Summary
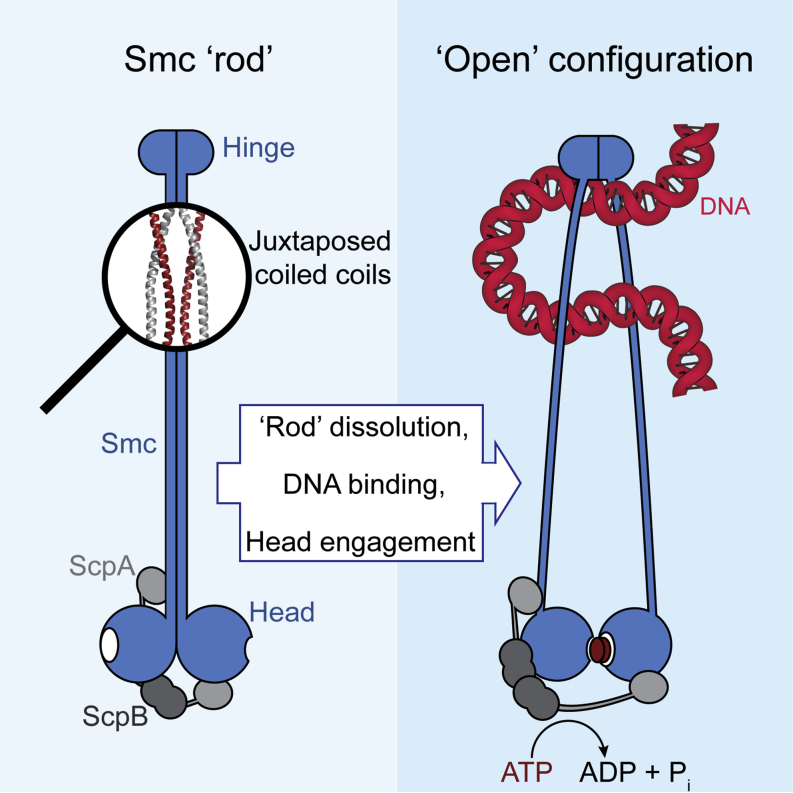
SMC condensin complexes are central modulators of chromosome superstructure in all branches of life. Their SMC subunits form a long intramolecular coiled coil, which connects a constitutive “hinge” dimerization domain with an ATP-regulated “head” dimerization module. Here, we address the structural arrangement of the long coiled coils in SMC complexes. We unequivocally show that prokaryotic Smc-ScpAB, eukaryotic condensin, and possibly also cohesin form rod-like structures, with their coiled coils being closely juxtaposed and accurately anchored to the hinge. Upon ATP-induced binding of DNA to the hinge, however, Smc switches to a more open configuration. Our data suggest that a long-distance structural transition is transmitted from the Smc head domains to regulate Smc-ScpAB’s association with DNA. These findings uncover a conserved architectural theme in SMC complexes, provide a mechanistic basis for Smc’s dynamic engagement with chromosomes, and offer a molecular explanation for defects in Cornelia de Lange syndrome.
Graphical Abstract
Highlights
-
•
Prokaryotic Smc-ScpAB complexes form rod-like structures
-
•
Binding of ATP and DNA induces a rod-to-ring transition in prokaryotic condensin
-
•
The condensin hinge is rigidly anchored to its coiled coil
-
•
The rod-like conformation is a conserved feature of SMC protein dimers
Soh et al. show that the rod-like conformation is a conserved architectural scheme of SMC complexes. Upon ATP-induced binding to DNA, the juxtaposed coiled coils of prokaryotic Smc-ScpAB adopt an open conformation to expose a DNA binding site at the inner surface of the hinge domain.
Introduction
Chromosome condensation takes place in all forms of life. It is essential for faithful partitioning of replicated chromosomes into nascent daughter cells during cell division. Multisubunit complexes, termed condensins, are key mediators of this process (Hirano, 2012; Thadani et al., 2012). They commonly have a large core subunit (>1,100 amino acids) that belongs to the family of structural maintenance of chromosome proteins (SMC; in capital letters, used as a generic term for protein family). Several types of condensins have been identified: three condensins in prokaryotes, Smc-ScpAB, MukBEF, and MksBEF; and two eukaryotic condensins, condensin I and condensin II (condensin I/II). Smc-ScpAB comprises a homodimer of Smc and the two non-SMC subunits ScpA and ScpB (Mascarenhas et al., 2002; Soppa et al., 2002), whereas MukBEF comprises a homodimer of the SMC subunit MukB and the two non-SMC subunits MukE and MukF (Woo et al., 2009; Yamazoe et al., 1999). Smc-ScpAB is nearly ubiquitous in prokaryotes and more closely related to condensin in eukaryotes than MukBEF, which is found in only some branches of γ-proteobacteria. Condensin I/II are composed of the same heterodimer of Smc2 and Smc4 (Smc2-4) and a different set of three non-SMC subunits (Onn et al., 2007).
The SMC subunit exhibits a peculiar folding pattern: the extreme N- and C-terminal segments together form an ABC-type nucleotide binding domain (also called SMC head), a middle segment folds into a so-called hinge domain, and the two intervening segments form an ∼50-nm-long antiparallel coiled coil connecting the two domains (Nolivos and Sherratt, 2014). The hinge domain is the interaction interface for the homo- or heterodimerization of SMC subunits (Haering et al., 2002). One of the non-SMC subunits generally belongs to a superfamily of proteins called kleisins, which bind and bridge the head domains of the SMC subunits (Schleiffer et al., 2003). In Smc-ScpAB condensin, the kleisin subunit ScpA binds two distinct interfaces on and near the Smc head domain to form a 1:1 asymmetric holocomplex between the Smc dimer and the ScpA1B2 subcomplex (Bürmann et al., 2013). Likewise, in condensin I/II, the head domains of the Smc2-4 heterodimer are presumably bridged by the kleisin subunit Cap-H/H2 that associates with two additional subunits, Cap-G/G2 and Cap-D2/D3 (Hirano, 2012).
The eukaryotic Smc1-Smc3 (Smc1-3) cohesin complex, which is evolutionarily related to condensin, is a chromosome concatenase that holds sister chromatid DNA within its closed ring structure (Nasmyth, 2011). Striking architectural similarities between different SMC complexes suggest that they all function using a fundamentally conserved mode of action (Bürmann et al., 2013). Consistent with this notion, eukaryotic condensin, like cohesin, associates with minichromosomes by entrapment of DNA within its ring (Cuylen et al., 2011).
The distant hinge and head domains are involved in the loading of SMC-kleisin complexes onto DNA. ATP hydrolysis by Smc1 and Smc3 head domains is essential for stable binding of cohesin to chromosomes, whereas the SMC hinge domains harbor affinity for DNA in cohesin, condensin, and Bacillus subtilis (Bs) Smc-ScpAB (Arumugam et al., 2003; Chiu et al., 2004; Griese et al., 2010; Hirano and Hirano, 2006; Weitzer et al., 2003). DNA binding stimulates ATP hydrolysis in Smc-ScpAB and condensin in vitro (Hirano and Hirano, 2006; Kimura and Hirano, 1997), and hinge opening appears to be required for loading DNA into cohesin rings (Gruber et al., 2006). Whether (and how) ATP binding and hydrolysis at the SMC heads might be mechanistically coordinated with DNA binding to the hinge and opening of the DNA entry gate is largely unclear. Conceivably, the coiled-coil arms could provide a mechanical link if they were somewhat stiff and rigidly connected to hinge and/or head domains. According to electron microscopy (EM) and atomic force microscopy, the coiled coils in SMC dimers and SMC holocomplexes are not in random conformations but mostly V or O shaped or juxtaposed onto each other over their entire length (Anderson et al., 2002; Fuentes-Perez et al., 2012; Haering et al., 2002; Matoba et al., 2005; Melby et al., 1998). In the crystal structure of the Thermotoga maritima (Tm) Smc hinge, two short coiled coils protrude from the hinge domain dimer in nearly opposite orientations (Haering et al., 2002). Similar coiled-coil configurations were found in crystal structures of the MukB hinge domain (Ku et al., 2010; Li et al., 2010; Vos et al., 2013). The observed variety in the conformations of SMC coiled coils might be partly, or entirely, due to (1) intrinsic structural flexibility, (2) structural differences between classes of SMC-kleisin complexes, or (3) experimental artifacts. Thus, it is largely unclear what configurations SMC-kleisin rings adopt on the chromosome, or in solution, and whether conformational changes are required during chromosomal loading and unloading cycles.
Using an integrative approach including crystallographic analyses of SMC protein fragments with long stretches of coiled coil, we demonstrate that both prokaryotic and eukaryotic condensins form rod-shaped holocomplexes with rigid and juxtaposed coiled coils. We further reveal that binding of DNA to Smc dimers is incompatible with the reported coiled-coil arrangement at the hinge and uncover an interplay between DNA and ATP binding in the dynamic control of Smc arm conformation. These findings allow us to propose a mechanism by which the ATPase head domains regulate DNA binding to the hinge via engagement and disengagement of Smc coiled coils.
Results
EM of Smc-ScpAB Holocomplexes
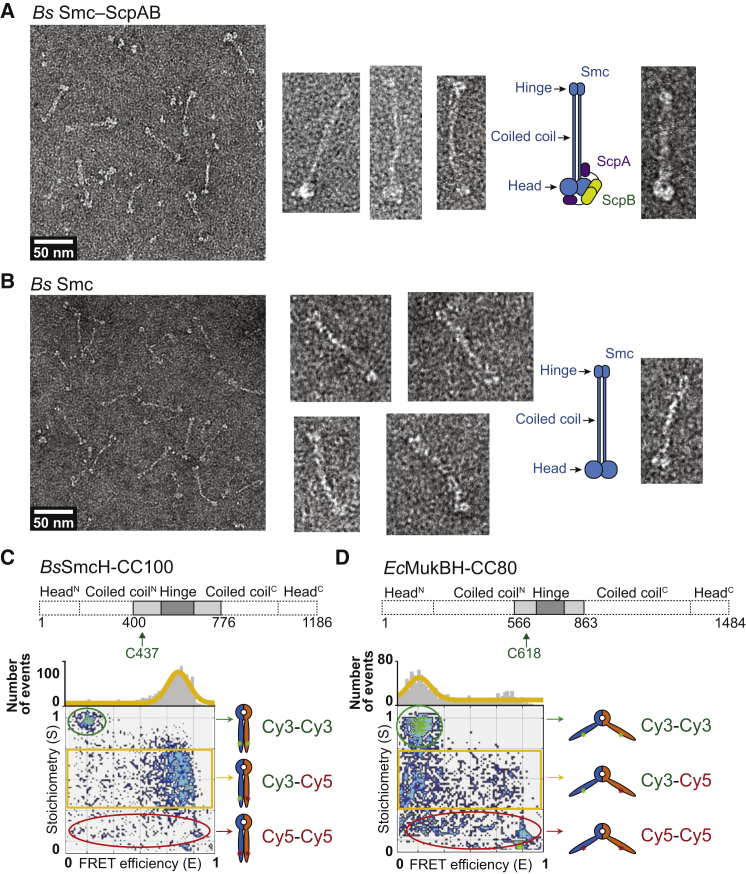
Crystallographic studies supported by biochemical and genetic data have provided detailed insights into the structures of all globular parts of prokaryotic Smc-ScpAB complexes. However, our understanding of the overall architecture of Smc-ScpAB and, in particular, the arrangement of the Smc coiled coils in the holocomplex of Smc-ScpAB has remained rather limited so far. Here, we have purified Bs Smc protein as well as Smc-ScpAB holocomplexes produced in Escherichia coli (Ec). As determined by size exclusion chromatography-multiangle light scattering (SEC-MALS), our preparations comprise near homogenous solutions with molecular weights fitting well to isolated Smc dimers and heteropentameric Smc2-ScpA1B2 complexes (Figure S1 available online). The proteins were negatively stained and visualized by EM (Figures 1A and 1B). Smc dimers and Smc-ScpAB holocomplexes were almost exclusively detected as straight objects comprising a single extended rod flanked by a small and a large globular density, which likely correspond to the Smc hinge and head domains with or without ScpAB. In good agreement with data obtained for Bs Smc protein by rotary shadowing experiments, these images suggest that the two Smc coiled coils are mostly aligned side by side (Melby et al., 1998).
Figure 1.
EM and FRET Analysis of Smc-ScpAB
(A) EM images of negatively stained Bs Smc-ScpAB. Selected objects are shown in high magnification (right).
(B) EM images of negatively stained Bs Smc protein.
(C) ALEX-FRET analysis of BsSmcH-CC100 (schematic drawing of construct on top) stochastically labeled at C437 with Cy3 and Cy5. In the FRET efficiency versus stoichiometry graph, each dot denotes a single BsSmcH-CC100 dimer. The green and red ellipses indicate dimers labeled by Cy3 or Cy5, respectively. Doubly labeled Cy3-BsSmcH-CC100-Cy5 dimers (yellow box) exhibit mostly high FRET.
(D) Same as in (C) using EcMukBH-CC80 labeled at C618.
See also Figure S1.
Juxtaposition of Smc Coiled Coils in Solution
We were concerned that the observed rod-like structure might arise during the harsh conditions used for EM sample preparation. Therefore, we probed the configuration of the Smc coiled coils under more physiological conditions by estimating the distance between symmetry-related positions on the two coiled coils in solution using fluorescence resonance energy transfer (FRET). We produced a Bs Smc fragment comprising the Smc hinge domain and a long stretch of coiled coil (∼100 residues), designated as BsSmcH-CC100 (Figure 1C). A single cysteine residue, Cys437, on the coiled coil was used for stochastic labeling with donor (Cy3) and acceptor (Cy5) dyes (Figure 1C). In order to discriminate the FRET pair (Cy3-Cy5 dimer) from any non-FRET pair (Cy3-Cy3 and Cy5-Cy5 dimers) in the sample, the single-molecule alternating-laser excitation FRET (ALEX-FRET) method was applied. The Cy3-Cy5 dimer species exhibited predominantly higher values of FRET efficiency, E, indicating that the two coiled coils are close to each other in most or all dimers of BsSmcH-CC100 (Figure 1C). Based on the observed FRET efficiency, the distance between Cy3 and Cy5 was estimated to be around 44 Å (E = 0.86) or approximately twice the diameter of a coiled coil. Similar experiments performed on a related fragment of the MukB protein, which exists in an open V conformation, demonstrated the validity of our FRET approach (Figure 1D). Furthermore, low real-time fluctuations in single-molecule total internal reflection fluorescence (TIRF)-FRET suggest that the coiled coils of Bs Smc hinge fragments are mostly or always closely juxtaposed (Figure S1).
Structure of Juxtaposed Smc Coiled Coils
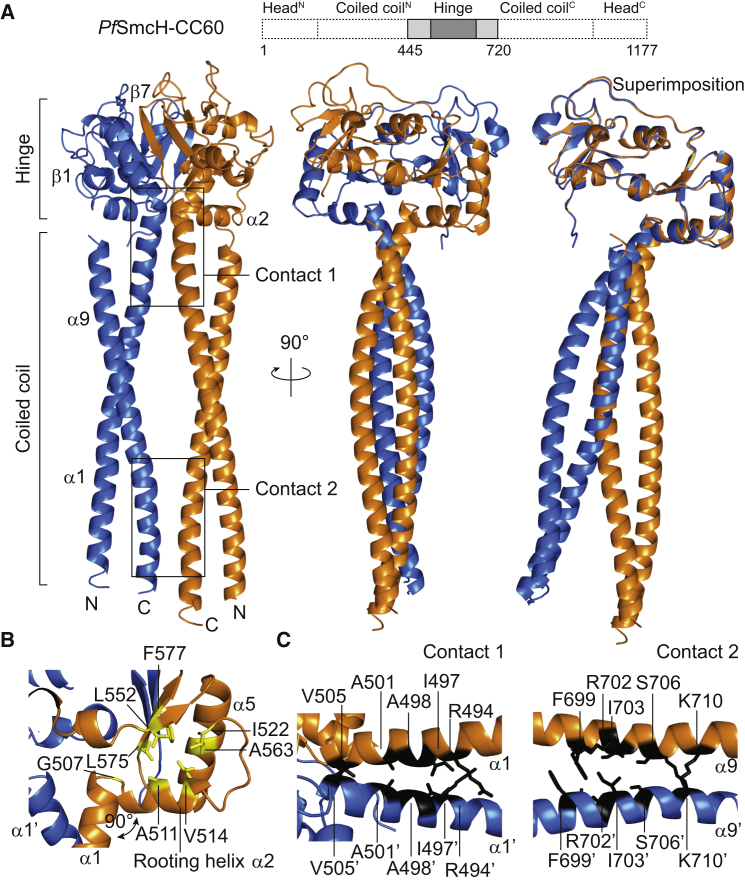
To elucidate the molecular basis for the alignment of Smc coiled coils, we determined the atomic structure of a fragment of Pyrococcus furiosus (Pf) Smc containing its hinge and a significant part of its coiled coil. After we screened several constructs with different lengths of coiled coil, crystals of a Pf Smc hinge domain with a 60-residue stretch of coiled coil, referred to as PfSmcH-CC60, were obtained, and the phase problem was solved by molecular replacement using the structure of an isolated Pf Smc hinge (Table 1) (Griese and Hopfner, 2011). The asymmetric unit of the crystal contained two copies of the PfSmcH-CC60 homodimer. As expected, the two hinge domains in the homodimer interact with each other at two identical interfaces to form a toroidal structure having a flat bottom and a central hole with the largest dimension of ∼16 Å (Figure 2A; Figure S2A). The two coiled coils are aligned in parallel and are closely juxtaposed onto each other, thereby forming a rod-like overall structure. The coiled coils emanate from the Smc hinge perpendicular to the bottom surface of the toroid, reminiscent of tentacles radiating from the body of a jellyfish. This shape is a result of a sharp ∼90° kink, governed by a single glycine residue (Gly507), at the junction between the N-terminal helix of the coiled coil and the following “rooting helix” α2, which interacts with a hydrophobic groove at the bottom of the toroid (Figure 2B). The presented architecture of the PfSmcH-CC60 homodimer is in sharp contrast with the V-shaped organization of the Tm Smc hinge with short coiled coils (discussed later) and of the Ec MukB hinge with long coiled coils (Figure S2B) (Li et al., 2010).
Table 1.
Data Collection and Structure Refinement Statistics
| Data Collection | PfSmcH-CC60 | ScSmc2H-CC110/ScSmc4H-CC110 |
|---|---|---|
| Crystal | Native | selenomethionine substituted |
| X-ray sourcea | 5C, PAL | BL17A, PF |
| Space group | P212121 | C2 |
| Unit cell dimensions | ||
| a, b, c (Å) | 101.92, 116.88, 145.493 | 185.26, 49.71, 154.28 |
| α, β, γ (°) | 90, 90, 90 | 90, 92.52, 90 |
| Wavelength (Å) | 1.0000 | 0.9789 |
| Resolution (Å) | 50.0–3.5 | 50.0–2.9 |
| Rsym (%) | 9.3 (28.2)b | 9.0 (33.8)b |
| I/σ(I) | 26.2(5.3) | 33.5(4.8) |
| Completeness (%) | 89.5 (74.7) | 90.2 (73.6) |
| Redundancy | 5.6 (2.9) | 4.4 (2.3) |
| Refinement | ||
| Resolution (Å) | 50.0–3.5 | 50.0–2.9 |
| Number of reflections | 20,012 | 49,139 |
| Rwork/Rfree (%) | 23.6/28.4 | 22.3/26.6 |
| Root-mean-square deviations | ||
| Bond (Å)/angle (°) | 0.003/0.78 | 0.010/1.29 |
| Average B values (Å2) | 47.08 | 84.58 |
| Ramachandran plot (%) | ||
| Most favored/favored | 88.2/11.3 | 86.7/13.1 |
| Generously allowed | 0.2 | 0.2 |
Beamline 5C at Pohang Accelerator Laboratory (PAL) and Beamline BL-17A at Photon Factory (PF).
The numbers in parentheses are the statistics from the highest resolution shell.
Figure 2.
Structure of the Pf Smc Hinge with Long Coiled Coils
(A) Crystal structure of a dimer of PfSmcH-CC60 shown in two perpendicular views (left and middle panels). Structural superimposition of the two monomers demonstrates slight asymmetry at the coils/hinge junction (right panel).
(B) Details of the coils/hinge interaction in Pf Smc. Conserved hydrophobic residues are displayed in stick representation in yellow. The arrow indicates a 90° kink at G507 between the coiled-coil helix, α1, and the rooting helix, α2.
(C) Structural view of the hinge-proximal (Contact 1, left panel) and hinge-distal coils/coils interface (Contact 2, right panel) in PfSmcH-CC60.
See also Figure S2.
Right below the bottom surface of the Pf Smc hinge, the two N-terminal coiled-coil helices of the homodimer pack against each other and engage in hydrophobic contacts (Figure 2C). In addition, ∼80 Å (or ∼50 residues) below the toroid, the two C-terminal coiled-coil helices are in contact with each other in a similar fashion (Figure 2C). This two-site interaction is found in both dimers of the asymmetric unit and appears to be responsible for holding the coiled coils together. Curiously, the two Smc coiled coils within a dimer display slightly distinct angles of attachment to the hinge, thus creating an asymmetric overall architecture (Figure 2A, right panel). This asymmetry might allow a more stable interaction to be formed at the coils/coils interface.
Smc-ScpAB Adopts a Rod-like Structure in B. subtilis
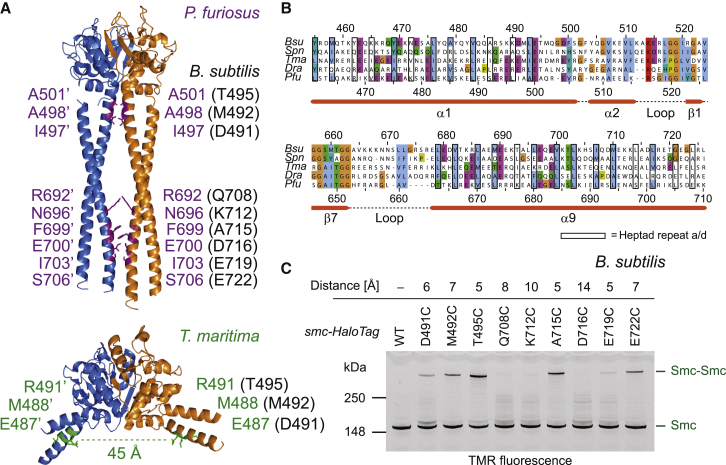
The coiled coils emanating from the Smc hinge adopt juxtaposed configurations in fragments of Bs and Pf Smc. However, multiple sequence alignments indicate that residues at the interfaces between the coiled coils in Pf Smc are not particularly well conserved (Figures 3A and 3B), raising the questions of whether this coils/coils interface is specific to archaeal Smc proteins or a general feature conserved through coevolution of pairs of residues. To test this, we probed the conformation of the coiled coils in endogenous holocomplexes of prokaryotic condensin using cysteine-specific crosslinking in living Bs cells. Based on the structure of PfSmcH-CC60, we engineered single-cysteine residues into the predicted coils/coils interfaces of Bs Smc. As the coils/coils interface is located along the 2-fold symmetry axis, crosslinking of cysteines by the thiol reactive compound BMOE will occur between symmetry-related Cys residues, which are in close proximity (<8 Å). To determine crosslinking efficiencies, we made use of a C-terminal HaloTag fusion to Smc permitting in-gel fluorescence detection of Smc species (Bürmann et al., 2013). Based on sequence alignments and coiled-coil predictions, three residues in the N-terminal helix of the Smc coiled coil were chosen to be mutated to cysteine (D491C, M492C, and T495C) (Figure 3A). All three mutant Smc proteins are functional, as judged by growth on rich medium (data not shown) (Gruber et al., 2014). They displayed significant levels of Smc-Smc crosslinking after incubation with BMOE, whereas a wild-type control showed little or no crosslinking (Figure 3C). Thus, the selected residues are located in close proximity of their symmetry mates, as predicted by the PfSmcH-CC60 but not the Tm Smc hinge structure (Figure 3A). The alignment of sequences in the C-terminal helix of the Smc coiled coil is more ambiguous; therefore, several residues were mutated to cysteines in a region about 45–60 residues from the Smc hinge domain. Three cysteine residues (Q708C, K712C, and D716C) showed very little Smc-Smc crosslinking—likely because their side chains are too far apart or facing opposite sides of the coils/coils structure. In stark contrast, residues A715C and E722C supported robust crosslinking of Smc (Figure 3C). In summary, efficient crosslinking by specific cysteine residues demonstrates that the coiled coils emanating from the Smc hinge are held together by a defined interface in endogenous Smc-ScpAB complexes.
Figure 3.
Juxtapositioning of the Smc Coiled Coils in Bs Smc-ScpAB
(A) Map of residues located at the coils/coils interface of PfSmcH-CC60 (top). Equivalent positions in Bs Smc were identified based on the sequence alignment shown in (B). In the Tm Smc hinge structure (PDB ID: 1GXL), these residues are distantly located from their symmetry mates (bottom panel). Labels for amino acids in Pf, Tm, and Bs Smc are shown in purple, green, and black, respectively.
(B) Alignment of the N- and C-terminal coils/hinge junctions (top and bottom panels, respectively) of four bacterial and one archaeal Smc protein sequence. (Bsu, Bs; Dra, D. radiodurans; Pfu, Pf; Spn, S. pneumoniae; Tma, Tm) Secondary structure elements are based on the structure of PfSmcH-CC60.
(C) In vivo crosslinking of cysteine mutants of Bs Smc-HaloTag with BMOE. Distances between symmetry-related Cys residues in Bs Smc were estimated according to the PfSmcH-CC60 structure. Cells were grown in Luria-Bertani medium. Strains: BSG1711, BSG1760–1765, and BSG1821–1823.
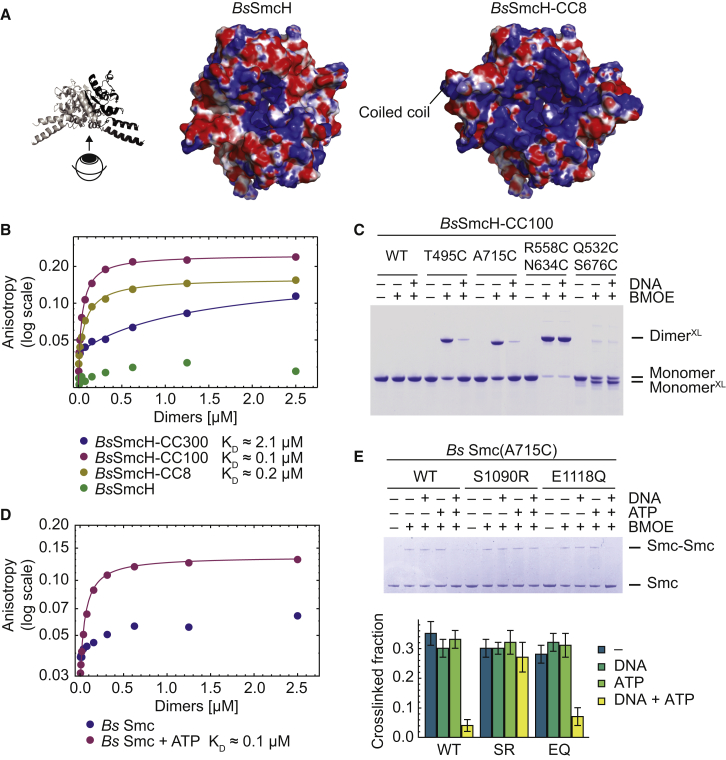
DNA Binding to the Hinge Facilitates Opening of Smc Arms
Given the fact that several SMC hinge domains display DNA binding affinity, we wondered whether the coiled-coil configuration would have any influence on the association of Smc with DNA. DNA binding activity of SMC hinges has been tentatively mapped to positively charged residues in the transition region between the hinge and the coiled coils in cohesin and Smc-ScpAB (Chiu et al., 2004; Hirano and Hirano, 2006). Accordingly, bound DNA might be located at the bottom surface of the hinge toroid (Figure 4A). Intriguingly, this area is obstructed in our Pf Smc structure by the presence of the aligned coiled coils, thus highlighting the possibility that a conformational change at the Smc hinge might control its association with DNA. Using fluorescence anisotropy measurements with short stretches of double-stranded DNA (dsDNA), we first confirmed that BsSmcH-CC100 displays affinity for DNA in the submicromolar range (dissociation constant, KD, ∼0.1 μM) (Figure 4B). Consistent with the notion that the transition region is involved in DNA binding, we found that an isolated Bs Smc hinge (BsSmcH) lacking this region fails to bind to DNA, whereas a slightly larger construct (BsSmcH-CC8) binds DNA (KD, ∼0.2 μM) with an affinity similar to that of BsSmcH-CC100. A Smc protein fragment harboring almost the entire Smc coiled coil attached to the hinge domain, designated as BsSmcH-CC300, associated with DNA only poorly, indicating that the long coiled coils interfere with efficient DNA binding at the hinge, possibly because of stable occlusion of the DNA binding site (Figure 4B). Next, we wondered whether the coiled coils are still juxtaposed when BsSmcH-CC100 is bound to DNA. To test this, we have purified BsSmcH-CC100 harboring T495C or A715C for crosslinking of Smc coiled coils. Intriguingly, formation of crosslinked dimers of BsSmcH-CC100 was strongly affected by the presence of DNA (Figure 4C). In contrast, DNA binding had no effect on the crosslinking of a pair of cysteines located at the hinge dimer interface (R558C/N634C) (Bürmann et al., 2013) (Figure 4C). Furthermore, a cysteine pair at the coils/hinge intersection (Q532C/S676C) displayed a modest but reproducible increase in intramolecular crosslinking in the presence of DNA (increased from 50% ± 1% to 60% ± 1%), implying that this cysteine pair might preferentially capture the more open conformation of the coiled coils (Figures 4C and S3D). We next repeated the crosslinking of A715C using bis-maleimide compounds having long linkers between the reactive groups (BM-PEG3, ∼30 Å; and BM-PEG11, ∼55 Å), which are able to capture more distant pairs of cysteines. BM-PEG3 and BM-PEG11 exhibited robust crosslinking of A715C in the absence of DNA but failed to do so in the presence of DNA (Figure S3G), implying that the A715C residues are too distantly located to be bridged by either BM-PEG3 or BM-PEG11 when BsSmcH-CC100 is bound to DNA. As positive control for long-distance crosslinking, we created a pair of cysteines (R516C, S597C) located about 30 Å apart from each other on the Smc hinge domain. As expected, this cysteine pair was efficiently crosslinked by BM-PEG11, but not by BM-PEG3 or BMOE, regardless of the presence or absence of DNA (Figure S3G). Together, these findings strongly suggest that DNA binding stabilizes an open conformation of the coiled coils at the SMC hinge. Intriguingly, this immediately implies a molecular model of regulated DNA binding by Smc: ATP binding or hydrolysis at the Smc heads might facilitate opening of Smc arms and thus expose the DNA binding site at the hinge. To test this, we purified full-length Bs Smc and cysless Smc(A715C) and performed DNA binding and crosslinking studies in the absence and presence of ATP. Without ATP, Smc imposed only a weak effect on the fluorescence anisotropy of DNA, indicating that the DNA binding site at the hinge is at least partly occluded in full-length Smc protein (Figure 4D). In the presence of ATP, however, the anisotropy response was substantial, producing an affinity (KD, ∼0.1 μM) similar to that of Smc hinge fragments. Exclusively under these conditions (i.e., with DNA and ATP), crosslinking of Smc arms at A715C was strongly reduced (Figure 4E). A hydrolysis-defective Smc mutant (E1118Q) displayed normal Smc arm opening, whereas a mutant blocked in Smc head engagement (S1090R) was locked in the rod-like state, suggesting that ATP-dependent head engagement drives dissolution of Smc rods (Figure 4E). Thus, Smc arms undergo an extended structural transition, which is cooperatively promoted by binding of DNA to the Smc hinge and ATP to the head domains.
Figure 4.
Structural Changes at the Coils/Hinge Junction upon DNA and ATP Binding
(A) Bottom view of electrostatic surface potential maps of Bs Smc hinge models (Kurze et al., 2011) based on the Tm Smc hinge structure (PDB ID: 1GXL). Left: isolated Bs Smc hinge. Right: hinge with short coiled coils.
(B) DNA binding of Bs Smc fragments measured by fluorescence anisotropy using fluorescein-labeled DNA (40 bp).
(C) Crosslinking of cysteine-bearing variants of BsSmcH-CC100 with and without DNA. XL denotes species crosslinked by BMOE. WT, wild-type.
(D) DNA binding of Bs Smc in the presence and absence of ATP measured by anisotropy using fluorescein-labeled DNA (40 bp).
(E) Crosslinking of Bs Smc(A715C) variants with and without mutations in the ABC signature and Walker B motif (S1090R [SR] and E1118Q [EQ], respectively). The four endogenous cysteines have been replaced by serines. Quantification of crosslinking efficiency is based on three independent replicates. Data are represented as mean ± SEM.
See also Figure S3.
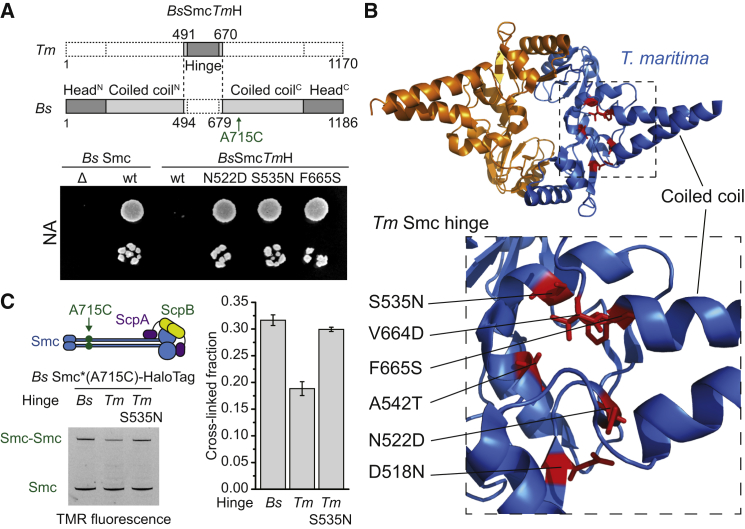
Artificial Opening of Smc Arms Is Detrimental
If the observed conformational change was physiologically relevant, then locking Smc-ScpAB in the open or closed configuration should jeopardize its functionality. Because of the extensive nature of the coils/coils interface and the rigid connection to the hinge, mutations in single residues are unlikely to have significant impact on the overall architecture. Thus, we decided to replace parts of the Bs Smc protein with homologous protein fragments, which might intrinsically bear higher propensity for one or the other conformation. The Tm hinge was chosen because it adopts an open, V-shaped organization in protein crystals. With the help of available structural information, a chimeric protein was constructed by splicing together N- and C-terminal sequences of Bs Smc with the central part of Tm Smc comprising its hinge domain and short stretches of the adjacent coiled coil (Figure 5A). The resulting BsSmcTmH protein was expressed from the endogenous locus in Bs. It accumulated at normal levels in vivo and efficiently formed Smc dimers according to crosslinking analysis, implying that protein folding was mostly unperturbed (data not shown). However, its functionality was severely compromised, as judged by colony formation assays (Figure 5A) (Gruber et al., 2014). To identify the underlying cause for this loss of function in BsSmcTmH, we mutagenized its Tm hinge moiety and isolated suppressor mutations that enabled normal growth on rich medium (Figure S4A). Most suppressor mutations mapped in the vicinity of the connection between the Tm Smc hinge and the adjacent coils, suggesting that these mutations might indeed provide increased structural flexibility at the coils/hinge interface (Figure 5B). Alternatively, these mutations could affect DNA binding to the Tm hinge. Although the Tm Smc hinge bound DNA in a more salt-sensitive manner than the Bs Smc hinge, the suppressor mutation S535N had no effect on the DNA binding of a chimeric Smc hinge fragment with long coiled coils (Figure S4B). Thus, defects in DNA binding at the Tm hinge are an unlikely explanation for the loss of functionality in BsSmcTmH. To measure the juxtapositioning of Smc arms in chimeric Smc proteins, we next fused a C-terminal HaloTag to BsSmcTmH and introduced the cysteine residue A715C for crosslinking. Crucially, the arms of the nonfunctional BsSmcTmH protein were only poorly crosslinked by BMOE, suggesting that the Tm hinge domain in BsSmcTmH promotes a more open coiled-coil arrangement, as suggested by its crystal structure (Figure 5C). Notably, the suppressed version, BsSmcTmH(S535N), displayed wild-type levels of Smc arm crosslinking. Thus, the ability to efficiently adopt the rod-shaped conformation appears crucial for Smc function. In combination with the observation that DNA binding stabilizes the open form, this finding strongly supports the notion that both open and closed conformations are crucially important for condensin function. We propose that transitions from rod-like to ring-like states and vice versa are essential for the biochemical action of Smc-ScpAB.
Figure 5.
Artificial Opening of Smc Arms at the Hinge Is Detrimental in B. subtilis
(A) Schematic drawing of the BsSmcTmH construct (top). Colony formation assay of strains of Bs encoding variants of Bs Smc or BsSmcTmH as the single source of Smc protein on nutrient-rich medium. Strains: BSG1001, BSG1007, BSG1363, BSG1365, BSG1368, and BSG1970.
(B) Mapping of suppressor mutations onto the Tm Smc hinge structure (PDB ID: 1GXL). Residues altered in BsSmcTmH suppressor mutants are highlighted in red as sticks.
(C) In vivo cysteine crosslinking of Bs Smc(A715C), BsSmcTmH(A715C) and its functional variant harboring the S535N mutation. All four endogenous cysteines have been replaced by serines. The graph shows means and SD from triplicate reactions. Cells were grown in SMG medium. Strains: BSG1921, BSG1932, and BSG1934.
See also Figure S4.
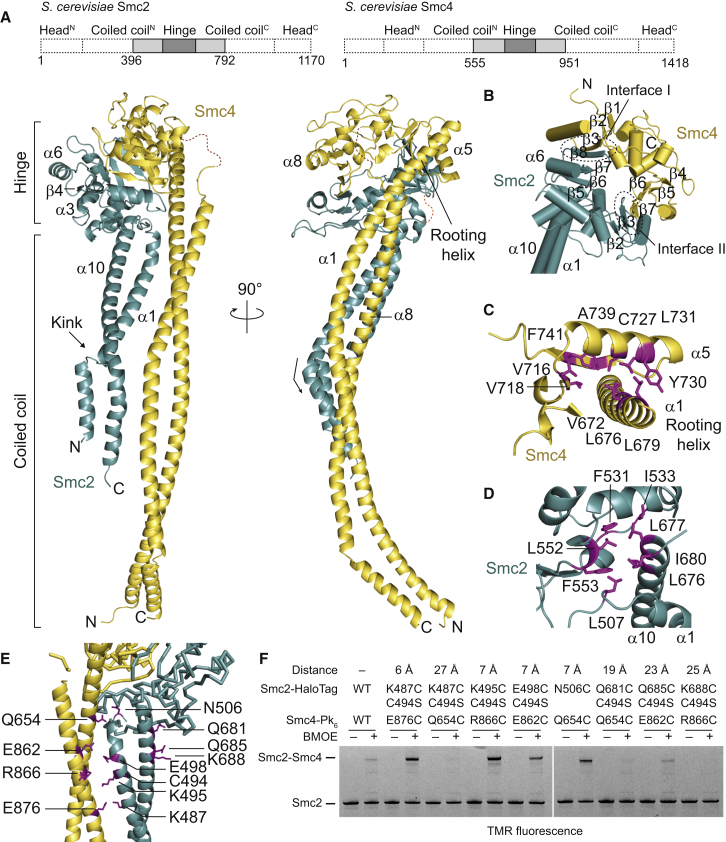
Structure of a Yeast Smc2-4 Hinge Heterodimer with Long Coiled Coils
Eukaryotic condensin has been observed as a rod-like structure by EM, suggesting that the architecture of the coils/hinge connection might be conserved between pro- and eukaryotic SMC complexes. The structure of a mouse Smc2-4 hinge heterodimer has recently been solved (Griese et al., 2010). However, because of the lack of coiled coils, no insight into their arrangement with respect to the hinge was gained. To address this, we generated a number of yeast Smc2 and Smc4 constructs containing the hinge domain and long stretches of coiled coil. Of these, Smc2 (residues 396–792) and Smc4 (residues 555–951), referred to as ScSmc2H-CC110 and ScSmc4H-CC110, respectively, were crystallized as a heterodimeric complex (Figure 6A). The coiled-coil stretches in these proteins correspond to about 150-Å-long α helices or approximately one third of the entire length of the Smc2 and Smc4 coiled coils.
Figure 6.
Structure of the Yeast Smc2-4 Hinge with Long Coiled Coils
(A) Crystal structure of a heterodimer of ScSmc2H-CC110 and ScSmc4H-CC110 proteins in cartoon representation in green and yellow, respectively, in two perpendicular views. The dotted lines indicate disordered segments in Smc2 and Smc4.
(B) The hinge domain toroid and the two interfaces between Smc2 and Smc4, each composed of two short β strands. The Smc4 coiled coil is omitted for clarity.
(C) Details of the Smc4 coils/hinge interface. Residues at the interface are shown in stick representation in pink.
(D) Smc2 coils/hinge interface.
(E) Coils/coils interface. Residues at the Smc2-4 coils/coils interface mutated to cysteine are indicated in stick representation in pink.
(F) Cysteine crosslinking of Smc2 and Smc4 coiled coils in holocomplexes of yeast condensin. Distances between pairs of Smc2 and Smc4 cysteine residues are predicted based on the crystal structure. Yeast condensin was immunoprecipitated with antibodies against the Pk6 epitope tag on Smc4 and crosslinked with BMOE. Smc2-HaloTag protein was fluorescently labeled and analyzed by in-gel detection. Strains: YSG81, YSG99–102, YSG158, and YSG192–194.
See also Figure S5.
The hinge domains of ScSmc2H-CC110 and ScSmc4H-CC110 together form a toroid structure having a central hole similar to the counterparts in Smc-ScpAB and cohesin (Figure 6B) (Haering et al., 2002; Kurze et al., 2011). Strikingly, however, the segments connected to the coiled coils are very different between the two subunits in their secondary structures and arrangement (Figures 6C and 6D). Opposite orientations of the coiled coils with regard to their hinge domain make them run in parallel upon heterodimerization of Smc2 and Smc4 to produce a highly asymmetric, folded rod-like overall structure.
The coiled coil of ScSmc4H-CC110 is entirely visible and extends out by about 150 Å. In case of ScSmc2H-CC110, about half of its coiled coil is visible in the electron density map (Figure 6A). The end of the Smc4 coiled coil is involved in crystal packing, whereas that of the Smc2 coiled coil is not.
Alignment of Smc2-4 Coiled Coils in the Condensin Holocomplex
Does the crystal structure faithfully reflect a conformation adopted by condensin holocomplexes isolated from yeast? To test this, we probed the complex by site-specific crosslinking with BMOE. We identified pairs of residues at the coils/coils interface of Smc2 and Smc4 that are in close proximity in the crystal structure and mutated them to cysteines (Figure 6E). For some cysteine combinations, the endogenous Cys494 in Smc2 was replaced by serine to prevent interference with the assay. Cysteine mutations were combined with a HaloTag on Smc2 and a Pk6 tag on Smc4 and introduced into the respective endogenous genetic loci of a haploid yeast strain. The modified genes were expressed as the sole source of SMC2 and SMC4 and supported viability, indicating that condensin remained functional. We then used antibodies against the Pk epitopes on Smc4 to immunoprecipitate holocomplexes from asynchronous cultures. Immobilized complexes were treated with the crosslinker BMOE and conjugated to the HaloTag-tetramethylrhodamine (TMR) substrate. Subsequently, crosslinked species of Smc2-HaloTag were detected by in-gel fluorescence (Figure 6F). When wild-type complexes were treated with BMOE, crosslinking of Smc2 to Smc4 was hardly detectable. Similarly, only insubstantial crosslinking was observed with cysteine pairs when their thiol group distance exceeded the ∼8 Å linker length of BMOE. In contrast, introduction of more closely positioned pairs of cysteines promoted robust crosslinking of Smc2 and Smc4 (Figure 6F). These data strongly suggest that the conformations of the coiled coils observed by X-ray crystallography are adopted by native condensin holocomplexes.
Structural Basis and Conservation of the Parallel Orientations of the Smc2-4 Coiled Coils
Quite extensive hydrophobic interactions are found at the coils/hinge interface in condensin, a feature that is much less pronounced or lacking in prokaryotic condensin. For description, we designate the N-terminal α-helix of the Smc2 coiled coil as N-αH2CC and the C-terminal α-helix as C-αH2CC; likewise, we designate those of the Smc4 coiled coil as N-αH4CC and C-αH4CC. In the case of the Smc4 coiled coil, N-αH4CC is longer than C-αH4CC, and the last part of N-αH4CC (residues 665–682) does not interact with C-αH4CC but with the Smc4 hinge domain. This α-helical segment of Smc4—which, in analogy to the Pf structure, we call “rooting α-helix”—has at least three hydrophobic residues (Val672, Leu676, and Leu679) that interact with a hydrophobic groove on the Smc4 hinge domain (Figure 6C). These interactions are quite extensive and appear to be responsible for fixing the position and the orientation of the Smc4 coiled coil. Notably, the key hydrophobic residues involved in the Smc4 hinge/coiled coil interaction are conserved throughout eukaryotic condensins. Consistent with this notion, we found that mutation of hydrophobic residues L676 or L731—located at the Smc4 hinge/coil interface—to aspartate rendered the protein nonfunctional in yeast (Figures S5A–S5F). The Smc2 coiled coil associates with the hinge heterodimer at a hydrophobic interface involving Leu676, Leu677, and Ile680 at the beginning of C-αH2CC and Leu507, Phe531, Ile533, Leu552, and Phe553 on the Smc2 hinge domain (Figure 6D). All these residues are also well conserved. However, single mutations in the residues in C-αH2CC did not result in any obvious growth defects (Figures S5G–S5I). Nevertheless, the high level of sequence conservation suggests that the observed conformations of the Smc2 and Smc4 coiled coils relative to the hinge domains are likely a general feature of condensin in eukaryotes.
In addition to the coils/hinge interactions, also coils/coils interactions are found. The Smc2 coiled coil is in contact with the Smc4 coiled coil at one site via three exposed hydrophilic interactions and one ring-to-ring stacking interaction (Figure S5K). The hydrophilic interactions are solvent exposed, and the contacting helices are not tightly packed against each other, unlike those observed in the PfSmcH-CC60 structure (compare Figures 2C and S5K). Therefore, these coils/coils interactions appear to be a result, rather than a cause, of the juxtaposition of the coiled coils, although they might reinforce the parallel orientations of the coiled coils.
The ScSmc2H-CC110/ScSmc4H-CC110 complex binds dsDNA with high affinity (KD ∼50 nM; measured by fluorescence anisotropy). However, the crosslinking of two pairs of cysteines [Smc2(K487C)-Smc4(E876C) and Smc2(K495C)-Smc4(E866C)] at its coils/coils interface is unaffected by the presence of short DNA molecules (40 base pairs [bp]) (data not shown). A surface potential map of the Smc2-4 structure features a prominently positively charged area on top of the Smc2-4 hinge (Figure S6E), suggesting that initial DNA contact might occur at the top hinge surface in condensin. Possibly, additional elements such as ATP binding to the Smc2-4 heads, non-SMC subunits, loading factors, or nucleosomes might control any binding of DNA to the bottom hinge surface and/or opening of condensin SMC arms. To address the first two possibilities, we purified endogenous yeast condensin from exponentially growing or mitotically arrested populations of cells harboring Smc2(K495C) and Smc4(E866C) mutations by immunoprecipitation or affinity tag purification. Purified fractions of yeast condensin were then incubated with BMOE crosslinker in the presence or absence of ATP and short DNA. However, no significant differences in crosslinking were detected under the various conditions (data not shown). Thus, these attempts failed to provide evidence for the opening of SMC arms in yeast condensin in vitro.
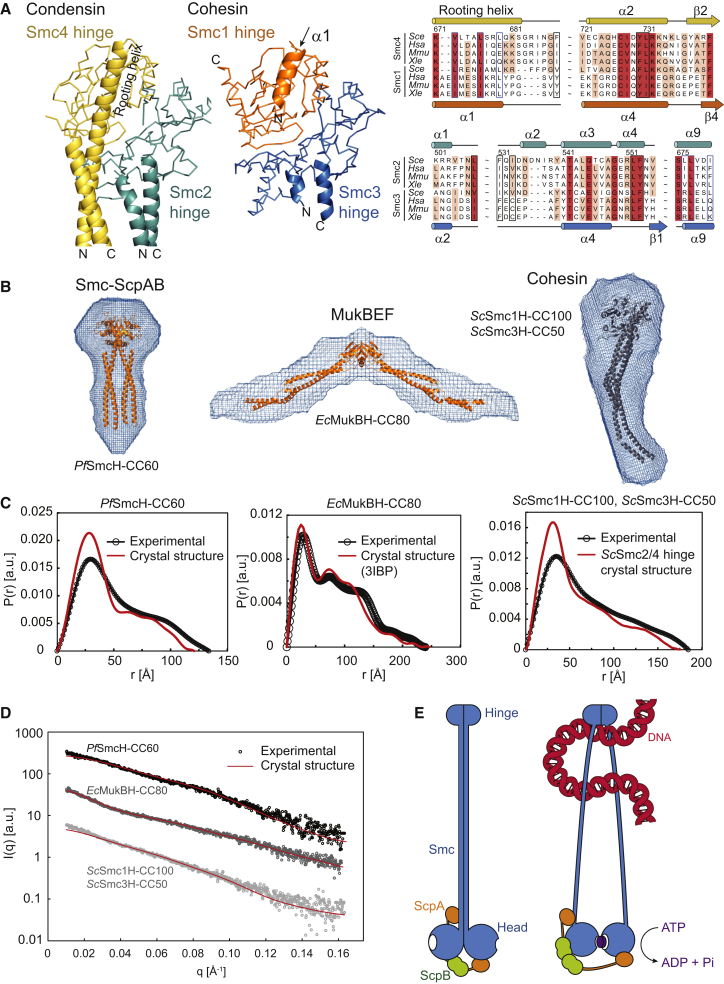
Open or Closed SMC Arms at the Cohesin Hinge?
Several EM studies depict cohesin and MukBEF in wide open conformations. In case of the latter, the open architecture is further supported by crystal structures of three isolated hinge fragments, all displaying diametrically opposed coiled coils (Ku et al., 2010; Li et al., 2010; Vos et al., 2013). Conceivably, cohesin and MukBEF might be fundamentally different in their structure from both condensin and Smc-ScpAB. Alternatively, all SMC complexes might require closed and open conformations during the course of action but might have distinct intrinsic preferences for these arrangements. To our surprise, we found that the coils/hinge junction in cohesin bears a strong resemblance to condensin. A crystal structure of the cohesin Smc1-3 hinge heterodimer was reported previously (Kurze et al., 2011). It contains a very short Smc3 coiled coil and a short α-helix of Smc1 that corresponds to the rooting α-helix of Smc4. Structural comparison with ScSmc2H-CC110/ScSmc4H-CC110 shows that the short Smc3 coiled coil is oriented similarly as the Smc2 coiled coil and that the short α-helix of Smc1 points in the same direction as the rooting α-helix of Smc4 (Figure 7A). Furthermore, most hydrophobic residues important for the coils/hinge interaction in the Smc2-4 heterodimer are conserved in Smc1 and Smc3 proteins. To address whether cohesin Smc1-3 complexes may indeed be able to adopt a rod shape, we generated a ScSmc1H-CC100/ScSmc3H-CC50 dimer and analyzed its structure using small-angle X-ray scattering (SAXS), a robust technique for characterization of macromolecular conformations in solution (Hura et al., 2009; Rambo and Tainer, 2010, 2013). As proof of principle, we initially performed SAXS analysis on PfSmcH-CC60 and an Ec MukB hinge fragment with long coiled coils, designated as EcMukBH-CC80. For both proteins, molecular envelopes fitted accurately to the rod-shaped or open V-shaped structures obtained by X-ray crystallography (Figures 7B–7D; Figure S6C; Table S3). These findings demonstrate that the coils/hinge junctions have a defined structure in solution as previously indicated by FRET analysis (Figures 1C and 1D). In addition, these experiments confirm the validity and suitability of SAXS for the study of SMC coils/hinge organization. Next, we analyzed the architecture of the ScSmc1H-CC100/ScSmc3H-CC50 dimer by SAXS. Remarkably, the SAXS envelopes derived for the cohesin fragment were clearly too short to accommodate an open V-shaped dimer as seen for EcMukBH-CC80 but were similar in size and shape to the condensin structure (Figures 7B–7D; Figure S6C), suggesting that Smc1-3 proteins fold into rods in solution. Two additional pieces of evidence support this surprising notion. First, a comprehensive lysine proximity map of purified human cohesin—based on the identification of crosslinked peptides by mass spectrometry by the Jan-Michael Peters laboratory—revealed 19 juxtaposed pairs of Smc1 and Smc3 coiled-coil residues (out of a total of 51 on cohesin) (Huis in ‘t Veld et al., 2014). Almost all these chemical crosslinks occurred between residues with similar position (plus or minus ten amino acids) along the length of the Smc1 and Smc3 coiled coils, being consistent with a well-defined, physical association between the two coiled coils (J.-M. Peters, personal communication). Second, we found that the affinity of the human cohesin hinge for DNA is reduced about 2-fold when 100 amino acids long coiled coils are attached to the Smc1 and Smc3 hinge domains, indicating that the DNA binding site at the cohesin hinge might at least be partially occluded (Figure S6D). Although further studies are clearly necessary, these initial observations provide an indication that all SMC complexes—with the possible exception of MukBEF—might at least transiently adopt a rod-like structure with juxtaposed coiled coils. Intriguingly, cohesin, condensin, and Smc hinge domains, but not the MukB hinge (Ku et al., 2010) (data not shown), display decent affinity for DNA, highlighting the possibility of a conserved functional connection between hinge architecture and regulated association with DNA.
Figure 7.
Organization of the Coils/Hinge Junction in Different SMC Complexes
(A) Side-by-side structural views of condensin hinge domains (ScSmc2H-CC110/ScSmc4H-CC110) (left) and cohesin hinge domains (PDB ID: 2WD5) (middle) reveals similarities in the attachment of coiled-coil helices onto the SMC hinge. Alignment of cohesin and condensin sequences at the coils/hinge junctions (right).
(B) SAXS envelopes for dimers of SMC hinge fragments with attached coiled coils of variable length: PfSmcH-CC60, EcMukBH-CC80, ScSmc1H-CC100/ScSmc3H-CC50.
(C) SAXS. Measured and calculated distance-distribution functions for different SMC hinge fragments (shown in B). a.u., arbitrary units.
(D) Experimental SAXS data for PfSmcH-CC60, EcMukBH-CC80, and ScSmc1H-CC100/ScSmc3H-CC50 and theoretical scattering curves calculated from the crystal structures of PfSmcH-CC60, EcMukBH-CC80, and ScSmc2H-CC110/ScSmc4H-CC110, respectively. For clarity, the curves are displayed with a y axis offset. Discrepancies (χ2) between the experimental and theoretical curves are PfSmcH-CC60 = 9.03, EcMukBH-CC80 = 4.22, and ScSmc1H-CC100/ScSmc3H-CC100 = 9.08. Relevant scattering derived parameters are shown in Table S3.
(E) Tentative model for a large structural transition in Smc-ScpAB upon binding to DNA.
Discussion
Little information is available on the arrangement of the two long coiled coils in SMC-kleisin complexes in vivo. Here, we start to fill this void by solving high-resolution X-ray structures of prokaryotic Smc-ScpAB and eukaryotic condensin and by performing subsequent biochemical and genetic characterization. We identify close juxtapositioning of SMC coiled coils at the hinge domain as a predominant architectural theme in SMC complexes and establish a functional link between hinge structure and DNA association.
Rod Formation in Prokaryotic Smc-ScpAB and Eukaryotic Condensin
Our work demonstrates how the long coiled coils in Bs Smc-ScpAB and yeast condensin are attached to their hinge domain dimers. The respective parts of these complexes share two striking structural features: the toroid-like hinge formed by homo- versus heterotypic interaction of two hinge domains and a four-helix bundle built by the intimate alignment of two SMC coiled coils. The most pronounced difference between the two structures, however, is the orientation of the coiled coils with respect to the hinge-domain toroid. Whereas the two coiled coils in the prokaryotic Smc hinge are virtually symmetric and perpendicular to the hinge toroid, those in the eukaryotic hinge are highly asymmetric and roughly parallel to the plane of the bottom surface of the hinge toroid (Figure S6A). The difference possibly reflects the homo- versus heterodimerization of these SMC proteins. The rooting α-helix of the yeast Smc4 hinge corresponds to α2 of the Pf Smc hinge. It can form a single straight α-helix with the preceding N-terminal Smc4 coiled-coil helix, as the Smc2 coiled coil keeps the required space free by adopting a different orientation. In contrast, α2 and the N-terminal coiled coil helix in the Pf Smc hinge cannot form a single continuous helix, because two such helices on the homodimer would inevitably clash with each other because of the molecular symmetry (Figure S6B). The ∼90° bending at the junction between the two helices avoids this scenario and, instead, allows the coiled coils to stretch out in an I shape from the hinge toroid and juxtapose onto each other. Apparently, as Smc2 diverged from prokaryotic Smc, it changed the orientation of its coiled coil drastically through a unique interaction between C-αH2CC and the hinge domain. It is important that, while eukaryotic SMC proteins broke the symmetry at the hinge, they retained the parallel and juxtaposed organization of the coiled coils, thus underscoring its functional importance.
The folded structure of prokaryotic Smc-ScpAB and eukaryotic condensins may be a way to limit the total number of entrapped DNA molecules within their circumference and/or to ensure that condensin would be occupied by selected DNA fibers only at defined moments in its catalytic cycle.
A Rod-to-Ring Transition in Smc-ScpAB—Regulating DNA Binding and Making a First Step toward Ring Opening?
Juxtapositioning of Smc coiled coils at the hinge is likely not a permanent feature. Rather, Smc-ScpAB complexes undergo marked transitions at the hinge between the folded rod and a more open ring-like configuration. The latter conformation is promoted by ATP binding to Smc heads and DNA binding to the hinge, whereas the former seems to be an intrinsically more favorable resting state. How could SMC proteins convert from one state to the other? EM images suggest that Smc arms are closely aligned along their entire length (Figure 1). During the ATP hydrolysis cycle, however, the head-proximal coiled coil might transiently become fixed in a conformation that is incompatible with coiled-coil juxtapositioning in this region (Haering et al., 2004). This, in turn, might promote the progression of coiled-coil disengagement up to the hinge, as seen in our BMOE crosslinking experiments (Figure 4E). We propose that the rod-like and ring-like configurations of Smc-ScpAB resemble, in structural and functional terms, the inward- and outward-facing conformations of the related ABC transmembrane transporters. In ABC transporters, the transitions between these conformations are controlled by the occupancy of the substrate binding pocket, thus ensuring unidirectional transport of substrates across membranes via a defined series of conformational states (Oldham et al., 2008). Our results give an indication as to how binding of the substrate molecule, DNA, might be restricted to the more open conformation of Smc by exposure of an otherwise occluded interface for DNA at the hinge. The hinge might thus serve as a sensor for DNA that links Smc arm architecture to the presence of DNA. Once DNA is bound to the hinge, it might stimulate hydrolysis of ATP by Smc (Hirano and Hirano, 2006) by simply promoting head engagement or through another long-range conformational change.
The location of the DNA binding site on the inner surface of the Smc hinge also has strong implications on how DNA might initially get in contact with Smc-ScpAB rings. Circular DNA molecules, such as bacterial chromosomes, need to form loops within the circumference of a SMC-kleisin ring so that DNA can fully engage with the binding site at the inner face of the hinge (Figure 7E). It will be exciting to determine the fate of the hinge-bound stretch of DNA—and the proposed DNA loop—upon completion of the loading reaction. Reformation of Smc rods will likely require the prior eviction of DNA from between Smc arms. We can think of two possible scenarios: (1) passage of DNA toward the head domains or (2) exit of DNA from the Smc-ScpA ring through a transiently opened hinge. The former could be related to processive extrusion of DNA loops from the Smc-ScpAB ring (Alipour and Marko, 2012; Nasmyth 2001), whereas the latter could create a topological interaction between circular DNA and Smc-ScpAB, as seen with cohesin and condensin, starting from a DNA loop. In this scenario, opening of the Smc-ScpAB ring might occur in two steps: by initial disengagement of Smc coiled coils and by subsequent hinge opening, possibly triggered by ATP hydrolysis.
Intriguingly, causative mutations in genes for cohesin subunits in Cornelia de Lange syndrome (CdLS) patients are mainly found in Smc1 and Smc3 coiled-coil sequences. In addition, several mutations that are located near the coils/hinge transition have been shown to increase DNA binding by Smc1-3 hinge heterodimers (Revenkova et al., 2009). Our data raise the exciting possibility that CdLS cohesin might be defective in SMC rod formation and, thus, display increased or misregulated association with DNA. Accordingly, cohesin in CdLS patients might have lost tight coordination between DNA binding at the hinge and the ATPase activity located at the SMC heads.
SMC proteins share their unusual architecture with Rad50, which uses a “zinc hook” rather than an SMC hinge domain for dimerization. Rad50 associates with Nbs1 and the nuclease Mre11 to form the MRN complex that is crucially important for efficient repair of DNA double-strand breaks and other DNA lesions (Williams et al., 2007). Simple binding of DNA to isolated Rad50 proteins has been suggested to convert ring-like dimers into straight rod-shaped structures (Moreno-Herrero et al., 2005). Thus, transitions between rod-like and ring-like states might be a conserved, albeit differently regulated, feature of all SMC-like proteins.
Experimental Procedures
Detailed methods can be found in the Supplemental Experimental Procedures.
Crystallization, X-Ray Data Collection, and Structure Determination
The PfSmcH-CC60 crystals grew from a precipitant solution containing 1 M sodium citrate, 0.1 M CHES (pH 9.5), and 8% glycerol; and the ScSmc2H-CC110/ScSmc4H-CC110 crystals grew from a solution of 16% polyethylene glycol 300, 0.1 M Na/K phosphate (pH 6.0), 8% glycerol, and 10 mM dithiothreitol (DTT). The structure of PfSmcH-CC60 was determined by molecular replacement using the structure of the coiled-coil-less Pf Smc hinge (Protein Data Bank [PDB] entry: 3NWC) as a search model. The ScSmc2H-CC110/ScSmc4H-CC110 structure was solved by the single isormorphous replacement with anomalous scattering method (Table 1).
SAXS Analysis
BL45XU of SPring-8 (Hyogo, Japan), 4C SAXS II beamline of Pohang Light Source II (Pohang, Korea) and a BioSAXS-1000 system (Rigaku) were used to collect SAXS intensity data. The data were processed and analyzed using the software applications embedded in the ATSAS package.
ALEX-FRET and TIRF-FRET Analyses
BsSmcH-CC100 was labeled with Cy3- and Cy5-maleimide (GE Healthcare). The LABVIEW software (National Instruments) was used to select fluorescent bursts induced by single molecules. The distance between Cy3 and Cy5 was estimated by the equation of R = R0(1/E − 1)1/6, with the R0 value of 6 nm for the Cy3-Cy5 pair.
Bs Strains and Crosslinking
All strains are derivatives of Bs 1A700 (Bacillus Genetic Stock Centre). They were constructed and grown as described by Bürmann et al. (2013). A list of strains is presented in Table S1. In vivo crosslinking was performed as detailed by Bürmann et al. (2013).
Yeast Strain Construction and Protein Crosslinking
Yeast strains are derivatives of Saccharomyces cerevisiae W303. Genetic modifications of SMC2 and SMC4 loci were performed by double crossover recombination. Genotypes are listed in Table S2. Yeast protein extracts were incubated with Dynabeads Protein G charged with monoclonal SV5-Pk1 antibody. Beads were washed, resuspended, and treated with BMOE (0.5 mM) and incubated for 10 min on ice before quenching with 2-mercaptoethanol (2-ME, 14 mM).
Anisotropy Titration Measurements
Fluorescence anisotropy titrations were performed at 25°C using a BioTek Neo plate reader in a buffer containing 50 mM Tris-HCl at pH 7.5, 50 mM NaCl, and 3 mM MgCl2 (plus 1 mM ATP) with 50 nM fluorescein-labeled dsDNA (40 bp).
Cysteine Crosslinking of Bs Smc and BsSmcH-CC100
BsSmcH-CC100 protein and double-stranded oligonucleotides (40 bp) were mixed at 4 μM and 20 μM, respectively, in 50 mM Tris, 50 mM NaCl, 2 mM MgCl2, 0.25 mM TCEP (pH 7.5)/23°C (final). After incubation at room temperature for 5 min, BMOE was added (0.5 mM final). Reactions were incubated for 1 min at room temperature and quenched with 2-mercaptoethanol (14 mM). BMOE crosslinking of wild-type and mutant Bs Smc(A715C) (at 1 μM) was performed for 5 min at room temperature in the same buffer but with 3 mM MgCl2, 10 μM DNA, with or without 1 mM ATP, and quenched with DTT in SDS loading buffer.
Author Contributions
Y.-M.S. and H.-C.S. performed protein purification, structure determination, and biochemical experiments; F.B., Sc and Bs strain constructions and cellular and biochemical experiments; K.S.J., T.O., and M.S., SAXS conception and experiments; C.K., H.L., and N.K.L., FRET conception and experiments; S.J.K. and H.M.K., electron microscopy; Y.-G.K., X-ray data collection; C.P.T., DNA-binding measurements; M.-S.K. and M.-L.D.-D., protein purification; and Y.-M.S., F.B., H.-C.S., S.G., and B.-H.O., conception of experiments and preparation of the manuscript.
Acknowledgments
The X-ray diffraction experiments used the Beamline 5C at the Pohang Accelerator Laboratory in Pohang, Korea, and the Beamline BL-17A at Photon Factory in Japan. We thank Dr. T. Hikima at SPring 8 for the help in SAXS data collection and Dr. M. Ikeguchi and Dr. Y. Kokabu for SAXS envelope models. We are grateful to J.-M. Peters for sharing results prior to publication and M. Dillingham for kindly providing expression plasmids and purification protocols for untagged Bs Smc protein. We thank Stefan Jentsch for sharing equipment and the Max Planck Institute of Biochemistry core facility for SEC-MALS analysis. This work was supported by the National Research Foundation of Korea (No. 2013-034955 to B.-H.O.), by the Intelligent Synthetic Biology Center of Global Frontier Project funded by the Ministry of Education, Science and Technology (No. 2011-0031955 to B.-H.O.), a European Research Council Starting Grant (DiseNtAngle #260853 to S.G.), and the Max Planck Society. The SAXS experiments were supported by the Platform for Drug Discovery, Informatics, and Structural Life Science from the Ministry of Education, Culture, Sports, Science and Technology, Japan.
Footnotes
This is an open access article under the CC BY-NC-ND license (http://creativecommons.org/licenses/by-nc-nd/3.0/).
Contributor Information
Byung-Ha Oh, Email: bhoh@kaist.ac.kr.
Stephan Gruber, Email: sgruber@biochem.mpg.de.
Accession Numbers
The coordinates of the structures together with the structure factors have been deposited in the PDB: PfSmcH-CC60 (4RSJ) and ScSmc2H-CC110/ScSmc4H-CC110 (4RSI).
Supplemental Information
References
- Alipour E., Marko J.F. Self-organization of domain structures by DNA-loop-extruding enzymes. Nucleic Acids Res. 2012;40:11202–11212. doi: 10.1093/nar/gks925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson D.E., Losada A., Erickson H.P., Hirano T. Condensin and cohesin display different arm conformations with characteristic hinge angles. J. Cell Biol. 2002;156:419–424. doi: 10.1083/jcb.200111002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arumugam P., Gruber S., Tanaka K., Haering C.H., Mechtler K., Nasmyth K. ATP hydrolysis is required for cohesin’s association with chromosomes. Current biology: CB. 2003;13:1941–1953. doi: 10.1016/j.cub.2003.10.036. [DOI] [PubMed] [Google Scholar]
- Bürmann F., Shin H.C., Basquin J., Soh Y.M., Giménez-Oya V., Kim Y.G., Oh B.H., Gruber S. An asymmetric SMC-kleisin bridge in prokaryotic condensin. Nat. Struct. Mol. Biol. 2013;20:371–379. doi: 10.1038/nsmb.2488. [DOI] [PubMed] [Google Scholar]
- Chiu A., Revenkova E., Jessberger R. DNA interaction and dimerization of eukaryotic SMC hinge domains. J. Biol. Chem. 2004;279:26233–26242. doi: 10.1074/jbc.M402439200. [DOI] [PubMed] [Google Scholar]
- Cuylen S., Metz J., Haering C.H. Condensin structures chromosomal DNA through topological links. Nat. Struct. Mol. Biol. 2011;18:894–901. doi: 10.1038/nsmb.2087. [DOI] [PubMed] [Google Scholar]
- Fuentes-Perez M.E., Gwynn E.J., Dillingham M.S., Moreno-Herrero F. Using DNA as a fiducial marker to study SMC complex interactions with the atomic force microscope. Biophys. J. 2012;102:839–848. doi: 10.1016/j.bpj.2012.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griese J.J., Hopfner K.P. Structure and DNA-binding activity of the Pyrococcus furiosus SMC protein hinge domain. Proteins. 2011;79:558–568. doi: 10.1002/prot.22903. [DOI] [PubMed] [Google Scholar]
- Griese J.J., Witte G., Hopfner K.P. Structure and DNA binding activity of the mouse condensin hinge domain highlight common and diverse features of SMC proteins. Nucleic Acids Res. 2010;38:3454–3465. doi: 10.1093/nar/gkq038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruber S., Arumugam P., Katou Y., Kuglitsch D., Helmhart W., Shirahige K., Nasmyth K. Evidence that loading of cohesin onto chromosomes involves opening of its SMC hinge. Cell. 2006;127:523–537. doi: 10.1016/j.cell.2006.08.048. [DOI] [PubMed] [Google Scholar]
- Gruber S., Veening J.W., Bach J., Blettinger M., Bramkamp M., Errington J. Interlinked sister chromosomes arise in the absence of condensin during fast replication in B. subtilis. Current biology: CB. 2014;24:293–298. doi: 10.1016/j.cub.2013.12.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haering C.H., Löwe J., Hochwagen A., Nasmyth K. Molecular architecture of SMC proteins and the yeast cohesin complex. Mol. Cell. 2002;9:773–788. doi: 10.1016/s1097-2765(02)00515-4. [DOI] [PubMed] [Google Scholar]
- Haering C.H., Schoffnegger D., Nishino T., Helmhart W., Nasmyth K., Löwe J. Structure and stability of cohesin’s Smc1-kleisin interaction. Mol. Cell. 2004;15:951–964. doi: 10.1016/j.molcel.2004.08.030. [DOI] [PubMed] [Google Scholar]
- Hirano T. Condensins: universal organizers of chromosomes with diverse functions. Genes Dev. 2012;26:1659–1678. doi: 10.1101/gad.194746.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano M., Hirano T. Opening closed arms: long-distance activation of SMC ATPase by hinge-DNA interactions. Mol. Cell. 2006;21:175–186. doi: 10.1016/j.molcel.2005.11.026. [DOI] [PubMed] [Google Scholar]
- Huis in ‘t Veld P.J., Herzog F., Ladurner R., Davidson I.F., Piric S., Kreidl E., Bhaskara V., Aebersold R., Peters J.M. Characterization of a DNA exit gate in the human cohesin ring. Science. 2014;346:968–972. doi: 10.1126/science.1256904. [DOI] [PubMed] [Google Scholar]
- Hura G.L., Menon A.L., Hammel M., Rambo R.P., Poole F.L., 2nd, Tsutakawa S.E., Jenney F.E., Jr., Classen S., Frankel K.A., Hopkins R.C. Robust, high-throughput solution structural analyses by small angle X-ray scattering (SAXS) Nat. Methods. 2009;6:606–612. doi: 10.1038/nmeth.1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura K., Hirano T. ATP-dependent positive supercoiling of DNA by 13S condensin: a biochemical implication for chromosome condensation. Cell. 1997;90:625–634. doi: 10.1016/s0092-8674(00)80524-3. [DOI] [PubMed] [Google Scholar]
- Ku B., Lim J.H., Shin H.C., Shin S.Y., Oh B.H. Crystal structure of the MukB hinge domain with coiled-coil stretches and its functional implications. Proteins. 2010;78:1483–1490. doi: 10.1002/prot.22664. [DOI] [PubMed] [Google Scholar]
- Kurze A., Michie K.A., Dixon S.E., Mishra A., Itoh T., Khalid S., Strmecki L., Shirahige K., Haering C.H., Löwe J., Nasmyth K. A positively charged channel within the Smc1/Smc3 hinge required for sister chromatid cohesion. EMBO J. 2011;30:364–378. doi: 10.1038/emboj.2010.315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Schoeffler A.J., Berger J.M., Oakley M.G. The crystal structure of the hinge domain of the Escherichia coli structural maintenance of chromosomes protein MukB. J. Mol. Biol. 2010;395:11–19. doi: 10.1016/j.jmb.2009.10.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mascarenhas J., Soppa J., Strunnikov A.V., Graumann P.L. Cell cycle-dependent localization of two novel prokaryotic chromosome segregation and condensation proteins in Bacillus subtilis that interact with SMC protein. EMBO J. 2002;21:3108–3118. doi: 10.1093/emboj/cdf314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matoba K., Yamazoe M., Mayanagi K., Morikawa K., Hiraga S. Comparison of MukB homodimer versus MukBEF complex molecular architectures by electron microscopy reveals a higher-order multimerization. Biochem. Biophys. Res. Commun. 2005;333:694–702. doi: 10.1016/j.bbrc.2005.05.163. [DOI] [PubMed] [Google Scholar]
- Melby T.E., Ciampaglio C.N., Briscoe G., Erickson H.P. The symmetrical structure of structural maintenance of chromosomes (SMC) and MukB proteins: long, antiparallel coiled coils, folded at a flexible hinge. J. Cell Biol. 1998;142:1595–1604. doi: 10.1083/jcb.142.6.1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno-Herrero F., de Jager M., Dekker N.H., Kanaar R., Wyman C., Dekker C. Mesoscale conformational changes in the DNA-repair complex Rad50/Mre11/Nbs1 upon binding DNA. Nature. 2005;437:440–443. doi: 10.1038/nature03927. [DOI] [PubMed] [Google Scholar]
- Nasmyth K. Disseminating the genome: joining, resolving, and separating sister chromatids during mitosis and meiosis. Annu. Rev. Genet. 2001;35:673–745. doi: 10.1146/annurev.genet.35.102401.091334. [DOI] [PubMed] [Google Scholar]
- Nasmyth K. Cohesin: a catenase with separate entry and exit gates? Nat. Cell Biol. 2011;13:1170–1177. doi: 10.1038/ncb2349. [DOI] [PubMed] [Google Scholar]
- Nolivos S., Sherratt D. The bacterial chromosome: architecture and action of bacterial SMC and SMC-like complexes. FEMS Microbiol. Rev. 2014;38:380–392. doi: 10.1111/1574-6976.12045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oldham M.L., Davidson A.L., Chen J. Structural insights into ABC transporter mechanism. Curr. Opin. Struct. Biol. 2008;18:726–733. doi: 10.1016/j.sbi.2008.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onn I., Aono N., Hirano M., Hirano T. Reconstitution and subunit geometry of human condensin complexes. EMBO J. 2007;26:1024–1034. doi: 10.1038/sj.emboj.7601562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rambo R.P., Tainer J.A. Bridging the solution divide: comprehensive structural analyses of dynamic RNA, DNA, and protein assemblies by small-angle X-ray scattering. Curr. Opin. Struct. Biol. 2010;20:128–137. doi: 10.1016/j.sbi.2009.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rambo R.P., Tainer J.A. Super-resolution in solution X-ray scattering and its applications to structural systems biology. Annual review of biophysics. 2013;42:415–441. doi: 10.1146/annurev-biophys-083012-130301. [DOI] [PubMed] [Google Scholar]
- Revenkova E., Focarelli M.L., Susani L., Paulis M., Bassi M.T., Mannini L., Frattini A., Delia D., Krantz I., Vezzoni P. Cornelia de Lange syndrome mutations in SMC1A or SMC3 affect binding to DNA. Hum. Mol. Genet. 2009;18:418–427. doi: 10.1093/hmg/ddn369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schleiffer A., Kaitna S., Maurer-Stroh S., Glotzer M., Nasmyth K., Eisenhaber F. Kleisins: a superfamily of bacterial and eukaryotic SMC protein partners. Mol. Cell. 2003;11:571–575. doi: 10.1016/s1097-2765(03)00108-4. [DOI] [PubMed] [Google Scholar]
- Soppa J., Kobayashi K., Noirot-Gros M.F., Oesterhelt D., Ehrlich S.D., Dervyn E., Ogasawara N., Moriya S. Discovery of two novel families of proteins that are proposed to interact with prokaryotic SMC proteins, and characterization of the Bacillus subtilis family members ScpA and ScpB. Mol. Microbiol. 2002;45:59–71. doi: 10.1046/j.1365-2958.2002.03012.x. [DOI] [PubMed] [Google Scholar]
- Thadani R., Uhlmann F., Heeger S. Condensin, chromatin crossbarring and chromosome condensation. Current biology: CB. 2012;22:R1012–R1021. doi: 10.1016/j.cub.2012.10.023. [DOI] [PubMed] [Google Scholar]
- Vos S.M., Stewart N.K., Oakley M.G., Berger J.M. Structural basis for the MukB-topoisomerase IV interaction and its functional implications in vivo. EMBO J. 2013;32:2950–2962. doi: 10.1038/emboj.2013.218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weitzer S., Lehane C., Uhlmann F. A model for ATP hydrolysis-dependent binding of cohesin to DNA. Curr. Biol. 2003;13:1930–1940. doi: 10.1016/j.cub.2003.10.030. [DOI] [PubMed] [Google Scholar]
- Williams R.S., Williams J.S., Tainer J.A. Mre11-Rad50-Nbs1 is a keystone complex connecting DNA repair machinery, double-strand break signaling, and the chromatin template. Biochemistry and cell biology. 2007;85:509–520. doi: 10.1139/O07-069. [DOI] [PubMed] [Google Scholar]
- Woo J.S., Lim J.H., Shin H.C., Suh M.K., Ku B., Lee K.H., Joo K., Robinson H., Lee J., Park S.Y. Structural studies of a bacterial condensin complex reveal ATP-dependent disruption of intersubunit interactions. Cell. 2009;136:85–96. doi: 10.1016/j.cell.2008.10.050. [DOI] [PubMed] [Google Scholar]
- Yamazoe M., Onogi T., Sunako Y., Niki H., Yamanaka K., Ichimura T., Hiraga S. Complex formation of MukB, MukE and MukF proteins involved in chromosome partitioning in Escherichia coli. EMBO J. 1999;18:5873–5884. doi: 10.1093/emboj/18.21.5873. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.