studies (I2=64%, P=0.0006). Therefore, a random effects model for pooled analysis was applied. The GG genotype was defined as the exposure factor, and the TT genotype was defined as the non-exposure factor. The meta-analysis of a recessive model showed a significant difference between the GG genotype and the GT+TT genotype in terms of leukemia risk [OR=1.47, 95% CI (1.11, 1.96), P=0.008] (Figure 1). The meta-analysis of a dominant model showed no significant difference between the GG+GT genotype and the TT genotype in terms of leukemia risk [OR=1.22, 95% CI (0.98, 1.52), P=0.07]. (Figure 2). The meta-analysis shows that people with the G allele had increased susceptibility to leukemia compared to people with the T allele [OR=1.24, 95% CI (1.06, 1.45), P=0.007] (Figure 3).
Figure 1.
Forest plot of leukemia susceptibility and SNP309 of MDM2 gene (a recessive model: GG vs. GT+TT), the horizontal lines correspond to the study-specific OR and 95% CI, respectively. The area of the squares reflects the study-specific weight. The diamond represents the pooled results of OR and 95%CI.
Figure 2.
Forest plot of leukemia susceptibility and SNP309 of MDM2 gene (a dominant model: TT vs. GG+GT), the horizontal lines correspond to the study-specific OR and 95% CI, respectively. The area of the squares reflects the study-specific weight. The diamond represents the pooled results of OR and 95%CI.
Figure 3.
Forest plot of leukemia susceptibility and SNP309 of MDM2 gene (an allele model: G vs. T), the horizontal lines correspond to the study-specific OR and 95% CI, respectively. The area of the squares reflects the study-specific weight. The diamond represents the pooled results of OR and 95%CI.
Sensitivity analysis
The individual exclusion method was used for the sensitivity analysis in both the GG and TT genotypes. The smallest combined OR after exclusion was 1.32 [95% CI (0.94, 1.96)], and the largest combined OR after exclusion was 1.62 [95% CI (1.14, 2.29)]. Comparisons of the meta-analysis results before and after this exclusion revealed no significant differences, indicating that the analysis exhibited relatively low sensitivity and that the analysis results are therefore relatively robust and credible.
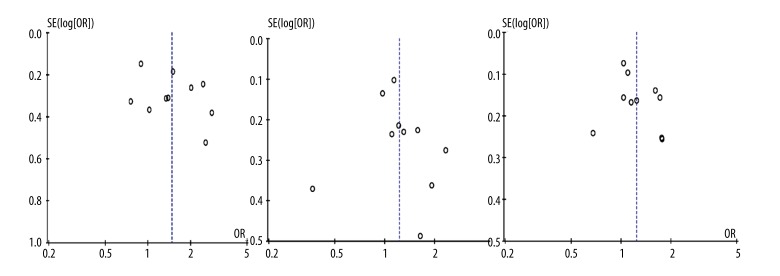
Publication bias
Funnel plot and Egger’s test were performed to assess the publication bias of the literature. Egger’s test further confirmed the absence of publication bias in this meta-analysis (P>0.05) (Figure 4).
Figure 4.
Begg’s funnel plot for publication bias tests. Each point represents a separate study for the indicated association. Log or represents natural logarithm of OR. Vertical line represents the mean effects size.
In this study, we performed a systematic review of the associations between MDM2 gene polymorphisms and susceptibility to leukemia. Our results indicate that at the MDM2 gene SNP309 polymorphism loci, the OR of the relative susceptibility to leukemia of the G allele compared to the T allele was 1.24 [95% CI (1.06, 1.45), P=0.007] and was statistically significant.
People who carried the homozygous GG allele had 1.47 times the risk of leukemia compared to people who carried the TT or GT genotype. These results indicate that the G allele significantly increases susceptibility to leukemia. The biological mechanisms of this susceptibility may be as follows. It is known that MDM2 has two promoters, namely, the Pl and P2 promoters [17–20]. The Pl promoter is a constitutive promoter that does not affect the expression levels of MDM2. The P2 promoter is a p53-dependent intronic promoter containing two adjacent p53 binding elements. P2 may affect the expression levels of MDM2. The MDM2 gene contains several polymorphic loci, among which SNP309T> G is located on the P2 promoter. Compared to the wild-type T allele, the G allele can significantly increase the affinity of MDM2 to transcriptional activator SPl. The transcriptional activity of MDM2 is increased, resulting in increased mRNA transcription and protein expression of MDM2. MDM2 directly inhibits the p53 tumor suppressor pathway [21], thereby increasing the susceptibility to leukemia.
The heterogeneity analysis of the included studies showed the presence of heterogeneity among the studies. In the present study we included studies involving both Caucasian and Asian populations; the genetic background may be a major cause of heterogeneity among studies. In addition, the different genotyping methods were used in the included studies, which may result in heterogeneity between each study.
Publication bias was not found using a funnel plot of the qualitative Begg’s test. Therefore, our study carries considerable credibility. However, our study may still have the following limitations. First, our study was restricted to published studies, and the languages were limited to Chinese and English. Therefore, potential publication and language biases could interfere with the meta-analysis. Second, most current research on the associations between MDM2 SNP309 polymorphisms and cancer susceptibility focus on solid tumors [22,23]. Studies on hematologic oncology are limited. Thus, only a limited number of studies were included in our study, resulting in a small number of patients and controls. The generalizability of our conclusions is potentially affected. Additionally, because our study only included genotype data on Asian and Caucasian populations, but not on other ethnic groups such as Africans, the results are not comprehensive.
Conclusions
In summary, MDM2 SNP309 polymorphism is associated with susceptibility to leukemia. The G allele may be a risk factor for leukemia. More multicenter case-control studies with large sample sizes and high homogeneity are expected in the future. These studies will more accurately evaluate the correlations between MDM2 polymorphisms and susceptibility to leukemia. More reliable evidence will be provided for both basic research and clinical treatment.
Footnotes
Source of support: Self financing
References
- 1.Satoh Y, Matsumura I, Tanaka H, et al. C-terminal mutation of RUNXl attenuates the DNA-damage repair response in hematopoietic stem cells. Leukemia. 2012;26:303–11. doi: 10.1038/leu.2011.202. [DOI] [PubMed] [Google Scholar]
- 2.Mühlbacher V, Zenger M, Schnittger S, et al. Acute lymphoblastic leukemia with low hypodiploid/near triploid karyotype is a specific clinical entity and exhibits a very high TP53 mutation frequency of 93% Genes Chromosomes Cancer. 2014;53:524–36. doi: 10.1002/gcc.22163. [DOI] [PubMed] [Google Scholar]
- 3.Chen J, Zhu B, Chen J, Li Y. Genetic variations in MDM2 and P53 genes confer risk for adult acute lymphoblastic leukemia in a Chinese population. DNA Cell Biol. 2013;32:414–19. doi: 10.1089/dna.2012.1900. [DOI] [PubMed] [Google Scholar]
- 4.Vogelstein B, Lane D, Levine AI. Surfingthe P53 network. Nature. 2000;408:307–10. doi: 10.1038/35042675. [DOI] [PubMed] [Google Scholar]
- 5.Michael D, Oren M. The p53-Mdm2 module and the ubiquitin system. Semin Cancer Biol. 2003;13:49–58. doi: 10.1016/s1044-579x(02)00099-8. [DOI] [PubMed] [Google Scholar]
- 6.Bond GL, Hu W, Bond EE, et al. A single nucleotide polymorphism in the MDM2 promoter attenuates the p53 tumor suppressor pathway and accelerates tumor formation in humans. Cell. 2004;119:591–602. doi: 10.1016/j.cell.2004.11.022. [DOI] [PubMed] [Google Scholar]
- 7.Wu XM, Shi D, Shi JP, et al. Meta-analysis of association of angiotensin-converting enzyme gene insert/deletion polymorphism with cerebral hemorrhage in Chinese Han population. Chinese Journal of Evidence-Based Medicine. 2009;9:1323–27. [Google Scholar]
- 8.Ellis NA, Huo D, Yildiz O, et al. MDM2 SNP309 and TP53 Arg72Pro interact to alter therapy-related acute myeloid leukemia susceptibility. Blood. 2008;112:741–49. doi: 10.1182/blood-2007-11-126508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zenz T, Habe S, Benner A, et al. The MDM2-309 T/G promoter single nucleotide polymorphism does not alter disease characteristics in chronic lymphocytic leukemia. Haematologica. 2008;93:1111–13. doi: 10.3324/haematol.12738. [DOI] [PubMed] [Google Scholar]
- 10.Phang BH, Linn YC, Li H, et al. MDM2 SNP309 G allele decreases risk but does not affect onset age or survival of Chinese leukaemia patients. Eur J Cancer. 2008;44:760–66. doi: 10.1016/j.ejca.2008.02.007. [DOI] [PubMed] [Google Scholar]
- 11.Chen YL, Chang YS, Chang JG, et al. Genotyping of single nucleotide polymorphism in MDM2 genes by universal fluorescence primer PCR and capillary electrophoresis. Anal Bioanal Chem. 2009;394:1291–97. doi: 10.1007/s00216-008-2416-y. [DOI] [PubMed] [Google Scholar]
- 12.Xiong X, Wang M, et al. Risk of MDM2 SNP309 alone or in combination with the p53 codon 72 polymorphism in acute myeloid leukemia. Leuk Res. 2009;33:1454–58. doi: 10.1016/j.leukres.2009.04.007. [DOI] [PubMed] [Google Scholar]
- 13.Do TN, Ucisik-Akkaya E, Davis CF, et al. TP53 R72P and MDM2 SNP309 polymorphisms in modification of childhood acute lymphoblastic leukemia susceptibility. Cancer Genet Cytogenet. 2009;195:31–36. doi: 10.1016/j.cancergencyto.2009.05.009. [DOI] [PubMed] [Google Scholar]
- 14.Phillips CL, Gerbing R, Alonzo T, et al. MDM2 polymorphism increases susceptibility to childhood acute myeloid leukemia: a report from the Children’s Oncology Group. Pediatr Blood Cancer. 2010;55:248–53. doi: 10.1002/pbc.22519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dong HI, Fang C, Fan L, et al. MDM2 promoter SNP309 is associated with an increased susceptibility to chronic lymphocytic leukemia and correlates with MDM2 mRNA expression in Chinese patients with CLL. Int Cancer. 2012;130:2054–61. doi: 10.1002/ijc.26222. [DOI] [PubMed] [Google Scholar]
- 16.Ebid GT, Sedhom IA, El-Gammal MM, Moneer MM. MDM2 T309G has a synergistic effect with P21 ser31arg single nucleotide polymorphisms on the risk of acute myeloid leukemia. Asian Pac J Cancer Prev. 2012;13:4315–20. doi: 10.7314/apjcp.2012.13.9.4315. [DOI] [PubMed] [Google Scholar]
- 17.Yi ZH. A meta analysis on expression of cyclooxygenase-2 in gastric carcinoma. Bulletin of Chinese Cancer. 2007;16:365–67. [Google Scholar]
- 18.Lalonde ME, Ouimet M, Larivière M, et al. Identification of functional DNA variants in the constitutive promoter region of MDM2. Hum Genomics. 2012;6:15. doi: 10.1186/1479-7364-6-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dimitriadi M, Poulogiannis G, Liu L, et al. p53-independent mechanisms regulate the P2-MDM2 promoter in adult astrocytic tumours. Br J Cancer. 2008;99:1144–52. doi: 10.1038/sj.bjc.6604643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liang H, Lunec J. Characteristics of a novel p53 down-regulated promoter in intron 3 of the human MDM2 oncogene. Gene. 2005;361:112–18. doi: 10.1016/j.gene.2005.07.018. [DOI] [PubMed] [Google Scholar]
- 21.Oberst A, Rossi M, Salomoni P, et al. Regulation of the p73 protein stability and degradation. Biochem Biophys Res Commun. 2005;331:707–12. doi: 10.1016/j.bbrc.2005.03.158. [DOI] [PubMed] [Google Scholar]
- 22.Dharel N, Kato N, Muroyama R, et al. MDM2 promoter SNP309 is associated with the risk of hepatocellular carcinoma in patients with chronic hepatitis C. Clin Cancer Res. 2006;12:4867–71. doi: 10.1158/1078-0432.CCR-06-0111. [DOI] [PubMed] [Google Scholar]
- 23.Onat OE, Tez M, Ozcelik T, et al. MDM2 T309G polymorphism is associated with bladder cancer. Anticancer Res. 2008;26:3473–75. [PubMed] [Google Scholar]