Abstract
Protein S-sulfhydration (forming -S-SH adducts from cysteine residues) is a newly defined oxidative posttranslational modification and plays an important role in H2S-mediated signaling pathways. In this study we report the first selective, “tag-switch” method which can directly label protein S-sulfhydrated residues by forming stable thioether conjugates. Furthermore we demonstrate that H2S alone cannot lead to S-sulfhydration and that the two possible physiological mechanisms include reaction with protein sulfenic acids (P-SOH) or the involvement of metal centers which would facilitate the oxidation of H2S to HSC.
Keywords: hydrogen sulfide, protein S-sulfhydration, signal transduction, tag-switch, thiols
Hydrogen sulfide (H2S) has been recently classified as a critical cell-signaling molecule.[1] Literature published in the past few years increasingly suggests that H2S is a mediator of many physiological and/or pathological processes.[2] Some of these effects are ascribed to the formation of protein persulfides, or protein S-sulfhydration (i.e. conversion of cysteine residues -SH to persulfides -S-SH). This has been defined as a new oxidative posttranslational modification (oxPTM).[3,4] Formation of persulfides is potentially significant because it provides a possible mechanism by which H2S alters the functions of a wide range of cellular proteins and enzymes.[5] To date, the underlying mechanisms of S-sulfhydration mediated by H2S are still unclear.[3,4] A significant challenge is that the persulfide group (-S-SH) shows reactivity akin to that of other sulfur species, especially thiols (-SH), which causes difficulties in developing selective detection methods for S-sulfhydration.[4]
So far two methods have been utilized in the detection of S-sulfhydration (Scheme 1). The first method is a modified biotin switch technique.[5a] It employs an alkylating agent S-methyl methanethiosulfonate (MMTS) to differentiate thiols and persulfides. Thiols (-SH) in proteins are first blocked by MMTS. Persulfides (-S-SH) are believed to remain unreacted and be available for subsequent conjugation to N-[6-(biotinamido)hexyl]-3′-(2′-pyridyldithio)propionamide (biotin-HPDP). Using this method, a large number of proteins were identified as targets for S-sulfhydration and the basal sulfhydration level of some proteins was estimated to be as high as 25 %. In the second method,[5c] it suggested that both -SH and -SSH units can be blocked by alkylating reagents like iodoacetic acid (IAA). Then the persulfide adducts can be reduced by dithiothreitol (DTT) to form free -SH groups, and subsequently labeled with iodoacetamide-linked biotin (IAP).
Scheme 1.
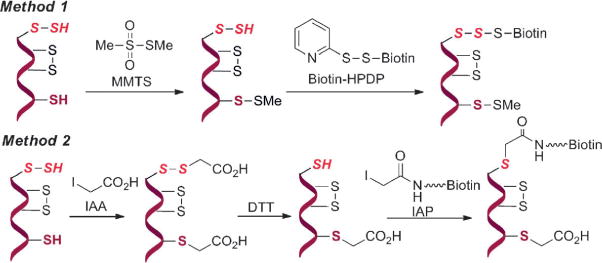
Current strategies for profiling protein S-sulfhydration.
From a chemistry perspective, both methods are problematic. In Method 1, the underlying mechanism of selectivity of MMTS for thiol versus persulfide is unclear. Studies have demonstrated that persulfides and thiols should have similar reactivity towards electrophiles such as MMTS.[4] In Method 2, it is unclear how DTT reduction would distinguish persulfide modifications from other DTT-reducible residues, such as disulfides and S-nitrosothiols.
Therefore, the chemical foundations of current methods are questionable, which may lead to erroneous results. Apparently more reliable methods for the detection of protein S-sulfhydration are needed. Having realized the very similar reactivity of both thiols and persulfides, we proposed a tag-switch technique to detect S-sulfhydration. Herein we report the development and application of this method.
As illustrated in Scheme 2, we proposed that S-sulfhydration can be selectively detected by the tag-switch method (i.e. using two reagents to label protein persulfides in two steps). In the first step a SH-blocking reagent will be introduced and it should tag both -SH and -SSH to form intermediate T. If an appropriate tag is employed, the disulfide bonds in persulfide adducts may show much enhanced reactivity to certain nucleophiles relative to the reactivity of common disulfides in proteins. Therefore we could introduce a tag-switching reagent (containing both the nucleophile and a reporting molecule such as biotin) to label only the persulfide adducts. It should be noted that thiol adducts from the first step are thioethers, which are not expected to react with the nucleophile.
Scheme 2.
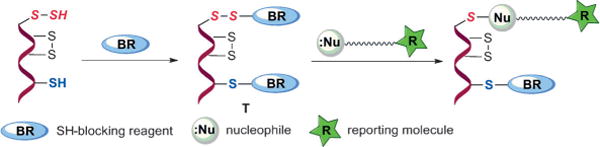
Proposed tag-switch technique for detecting S-sulfhydration.
A major challenge in this technology is whether the newly generated disulfide linkage from persulfide moieties can display a unique reactivity for a suitable nucleophile to an extent that distinguishes them from common disulfides. SH-blocking reagents are well known.[6] However, those fulfilling the criteria for this assay are limited. For example, irreversible thiol-blocking reagents such as maleimides and iodoacetamides displayed good selectivity and fast reactivity for thiols.[6] If such reagents react with persulfides, alkyl disulfide adducts are produced and their reactivity should not differ from that of cysteine or glutathionylated protein disulfides. Therefore, these reagents are not suitable for tag-switch. We envisioned that a reagent, upon reaction with persulfides to give a mixed aromatic disulfide linkage, could meet the reactivity criteria. One potential candidate is methylsulfonyl benzothiazole (MSBT), a thiol-blocking reagent recently developed by our group.[7] We expected the disulfides generated from MSBT and persulfides should be highly activated and exert a unique reactivity with certain nucleophiles, in particular, enolates.[8]
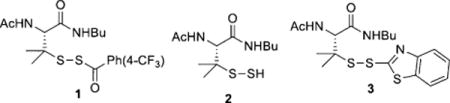
With this idea in mind, we first tested the reaction between MSBT and persulfide substrates. Since MSBT is a very effective SH-blocking reagent[7] and persulfides (-S-SH) are known to have very similar reactivity to thiols,[4] we expected MSBT should effectively block persulfides. However, it is known that small-molecule persulfides are very unstable species.[9] We could not use purified/isolated persulfides in the experiments. Instead we attempted several approaches to generate persulfides in situ from precursors like 1 (Scheme S1 in the Supporting Information) and used the persulfide intermediates directly in MSBT blocking. Indeed we obtained the desired product 3, although in low yield (13%). This result demonstrated that MSBT can react with persulfides to form R-S-S-BT adducts. The major product in the reaction was found to be polysulfides derived from persulfide 2. This should not be a concern in the case of protein persulfides because polysulfide formation is not expected to occur easily on protein persulfides.
We next used a cysteine substrate 4 to screen the appropriate nucleophile for the tag-switch step (Table 1). The preparation of 4 is shown in Scheme S2 in the Supporting Information. It should be noted that R-S-S-BT products like 4 are quite stable. They do not react with potential nucleophilic groups such as -OH and -NH2 (Scheme S3 in the Supporting Information). We screened a series of carbon-based nucleophiles as potential candidates. As shown in Table 1, three reagents (dimedone, malononitrile, and methyl cyanoacetate) proved to be effective and the corresponding products 5 b, 5 e, and 5 f were obtained in valuable yields. The reactions were also found to be fast (completing within 20 min). Among these candidates, methyl cyanoacetate (MCA; Table 1, entry 6) was particularly attractive as the ester group could allow easy installation of reporting molecules. Therefore MCA was selected in subsequent studies.
Table 1.
Screening nucleophiles for the tag-switch step.

| ||
|---|---|---|
| Entry | Nucleophile | Product (yield) |
| 1 |
|
|
| 2 |
|
|
| 3 |
|
|
| 4 |
|
|
| 5 |
|
|
| 6 |
|
|
Bn = Benzyl.
Given the dramatic structure changes in protein persulfide substrates, we wondered whether MCA could effectively react with different R-S-S-BT substrates. The reaction scope was then studied using a series of cysteine-S-S-BT derivatives (Scheme 3). Pleasingly, the reaction was found to be highly effective. In all cases the substitution products were afforded in good yields.
Scheme 3.
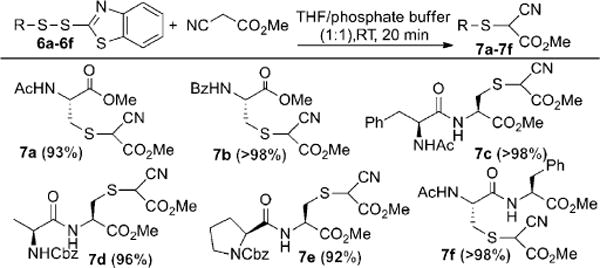
Scope of the reaction of MCA with R-S-S-BT derivatives. Cbz = carbobenzyloxy.
If MCA is used to specifically label protein persulfide derived R-S-S-BT moieties, it is critical to prove that MCA is inert towards common disulfides. We thus carried out several control experiments (Scheme S5 in the Supporting Information). We first examined the reactivity of MCA against Cys disulfide 8 (Boc = tert-butyloxycarbonyl). Under the tag-switch reaction conditions the corresponding product was not observed, even after hours. We also checked the reactivity of MCA toward S-nitrosothiol 9, which represents another well-known thiol modification in proteins. Again, no reaction was observed. Finally a crossover experiment using both R-S-BT 10 (derived from thiols) and R-S-S-BT 4 (derived from persulfides) was tested. We only observed product 5 f (from 4). The thiol-derived substrate 10 was unreactive and could be fully recovered. These results suggested that the proposed tag-switch method was selective for persulfides.
The results shown above demonstrate the chemical foundation of the tag-switch method. We then tested it in protein samples. Gpx3, an established protein-S-SH model,[4] was used in this experiment. Freshly prepared Gpx3 persulfide was treated with MSBT-A, a water-soluble MSBT derivative,[7] followed by the addition of cyanoacetate. The protein was then purified and analyzed by LC-MS. As shown in Figure 1B, cyanoacetate-labeled protein was clearly identified by MS. In the control (Figure 1A, without MSBT-A), we did not observe the peak for the cyanoacetate-labeled protein. An oxidative byproduct (P-S-SO3H) was observed in both samples and this is common for Gpx3 based on our previous experience.[4]
Figure 1.
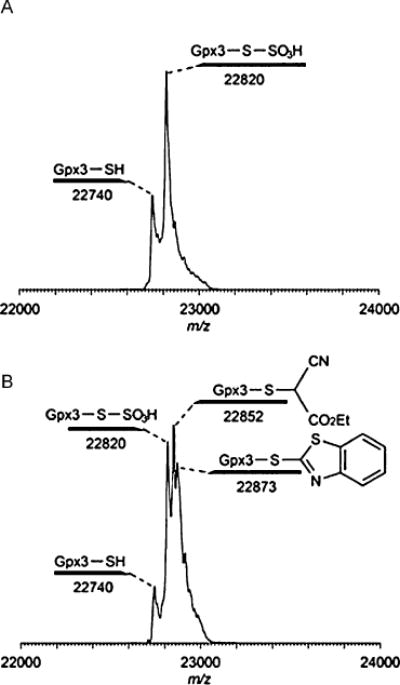
MS analysis of the tag-switch assay with Gpx3-persulfide. A) The control reaction between Gpx3 persulfide and ethyl cyanoacetate (without MSBT-A). B) The reaction between Gpx3 persulfide and ethyl cyanoacetate (with MSBT-A).
We next tested the selectivity of tag-switch assay towards different oxPTMs. A biotin-linked cyanoacetate (CN-biotin) was prepared and used in this study. A relatively stable sulfenic acid derivative of bovine serum albumin (BSA-SOH) was prepared[10] and in its reactions with glutathione[10,11] and H2S the corresponding glutathionylated and S-sulfhydrated derivatives were generated (Figure 2 A,B). Neither BSA-SH, BSA-SOH, nor BSA-SSG gave positive signals in the tag-switch assay. Only in the case of BSA-SSH could biotinylated product be pulled down by streptavidin agarose beads and detected by dot blot or ESI-TOF MS (Figure 2C and Figure S1 in the Supporting Information).
Figure 2.
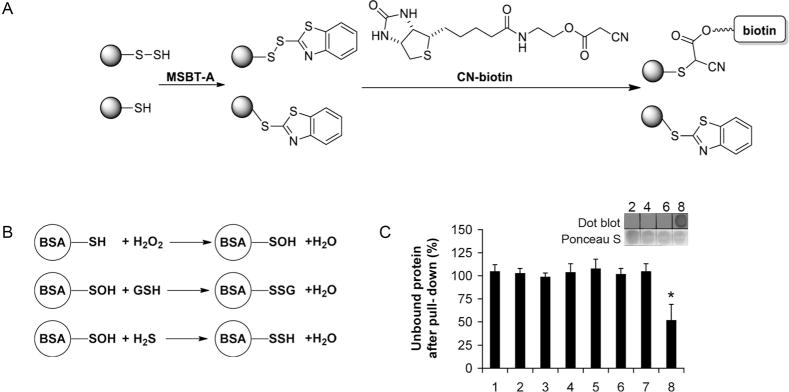
Testing the selectivity of the tag-switch assay. A) Schematic description of the tag-switch assay. B) Preparative procedure for generating different oxPTMs of BSA. C) Percentage of the unbound protein after treatment with streptavidin agarose beads. Samples 1, 3, 5, and 7 are untreated samples of BSA-SH, BSA-SOH, BSA-SSG, and BSA-SSH, respectively. Samples 2, 4, 6, and 8 are BSA-SH, BSA-SOH, BSA-SSG and BSA-SSH, respectively, treated with tag-switch reagents. n = 3, *p < 0.001. Inset shows the dot blot detection of successful biotinylation.
Although a few studies have suggested S-sulfhydration to be a potential posttranslational modification mediated by H2S that could regulate protein function,[5] there is no information, to date, dealing with the mechanism(s) underlying its formation. From a chemistry perspective, the direct reaction of protein thiols (-SH) with H2S would not be possible. However, the intermediate role of oxygen and metal centers as well as the reactions with other posttranslational modifications of cysteine could be possible.
Based on the mechanistic studies for S-glutathionylation[3,11] we addressed the following hypothetical reactions, as potential paths for P-SSH/PSS− generation under physiological conditions [Eq. (1)–(6)].
| (1) |
| (2) |
| (3) |
| (4) |
| (5) |
| (6) |
The reaction shown in Equation (1) is only the sum of the multiple reaction steps that could occur during the spontaneous oxidation of H2S where HS• is a possible intermediate[12] that would then lead to formation of S-sulfhydrated protein by means of the reaction shown in Equation (6).
The reduction of a disulfide bond by H2S [Eq. (2)] is thermodynamically unfavorable.[13] Based on the calculation of the bond energies of GSSG and GSSH, the bonding energy in the latter is roughly 18 kJmol−1 lower.[14] The reaction with sulfenic acids [Eq. (3)] does occur, as demonstrated in Figure 2B. S-Nitrosothiols would react with H2S to give HSNO, rather than to form HNO and the corresponding S-sulfhydrated protein as we previously demonstrated (with ∆rxn1G° =+ 40 kJmol−1).[14] However, a recent computational study pointed out that the surrounding of the S–NO bond could significantly affect the thermodynamic feasibility of the reaction shown in Equation (4), making the reaction possible for certain proteins that have positively charged amino acids in close proximity to the S–NO bond.[15]
The reaction of nitroxyl (HNO), a redox sibling of NO with distinct signaling pathways,[16] with a protein thiol leads to the formation of a (hydroxyamino)sulfanyl derivative. It is that this (hydroxyamino)sulfanyl derivative then reacts with other thiols with elimination of hydroxylamine as shown in Equation (5).[16]
Finally, a possible way to form S-sulfhydrated proteina is the reaction of HS• with protein thiols, where initially PSSH• is formed, which subsequently reacts quickly with O2 to give O2•− and PSSH [Eq. (6)], as observed in the formation of S-glutathionylated proteins.[11] Formation of HS• under physiological conditions would require interaction with oxidized metal centers, such as those in ferrioc heme porphyrins, which would be reduced forming HS• by means of inner-sphere electron transfer.[17]
We tested the reaction pathways (1), (3), (5), and (6) using glyceraldehyde 3-phosphate dehydrogenase (GAPDH) as a model. Angeli’s salt was used as a donor of HNO[18] to form the (hydroxyamino)sulfanyl derivative. Formation of P-SOH was induced by reaction with H2O2. Proteins were also treated with supraphysiological/pharamcological concentrations of H2S alone and H2S in combination with rigorous shaking to increase the oxygenation of the solution. As a source of HS• we used a combination of water-soluble ferric porphyrin and H2S.[19] Only when H2S reacted with P-SOH or when HS• was generated with the iron center was PS-SH detectable (Figure S2 in the Supporting Information). When BSA was used as a model of the protein with intramolecular disulfide bonds, no S-sulfhydration was observed upon addition of H2S (data not shown), confirming the low reducing power of free H2S. It is worth mentioning that an alternative mechanism could lead to protein persulfide formation, such as the reaction with polysulfides,[5g] although their physiological relevance remains to be elucidated.
As a proof of concept that the method can be applied to more complex systems such as the intracellular environment, we attempted to label proteins by the tag-switch technique in cell extracts. Protein extracts from control and Jurkat cells treated with 200 μM H2S (using Na2S as the equivalent) for 30 min at 37°C were labeled by the tag-switch method, resolved by SDS-PAGE, transferred to a nitrocellulose membrane, and identified by anti-biotin horse radish peroxidase (HRP)-conjugated antibodies. Representative Western blots (Figure 3A–C) demonstrated that a small number of proteins showed positive signals, confirming the existence of endogenous S-sulfhydration. The treatment with Na2S increased the signal intensity, but did not significantly affect the total number of S-sulfhydrated proteins. Importantly, when cell lysates were treated with streptavidin agarose beads and subsequently analyzed, no biotinylated proteins could be detected, suggesting that all modified proteins could be pulled down and the method used for the further proteomic analysis (for example, see Figure S3A in the Supporting Information).
Figure 3.
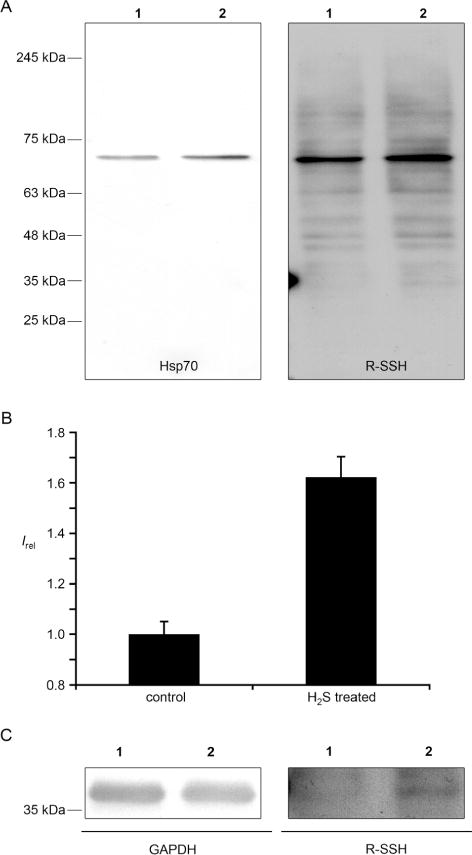
Detection of protein S-sulfhydration in cell lysates by the tag-switch assay. A) Jurkat cell lysates of the control (lane 1) and H2S-treated cells (lane 2) (200 μm Na2S, 30 min, 37°C) analyzed by the tag-switch assay. In parallel the same cell extracts were tested by Western blot analysis for the presence of Hsp70, which appeared at exactly the same position as the strongest S-sulfhydrated band. B) Quantification of the S-sulfhydration levels in the control and H2S-treated cells based on the intensity of the band at 70 kDa. C) Detection of GAPDH as a standard S-sulfhydrated protein.
It has been reported previously that GAPDH could be one of the major targets for S-sulfhydration.[5ab] Indeed, when we attempted to identify GAPDH with specific antibodies we found that this protein was endogenously S-sulfhydrated, although the signal was much stronger after the treatment with H2S (Figure 3C).
The most prominent S-sulfhydration was detected on a protein with a molecular weight of roughly 70 kDa. Using antibodies specific for heat shock protein 70 (Hsp70), we demonstrated that this protein is most likely Hsp70 (Figure 3A). Hsp70 is of great pharmacological interest[20a] and recent studies showed that it could serve as a redox sensor through oxidation of its cysteines by sulfenylation (P-SOH),[20b] which can also explain how Hsp70 could form persulfides [Eq. (3)].
As we suggested in Figure S2 in the Supporting Information and in Equation (6), metal-assisted generation of P-SSH could be the predominant way for forming P-SSH, but also a source of its artificial formation. Indeed when Jurkat cell lysates used in the experiments shown in Figure 3 were additionally exposed to 200 μM H2S, much stronger overall sulfhydration was detected than when living cells were treated (Figure S3B in the Supporting Information).
Finally, in a preliminary set of experiments we tried to adapt the tag-switch technique for the in situ labeling of methanol- or paraformaldehyde-fixed cells. Human umbilical vein cells (HUVECs), previously exposed to 100 μM Na2S for 30 min or to 2 mM propargylglycine, a cystathionine gamma lyase inhibitor,[14] for 2 h, were fixed with ice-cold methanol. Free thiols were blocked with MSBT-A and the protein S-sulfhydration (protein persulfides) tagged with CN-biotin. Finally, the cells were exposed to fluorescein-labeled streptavidin. A similar protocol was used with cells treated with Na2S or 2-ketobutyric acid (inhibitor of mercaptopiruvate S-transferase, a mitochondrial enzyme for H2S production), but with the initial difference that they were fixed with paraformaldehyde.
As shown in Figure 4 A,B, detectable S-sulfhydrated proteins increased in HUVECs treated with H2S. Partial inhibition, relative to the control, was achieved by treatment with propargylglycine but it was almost completely abolished by the use of 2-ketobutyric acid, implying the essential role of mitochondrially produced H2S, as suggested previously.[21] Almost complete absence of the signal in 2-ketobutyric acid treated cells also confirms that the nonselective binding of the fluorescent probe, incomplete blocking of free thiols, and/or unselective background fluorescence are not contributors of the main fluorescence signal. In addition, the pretreatment of the fixed cells with dimedone did not affect the detection of intracellular persulfides (Figure S4 in the Supporting Information). These data suggest that the tag-switch assay is selective for P-S-SH in the presence of P-S-OH. It should be noted that the reactivity of sulfenic acids toward carbon nucleophiles like cyanoacetate may change depending on the protein enviroment. However, even if certain P-S-OH groups would react with cyanoacetate, samples could always be pretreated with dimedone to remove the false signals from P-S-OH, as we previously demonstrated that dimedone reacts with P-S-OH but not with P-S-SH.[4]
Figure 4.
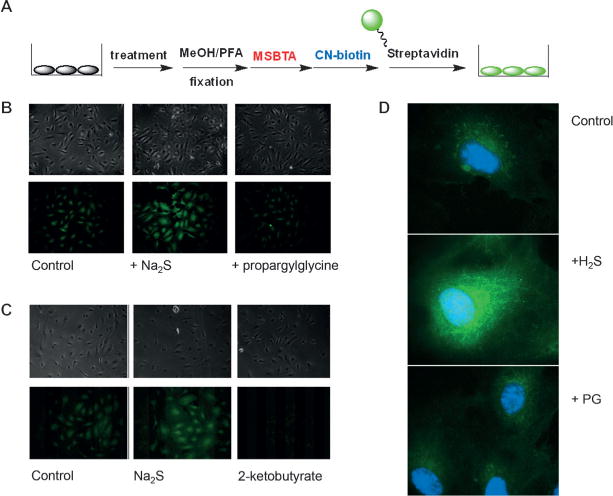
In situ fluorescence detection of intracellular S-sulfhydration of proteins. A) Schematic representation of the protocol used for intracellular labeling of S-sulfhydration. B,C) Phase contrast and fluorescence micrographs of cells fixed with methanol (MeOH) (B) and paraformaldehyde (PFA) (C). Untreated cells were used as a control. Treatments were as follows: 100 μm Na2S (30 min, 37°C), 2 mm propargylglycine (PG; 2 h, 37°C), or 2 mm 2-ketobutyric acid (2 h, 37°C). D) Fluorescent micrographs recorded at 100-fold magnification. Nuclei were stained with 4′,6-diamidino-2-phenylindole (DAPI).
Higher magnification microscopy (100 ×) gave us some indication of the intracellular distribution of the signal (Figure 4D). The perinuclear localization of the signal is indicative of localization in the mitochondrion and/or endoplasmic reticulum (ER). Since the signal in the cells was less diffused with methanol fixation we used it for co-localization studies. Mitochondrial and ER trackers proved that most of the detected persulfide signal is localized within these two organelles, within the ER predominantly (Figure S5 in the Supporting Information).
Taken together, these data offer a new, selective method for the detection of protein S-sulfhydration. Our results demonstrate that carbon-based nucleophiles such as cyanoacetate do not react with common disulfides in proteins, but with highly chemically activated disulfide species. Some mechanistic insight into physiological mechanisms for the formation of protein S-sulfhydration is presented, suggesting that the metal-center-assisted oxidation of H2S could be a predominant mechanism along with the reaction of H2S with cysteine residues which are oxidized to sulfenic acids.
Regardless of the teleological basis for the reaction, the data indeed suggest that S-sulfhydration could be a form of posttranslational modification of the mammalian proteome. The detailed mechanistic studies for P-SSH generation and the functional effects this modification can have on specific targets serve as the basis for ongoing study.
Supplementary Material
Footnotes
M.X. thanks the NSF-CAREER Program (0844931) and the American Chemical Society (Teva USA Scholar Award). M.R.F. and I.M. thank the University of Erlangen-Nuremberg within Emerging Field Initiative (EFI-MRIC) for support. K.S.C. thanks NIH R01 GM102187.
Contributor Information
Dehui Zhang, Department of Chemistry, Washington State University Pullman, WA 99164 (USA).
Igor Macinkovic, Department of Chemistry and Pharmacy, University of Erlangen-Nuremberg, Erlangen (Germany).
Nelmi O. Devarie-Baez, Department of Chemistry, Washington State University Pullman, WA 99164 (USA)
Jia Pan, Department of Chemistry, The Scripps Research Institute, Jupiter, FL 33458 (USA).
Chung-Min Park, Department of Chemistry, Washington State University Pullman, WA 99164 (USA).
Kate S. Carroll, Department of Chemistry, The Scripps Research Institute, Jupiter, FL 33458 (USA)
Milos R. Filipovic, Email: milos.filipovic@fau.de, Department of Chemistry and Pharmacy, University of Erlangen-Nuremberg, Erlangen (Germany).
Ming Xian, Email: mxian@wsu.edu, Department of Chemistry, Washington State University Pullman, WA 99164 (USA).
References
- 1.a) Li L, Rose P, Moore PK. Annu Rev Pharmacol Toxicol. 2011;51:169–187. doi: 10.1146/annurev-pharmtox-010510-100505. [DOI] [PubMed] [Google Scholar]; b) Kabil O, Banerjee R. J Biol Chem. 2010;285:21903–21907. doi: 10.1074/jbc.R110.128363. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Szabó C. Nat Rev Drug Discovery. 2007;6:917–935. doi: 10.1038/nrd2425. [DOI] [PubMed] [Google Scholar]
- 2.a) Abe K, Kimura H. J Neurosci. 1996;16:1066–1071. doi: 10.1523/JNEUROSCI.16-03-01066.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Zhao W, Zhang J, Lu Y, Wang R. EMBO J. 2001;20:6008–6016. doi: 10.1093/emboj/20.21.6008. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Yang G, Wu L, Jiang B, Yang W, Qi J, Cao K, Meng Q, Mustafa AK, Mu W, Zhang S, Snyder SH, Wang R. Science. 2008;322:587–590. doi: 10.1126/science.1162667. [DOI] [PMC free article] [PubMed] [Google Scholar]; d) Mustafa AK, Sikka G, Gazi SK, Steppan J, Jung SM, Bhunia AK, Barodka VM, Gazi FK, Barrow RK, Wang R, Amzel LM, Berkowitz DE, Snyder SH. Circ Res. 2011;109:1259–1268. doi: 10.1161/CIRCRESAHA.111.240242. [DOI] [PMC free article] [PubMed] [Google Scholar]; e) Elrod JW, Calvert JW, Morrison J, Doeller JE, Kraus DW, Tao L, Jiao X, Scalia R, Kiss L, Szabo C, Kimura H, Chow CW, Lefer DJ. Proc Natl Acad Sci USA. 2007;104:15560–15565. doi: 10.1073/pnas.0705891104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Paulsen CE, Carroll KS. Chem Rev. 2013;113:4633–4679. doi: 10.1021/cr300163e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pan J, Carroll KS. ACS Chem Biol. 2013;8:1110–1116. doi: 10.1021/cb4001052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.a) Mustafa AK, Gadalla MM, Sen N, Kim S, Mu W, Gazi SK, Barrow RK, Yang G, Wang R, Snyder SH. Sci Signaling. 2009;2:ra72. doi: 10.1126/scisignal.2000464. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Sen N, Paul BD, Gadalla MM, Mustafa AK, Sen T, Xu R, Kim S, Snyder SH. Mol Cell. 2012;45:13–24. doi: 10.1016/j.molcel.2011.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Krishnan N, Fu C, Pappin DJ, Tonks NK. Sci Signaling. 2011;4:ra86. doi: 10.1126/scisignal.2002329. [DOI] [PMC free article] [PubMed] [Google Scholar]; d) Vandiver MS, Paul BD, Xu R, Karuppagounder S, Rao F, Snowman AM, Ko HS, Lee YI, Dawson VL, Dawson TM, Sen N, Snyder SH. Nat Commun. 2013 doi: 10.1038/ncomms2623. [DOI] [PMC free article] [PubMed] [Google Scholar]; e) Paul BD, Snyder SH. Nat Rev Mol Cell Biol. 2012;13:499–507. doi: 10.1038/nrm3391. [DOI] [PubMed] [Google Scholar]; f) Yang G, Zhao K, Ju Y, Mani S, Cao Q, Puukila S, Khaper N, Wu L, Wang R. Antioxid Redox Signaling. 2013;15:1906–1919. doi: 10.1089/ars.2012.4645. [DOI] [PubMed] [Google Scholar]; g) Greiner R, Pálinkás Z, Bäsell K, Becher D, Antelmann H, Nagy P, Dick TP. Antioxid Redox Signaling. 2013 doi: 10.1089/ars.2012.5041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.a) Lane J, Spence MTZ. Molecular Probes Handbook, A Guide to Fluorescent Probes and Labeling Technologies. 11. Life Technologies, Inc.; Eugene: 2010. pp. 97–116. chap. 2. [Google Scholar]; b) Hermanson GT. Bioconjugate Techniques. 2. Elsiever; Amsterdam: 2008. [Google Scholar]
- 7.Zhang D, Devarie-Baez NO, Li Q, Lancaster JR, Jr, Xian M. Org Lett. 2012;14:3396–3399. doi: 10.1021/ol301370s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pan J, Xian M. Chem Commun. 2011;47:352–354. doi: 10.1039/c0cc02076a. [DOI] [PubMed] [Google Scholar]
- 9.Heimer NE, Field L, Neal RA. J Org Chem. 1981;46:1374–1377. [Google Scholar]
- 10.Carballal S, Radi R, Kirk MC, Barnes S, Freeman BA, Alvarez B. Biochemistry. 2003;42:9906–9914. doi: 10.1021/bi027434m. [DOI] [PubMed] [Google Scholar]
- 11.a) Dalle-Donne I, Rossi R, Giustarini D, Colombo R, Milzani A. Free Radical Biol Med. 2007;43:883–898. doi: 10.1016/j.freeradbiomed.2007.06.014. [DOI] [PubMed] [Google Scholar]; b) Cai Z, Yan LJ. J Biochem Pharmacol Res. 2013;1:15–26. [PMC free article] [PubMed] [Google Scholar]; c) Dalle-Donne I, Milzani A, Gagliano N, Colombo R, Giustarini D, Rossi R. Antioxid Redox Signaling. 2008;10:445–473. doi: 10.1089/ars.2007.1716. [DOI] [PubMed] [Google Scholar]
- 12.“Removal of Hydrogen Sulphide (H2S): Catalytic oxidation of sulphide species”: I. Ivanovic-Burmazovic, M. R. Filipovic, WO 2012/175630, 2012.
- 13.Cavallini D, Federici G, Barboni E. Eur J Biochem. 1970;14:169–174. doi: 10.1111/j.1432-1033.1970.tb00275.x. [DOI] [PubMed] [Google Scholar]
- 14.Filipovic MR, Miljkovic JL, Nauser T, Royzen M, Klos K, Shubina T, Koppenol WH, Lippard SJ, Ivanovic-Burmazovic I. J Am Chem Soc. 2012;134:12016–12027. doi: 10.1021/ja3009693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Timerghazin QK, Talipov MR. J Phys Chem Lett. 2013;4:1034–1038. doi: 10.1021/jz400354m. [DOI] [PubMed] [Google Scholar]
- 16.a) Flores-Santana W, Salmon DJ, Donzelli S, Switzer CH, Basudhar D, Ridnour L, Cheng R, Glynn SA, Paolocci N, Fukuto JM, Miranda KM, Wink DA. Antioxid Redox Signaling. 2011;13:1659–1674. doi: 10.1089/ars.2010.3841. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Fukuto JM, Carrington SJ. Antioxid Redox Signaling. 2011;13:1649–1657. doi: 10.1089/ars.2010.3855. [DOI] [PubMed] [Google Scholar]; c) Fukuto JM, Switzer CH, Miranda KM, Wink DA. Annu Rev Pharmacol Toxicol. 2005;45:335–355. doi: 10.1146/annurev.pharmtox.45.120403.095959. [DOI] [PubMed] [Google Scholar]
- 17.Pavlik JW, Noll BC, Oliver AG, Schulz CE, Scheidt WR. Inorg Chem. 2010;49:1017–1026. doi: 10.1021/ic901853p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Miranda KM, Dutton AS, Ridnour LA, Foreman CA, Ford P, Paolocci N, Katori T, Tocchetti CG, Mancardi D, Thomas DD, Espey MG, Houk KN, Fukuto JM, Wink DA. J Am Chem Soc. 2005;127:722–731. doi: 10.1021/ja045480z. [DOI] [PubMed] [Google Scholar]
- 19.Miljkovic JL, Kenkell I, Ivanovic-Burmazovic I, Filipovic MR. Angew Chem. 2013 doi: 10.1002/ange.201305669. [DOI] [Google Scholar]; Angew Chem Int Ed. 2013 doi: 10.1002/anie.201305669. [DOI] [Google Scholar]
- 20.a) Liu T, Daniels CK, Cao S. Pharmacol Ther. 2012;136:354–374. doi: 10.1016/j.pharmthera.2012.08.014. [DOI] [PubMed] [Google Scholar]; b) Miyata Y, Rauch JN, Jinwal UK, Thompson AD, Srinivasan S, Dickey CA, Gestwicki JE. Chem Biol. 2012;19:1391–1399. doi: 10.1016/j.chembiol.2012.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.a) Shibuya N, Mikami Y, Kimura Y, Nagahara N, Kimura H. J Biochem. 2009;146:623–626. doi: 10.1093/jb/mvp111. [DOI] [PubMed] [Google Scholar]; b) Módis K, Coletta K, Erdelyi K, Papapetropoulos A, Szabo C. FASEB J. 2013;27:601–611. doi: 10.1096/fj.12-216507. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


