Abstract
Aim
To describe practicing physicians’ perceived clinical utility of genome sequencing.
Materials & methods
We conducted a mixed-methods analysis of data from 18 primary care physicians and cardiologists in a study of the clinical integration of whole-genome sequencing. Physicians underwent brief genomics continuing medical education before completing surveys and semi-structured interviews.
Results
Physicians described sequencing as currently lacking clinical utility because of its uncertain interpretation and limited impact on clinical decision-making, but they expressed the idea that its clinical integration was inevitable. Potential clinical uses for sequencing included complementing other clinical information, risk stratification, motivating patient behavior change and pharmacogenetics.
Conclusion
Physicians given genomics continuing medical education use the language of both evidence-based and personalized medicine in describing the utility of genome-wide testing in patient care.
Keywords: genomics, high-throughput nucleotide sequencing, pharmacogenetics, physician’s practice patterns, qualitative research
The claim that genomics will revolutionize the practice of medicine has been the subject of hope and hype, promise and skepticism [1–3]. Routine testing for genetic conditions such as phenylketonuria has been a part of clinical care since the 1960s, and today many general practitioners have at least some experience with targeted genetic testing for conditions such as cystic fibrosis, hemochromatosis and factor V Leiden thrombophilia [4–6]. However, the sequencing of the human genome in 2003 enabled genome-wide analyses of unprecedented scope and resolution, such that most variation in the human genome can now be analyzed with a single test. Genome sequencing has demonstrated clinical utility in specialized settings, including the diagnosis and management of rare diseases, infectious disease outbreaks and cancer [7–11]. However, the widespread uptake of genomics into general practice has not yet occurred, in part because of an absence of evidence for its clinical validity (how accurately the test detects or predicts a health outcome) and clinical utility (how likely the test is to improve medical decision-making and health outcomes) [12–17], particularly among generally healthy patients.
At the same time, some thought leaders as well as industry and patient advocacy representatives have voiced the promise that genomics may usher in an era of personalized medicine [2,18–20], in which even healthy individuals undergo sequencing as a part of routine preventive medicine and medical care. Many of the articulated values overlap with movements calling for increased patient engagement in healthcare, including giving patients greater access to their health information and more active roles in medical decision-making [21,22]. Because genome-wide testing in these settings has not yet met the standards for clinical use generally required by clinicians, professional organizations and payers, some individuals have bypassed the healthcare system and turned to direct-to-consumer (DTC) products such as 23 and-Me’s Personal Genome Service (PGS) to learn more about the health implications of their genomes [23–25]. These products have presented clinical challenges for providers and regulatory challenges for the US FDA, which in November 2013 instructed 23andMe to stop marketing Personal Genome Service as a health-related product, citing a lack of documentation of its analytic and clinical validity for this purpose [24,26].
DTC genomics products often use arrays of only hundreds of thousands of preselected common variants, called single-nucleotide polymorphisms (SNPs), some of which have association with human disease. Whole-genome sequencing (WGS), which reads more than 99% of the three billion base pairs in an individual’s genome, has the capacity to reveal uncommon or even unique variants, much of which have uncertain clinical significance for an individual [17,27–28], but some of which can be highly diagnostic. The cost of sequencing has fallen considerably, making it feasible for WGS to replace more targeted genetic testing in the near future. However, the evidence for its clinical utility and cost–effectiveness in general patient populations lags behind [29]. Many health insurers are not reimbursing the costs of sequencing even for individuals with suspected rare genetic conditions, citing its uncertain impact on patient management [30]. To a large extent, the clinical integration of these technologies will depend on how useful practicing physicians perceive them to be for improving patient care. In a climate of clinical and regulatory uncertainty for the future of genomics, the voices of practicing clinicians will therefore be important for shaping policy and setting research priorities. To that end, we asked physicians participating in a research study of the clinical integration of genomics to describe their perceptions of the clinical utility that widespread uptake of genome sequencing might have for patient care.
Materials & methods
Study overview
We report here data from the MedSeq Project, a pair of randomized controlled trials of WGS in the clinical care of healthy adult primary care patients and cardiomyopathy patients [31]. In this study, both physicians and patients were research subjects who were studied with serial surveys and interviews during the project. In this report, we use mixed methods to describe the physician participants’ perceived utility of WGS for patient care at enrollment. The Partners Human Research Committee and the Baylor College of Medicine Institutional Review Board approved this study.
Setting, study design & participants
Eligible MedSeq Project physician participants included any primary care physician (PCP) or cardiologist actively seeing patients in a clinic setting in a single large urban network of academic hospitals and outpatient practices. Physicians were invited to enroll in the MedSeq Project as described previously [31]. Physicians were told that they would each be asked to identify potentially eligible patients and that research study staff would aim to recruit 8–12 patients per physician. For the primary care trial, eligible patient participants were generally healthy adults aged 40–65 years without an indication for genetic testing. The PCPs used their judgment to determine whether each patient was ‘generally healthy,’ although patients with cardiovascular disease or diabetes were specifically excluded. For the cardiology trial, eligible patient participants had a diagnosis of hypertrophic or dilated cardiomyopathy and may have undergone prior or concurrent targeted genetic testing for their condition. All patient participants were to undergo a family history assessment; half were randomly assigned to additionally undergo WGS and have the results interpreted on a genome report delivered to their physicians. A disclosure visit between each patient participant and his or her physician participant allowed the two to discuss the findings of the family history report alone or the combination of the family history and WGS reports.
After physician enrollment but before patient enrollment, the MedSeq Project PCP and cardiologist physician participants underwent a genomics educational curriculum comparable in duration and scope to other continuing medical education (CME) offerings required for maintenance of certification. This curriculum consisted of two 1-h in-person group classes taught by medical geneticists and genetic counselors and 12 self-paced online modules, designed to take about 4 h total. The curriculum used case-based examples to cover general genetics concepts such as inheritance patterns, an overview of Mendelian conditions, genome-wide association studies and pharmacogenomics. Diseases illustrating key concepts included breast cancer, cystic fibrosis, familial hyperlipidemia and atrial fibrillation. Potential future clinical uses of WGS were not specifically discussed. Physician participants received 6 h of CME credits for completing the curriculum and financial compensation for study participation [31].
Measurements
The MedSeq Project collected data on many patient and physician outcomes, including attitudes and preferences, psychological and behavioral impact and healthcare utilization, assessed at multiple time points during the study [31]. Here, we present data from two instruments administered to the MedSeq Project physician participants at enrollment. After the educational curriculum, physician participants completed a survey measuring their perceived utility of family history and genome sequencing information for clinical care. Specifically, physicians were asked to rate on a scale from 1 to 10 (‘Not at all useful’ to ‘Extremely useful’) how useful they thought family history and sequencing information would be for ‘managing [their] patients’ health’ at two times: ‘now’ and ‘in the future.’ Also after the educational curriculum, but before any disclosure visits with their patients, physician participants underwent in-depth individual semi-structured interviews conducted by nonphysician interviewers trained in qualitative methods. The interview guide included domains about physicians’ motivations for study participation, attitudes and expectations for genome sequencing and its utility for clinical care. Question prompts included how physicians felt about ‘WGS becoming a part of clinical care’ and how WGS results might ‘influence [their] approach to patient care.’ Interviews lasted about 45 min and occurred between March and December 2013. Interviews were audio-recorded and transcribed verbatim.
Analysis
We used mixed methods to describe the physicians’ perceptions of the utility of WGS and family history information for clinical care. We used paired t-tests to compare the present and future utility ratings of WGS and family history information from the physician surveys. We analyzed interview data using thematic analysis [32], following standardized procedures for team-based qualitative analysis [33] and consensus-coding [34,35]. Interview data were stored and managed using ATLAS.ti version 7 software. A comparison of PCPs and cardiologists was not a focus of these analyses.
Results
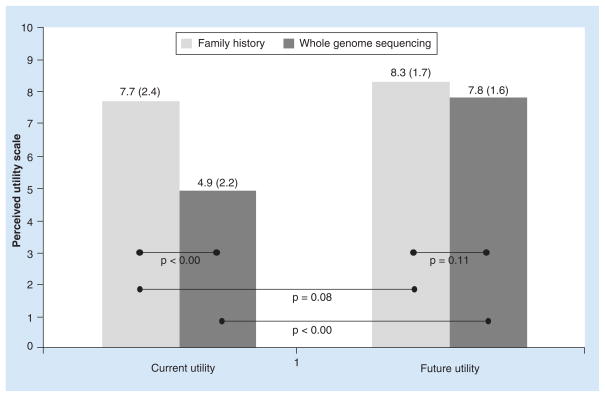
Eighteen physician participants were recruited to the MedSeq Project (Table 1). Physicians rated family history information to have higher utility than WGS now, but they rated the two to have similar utility in the future (Figure 1). They anticipated that the utility of WGS would increase in the future (Δ = 2.9, p < 0.001) but the utility of family history information would remain stable (Δ = 0.6, p = 0.08).
Table 1.
Characteristics of physician participants recruited to the MedSeq Project.
| PCPs (n = 10) | Cardiologists (n = 8) | Total (n = 18) | |
|---|---|---|---|
| Mean age, years (SD) | 53 (9) | 50 (9) | 52 (9) |
| Women (n) | 5 | 2 | 7 |
| Nonwhite race/ethnicity (n) | 2 | 2 | 4 |
| Genetics training† (n) | 1 | 4 | 5 |
Number responding ‘yes’ to the question “Have you ever had genetics training beyond the typical medical school curriculum?”
PCP: Primary care physician.
Figure 1. Physician perception of present and future clinical utility of family history and whole genome sequencing, rated on a scale from 1 to 10 (‘Not at all useful’ to ‘Extremely useful’).
Values shown are mean (SD) utility ratings. p-values correspond to paired t-tests.
From interview data, we identified three major themes physicians discussed in describing their perceived utility of WGS for the practice of medicine: its lack of current clinical utility, its inevitability and contexts in which WGS might achieve utility. Table 2 shows quotes illustrating these themes. In expressing these perceptions, physicians often used analogies to current medical testing or more familiar clinical scenarios.
Table 2.
Illustrative quotes from MedSeq Project physician participants about their perception of the utility of whole-genome sequencing in patient care.
| Domain | Illustrative physician quotes |
|---|---|
| Lack of clinical utility | |
| Nature of WGS data | “Nobody knows really what to do with the information and knows what it is and what it isn’t…” (P17) “I think there’s so many mutations with unknown consequence. There’s so much variability in the expression of some of the gene mutations so that even if you have them you may or may not have disease outcome with it. There are so many modifiers to it.” (P15) |
| Potential for harm | “It’s like doing a CT scan and finding a nodule and not knowing what it means. And the next thing you know you have a biopsy, and then you have a complication from the biopsy. Then the results from the biopsy are ambiguous again.” (C07) “Like PSA testing and mammography, you might end up doing tests and procedures and treatments that are really not indicated because we don’t really understand what it means.” (P01) “You may tell the patient that he’s not at high risk of something when he may be…so I am concerned we can make serious mistakes that may haunt us in the future because we misinterpreted something.” (P10) |
| Inevitability | “I don’t know when, but I’m sure sometime in the future it will become very much a part of what primary care physicians do. It will be just one of the tools in their toolbox that’s available for taking care of patients.” (P19) |
| Contexts of utility | |
| Complementing other clinical information | “We tell patients, ‘Don’t smoke, let’s lose weight. Let’s look back at that cholesterol—it’s average. But, given the genetics… And then let’s look at your family history. There was this one aunt.’ When you pull it all together, you say, ‘You know what? Maybe go on a statin. Maybe go on a little bit of aspirin.’” (P13) “I would take the genetic sequencing information and definitely correlate it to the family history… Separate from that I think that it can actually be dangerous. I think if you look through family histories, that’s where the information carries its greatest value.” (P15) |
| Risk stratification | “If it turns out that we don’t have to do colonoscopies every 10 years on 100% of the people, maybe that will save a lot of money. If we know who we really should be doing PSA’s on, or mammograms on, instead of doing them on everybody, we can stop creating a lot of harm and disability.” (P01) “I look at this just like I look at risk in general. It’s one other thing that might push me to be more aggressive in protecting someone against cardiac disease, Alzheimer’s. For what impact we can have on those diseases that is, which is probably modest. But if someone were to have risk factors that suggested a higher risk for heart disease, I’d probably be more likely to put them on a statin, on an aspirin, do a stress test—do things that would be more proactive.” (P15) “Whether it’s from cholesterol, coronary artery disease, arrhythmias, hypertension, heart failure – I think there’s going to be a wealth of areas that I deal with on a daily basis, that we will be able to focus our treatments to people who really need it and be able to reassure people who really don’t.” (C09) |
| Motivating patient health behaviors | “People today want to be more in control of their health. And [WGS] will enable them to do that. They’ll be able to take something to their doctor and say, in addition to ‘I had my appendix out, and I have chronic irritable bowel syndrome, and I get a lot of headaches,’ ‘By the way, this is my whole genome sequence, and these are the ones I was positive for. I need you to keep an eye out for these.’ So it gives a patient a lot more power and control, which I think is very important.” (P01) “You get a lot of information that may not be immediately useful and could potentially make some patients anxious. Although, you can always turn that around and use it to prod them to alter their behavior in a more healthy way. So I suppose one could leverage fear into something constructive.” (P11) “I think maybe showing something concrete sometimes does work for the patients. I think that seeing results, seeing a paper, may make them change and be more compliant to come and get their mammograms and colonoscopies, which they have postponed and don’t want to have done.” (P10) |
| Pharmacogenomics | “One thing I do look forward to is knowing more about the pharmacogenomics because with basically everything we prescribe—blood pressure medications, anti-depressants, all this stuff— you’re just guessing. And you’re starting with the basic first-step evidence-based stuff, but you don’t know. It would be nice if you just knew this is a patient who needs this SSRI because we know that it’s a waste to try the first three steps—just go to the one that you know would work.” (P17) “I’m sure many of the drugs that the future doctor uses will be throughput onto their genome to figure out the dose and the drug that’s going to work best. That’s very exciting, but it’s a long way away.” (P05) “I wonder about the usefulness of some of the pharmacogenomics associations, because I don’t see them. The studies that have been done looking at the pharmacogenomics for warfarin dosing have been at best inconsistent, at worst disappointing, and so it really isn’t endorsed as being useful.” (C18) |
Primary care physicians and cardiologists are designated by ‘P’ and ‘C,’ respectively.
PSA: Prostate-specific antigen; SSRI: Selective serotonin-reuptake inhibitor; WGS: Whole-genome sequencing.
Lack of current clinical utility
Physicians did not think that the widespread clinical integration of WGS was appropriate at present, citing a general perception that it would not change their clinical care of the majority of their patients. The nature of WGS data was often given as a reason for its limited clinical utility: it was seen to have uncertain disease associations with limited implications for clinical management. Physicians also described WGS results as probabilistic, although they acknowledged that this quality was not unique to WGS. One commented,
“Medicine is used to using imperfect information – it does it all the time. In fact, I don’t think there’s any such thing as a perfect set of data in medicine because you are trying to take the general and apply it in the individual. And that is always fraught because there’s no such thing as a probability when the individual is concerned. You have to actually make a binary decision. You either have to treat them or not treat them, take the lesion out or leave the lesion in” (C08).
Physicians cited the potential for patient harm as another limitation to the clinical utility of WGS, often invoking the concern that its uncertain interpretation would lead to a cascade of further diagnostic tests carrying their own risks and costs.
Inevitability
Despite its current limitations, physicians generally expressed a sense that the widespread clinical integration of WGS was inevitable in the near future. When asked his opinion about WGS becoming a routine part of clinical care, one physician responded, “Well, that’s sort of like saying, ‘So what do you think about getting cholesterol tests on people?’ I mean, it’s a reality, and I think it’s going to happen” (C18). Physicians did not generally elaborate on why they thought WGS would inevitably be a part of clinical care.
Contexts of potential clinical utility
Addressing the limitations above was seen to be a prerequisite for the clinical integration of WGS. Mirroring the quantitative survey results, one physician commented, “I think for a long time there will be too many genes and not enough correlation to disease, and that ratio will have to gradually change so that we understand more about the specifics” (P05). Physicians described four clinical contexts in which WGS might be useful for patient care: complementing other clinical information, risk stratification, motivating patient health behaviors and pharmacogenomics.
Complementing other clinical information
Physicians envisioned using WGS results to complement family history and other clinical information about individual patients. They described its potential to explain disease patterns within families, but they felt that they might minimize the clinical significance of WGS results not supported by observed family history. Many emphasized the importance of contextualizing WGS findings to the specific circumstances of individual patients, including their ages, health behaviors, family histories and other lab test results.
Risk stratification
Physicians would find WGS clinically useful for widespread implementation if it helped them distinguish low-risk from high-risk patients and if that stratification resulted in specific changes in clinical management, such as disease surveillance frequency and therapeutic choice. Physicians often described this concept in the setting of current health screening tests, such as colonoscopy and mammography.
Motivating patient health behavior
Physicians responded that WGS results might motivate patients to adopt healthier lifestyle habits, undergo screening tests already recommended for them and be more adherent to pharmacotherapy. Some physicians described WGS as empowering to patients in this regard, while others described WGS as a tool for physicians to use in persuading patients to adopt healthy behaviors. One physician responded that the low disease variance generally explained by genetics would paradoxically lead to an emphasis on non-genetic causes and increase patient motivation, stating,
“When you start talking about the diseases that I deal with most commonly—atrial fibrillation, coronary heart disease, hypertension—and we have these genetic risks that are a limited percentage of the variation that gets explained, it really just emphasizes the importance of all the lifestyle modifications” (C18).
Pharmacogenomics
Physicians articulated a hope that WGS information would lessen the inefficient trial-and-error nature of pharmacotherapy, improving efficacy and minimizing adverse events. Still, some reported that the field of pharmacogenomics has the same limitations as genomic medicine overall, with unclear significance for medical decision-making.
Discussion
Physician participants of a study of WGS cited the complex and uncertain nature of its data, coupled with its potential for harm, as reasons for why genome sequencing is not yet ready for widespread clinical integration. Although they did not use the terms explicitly, physicians often invoked the uncertain clinical validity and utility of sequencing for use in their general patient populations. These physicians related WGS to common primary care scenarios in envisioning the future clinical utility of genome sequencing. They acknowledged its potential use as a screening test in routine health maintenance among asymptomatic individuals, as a complement to or replacement for current modalities such as colonoscopies or prostate-specific antigen (PSA) testing. They anticipated that such screening might one day help them target more intensive monitoring or treatment to patients at highest disease risk. They expressed a hope that sequence results might motivate healthy behaviors among their patients. They also looked forward to a pharmacogenomics-based body of evidence that would guide more effective pharmacotherapy, although they had the sense that such evidence does not yet exist. Despite the current limitations of genome-wide testing in healthy adults, physicians were generally pragmatic about its use. Many saw genome sequencing as just one of many tools for use in medical decision-making that might complement or even be superseded by other clinical information or by the larger context of an individual patient.
To our knowledge, this is the first study examining practicing physicians’ views on the widespread use of genome sequencing in clinical medicine, but our findings are consistent with prior work around more limited genome-wide technologies. Concerns about the clinical utility of SNP-based arrays have been reported in surveys and focus groups of general practitioners [4,36–40], including a group of PCPs who had undergone this testing themselves [41]. Early focus groups of family physicians expressed concerns about the complexity and uncertain clinical management of genetic cancer susceptibility testing [36], and focus groups a decade later echoed similar concerns about personalized genomic medicine more broadly [40]. The potential uses for WGS elicited in our interviews mirrored those anticipated for SNP array testing, such as risk stratification to inform disease surveillance frequency and treatment intensity [4,39,42]. Survey data suggest that physicians see genome-wide testing as one possible way to motivate their patients to make health behavior changes but, at the same time, are uncertain about its efficacy in doing so [4,43–44]. Of note, randomized trial evidence to date generally suggests that testing for genetic susceptibility to diseases such as diabetes and lung cancer does not improve health behaviors such as dietary habits, physical activity and smoking cessation [45–48]. The study physicians similarly expressed an enthusiasm for pharmacogenomics seen in prior surveys [41,49] but also skepticism for its clinical utility, this even before the high-profile publication of studies calling into question the clinical utility of warfarin pharmacogenomics [50]. Nonetheless, physicians in this and previous studies predict that genomics will become increasingly important for general medical practice in the future [4,42].
This study is limited to one network of academic primary care and cardiology practices, although the physicians represent a diverse range of demographics and training. These analyses are derived from a research study setting; however, at present, that is the only experience that most physicians, and particularly PCPs, have with WGS [17]. Moreover, the MedSeq Project aims to model the real-world integration of genome sequencing into general medicine. Specifically, we provided genomics education similar in duration to other CME opportunities and then studied how physicians perceived WGS. Our interviews demonstrate that these physicians are not necessarily early-adopting genome sequencing enthusiasts, but, rather, pragmatic clinicians who might represent the future of genomic medicine in patient care. The specific CME we provided almost certainly influenced the physicians’ responses, but given its brief nature, we argue that these physicians may represent the future genomic medicine practitioner: not a genetics specialist but a physician given an introduction to genomics through both didactic and practical experience. Although our sample size limits the conclusions that can be drawn from our quantitative physician surveys, it was sufficiently large to enable the richness that is the strength of qualitative data [51]. However, our findings may not be generalizable to other physicians and other practice settings, particularly outside the research context. Although we provide a description of the experience of one group of physicians with genomic medicine, additional research in more diverse practice settings will be necessary to inform the future of genomics education, practice and research.
The recent conflict between the FDA and 23andMe illustrates the importance that policy-makers place on the clinical validity and utility of new genomic technologies. WGS will likely be held to the same standards. In late 2013, insurer Blue Cross and Blue Shield concluded that exome sequencing can be diagnostic for certain patients but still not have an impact on disease treatment or outcomes [52]. In January 2014, Blue Cross and Blue Shield of North Carolina issued a policy that it would not cover WGS, considering it investigational and lacking in utility. The rise of DTC genomics products, on the other hand, reflects the growing interest of some patients in personalized medicine and engaging in their own healthcare. This tension between evidence-based and personalized approaches is not unique to genomic medicine, but nowhere is the uniqueness of each patient made more concrete than in his or her DNA sequence. To inform this debate, our study gives insight into the potential benefits and pitfalls that physician stakeholders perceive in increasingly integrating genome-wide testing into clinical care. Even before the FDA letter to 23andMe in November 2013, these physicians expressed concerns about the clinical validity and utility of the broad application of WGS.
In the policy debate around genome-wide testing, it is important to recognize that physicians still use the language of evidence-based medicine to describe the application of genome sequencing to patient care. Given the unprecedented scope and uncertainty of WGS results, applying an evidentiary framework based on traditional comparative effectiveness metrics presents considerable challenges. At the same time, physicians describe a kind of personalized medicine when talking about the utility of genome sequencing, although this means interpreting a patient’s WGS results in the context of their family history and other risk factors at least as often as it means medical decision-making based on one’s genomic profile alone. It is unlikely that physicians will widely adopt genome sequencing unless policymakers can develop a framework that balances the competing demands of evidence-based and personalized medicine. In addition to framing the policy debate, the recognition that practicing physicians feel this tension when envisioning the future of genomic medicine will also shape a research agenda to develop the appropriate methodologies, metrics and standards to determine the role that genome sequencing should play in clinical medicine [16,53]. In contributing their perspectives to defining the thresholds that genome-wide testing should meet for clinical integration, physicians will minimize their risk of being excluded from the relationships between patients, their genomes and their health.
Conclusion
Practicing physicians given CME-level training in genomic medicine use the language of both evidence-based medicine and personalized medicine in describing the utility of genome-wide testing in patient care. The promise and limitations of genome sequencing identified by these potential users of this new technology should help shape the policy and research agendas around its integration into clinical practice.
Future perspective
Rapid advances in genomic technology and discovery in the last few years will almost certainly change the way that medicine is practiced in the next decade. However, the nature and extent of that impact remains to be defined, including the specific clinical contexts in which physicians find genome sequencing to improve medical decision-making and improve the care of their patients. Determining the clinical utility of genome sequencing across the spectrum of healthcare settings remains a next frontier in genomic medicine.
Executive Summary.
After brief continuing medical education (CME) in genetics and genomics, primary care physicians and cardiologists without specialized genetics training rated genome sequencing to have lower utility for current patient care compared with family history assessment.
Physicians perceived the current limitations in the clinical utility of sequencing to include the uncertain interpretation of sequencing data and its limited ability to change clinical decision-making.
Nonetheless, physicians expected the clinical utility of genome sequencing to increase in the future, matching that of family history.
Physicians perceived the potential uses of sequencing to include complementing other clinical information; improved risk stratification of patients, leading to better preventive efforts, motivating patient behavior change and tailoring pharmacotherapy to patient genotype.
Practicing physicians given CME-level training in genomic medicine use the language of both evidence-based medicine and personalized medicine in describing the utility of genome-wide testing in patient care.
The promise and limitations of genome sequencing identified by these potential users of this new technology should be added to those of other stakeholders to help shape the policy and research agendas around its integration into clinical practice.
Footnotes
Ethical conduct
The authors state that they have obtained appropriate institutional review board approval or have followed the principles outlined in the Declaration of Helsinki for all human or animal experimental investigations. In addition, for investigations involving human subjects, informed consent has been obtained from the participants involved.
For reprint orders, please contact: reprints@futuremedicine.com
Financial & competing interests disclosure
This work was supported the US National Institutes of Health (U01-HG006500, U19-HD077671, L30-DK089597, F32-HG006993). The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
References
Papers of special note have been highlighted as:
• of interest;
•• of considerable interest
- 1.Green ED, Guyer MS. Charting a course for genomic medicine from base pairs to bedside. Nature. 2011;470(7333):204–213. doi: 10.1038/nature09764. [DOI] [PubMed] [Google Scholar]
- 2.Evans JP, Meslin EM, Marteau TM, Caulfield T. Genomics. Deflating the genomic bubble. Science. 2011;331(6019):861–862. doi: 10.1126/science.1198039. [DOI] [PubMed] [Google Scholar]
- 3.Evaluation of Genomic Applications in Practice and Prevention (Egapp) Working Group. The EGAPP initiative: lessons learned. Genet Med. 2014;16(3):217–224. doi: 10.1097/GIM.0b013e318184137c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bernhardt BA, Zayac C, Gordon ES, Wawak L, Pyeritz RE, Gollust SE. Incorporating direct-to-consumer genomic information into patient care: attitudes and experiences of primary care physicians. Per Med. 2012;9(7):683–692. doi: 10.2217/pme.12.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Klitzman R, Chung W, Marder K, et al. Attitudes and practices among internists concerning genetic testing. J Genet Couns. 2013;22(1):90–100. doi: 10.1007/s10897-012-9504-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Krousel-Wood M, Andersson HC, Rice J, Jackson KE, Rosner ER, Lubin IM. Physicians’ perceived usefulness of and satisfaction with test reports for cystic fibrosis (DeltaF508) and factor V Leiden. Genet Med. 2003;5(3):166–171. doi: 10.1097/01.GIM.0000066797.05220.85. [DOI] [PubMed] [Google Scholar]
- 7.Bainbridge MN, Wiszniewski W, Murdock DR, et al. Whole-genome sequencing for optimized patient management. Sci Transl Med. 2011;3(87):87re83. doi: 10.1126/scitranslmed.3002243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Worthey EA, Mayer AN, Syverson GD, et al. Making a definitive diagnosis: successful clinical application of whole exome sequencing in a child with intractable inflammatory bowel disease. Genet Med. 2011;13(3):255–262. doi: 10.1097/GIM.0b013e3182088158. [DOI] [PubMed] [Google Scholar]
- 9.Yang Y, Muzny DM, Reid JG, et al. Clinical whole-exome sequencing for the diagnosis of Mendelian disorders. N Engl J Med. 2013;369(16):1502–1511. doi: 10.1056/NEJMoa1306555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Koser CU, Holden MT, Ellington MJ, et al. Rapid whole-genome sequencing for investigation of a neonatal MRSA outbreak. N Engl J Med. 2012;366(24):2267–2275. doi: 10.1056/NEJMoa1109910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11••.Green RC, Biesecker LG. Diagnostic clinical genome and exome sequencing. N Engl J Med. 2014;370(25):2418–2425. doi: 10.1056/NEJMra1312543. Recent description of the current use of next-generation sequencing in clinical diagnosis. [DOI] [PubMed] [Google Scholar]
- 12••.Haddow J, Palomaki G. ACCE. A Model Process for Evaluating Data on Emerging Genetic Tests. In: Khoury M, Little J, Burke W, editors. Human Genome Epidemiology. Oxford University Press; Oxford: 2004. pp. 217–233. Description of a widely used approach to evaluating a genetic test according to its analytic validity, clinical validity, clinical utility and associated ethical, legal and social implications. [Google Scholar]
- 13.Khoury MJ, Janssens AC, Ransohoff DF. How can polygenic inheritance be used in population screening for common diseases? Genet Med. 2013;15(6):437–443. doi: 10.1038/gim.2012.182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Grosse SD, Rogowski WH, Ross LF, Cornel MC, Dondorp WJ, Khoury MJ. Population screening for genetic disorders in the 21st century: evidence, economics, and ethics. Public Health Genomics. 2010;13(2):106–115. doi: 10.1159/000226594. [DOI] [PubMed] [Google Scholar]
- 15.European Society of Human Genetics. Statement of the ESHG on direct-to-consumer genetic testing for health-related purposes. Eur J Hum Genet. 2010;18(12):1271–1273. doi: 10.1038/ejhg.2010.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Garber AM, Tunis SR. Does comparative-effectiveness research threaten personalized medicine? N Engl J Med. 2009;360(19):1925–1927. doi: 10.1056/NEJMp0901355. [DOI] [PubMed] [Google Scholar]
- 17.Dewey FE, Grove ME, Pan C, et al. Clinical interpretation and implications of whole-genome sequencing. JAMA. 2014;311(10):1035–1045. doi: 10.1001/jama.2014.1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen R, Mias GI, Li-Pook-Than J, et al. Personal omics profiling reveals dynamic molecular and medical phenotypes. Cell. 2012;148(6):1293–1307. doi: 10.1016/j.cell.2012.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen R, Mias GI, Li-Pook-Than J, et al. Comment on “The predictive capacity of personal genome sequencing”. Sci Transl Med. 2012;4(135):135le135. author reply 135lr133. [Google Scholar]
- 20.Steenhuysen J. The dawning of the age of genomic medicine, finally. Retuers. 2014 [Google Scholar]
- 21.Carman KL, Dardess P, Maurer M, et al. Patient and family engagement: a framework for understanding the elements and developing interventions and policies. Health Aff (Millwood) 2013;32(2):223–231. doi: 10.1377/hlthaff.2012.1133. [DOI] [PubMed] [Google Scholar]
- 22.Delbanco T, Walker J, Bell SK, et al. Inviting patients to read their doctors’ notes: a quasi-experimental study and a look ahead. Ann Intern Med. 2012;157(7):461–470. doi: 10.7326/0003-4819-157-7-201210020-00002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Allison M. Direct-to-consumer genomics reinvents itself. Nat Biotechnol. 2012;30(11):1027–1029. doi: 10.1038/nbt.2409. [DOI] [PubMed] [Google Scholar]
- 24.Murray MF. Why We Should Care About What You Get for “Only $99” From a Personal Genomic Service. Ann Intern Med. 2014;160(7):507–508. doi: 10.7326/M13-2804. [DOI] [PubMed] [Google Scholar]
- 25.Murphy E. Inside 23andMe Founder Anne Wojcicki’s $99 DNA Revolution. Fast Company. 2013;(180) www.fastcompany.com/3018598/for-99-this-ceo-can-tell-you-what-might-kill-you-inside-23andme-founder-anne-wojcickis-dna-r.
- 26.Green RC, Farahany NA. Regulation: the FDA is overcautious on consumer genomics. Nature. 2014;505(7483):286–287. doi: 10.1038/505286a. [DOI] [PubMed] [Google Scholar]
- 27.Boone PM, Soens ZT, Campbell IM, et al. Incidental copy-number variants identified by routine genome testing in a clinical population. Genet Med. 2013;15(1):45–54. doi: 10.1038/gim.2012.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28••.Teutsch SM, Bradley LA, Palomaki GE, et al. The Evaluation of Genomic Applications in Practice and Prevention (EGAPP) Initiative: methods of the EGAPP Working Group. Genet Med. 2009;11(1):3–14. doi: 10.1097/GIM.0b013e318184137c. Description of the methodology used by the US Centers for Disease Control and Prevention to evalaute the clinical utility of specific genomic tests. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Grosse SD. Economic analyses of genetic tests in personalized medicine: clinical utility first, then cost utility. Genet Med. 2014;16(3):225–227. doi: 10.1038/gim.2013.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Steenhuysen J. As sequencing moves into clinical use, insurers balk. Reuters. 2014 Jun 19; [Google Scholar]
- 31.Vassy JL, Lautenbach DM, Mclaughlin HM, et al. The MedSeq Project: a randomized trial of integrating whole genome sequencing into clinical medicine. Trials. 2014;15(1):85. doi: 10.1186/1745-6215-15-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Creswell JW. Qualitative Inquiry and Research Design: Choosing among Five Approaches. 2. SAGE Publications; Thousand Oaks, CA, USA: 2007. [Google Scholar]
- 33.Guest GS, Macqueen KM, Namey EE. Applied Thematic Analysis. SAGE Publications; Thousand Oaks, CA, USA: 2012. [Google Scholar]
- 34.Carey JW, Gelaude D. Systematic methods for collecting and analyzing multidisciplinary team based qualitative data. In: Guest GS, Macqueen KM, editors. Handbook for Team-Based Qualitative Research. AltaMira; Lanham, MD, USA: 2008. pp. 227–274. [Google Scholar]
- 35.Macqueen KM, Mclellan E, Kay K, Milstein B. Codebook development for team-based qualitative analysis. Cult Anthropol Meth. 1998;10(2):31–36. [Google Scholar]
- 36•.Carroll JC, Brown JB, Blaine S, Glendon G, Pugh P, Medved W. Genetic susceptibility to cancer: family physicians’ experience. Can Fam Phys. 2003;49:45–52. Early focus groups among Canadian family physicians about cancer susceptibility testing. [PMC free article] [PubMed] [Google Scholar]
- 37.Levy DE, Youatt EJ, Shields AE. Primary care physicians’ concerns about offering a genetic test to tailor smoking cessation treatment. Genet Med. 2007;9(12):842–849. doi: 10.1097/gim.0b013e31815bf953. [DOI] [PubMed] [Google Scholar]
- 38••.Scheuner MT, Sieverding P, Shekelle PG. Delivery of genomic medicine for common chronic adult diseases: a systematic review. JAMA. 2008;299(11):1320–1334. doi: 10.1001/jama.299.11.1320. Comprehensive review of barriers to and physician use of genomic medicine. [DOI] [PubMed] [Google Scholar]
- 39.Powell KP, Cogswell WA, Christianson CA, et al. Primary care physicians’ awareness, experience and opinions of direct-to-consumer genetic testing. J Genet Couns. 2012;21(1):113–126. doi: 10.1007/s10897-011-9390-9. [DOI] [PubMed] [Google Scholar]
- 40.Najafzadeh M, Davis JC, Joshi P, Marra C. Barriers for integrating personalized medicine into clinical practice: a qualitative analysis. Am J Med Genet A. 2013;161A(4):758–763. doi: 10.1002/ajmg.a.35811. [DOI] [PubMed] [Google Scholar]
- 41•.Haga SB, Carrig MM, O’daniel JM, et al. Genomic risk profiling: attitudes and use in personal and clinical care of primary care physicians who offer risk profiling. J Gen Intern Med. 2011;26(8):834–840. doi: 10.1007/s11606-011-1651-7. Survey of physicians who offer genomic testing. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mainous AG, 3rd, Johnson SP, Chirina S, Baker R. Academic family physicians’ perception of genetic testing and integration into practice: a CERA study. Fam Med. 2013;45(4):257–262. [PubMed] [Google Scholar]
- 43.Gramling R, Nash J, Siren K, Culpepper L. Predictive genetics in primary care: expectations for the motivational impact of genetic testing affects the importance family physicians place on screening for familial cancer risk. Genet Med. 2003;5(3):172–175. doi: 10.1097/01.GIM.0000068986.03217.BB. [DOI] [PubMed] [Google Scholar]
- 44.Grant RW, Hivert M, Pandiscio JC, Florez JC, Nathan DM, Meigs JB. The clinical application of genetic testing in type 2 diabetes: a patient and physician survey. Diabetologia. 2009;52(11):2299–2305. doi: 10.1007/s00125-009-1512-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Grant RW, O’brien KE, Waxler JL, et al. Personalized genetic risk counseling to motivate diabetes prevention: a randomized trial. Diabetes Care. 2013;36(1):13–19. doi: 10.2337/dc12-0884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mcbride CM, Koehly LM, Sanderson SC, Kaphingst KA. The behavioral response to personalized genetic information: will genetic risk profiles motivate individuals and families to choose more healthful behaviors? Annu Rev Public Health. 2010;31:89–103. doi: 10.1146/annurev.publhealth.012809.103532. [DOI] [PubMed] [Google Scholar]
- 47.Marteau TM, French DP, Griffin SJ, et al. Effects of communicating DNA-based disease risk estimates on risk-reducing behaviours. Cochrane Database Syst Rev. 2010;(10):CD007275. doi: 10.1002/14651858.CD007275.pub2. [DOI] [PubMed] [Google Scholar]
- 48.Sanderson SC, Humphries SE, Hubbart C, Hughes E, Jarvis MJ, Wardle J. Psychological and behavioural impact of genetic testing smokers for lung cancer risk: a phase II exploratory trial. J Health Psychol. 2008;13(4):481–494. doi: 10.1177/1359105308088519. [DOI] [PubMed] [Google Scholar]
- 49.Stanek EJ, Sanders CL, Taber KA, et al. Adoption of pharmacogenomic testing by US physicians: results of a nationwide survey. Clin Pharmacol Ther. 2012;91(3):450–458. doi: 10.1038/clpt.2011.306. [DOI] [PubMed] [Google Scholar]
- 50.Cavallari LH, Kittles RA, Perera MA. Genotype-guided dosing of vitamin K antagonists. N Engl J Med. 2014;370(18):1763. doi: 10.1056/NEJMc1402521. [DOI] [PubMed] [Google Scholar]
- 51.Guest G, Bunce A, Johnson L. How many interviews are enough?: an experiment with data saturation and validity. Field Methods. 2006;18(1):59–82. [Google Scholar]
- 52.Blue Cross and Blue Shield Association: special report: exome sequencing for clinical diagnosis of patients with suspected genetic disorders. Technol Eval Cent Assess Program Exec Summ. 2013;28(3):1–4. [PubMed] [Google Scholar]
- 53.Veenstra DL, Piper M, Haddow JE, et al. Improving the efficiency and relevance of evidence-based recommendations in the era of whole-genome sequencing: an EGAPP methods update. Genet Med. 2013;15(1):14–24. doi: 10.1038/gim.2012.106. [DOI] [PMC free article] [PubMed] [Google Scholar]