Abstract
Aims
To assess strains of lactobacilli for their capacity to produce functional fatty acid-conjugated linoleic acid. To assess the linoleate isomerase for CLA production in the most efficient CLA producer.
Methods and Results
In this study, strains of food-derived lactobacilli were cultured in media with linoleic acid and CLA production was assessed. Most of the selected strains produced CLA at different levels, with Lactobacillus plantarum ZS2058 being the most efficient CLA producer converting over 50% of linoleic acid to c9, t11-CLA and t9, t11-CLA. Some intermediates 10-hydroxy-cis-12-octadecenoic acid, 10-oxo-cis-12-octadecenoic acid and 10-oxo-trans-11-octadecenoic acid were determined via GC-MS. The genes coding the multicomponent linoleate isomerase containing myosin-cross-reactive antigen, short-chain dehydrogenase/oxidoreductase and acetoacetate decarboxylase for CLA production in Lact. plantarum ZS2058 were cloned and expressed in Escherichia coli. With the mixture of recombinant E. coli, c9, t11-CLA and three kinds of intermediates were produced from linoleic acid, which were in line with those in the lactobacilli.
Conclusions
The ability for CLA production by lactobacilli exhibited variation. Lactobacillus plantarum and Lact. bulgaricus were the most efficient producers in the selected strains. Lact. plantarum ZS2058 converted linoleic acid to CLAs with 10-hydroxy-cis-12-octadecenoic acid, 10-oxo-cis-12-octadecenoic acid and 10-oxo-trans-11-octadecenoic acid as intermediates. The multiple-step reactions for CLA production catalysed by multicomponent linoleate isomerase in Lact. plantarum ZS2058 were confirmed successfully.
Significance and Impact of the study
Multicomponent linoleate isomerase provides important results for the illustration of the mechanism for CLA production in lactic acid bacteria. Food-derived lactobacilli with CLA production ability offers novel opportunities for functional foods development.
Keywords: conjugated linoleic acid, lactic acid bacteria, Lactobacillus plantarum, linoleate isomerase
Introduction
Conjugated linoleic acid (CLA) refers to a group of positional and geometric isomers of linoleic acid (LA, 18:2 n-6) with conjugated double bonds. In the past several decades, CLA has attracted great interest because of its heath-associated activities including anticarcinogenic (Ip et al. 1991, 1994; Field and Schley 2004; Shen et al. 2013), anti-atherogenic (Lee et al. 1994; Valeille et al. 2004; McClelland et al. 2010), antidiabetic (Moloney et al. 2007; Castro-Webb et al. 2012; Rungapamestry et al. 2012), anti-inflammatory (Sugano et al. 1998; Coakley et al. 2006) and anti-obesity (Noone et al. 2002; Park et al. 2004; Sluijs et al. 2010).
CLA isomers occur naturally in ruminant meat and a variety of dairy food derived from ruminants as a minor component of the lipid fraction. CLAs are formed as intermediates during linoleic acid biohydrogenation to stearic acid by the anaerobic rumen bacteria. The complete biohydrogenation of linoleic acid by the anaerobic rumen bacterium (such as Butyrivibrio fibrisolvens) is a multi-step process (Kepler and Tove 1967). The first reaction occurs rapidly by linoleate isomerase, the conversion of linoleic acid to c9, t11-CLA, followed by the slower conversion to trans-11 vaccenic acid (Kepler et al. 1966). Vaccenic acid is known to be reduced to stearic acid by microbial activity other than that of B. fibrisolvens in the rumen. Vaccenic acid can also be converted to c9, t11-CLA by the delta-9 desaturase in the mammary tissue itself, providing another mechanism for its formation in milk (Griinari and Bauman 1999).
Additionally, it has also demonstrated that certain strains used in food fermentation possess the capacity to generate c9, t11-CLA. Two strains of Propionibacterium freudenreichii subsp. freudenreichii and one strain of P. freudenreichii subsp. shermanii can convert free LA to c9, t11-CLA (Jiang et al. 1998). Several bifidobacteria, isolated from the human gut and other sources, can produce c9, t11-CLA with LA in the medium (Coakley et al. 2003; Rosberg-Cody et al. 2004; Gorissen et al. 2010). Furthermore, several studies have reported the production of CLA isomers from LA by different lactic acid bacteria grown in MRS, skim milk and cheddar cheese (Lin et al. 1999; Alonso et al. 2003; Mohan et al. 2013; Ye et al. 2013).
To date, only three linoleate isomerases derived from Lactobacillus reuteri PYR8 (Rosson et al. 2004), Clostridium sporogenes (Peng et al. 2007) and Propionibacterium acnes (Liavonchanka et al. 2006) have been characterized. The linoleate isomerase from Lact. reuteri PYR8 was a myosin-cross-reactive antigen (MCRA), which was originally found in Streptococcus pyogenes and predicted to have a polyunsaturated fatty acid isomerase function (Kil et al. 1994). Several putative linoleate isomerases, which were highly homologous to that from Lact. reuteri PYR8, were expressed in E. coli; unfortunately, none can produce CLA (Volkov et al. 2010; Rosberg-Cody et al. 2011), and instead of CLA, 10-hydroxy-cis-12-octadecenoic acid (10-HOE) was produced. A multiple-fraction linoleate isomerase was purified from Lact. plantarum AKU 1009a, which produced c9, t11-CLA, t10, c12-CLA and t9, t11-CLA, but no detailed results for the enzyme were reported (Kishino et al. 2011a). In a later study, the genes encoding the multicomponent enzyme machinery catalysing double bond migration in Lact. plantrum AKU 1009a were illustrated (Kishino et al. 2011b), with the transformed E. coli as the catalysts, t9, t11-CLA was produced at a significant level with c9, t11-CLA and 10-HOE. A multiple-step reaction for CLA production in Lactobacillus was hypothesized but without evidences for the putative intermediates. Recently, the mass spectra and NMR data for 10-hydroxyl-cis-12-octadecenoic acid, 10-oxo-cis-12-octadecenoic acid and 10-oxo-trans-11-octadecenoic acid, intermediates in CLA bioconversion catalysed by the multicomponent linoleate isomerase, were further demonstrated (Kishino et al. 2013).
In our previous study, MCRAs from several lactic acid bacteria were confirmed as fatty acid hydratase (Yang et al. 2013). In the present study, a selection of strains including different food-derived lactobacilli was assessed for CLA production from free linoleic acid. The genetic determinants for CLA production in the most efficient producer, Lact. plantarum ZS2058, were cloned in isolation in E. coli, and the ability of the resultant strains was then assessed for CLA production successfully.
Materials and methods
Strains, media and growth conditions
The lactic acid bacteria strains used in this study are detailed in Table 1. Strains of lactobacilli were cultured in de Man, Rogosa and Sharpe (MRS) medium consisting of 1·0% tryptone, 1·0% meat extract, 0·5% yeast extract, 2·0% glucose, 0·1% Tween 80, 0·2% K2HPO4, 0·5% sodium acetate, 0·2% diammonium citrate, 0·02% MgSO4·7H2O and 0·005% MnSO4·H2O (pH 6·5) under anaerobic conditions at 37°C for 48 h. When solid media were required, 2·0% agar was added to the MRS medium. For growth of bifidobacteria, 0·05% (w/v) L-cysteine hydrochloride was added to the MRS medium and cultured at 37°C for 48 h. Escherichia coli BL21 (DE3) carrying the plasmid pET28a was routinely cultured aerobically in Luria–Bertani (LB) medium (10 g l−1 tryptone, 5 g l−1 yeast extract, 10 g l−1 NaCl) at 37°C in the presence of kanamycin (50 μg ml−1) as a selective marker.
Table 1.
List of selected strains in the present study
| Species | Strain | Biological Origin | CLA production from 0·55 mg ml−1 linoleic acid
|
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| c9,t11 | S.D. | % converted | t10,c12 | S.D. | % converted | t9,t11 | S.D. | % converted | |||
| Lactobacillus acidophilus | CCFM1006 | Unknown | 0·0039 | 0·0006 | 0·71 | 0·0035 | 0·0004 | 0·64 | 0·0082 | 0·0005 | 1·49 |
| CCFM1137 | Unknown | 0·0031 | 0·0001 | 0·56 | 0·0028 | 0·0003 | 0·51 | 0·0105 | 0·0008 | 1·91 | |
| NCFM | Human GIT | 0·0033 | 0·0003 | 0·60 | 0·0033 | 0·0002 | 0·60 | 0·0106 | 0·003 | 1·93 | |
| Lact. brevis | CCFM2135 | Unknown | 0·0035 | 0·0003 | 0·64 | 0·0026 | 0·0008 | 0·47 | 0·0127 | 0·001 | 2·31 |
| ATCC14869 | Unknown | 0·0046 | 0·0001 | 0·84 | 0·0038 | 0·0001 | 0·69 | 0·0081 | 0·0008 | 1·47 | |
| Lact. bulgaricus | CCFM3004 | Yoghurt | 0·0373 | 0·0009 | 6·78 | 0·0254 | 0·0001 | 4·62 | 0·0546 | 0·009 | 9·93 |
| CCFM3029 | Unknown | 0·0415 | 0·0001 | 7·55 | 0·0349 | 0·0004 | 6·35 | 0·0494 | 0·0007 | 8·98 | |
| Lact. casei | BD-II | Koumiss | 0·0254 | 0·0009 | 4·62 | 0·0233 | 0·0008 | 4·24 | 0·0309 | 0·014 | 5·62 |
| CCFM7030 | Raw milk | 0·0229 | 0·0015 | 4·16 | 0·0211 | 0·0014 | 3·84 | 0·0336 | 0·003 | 6·11 | |
| str. Zhang | Koumiss | 0·0036 | 0·0005 | 0·65 | 0·0036 | 0·0001 | 0·65 | 0·0104 | 0·0003 | 1·89 | |
| CCFM8236 | Sauerkraut | 0·0161 | 0·0002 | 2·93 | 0·016 | 0·0003 | 2·91 | 0·0355 | 0·007 | 6·45 | |
| Lact. crispatus | CCFM5136 | Unknown | 0·0165 | 0·0003 | 3·00 | 0·0151 | 0·0006 | 2·75 | 0·0307 | 0·0002 | 5·58 |
| Lact. gasseri | CCFM5115 | Yoghurt | 0·015 | 0·0007 | 2·73 | 0·0135 | 0·0012 | 2·45 | 0·0331 | 0·0017 | 6·02 |
| Lact. helveticus | CCFM8310 | Yoghurt | 0·0193 | 0·0017 | 3·51 | 0·0194 | 0·0003 | 3·53 | 0·0392 | 0·0015 | 7·13 |
| Lact. plantarum | STIII | Sauerkraut | 0·0023 | 0·0001 | 0·42 | 0·0026 | 0·0001 | 0·47 | 0·0069 | 0·0001 | 1·25 |
| ZS2058 | Sauerkraut | 0·2078 | 0·0033 | 37·78 | 0·0139 | 0·0001 | 2·54 | 0·0911 | 0·0026 | 16·57 | |
| CCFM9047 | Sauerkraut | 0·0545 | 0·0006 | 9·91 | 0·0290 | 0·0002 | 5·27 | 0·0974 | 0·0005 | 17·71 | |
| CCFM9187 | Sauerkraut | 0·0188 | 0·0004 | 3·42 | 0·0187 | 0·0005 | 3·40 | 0·0412 | 0·0005 | 7·49 | |
| CCFM9225 | Tibet Kefir | 0·02 | 0·0011 | 3·64 | 0·02 | 0·0009 | 3·64 | 0·0383 | 0·0011 | 6·96 | |
| CCFM9232 | Sauerkraut | 0·0326 | 0·0018 | 5·93 | 0·0239 | 0·0016 | 4·35 | 0·0672 | 0·0033 | 12·22 | |
| CCFM9235 | Sourdough | 0·0264 | 0·0012 | 4·80 | 0·0241 | 0·0003 | 4·38 | 0·0464 | 0·0037 | 8·44 | |
| CCFM9240 | Unknown | 0·0307 | 0·0014 | 5·58 | 0·0256 | 0·0015 | 4·65 | 0·049 | 0·0015 | 8·91 | |
| CCFM9307 | Milk Granule | 0·0185 | 0·0009 | 3·36 | 0·0177 | 0·0004 | 3·22 | 0·0343 | 0·008 | 6·24 | |
| Lact. reuteri | CCFM4014 | Tibet cheese | 0·0317 | 0·0003 | 5·76 | 0·024 | 0·0009 | 4·36 | 0·0361 | 0·0004 | 6·56 |
| ATCC55739 | Rat | 0·3282 | 0·0048 | 59·68 | 0·0095 | 0·0028 | 1·74 | 0·0146 | 0·004 | 2·66 | |
| Lact. rhamnosus | GG | Human GIT | 0·0033 | 0·0006 | 0·60 | 0·0032 | 0·0004 | 0·58 | 0·0072 | 0·0009 | 1·31 |
| Bif. animalis | BB12 | Unknown | 0·1106 | 0·0005 | 20·10 | 0·0047 | 0·001 | 0·85 | 0·0587 | 0·0002 | 10·68 |
S.D., trial was carried out in triplicate; CCFM, Collection Centre of Food Microbiology, Jiangnan University, Wuxi, China; ATCC, American Type Culture Collection, Manassas, VA, USA.
Fat extraction from bacterial supernatant fluids and pellets
Prior to examination of the strains for CLA production, each was subcultured twice in MRS broth. All strains were then cultured (1%) in broth spiked with 0·55 mg ml−1 free linoleic acid (99% purity; Sigma, St. Louis, MO). The linoleic acid was added as a 30 mg ml−1 stock solution containing 2% (v/v) Tween 80 and was previously filter sterilized through a 0·45-μm Minisart filter (Sigma) and stored in the dark at −20°C. The strains were incubated anaerobically at 37°C in a modular atmosphere-controlled system (Whitley DG250 anaerobic workstation; Don Whitley Scientific, West Yorkshire, UK) that was continuously sparged with a mixture of 80% nitrogen, 10% carbon dioxide and 10% hydrogen.
Lipid extraction and methylation
After 48-h incubation, the cultures were centrifuged at 5000 g for 10 min at room temperature. The fat was extracted from the culture supernatant fluid as follows. The internal standard, C17:0 heptadecanoic acid (99% pure; Sigma), was added to 4 ml of the supernatant fluid to give a final concentration of 217 μg internal standard per sample. Four millilitre of isopropanol was added to the supernatant fluid, and the samples were vortexed for 30 s. Four millilitre of n-hexane was added to this mixture, vortexed and centrifuged at 5000 g for 5 min. The resultant hexane layer (containing lipids) was dried off under a stream of nitrogen.
The fat was extracted from the bacterial pellet as follows. The pellet from 10 ml of bacterial culture was washed in 2 ml saline solution (0·137 mol l−1 NaCl, 7·0 mmol l−1 K2HPO4 and 2·5 mmol l−1 KH2PO4). The cells were vortexed and centrifuged at 5000 g for 10 min and the washing step repeated twice. The cells were suspended in 2 ml saline solution, and then the samples were extracted completely as described above for the bacterial supernatant fluid.
Preparation of fatty acid methyl esters and gas chromatography analysis
Fatty acids were converted to corresponding methyl esters with (trimethylsilyl)-diazomethane (Sigma) as described previously (Yang et al. 2013). The FAMEs were extracted in n-hexane and separated on a Rtx-2560 column (100 m × 0·25 mm × 0·25 μm) using a gas chromatograph (GC2010 plus, Shimadzu, Kyoto, Japan) fitted with a QP2010 ultra mass spectrometer. Injections of 1 μl were administered automatically at a split ratio of 10 : 1. Helium was used as the carrier gas. The column temperature was set initially at 150°C, then increased to 200°C in increments of 5°C min−1 and maintained for 10 min, and finally increased to 240°C in a rate of 4°C min−1. The 240°C was maintained for 10 min. The injector and detector were operated at 240°C. Electron energy of 70 eV and ion source temperature of 220°C were used. The CLA isomers were identified by retention time with reference to CLA standard mix (Sigma). The percentage conversion to CLA isomers was calculated by dividing the amount of CLA present in the broth after inoculation/ incubation with the amount of linoleic acid present in the spiked broth before incubation.
Cloning and expression of recombinant protein in Escherichia coli
Genomic DNAs were isolated rapidly from Lact. plantarum ZS2058 as described by Hoffman and Winston (1987). According to previous result (Kishino et al. 2011b), myosin-cross-reactive antigen (mcra, GenBank: JF747255.1), short-chain dehydrogenase/oxidoreductase (dh, GenBank: KJ019513) and acetoacetate decarboxylase (dc, GenBank: KJ019514) were amplified from Lact. plantarum ZS2058 genomic DNA with specific primers according to their homologous gene in Lact. plantarum WCFS1 (GenBank: NC_004567). PCR was performed with KOD-plus DNA polymerase (Toyobo, Osaka, Japan) according to the manufacturer’s protocol. PCR conditions were as follows: 30 cycles of 45 s of denaturation (94°C), 30 s of annealing (55°C) and 2 min of elongation (68°C). For expression in E. coli, the three genes were amplified with the primers listed in Table 2 and cloned into the pET28a expression vector (Novagen, Darmstadt, Germany), yielding plasmids pET28a-mcra, pET28a-dh and pET28a-dc (N-His-tagged version).
Table 2.
Primers used in this study
| Name | Sequence (5′–3′)* | Restriction site |
|---|---|---|
| mcra-F | GTTCTCGAGAAAAGA-ATGGGGGCGTTATTTATG | Xho I |
| mcra-R | CGGCGGCCGCTTATCA-ATCAAACATCTTCTTAGTTGC | Not I |
| dh-F | CCGGAATTCATGAAAGATTTTAAAGATAAAGTTATGTTTATCACG | EcoR I |
| dh-R | CCCAAGCTTTTACATGATACCGTCCATGATGTGCA | Hind III |
| dc-F | CCGGAATTCATGGCAAGTTTTATTGCAAGTGATCA | EcoR I |
| dc-R | CCCAAGCTTCTAAATAATGTAAGTCGCTGCCTTGG | Hind III |
Restriction sites are underlined.
For protein production, E. coli BL21 (DE3) Star strain (Invitrogen, Carlsbad, CA) harbouring pET28a-mcra, pET28a-dh and pET28a-dc plasmids, separately, was used. Bacteria were cultivated in LB medium supplied with kanamycin at 37°C until OD600 reached 0·6. At that point, IPTG was added to a final concentration of 0·05 mmol l−1, and the culture was placed at 18°C for 10-h induction of protein expression. pET28a vector-inserted E. coli was used as negative control. Following induction, cells were harvested by centrifugation, washed with 20 mmol l−1 potassium phosphate buffer (KPB) (pH 6·5) and sonicated (Uilbra-Cell VCX500; Sonics & Materials Inc., Newtown, CT). The cell debris was removed by centrifugation, and the supernatant containing soluble proteins was collected. Ten micrograms of protein was subjected to SDS-PAGE (12% SDS-PAGE) followed by protein transfer to a PVDF membrane (Amersham Pharmacia Biotech, Amersham, UK); the immunoblots were developed with the use of anti-His antibody at a dilution of 1 : 2000 (Tiangen, Beijing, China). Horseradish peroxidase (HRP)-conjugated anti-mouse IgG (Tiangen) secondary antibody diluted at 1 : 500 was used for the detection of specific antibody binding. The bands were visualized with enhanced chemiluminescence reagents (Kangwei, Beijing, China) according to the manufacturer instructions.
Activity assay and fatty acid analysis
For activity assay, the reaction was performed in a screw tube that contained 1 ml of reaction mixture (20 mmol l−1 KPB, pH 6·5) with 5 mmol l−1 NADH and 0·1 mmol l−1 FAD as cofactors, 0·5 mg ml−1 linoleic acid complexed with BSA as substrate and the recombinant E. coli cells under microaerobic conditions in a sealed chamber filled with N2 and shaken (180 strokes min−1) at 37°C for 6 h. All the transformed E. coli were suspended in KPB at a final concentration of 0·5 mg ml−1 wet cells. Fifty microlitre of each transformed E. coli suspension was used as catalyst in various combinations. The following reactions were carried out: (i) E. coli BL21(DE3)/pET28a-mcra recombinant suspension plus free linoleic acid, FAD and NADH; (ii) E. coli BL21(DE3)/pET28a-mcra recombinant suspension with FAD and NADH without linoleic acid; (iii) E. coli BL21(DE3)/pET28a-mcra recombinant suspension and E. coli BL21(DE3)/pET28a-dh recombinant suspension with substrate, FAD and NADH; (iv) E. coli BL21(DE3)/pET28a-mcra recombinant suspension and E. coli BL21(DE3)/pET28a-dh recombinant suspension without substrate but with cofactors; (v) mixture of E. coli BL21(DE3)/pET28a-mcra suspension, E. coli BL21(DE3)/pET28a-dh suspension and E. coli BL21(DE3)/pET28a-dc suspension plus linoleic acid, FAD and NADH; and (vi) all the three transformed E. coli suspension mixed with FAD and NADH without substrate. Following reaction, fatty acids were extracted, methylated and analysed as above.
Results
Screening of lactobacilli for conjugated linoleic acid production
In this study, a number of strains of lactobacilli (see Table 1) were assessed for the ability to generate CLA from free linoleic acid. The origin of these strains varied in vegetable fermentation and dairy fermentations to human intestinal isolates. Bifidobacteria previously reported to synthesize c9, t11-CLA from free linoleic acid, Bifidobacterium animalis subsp. lactis BB-12 (Coakley et al. 2003) and Lact. reuteri ATCC55739 (Rosson et al. 2004) were used as positive controls in this study.
Twenty-five selected strains were investigated for their ability to convert free linoleic acid (0·55 mg ml−1) to CLA. The results demonstrated that c9, t11-CLA, t10, c12-CLA and t9, t11-CLA could be generated, but to varying levels ranging from 3 to 56% for CLA production. Most CLA isomers were found in the supernatant from the cultures rather than in the cell pellets. All nine strains of Lact. plantarum tested, except Lact. plantarum STIII, produced CLA. Lact. plantarum ZS2058 was the most efficient of the lactobacilli strains tested for the conversion of linoleic acid to CLA. There was a 37·78% conversion of linoleic acid into c9, t11-CLA and 16·57% conversion to t9, t11-CLA. Lact. bulgaricus CCFM3004, Lact. bulgaricus CCFM3029, Lact. crispatus CCFM5136, Lact. gasseri CCFM5115 and Lact. helveticus CCFM8310 converted 10–20% linoleic acid into three CLA isomers. Of the remaining strains in Table 1, some did not convert linoleic acid to CLA at any significant level, including Lact. acidophilus, Lact. brevis, Lact. rhamnosus, one strain of Lact. casei and one strain of Lact. plantarum. Although the concentration of 10-HOE differed (data not shown), it was produced in all the assessed strains.
Conjugated linoleic acid production by Lactobacillus plantarum ZS2058 in the presence of linoleic acid
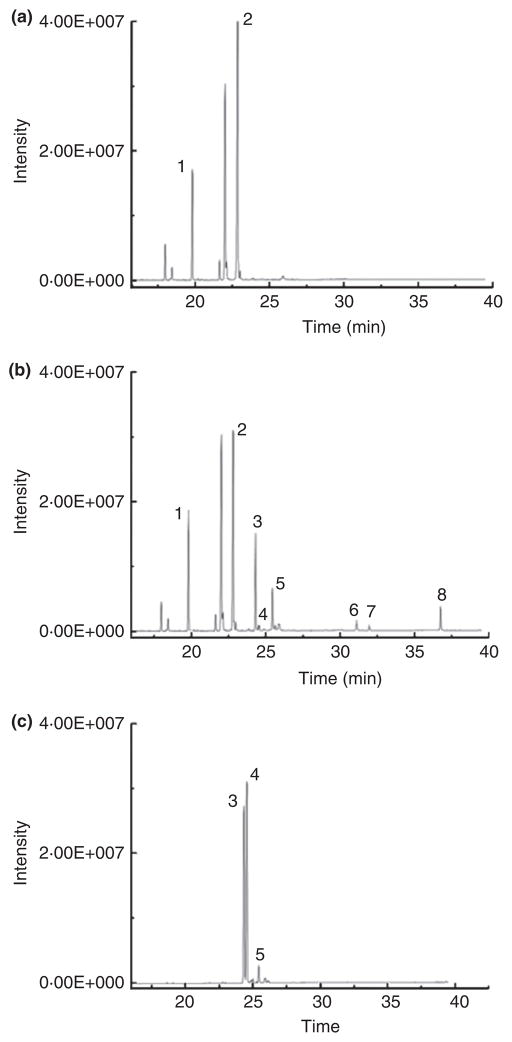
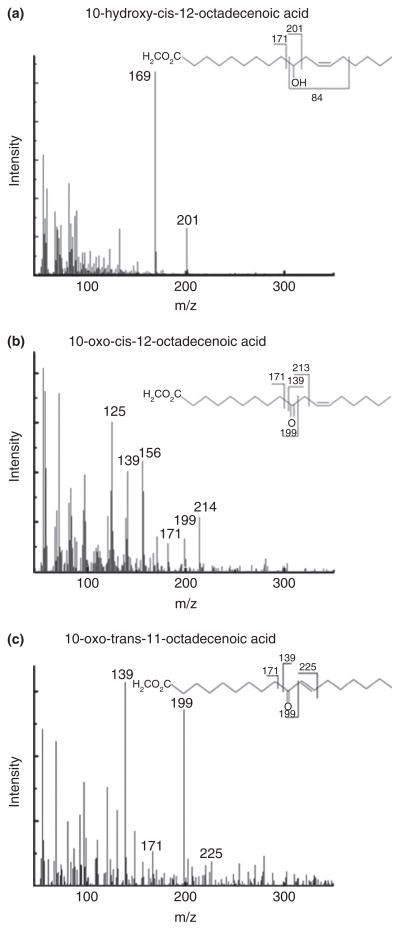
As Lact. plantarum ZS2058 was the most efficient strain for linoleic acid conversion in this study, this strain was then studied in a lot more detail. Following culturing with linoleic acid, both c9, t11-CLA and t9, t11-CLA isomers were mainly found in cell supernatant, with very low concentration being recovered in the cell pellet (data not shown). Interestingly, t10, c12-CLA was not significantly produced in this strain. Indeed, c9, t11-CLA was the major isomer and represented over 65% in total CLAs. During CLA production, three kinds of intermediates were produced (Fig. 1), based on previous result (Yang et al. 2013), and the mass spectra in this study, the intermediate with retention time at 36·3 min, with significant mass fractions of 169 and 201 m/z were identified as 10-HOE (Figs 1 and 2a). As all the intermediates in the reaction were out of commercial standards, according to previous hypothesis (Kishino et al. 2011b), the fragmentation patterns of each intermediate were analysed. As structures of the two intermediates, 10-oxo-cis-12-octadecenoic acid and 10-oxo-trans-11-octadecenoic acid, were of high similarity, amount of mass fractions were identical, such as 171, 139 and 199 m/z (Fig. 2b, c). The significant difference between the two intermediates was the position of carbon double bond; therefore, 213 m/z was more notable in 10-oxo-cis-12-octadecenoic acid instead of 225 m/z in 10-oxo-trans-11-octadecenoic acid (Fig. 2b, c).
Figure 1.
GC-MS total ion chromatograms of the products from fatty acid profile of Lactobacillus plantarum ZS2058 grown in MRS plus 0·55 mg ml−1 linoleic acid. (a) Lact. plantarum ZS2058 at 0 h; (b) Lact. plantarum ZS2058 at 48 h; (c) CLA standard. (1) C17:0 (Internal standard); (2) linoleic acid; (3) c9, t11-CLA; (4) t10, c12-CLA; (5) t9, t11-CLA; (6) 10-oxo-cis-12-octadecenoic acid; (7)10-oxo-trans-11-octadecenoic acid; and (8) 10-hydroxyl-cis-12-octadecenoic acid.
Figure 2.
Mass fractions of the intermediate products. (a) Mass spectra of 10-HOE and its fragmentation pattern. (b) Mass spectra of 10-oxo-cis-12-octadecenoic acid and its fragmentation pattern. (c) Mass spectra of 10-oxo-trans-11-octadecenoic acid and its fragmentation pattern.
Expression and activity of the recombinant proteins
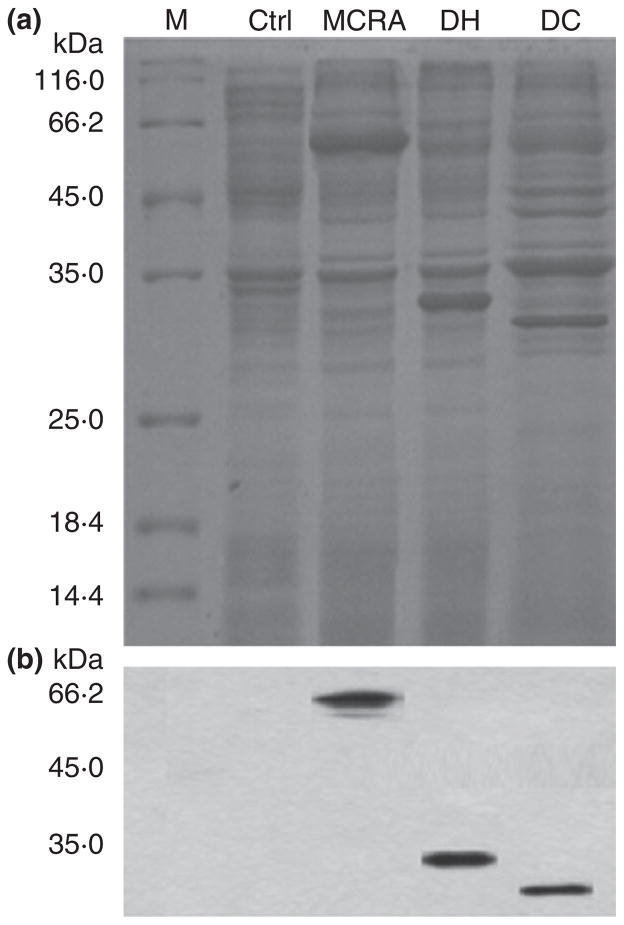
Based on the previous hypothesis (Kishino et al. 2011b), the three genes encoding myosin-cross-reactive antigen (mcra), short-chain dehydrogenase/oxidoreductase (dh) and acetoacetate decarboxylase (dc) were cloned from Lact. plantarum ZS2058. These three genes were overexpressed separately in E. coli BL21 (DE3) (Fig. 3a). Major bands with apparent molecular weights of 64·76 kDa (MCRA), 32·10 kDa (DH) and 30·71 kDa (DC) were visualized by SDS-PAGE and these corresponded to the expected molecular weights of the three proteins, which were absent in pET28a vector inserts. Immunoblotting with anti-His-tag antibodies confirmed that each single band contained a 6× His tag (Fig. 3b).
Figure 3.
SDS-PAGE and Western blot analyses of the recombinant proteins. (a) SDS-PAGE of control, MCRA, DH and DC; (b) Western blot of control, MCRA, DH and DC. Lane: M: Marker; Ctrl: E. coli/pET28a; MCRA; DH and DC.
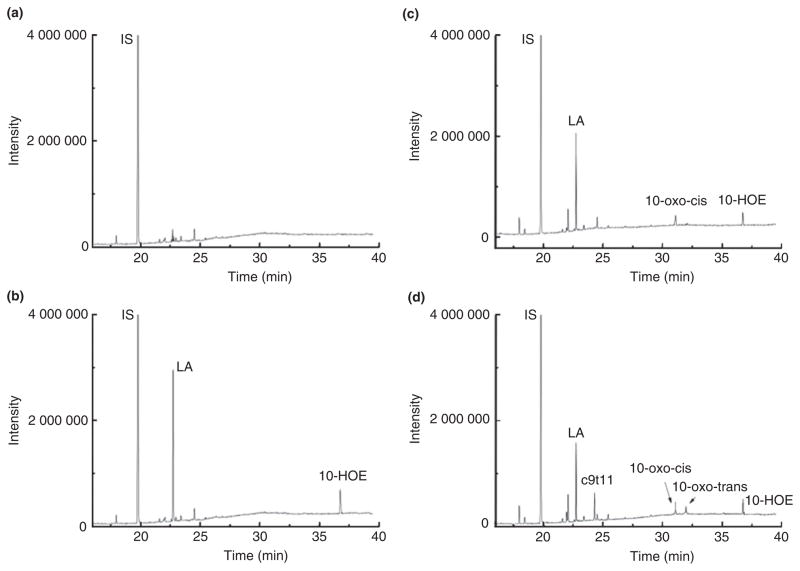
To assess the activity of the three recombinant proteins, different combinations of the resultant cells were tested for conversion of linoleic acid. In the case where the MCRA recombinant was used for reaction, linoleic acid was only converted to 10-HOE according to the retention time and mass fraction compared with that in Lact. plantarum ZS2058 (Fig. 4b). Addition of the E. coli cells containing DH protein resulted in the production of both 10-HOE and 10-oxo-cis-12-octadecenoic acid (Fig. 4c). While combination of all the three recombinant E. coli resulted in 10-HOE, 10-oxo-cis-octadecenoic acid, 10-oxo-trans-11-octadecenoic acid and c9, t11-CLA (Fig. 4d), about 13·66% of LA was converted to CLA by the recombinant E. coli.
Figure 4.
Fatty acid profile after reaction of recombinant E. coli suspension in various combinations. (a) E. coli/pET28a-mcra, E. coli/pET28a-dh and E. coli/pET28a-dc plus FAD and NADH without linoleic acid. (b) E. coli/pET28a-mcra plus FAD and NADH with linoleic acid. (c) E. coli/pET28a-mcra and E. coli/pET28a-dh plus FAD and NADH with linoleic acid. (d) E. coli/pET28a-mcra, E. coli/pET28a-dh and E. coli/pET28a-dc plus FAD and NADH with linoleic acid. IS: internal standard (C17:0); LA: linoleic acid; 10-HOE: 10-hydroxy-cis-12-octadecenoic acid; 10-oxo-cis: 10-oxo-cis-12-octadecenoic acid; 10-oxo-trans: 10-oxo-trans-11-octadecenoic acid; c9, t11: c9, t11-CLA.
Discussion
The reason for bacteria converting linoleic acid to CLA is unclear, and it remains uncertain as to why some strains of lactobacilli, in particular Lact. plantarum, exert superior CLA producers while some other do not produce CLA at a significant level. It has been reported that bioconversion of linoleic acid to CLA might be a key step for fatty acid detoxicification in bacteria (Jiang et al. 1998; Maia et al. 2007, 2010). In the present study, the ability of 25 strains of different food-derived lactobacilli to produce CLA from free linoleic acid was investigated. None of the Lact. acidophilus, Lact. brevis and Lact. rhamnosus tested converted linoleic acid to CLA at a significant level, while the range of Lact. plantarum exhibited considerable CLA production ability. From those strains, Lact. plantarum ZS2058, isolated from Chinese traditional fermented sauerkraut (Niu et al. 2007), was the most efficient producer of CLA, in which over 50% linoleic acid was converted to c9, t11-CLA and t9, t11-CLA as dominant isomers. The conversion percentages reported in the present study are in agreement with earlier studies (Alonso et al. 2003; Zeng et al. 2009; Li et al. 2012). Considering the potential health promotion of CLA, the discovery of food-grade lactobacilli with high ability to synthesize CLA, such as Lact. plantarum and Lact. bulgaricus strains in this study, may offer novel opportunities for developing health-promoting functional food safely with the multiple benefits of CLA and probiotics.
Following culturing with free linoleic acid in MRS medium, Lact. plantarum ZS2058 converted more than 50% linoleic acid to c9, t11-CLA and t9, t11-CLA. In addition, 10-HOE was produced during CLA production. Ogawa et al. (2001) firstly reported the 10-HOE accumulation in CLA production in Lact. acidophilus. With Lact. acidophilus AKU1137 grown in MRS medium containing free linoleic acid for four days, 10-HOE was significantly accumulated from linoleic acid in the first two days, while CLA was produced slowly. Moreover, when the concentration of 10-HOE reached a high level, it was converted into CLA rapidly. Volkov et al. (2010) firstly reported the MCRA from S. pypgenes as a fatty acid double-bond hydratase, which converted linoleic acid into 10-HOE and 10, 13-dihydroxy-octadacenoic acid. MCRA from Bifidobacterium breve NCIMB 702258, a high CLA producer, was reported as a FAD-dependent fatty acid hydratase (Rosberg-Cody et al. 2011), which has a function in stress protection. Kishino et al. (2011a) reported the multiple-component enzymes for CLA production from Lact. plantarum AKU1009a. With different fractions from ultracentrifugation, 10-HOE was produced by a membrane-bound protein; further then c9, t11-CLA was produced while the membrane fraction was mixed with the other two unknown proteins from soluble fractions. Later, more MCRAs were reported as fatty acid hydratases from different bacteria (Joo et al. 2012; Kim et al. 2012; Yang et al. 2013).
Following culturing with free linoleic acid, 10-HOE, 10-oxo-cis-12-octadecenoic acid and 10-oxo-trans-11-octadecenoic acid combined with c9, t11-CLA and t9, t11-CLA were produced at different levels in Lact. plantarum ZS2058. The conversion of linoleic acid to 10-HOE and 10-oxo-octadecenoic acid derivatives was in agreement with previous hypothesis (Kishino et al. 2011b), in which CLA production was a multiple-step reaction: firstly, linoleic acid was converted to 10-HOE, then dehydrated and double-bond isomerized to 10-oxo-trans-11-octadecenoic acid, followed by rehydration and converted to CLA as the end product.
In the present study, MCRA was amplified and confirmed as fatty acid hydratase and played a role in linoleic acid hydration to 10-HOE. The latter two genes, dh and dc, were also expressed in E. coli. With the mixture of MCRA and DH recombinants, both 10-HOE and 10-oxo-cis-12-octadecenoic acid were converted, while the two recombinants were mixed with DC recombinant, all 10-HOE, 10-oxo-cis-12-octadecenoic acid, 10-oxo-trans-11-octadecenoic acid and c9, t11-CLA were produced. The present results were highly in agreement with a newest result (Kishino et al. 2013), in which cla-hy, cla-dh and cla-dc from Lact. plantarum AKU1009a were the genes in charge of linoleic acid bioconversion to CLA. Those results indicated that the mechanism for CLA production might be shared by different Lactobacillus species. The lactobacilli strain consisting the mcra, dh and dc genes for multiple-component linoleate isomerase genome might have the ability for CLA production, which needs further investigation.
Acknowledgments
This work was supported by the Program for New Century Excellent Talents (NCET-13-0831), the National Natural Science Foundation of China (No. 21276108), the National High Technology Research and Development Program of China (2011AA100905, 2011AA100901), the National Science Fund for Distinguished Young Scholars (31125021), the National Basic Research Program 973 of China (2012CB720802), the 111 project B07029 and the Fundamental Research Funds for the Central Universities (No. JUSRP51320B).
Footnotes
Conflict of interest
We declare that we have no conflict of interest.
References
- Alonso L, Cuesta EP, Gilliland SE. Production of free conjugated linoleic acid by Lactobacillus acidophilus and Lactobacillus casei of human intestinal origin. J Dairy Sci. 2003;86:1941–1946. doi: 10.3168/jds.S0022-0302(03)73781-3. [DOI] [PubMed] [Google Scholar]
- Castro-Webb N, Ruiz-Narvaez EA, Campos H. Cross-sectional study of conjugated linoleic acid in adipose tissue and risk of diabetes. Am J Clin Nutr. 2012;96:175–181. doi: 10.3945/ajcn.111.011858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coakley M, Ross RP, Nordgren M, Fitzgerald G, Devery R, Stanton C. Conjugated linoleic acid biosynthesis by human-derived Bifidobacterium species. J Appl Microbiol. 2003;94:138–145. doi: 10.1046/j.1365-2672.2003.01814.x. [DOI] [PubMed] [Google Scholar]
- Coakley M, Johnson MC, McGrath E, Rahman S, Ross RP, Fitzgerald GF, Devery R, Stanton C. Intestinal bifidobacteria that produce trans-9, trans-11 conjugated linoleic acid: a fatty acid with antiproliferative activity against human colon SW480 and HT-29 cancer cells. Nutr Cancer. 2006;56:95–102. doi: 10.1207/s15327914nc5601_13. [DOI] [PubMed] [Google Scholar]
- Field CJ, Schley PD. Evidence for potential mechanism for the effect of conjugated linoleic acid on tumor metabolism and immune function: lessons from n-3 fatty acids. Am J Clin Nutr. 2004;79:1190–1198. doi: 10.1093/ajcn/79.6.1190S. [DOI] [PubMed] [Google Scholar]
- Gorissen L, Raes K, Weckx S, Dannenberger D, Leroy F, De Vuyst L, De Smet S. Production of conjugated linoleic acid and conjugated linolenic acid isomers by Bifidobacterium species. Appl Microbiol Biotechnol. 2010;87:2257–2266. doi: 10.1007/s00253-010-2713-1. [DOI] [PubMed] [Google Scholar]
- Griinari JM, Bauman DE. Biosynthesis of conjugated linoleic acid and its incorporation into meat and milk in ruminants. In: Yurawecz MP, Mossoba MM, Kramer JKG, Pariza MW, Nelson GJ, editors. Advances in Conjugated Linoleic Acid Research. Champaign, IL: AOCS Press; 1999. pp. 180–200. [Google Scholar]
- Hoffman CS, Winston F. A ten-minute DNA preparation from yeast efficiently releases autonomous plasmids for transformation of Escherichia coli. Gene. 1987;57:267–272. doi: 10.1016/0378-1119(87)90131-4. [DOI] [PubMed] [Google Scholar]
- Ip C, Chin SF, Scimeca JA, Pariza MW. Mammary cancer prevention by conjugated dienoic derivative of linoleic acid. Cancer Res. 1991;51:6118–6124. [PubMed] [Google Scholar]
- Ip C, Scimeca JA, Thompson HJ. Conjugated linoleic acid. A powerful anticarcinogen from animal fat sources. Cancer. 1994;74:1050–1054. doi: 10.1002/1097-0142(19940801)74:3+<1050::aid-cncr2820741512>3.0.co;2-i. [DOI] [PubMed] [Google Scholar]
- Jiang J, Bjorck L, Fonden R. Production of conjugated linoleic acid by dairy starter cultures. J Appl Microbiol. 1998;85:95–102. doi: 10.1046/j.1365-2672.1998.00481.x. [DOI] [PubMed] [Google Scholar]
- Joo YC, Seo ES, Kim YS, Kim KR, Park JB, Oh DK. Production of 10-hydroxystearic acid from oleic acid by whole cells of recombinant Escherichia coli containing oleate hydratase from Stenotrophomonas maltophilia. J Biotechnol. 2012;158:17–23. doi: 10.1016/j.jbiotec.2012.01.002. [DOI] [PubMed] [Google Scholar]
- Kepler CR, Tove SB. Biohydrogenation of unsaturated fatty acids. J Biol Chem. 1967;242:5686–5692. [PubMed] [Google Scholar]
- Kepler CR, Hirons KP, McNeill JJ, Tove SB. Intermediates and products of the biohydrogenation of linoleic acid by Butyrinvibrio fibrisolvens. J Biol Chem. 1966;241:1350–1354. [PubMed] [Google Scholar]
- Kil KS, Cunningham MW, Barnett LA. Cloning and sequence analysis of a gene encoding a 67-kilodalton myosin-cross-reactive antigen of Streptococcus pyogenes reveals its similarity with class II major histocompatibility antigens. Infect Immun. 1994;62:2440–2449. doi: 10.1128/iai.62.6.2440-2449.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim BN, Joo YC, Kim YS, Kim KR, Oh DK. Production of 10-hydroxystearic acid from oleic acid and olive oil hydrolyzate by an oleate hydratase from Lysinibacillus fusiformis. Appl Microbiol Biotechnol. 2012;95:929–937. doi: 10.1007/s00253-011-3805-2. [DOI] [PubMed] [Google Scholar]
- Kishino S, Ogawa J, Yokozeki K, Shimizu S. Linoleic acid isomerase in Lactobacillus plantarum AKU1009a proved to be a multi-component enzyme system requiring oxidoreduction cofactors. Biosci Biotechnol Biochem. 2011a;75:318–322. doi: 10.1271/bbb.100699. [DOI] [PubMed] [Google Scholar]
- Kishino S, Park SB, Takeuchi M, Yokozeki K, Shimizu S, Ogawa J. Novel multi-component enzyme machinery in lactic acid bacteria catalyzing C=C double bond migration useful for conjugated fatty acid synthesis. Biochem Biophys Res Commun. 2011b;416:188–193. doi: 10.1016/j.bbrc.2011.11.022. [DOI] [PubMed] [Google Scholar]
- Kishino S, Takeuchi M, Park SB, Hirata A, Kitamura N, Kunisawa J, Kiyono H, Iwamoto R, et al. Polyunsaturated fatty acid saturation by gut lactic acid bacteria affecting host lipid composition. Proc Natl Acad Sci USA. 2013;110:17808–17813. doi: 10.1073/pnas.1312937110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee KN, Kritchevsky D, Pariza MW. Conjugated linoleic acid and atherosclerosis in rabbits. Atherosclerosis. 1994;108:19–25. doi: 10.1016/0021-9150(94)90034-5. [DOI] [PubMed] [Google Scholar]
- Li H, Liu Y, Bao Y, Liu X, Zhang H. Conjugated linoleic acid conversion by six Lactobacillus plantarum strains cultured in MRS broth supplemented with sunflower oil and soymilk. J Food Sci. 2012;77:330–336. doi: 10.1111/j.1750-3841.2012.02723.x. [DOI] [PubMed] [Google Scholar]
- Liavonchanka A, Hornung E, Feussner I, Rudolph MG. Structure and mechanism of the Propionibacterium acnes polyunsaturated fatty acid isomerase. Proc Natl Acad Sci USA. 2006;103:2576–2581. doi: 10.1073/pnas.0510144103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin TY, Lin CW, Lee CH. Conjugated linoleic acid concentration as affected by lactic cultures and added linoleic acid. Food Chem. 1999;67:1–5. [Google Scholar]
- Maia MR, Chaudhary LC, Figueres L, Wallace RJ. Metabolism of polyunsaturated fatty acids and their toxicity to the microflora of the rumen. Antonie Van Leeuwenhoek. 2007;91:303–314. doi: 10.1007/s10482-006-9118-2. [DOI] [PubMed] [Google Scholar]
- Maia MR, Chaudhary LC, Bestwick CS, Richardson AJ, McKain N, Larson TR, Graham IA, Wallace RJ. Toxicity of unsaturated fatty acids to the biohydrogenating ruminal bacterium, Butyrivibrio fibrisolvens. BMC Microbiol. 2010;10:52. doi: 10.1186/1471-2180-10-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClelland S, Cox C, O’Connor R, de Gaetano M, McCarthy C, Cryan L, Fitzgerald D, Belton O. Conjugated linoleic acid suppresses the migratory and inflammatory phenotype of the monocyte/macrophage cell. Atherosclerosis. 2010;211:96–102. doi: 10.1016/j.atherosclerosis.2010.02.003. [DOI] [PubMed] [Google Scholar]
- Mohan MS, Anand S, Kalscheur KF, Hassan AN, Hippen AR. Starter cultures and cattle feed manipulation enhance conjugated linoleic acid concentrations in cheddar cheese. J Dairy Sci. 2013;96:2081–2094. doi: 10.3168/jds.2012-6101. [DOI] [PubMed] [Google Scholar]
- Moloney F, Toomey S, Noone E, Nugent A, Allan B, Loscher CE, Roche HM. Antidiabetic effects of cis-9, trans-11-conjugated linoleic acid may be mediated via anti-inflammatory effects in white adipose tissue. Diabetes. 2007;56:574–582. doi: 10.2337/db06-0384. [DOI] [PubMed] [Google Scholar]
- Niu XY, Chen W, Tian FW, Zhao JX, Zhang H. Bioconversion of conjugated linoleic acid by resting cells of Lactobacillus plantarum ZS2058 in potassium phosphate buffer system. [Article in Chinese] Wei Sheng Wu Xue Bao. 2007;47:244–248. [PubMed] [Google Scholar]
- Noone EJ, Roche HM, Nugent AP, Gibney MJ. The effect of dietary supplementation using isomeric blends of conjugated linoleic acid on lipid metabolism in healthy human subjects. Br J Nutr. 2002;88:243–251. doi: 10.1079/BJN2002615. [DOI] [PubMed] [Google Scholar]
- Ogawa J, Matsumura K, Kishino S, Omura Y, Shimizu S. Conjugated linoleic acid accumulation via 10-hydroxy-12-octadecaenoic acid during microaerobic transformation of linoleic acid by Lactobacillus acidophilus. Appl Environ Microbiol. 2001;67:1246–1252. doi: 10.1128/AEM.67.3.1246-1252.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park Y, Storkson JM, Liu W, Albright KJ, Cook ME, Pariza MW. Structure-activity relationship of conjugated linoleic acid ans its cognates in inhibiting heparin-releasable lipoprotein lipase and glycerol release from fully differentiated 3T3-L1 adipocytes. J Nutr Biochem. 2004;15:561–569. doi: 10.1016/j.jnutbio.2004.04.004. [DOI] [PubMed] [Google Scholar]
- Peng SS, Deng MD, Grund AD, Rosson RA. Purification and characterization of a membrane-bound linoleic acid isomerase from Clostridium sporogenes. Enzyme Microb Technol. 2007;40:831–839. [Google Scholar]
- Rosberg-Cody E, Ross RP, Hussey S, Ryan CA, Murphy BP, Fitzgerald GF, Devery R, Stanton C. Mining the microbiota of the neonatal gastrointestinal tract for conjugated linoleic acid-producing bifidobacteria. Appl Environ Microbiol. 2004;70:4635–4641. doi: 10.1128/AEM.70.8.4635-4641.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosberg-Cody E, Liavonchanka A, Göbel C, Ross RP, O’Sullivan O, Fitzgerald GF, Feussner I, Stanton C. Myosin-cross-reactive antigen (MCRA) protein from Bifidobacterium breve is a FAD-dependent fatty acid hydratase which has a function in stress protection. BMC Biochem. 2011;12:9. doi: 10.1186/1471-2091-12-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosson RA, Grund AD, Deng MD, Sanchez-Riera F. Linoleate isomerase. 6743609. United States Patent. 2004
- Rungapamestry V, McMonagle J, Reynolds C, Rucklidge G, Reid M, Duncan G, Ross K, Horgan G, et al. Inter-organ proteomic analysis reveals insights into the molecular mechanisms underlying the anti-diabetic effects of cis-9, trans-11-conjugated linoleic acid in ob/ob mice. Proteomics. 2012;12:461–476. doi: 10.1002/pmic.201100312. [DOI] [PubMed] [Google Scholar]
- Shen W, Chuang CC, Martinez K, Reid T, Brown JM, Xi L, Hixson L, Hopkins R, et al. Conjugated linoleic acid reduces adiposity and increases markers of browning and inflammation in white adipose tissue of mice. J Lipid Res. 2013;54:909–922. doi: 10.1194/jlr.M030924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sluijs I, Plantinga Y, de Roos B, Mennen LI, Bots ML. Dietary supplementation with cis-9, trans-11 conjugated linoleic acid and aortic stiffness in overweight and obese adults. Am J Clin Nutr. 2010;91:175–183. doi: 10.3945/ajcn.2009.28192. [DOI] [PubMed] [Google Scholar]
- Sugano M, Tsujita A, Yamasaki M, Noguchi M, Yamada K. Conjugated linoleic acid modulates tissue levels of chemical mediators and immunoglobulins in rats. Lipids. 1998;33:521–527. doi: 10.1007/s11745-998-0236-4. [DOI] [PubMed] [Google Scholar]
- Valeille K, Gripois D, Blouquit MF, Souidi M, Riottot M, Bouthegourd JC, Sérougne C, Martin JC. Lipid atherogenic risk markers can be more favourably influenced by the cis-9, trans-11-octadecadienoate isomer than a conjugated linoleic acid mixture or fish oil in hamsters. Br J Nutr. 2004;91:191–199. doi: 10.1079/BJN20031057. [DOI] [PubMed] [Google Scholar]
- Volkov A, Liavonchanka A, Kamneva O, Fiedler T, Goebel C, Kreikemeyer B, Feussner I. Myosin cross-reactive antigen of Streptococcus pyogenes M49 encodes a fatty acid double bond hydratase that plays a role in oleic acid detoxification and bacterial virulence. J Biol Chem. 2010;285:10353–10361. doi: 10.1074/jbc.M109.081851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang B, Chen H, Song Y, Chen YQ, Zhang H, Chen W. Myosin-cross-reactive antigens from four different lactic acid bacteria are fatty acid hydratases. Biotechnol Lett. 2013;35:75–81. doi: 10.1007/s10529-012-1044-y. [DOI] [PubMed] [Google Scholar]
- Ye SH, Yu T, Yang H, Li L, Wang H, Xiao S, Wang JH. Optimal culture conditions for producing conjugated linoleic acid in skim-milk by coculture of different Lactobacillus strains. Ann Microbiol. 2013;63:707–717. [Google Scholar]
- Zeng Z, Lin J, Gong D. Identification of lactic acid bacterial strains with high conjugated linoleic acid-producing ability from natural sauerkraut fermentations. J Food Sci. 2009;74:154–158. doi: 10.1111/j.1750-3841.2009.01123.x. [DOI] [PubMed] [Google Scholar]