Abstract
Mounting evidence suggests that microRNA (miR) dysregulation contributes to neurodegenerative disorders including Parkinson’s disease (PD). MiR-34b and miR-34c have been previously shown to be down-regulated in the brains of patients with PD. Here, we demonstrate that miR-34b and miR-34c repress the expression of α-synuclein (α-SYN), a key protein in PD pathogenesis. Inhibition of miR-34b and miR-34c expression in human dopaminergic SH-SY5Y cells increased α-SYN levels and stimulated aggregate formation. Additionally, a single nucleotide polymorphism (SNP) in the 3’-UTR of α-SYN was found to lower the miR-34b-mediated repression of the protein. Our results suggest that down-regulation of miR-34b and miR-34c in the brain, as well as an SNP in the 3’-UTR of α-SYN can increase α-SYN expression, possibly contributing to PD pathogenesis.
Keywords: microRNA-34b, microRNA-34c, α-synuclein, Parkinson’s disease
Introduction
Parkinson’s disease (PD) is the second most common neurodegenerative disorder affecting 1–2% of the population over the age of 65. PD is characterized by the presence of cytoplasmic inclusions in the surviving neurons called Lewy bodies (LBs) and Lewy neurites, which consist mainly of α-synuclein (α-syn)1, 2. Point mutations in the α-syn gene are linked to dominantly inherited PD2, and individuals with multiplication of this gene locus develop PD with an early-onset age and phenotypic severity that are consistent with a gene dosage effect3, 4. Additionally, a polymorphism in the α-syn gene promoter region results in enhanced transcription and is associated with increased risk of PD5. Even in sporadic PD patients, α-syn mRNA has been found to be significantly upregulated as assessed by quantitative RT-PCR using a pool of individual dopaminergic neurons isolated by UV-laser microdissection microscopy from postmortem brains6, 7. Therefore, increased expression of α-syn appears to be a key factor underlying the pathogenesis of PD. Here, we provide experimental evidence to suggest a possible mechanism leading to increased α-syn expression through dysregulation of α-syn targeting microRNAs (miRs), miR-34b and miR-34c, in the brains of PD patients.
miRs are small non-coding RNAs that regulate post-transcriptional gene expression by binding to the 3’-UTR of the mRNA. Dysregulation of miR expression and/or function has been associated with several neurodegenerative diseases including PD8–11. Previously, miR-7 and miR-153 were reported to target and downregulate α-syn expression12, 13. However, it is not known whether these two miRs are altered in PD brains. Interestingly, miR-34b (miR-34b-3p) and miR-34c (miR-34c-5p) are reportedly downregulated in the brain areas of PD patients including the amygdala, frontal cortex, substantia nigra and cerebellum, suggesting their possible mechanistic role in PD pathogenesis14, 15. Reducing the levels of miR-34b and miR-34c recapitulates several biochemical and cell biological alterations observed in PD brains including oxidative stress, impaired mitochondrial function and decreased viability of dopaminergic neuronal cells14. In the present study, we show that miR-34b and miR-34c repress α-syn expression through its 3’-UTR, while inhibition of miR-34b and miR-34c elevates α-syn level and leads to the formation of α-syn-containing aggregates. We suggest that decreased expression of miR-34b and miR-34c found in PD brains can result in elevated α-syn level and aggregate formation, which might contribute to PD pathogenesis.
Materials and Methods
Materials
All pre-miRs and anti-miRs were purchased from Ambion.
Plasmids
The firefly luciferase reporter construct containing human α-syn 3’-UTR was previously described12. Mutations of the miR-34b and miR-34c predicted target site in the α-syn 3′-UTR were created using the QuikChange site-directed mutagenesis kit (Stratagene) with primers 5’ - GAAGTCTTCCATCAGCAGTCTTTGAAGTATCTGTACCTG-3’ and 5’- CAGGTACAGATACTTCAAAGACTGCTGATGGAAGACTTC-3’ for miR-34b site 1, 5’- GTTGTTCAGAAGTTGTTAGTCTTTTGCTATCATATATTATAAG-3’ and 5’- CTTATAATATATGATAGCAAAAGACTAACAACTTCTGAACAAC-3’ for miR-34b site 2, and 5’- GAATTCCCTGAAGCAACAGACCCAGAAGTGTGTTTTGG-3’ and 5’- CCAAAACACACTTCTGGGTCTGTTGCTTCAGGGAATTC-3’ for miR-34c site. The minor allele sequence of SNP rs10024743 was introduced in the α-syn 3’-UTR reporter plasmid using primers 5’- CAGCAGTGATTGAAGTATCTG-3’ and 5’- CAGATACTTCAATCACTGCTG-3’.
Cell cultures and Transfections
Human neuroblastoma cell line, SH-SY5Y, was purchased from American Type Culture Collection. Cells were maintained in Dulbecco’s Modified Eagle Medium (Invitrogen) supplemented with 10% fetal bovine serum (Invitrogen). Cells were transfected with anti-miRs and pre-miRs at the concentration of 50 nM using Lipofectamine RNAiMax (Invitrogen) for 48 hours before harvest.
Reporter gene assay
Cells were cotransfected with luciferase reporter constructs and internal control construct pSV-β-galactosidase in the absence or presence of pre-miR-34b and pre-miR-34c. After cell lysis with Glo Lysis Buffer (Promega), luciferase activity was measured with Steady-Glo Luciferase Assay System (Promega) using a Wallac 1420 Multilabel Counter (PerkinElmer). β-Galactosidase activity was measured using chlorophenol red-β-D-galactopyranoside (CPRG)(Roche) in a reaction buffer (60 mM Na2HPO4, 40 mM NaH2PO4, 10 mM KCl, 1 mM MgSO4, 50 mM 2-mercaptoethanol, pH 7.0) to normalize luciferase activity. Experiments were performed in triplicates.
Western Blot Analysis
Cell lysates were analyzed by Western blotting as described previously12 using mouse monoclonal anti-α-syn antibody SYN-1 (Invitrogen) and monoclonal β-actin antibody (Sigma). Band densities were measured using National Institutes of Health (NIH) Image J.
Quantitative Real-time PCR
To quantify α-syn mRNA levels, total RNA was prepared from SH-SY5Y cells using Trizol reagent (Invitrogen) according to manufacturer’s instructions. Reverse Transcription was performed with 2 µg total RNA in 20µL reaction. Real-time PCR was performed in triplicates with SYBR® Select Master Mix (Applied Biosystems) in an Applied Biosystems 7500 Real-Time PCR System. PCR primer sequences used were as follows: 18S rRNA (5’- CGGCTACCACATCCAAGGAA-3’ and 5’- GCTGGAATTACCGCGGCT-3’), α-syn (5’- ACCAAACAGGGTGTGGCAGAAG-3’ and 5’- CTTGCTCTTTGGTCTTCTCAGCC-3’) and β-syn (5’-AAGCAAGACCCGAGAAGGTGTG-3’ and 5’-ATTCCTCCCTCTTCACCAGTCC-3’). Relative mRNA expression level was calculated by the 2ΔΔCt method.
Immunocytochemistry
Cells were fixed in 4% paraformaldehyde in PBS for 20 min, washed with PBS three times, and permeabilized with 0.5% Triton X-100 in PBS for 10 min. After washing with PBS again and blocking with 5% donkey serum for 20 min, cells were incubated with anti-α-syn antibody (SYN-1) diluted in 1% donkey serum at room temperature for 1 h and washed with PBS and incubated with DyLight 488-conjugated anti-mouse IgG (Jackson immunoresearch) diluted in PBS containing 1% donkey serum for 1 hour at room temperature. For nuclear staining, cells were incubated with 1 µg/mL 4’,6’-diamidino-2-phenylindole dihydrochloride (DAPI; Sigma-Aldrich) in PBS for 1 min at room temperature. Cells were washed five times with PBS and analyzed under a fluorescence microscope (Axiovert 200, Zeiss). For quantification of aggregates, 6 microscopic fields containing at least 100 cells per group were randomly selected, and the average number of aggregates per 100 cells was calculated.
Statistical analysis
All experiments were done in triplicates, and statistical significance between control and experimental values was determined using Student’s t test (paired, two-tailed). All data are expressed as means ± standard deviations (SD) or mean ± standard error (SEM) as described.
Results
Inhibition of miR-34b or miR-34c increases α-syn expression
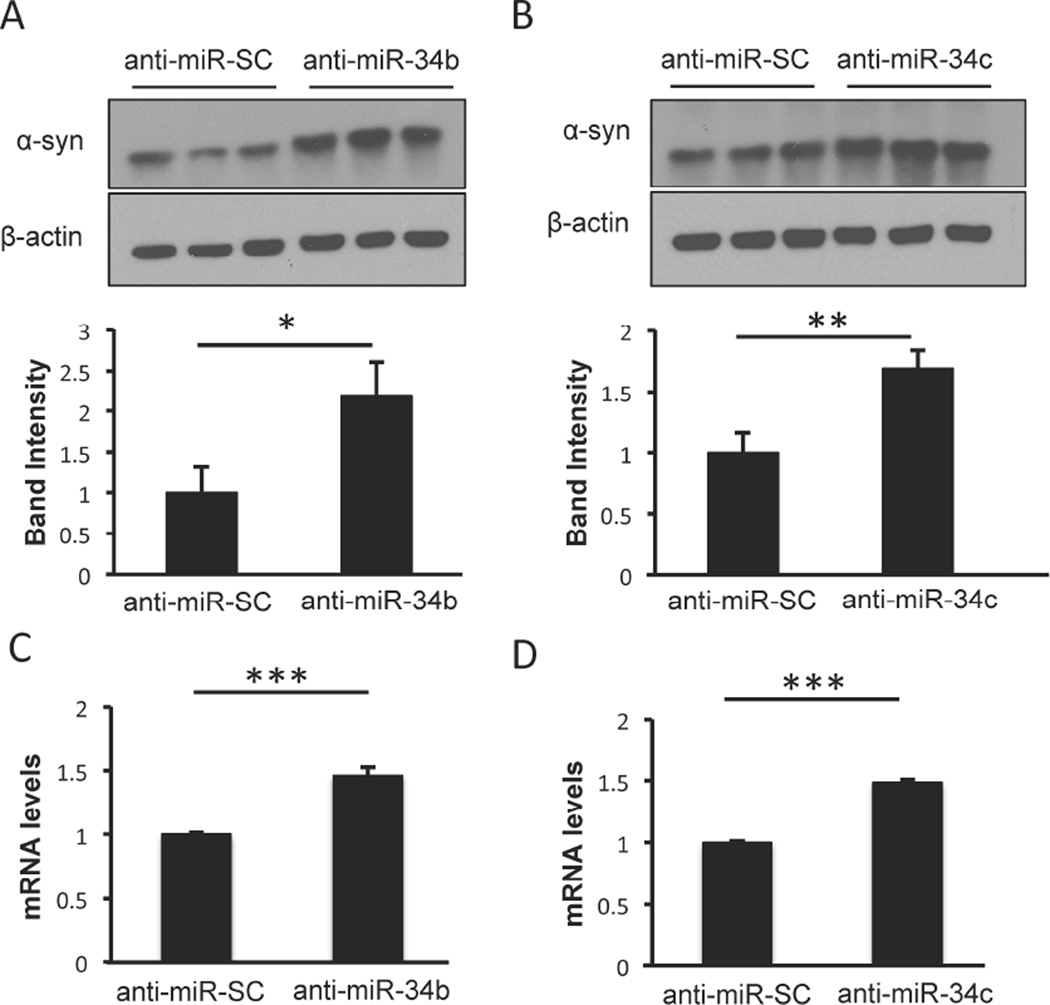
The effect of reduced levels of miR-34b and miR-34c in PD patients was recapitulated in human dopaminergic SH-SY5Y cells by utilizing anti-miRs, which are designed to specifically bind to and inhibit corresponding endogenous mature miRs. To examine whether inhibiting these miRs leads to increased α-syn protein level, lysates from cells transfected with anti-miR-34b or anti-miR-34c were examined using Western blot analysis. Indeed, inhibiting miR-34b (Fig. 1A) or miR-34c (Fig. 1B) significantly increased α-syn protein expression by 2.2-fold and 1.7-fold, respectively. Also, quantitative real time PCR analysis revealed that α-syn mRNA level was significantly elevated by 1.5 fold (Fig. 1C and 1D) upon inhibiting either of these miRs, suggesting the regulation of α-syn mRNA by miR-34b and miR-34c in SH-SY5Y cells.
Fig. 1. Inhibition of miR-34b and miR-34c increases α-syn expression.
(A–B) SH-SY5Y cells were transfected with anti-miR-34b and anti-miR-34c for 48 h followed by Western blot analysis using anti-α-syn antibody and β-actin antibody. Relative protein levels were normalized to β-actin and calculated based on band intensity using NIH Image J Software. Anti-miR-SC denotes anti-miR-scrambled sequence. (C–D) Quantitative RT-PCR of α-syn mRNA levels in cells transfected as in panels A–B. Values are normalized to 18S rRNA. Data are shown as mean ± SD. *p<0.05, **p<0.01 and ***p<0.001.
miR-34b and miR-34c reduce α-syn expression
By performing sequence alignment using TargetScan, we identified potential miR-34b and miR-34c binding sites in the 3’-UTR of α-syn mRNA, which are conserved in humans, chimpanzee and rhesus (Fig. 2A and 2B). Computational analysis using RNAhybrid algorithm16 predicted base pairing with two target sites for miR-34b and one target site for miR-34c within the α-syn mRNA. The minimal free energy is −14.3 kcal/mol for miR-34b site 1, −18.6 kcal/mol for miR-34b site 2, and −22.9 kcal/mol for miR-34c site, suggesting favorable interactions between these miRs and their respective sites. To examine the effect of miR-34b and miR-34c on the expression of α-syn, SH-SY5Y cells were transfected with pre-miR-34b and pre-miR-34c followed by quantitative real time PCR analysis. Overexpression of miR-34b or miR-34c resulted in significant reduction in α-syn mRNA expression (Fig. 2C and 2D) and α-syn protein level (Fig. 2E, and 2F), suggesting that miR-34b and miR-34c target α-syn mRNA. However, miR-34b and miR-34c were unable to repress β-synuclein (β-syn) expression, a highly homologous protein to α-syn (Supplementary Fig. 1). Rather, miR-34c increases β-syn expression up to 2.3 fold through an unknown mechanism (Supplementary Fig. 1). Taken together, we conclude that miR-34b and miR-34c repress α-syn expression, but not β-syn expression. Further, comparison of miR-34b and miR-34c to previously identified miRs that target α-syn 3’UTR, i.e., miR-7 and miR-153, revealed that all the tested miRs downregulate α-syn protein level to a similar extent (Supplementary Fig. 2).
Fig. 2. miR-34b and miR-34c target α-syn expression.
Schematic representation of the two predicted binding sites (#1 and #2) for miR-34b (A) and one site for miR-34c (B) in the 3’-UTR of α-syn. Sites are conserved in human, chimpanzee and rhesus monkey. Quantitative RT-PCR analysis of α-syn mRNA levels was performed on SH-SY5Y cells transfected with miR-34b (C) and miR-34c (D) for 48 h. Levels of α-syn mRNA are normalized to 18S rRNA. SH-SY5Y cells were transfected with pre-miR-34b (E) and pre-miR-34c (F) for 48h followed by Western Blot analysis using anti-α-syn antibody. Band intensities are quantified and shown below the respective blots. Data are shown as mean ± SD. **p<0.01 and ***p<0.001.
Confirmation of miR-34b and miR-34c target sites in the 3’-UTR of α-syn mRNA
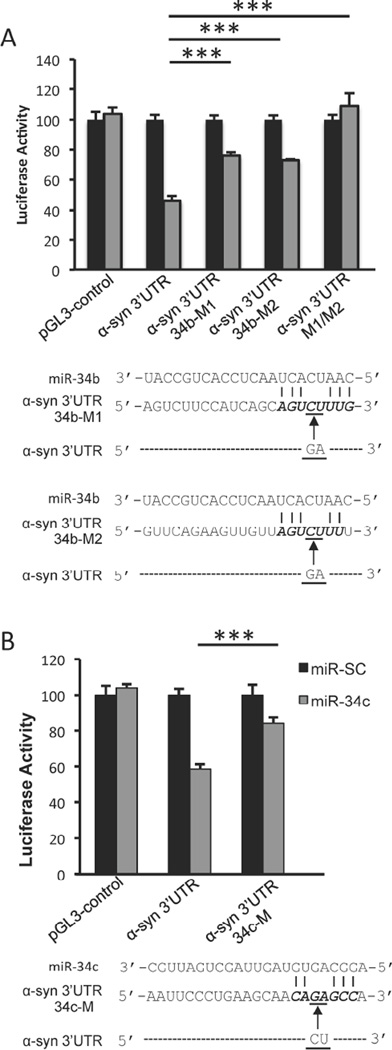
To verify whether miR-34b and miR-34c directly target the 3′-UTR of α-Syn mRNA, we utilized a plasmid construct expressing full-length α-syn 3′-UTR downstream of the firefly luciferase reporter gene. Co-transfection of miR-34b or miR-34c along with this reporter construct significantly decreased luciferase activity, but did not affect the control vector pGL3 devoid of the α-syn 3’-UTR (Fig. 3A and 3B), indicating that miR-34b and miR-34c work through the 3’-UTR of α-syn. To ensure that the predicted target sites of miR-34b and miR-34c in the α-Syn 3′-UTR are functional, these sites were mutated as shown in Fig. 3A and 3B. Compared with 54% repression of luciferase activity from the wild-type α-syn 3’-UTR construct upon co-transfection with pre-miR-34b, the α-syn 3’-UTR construct mutated at the predicted miR-34b site 1 (34b-M1) or site 2 (34b-M2) could be repressed by only 24% and 27%, respectively, (Fig. 3A). Further, mutating both miR-34b binding sites (34b-M1/M2) in the 3’UTR completely abrogated its suppression (Fig. 3A). Similarly, miR-34c was able to repress the expression of the reporter gene by only 16% from the α-syn 3’-UTR construct containing 34c-M mutated site compared to 41% repression from the construct containing the predicted wild-type sequence (Fig. 3B). These results indicate that the predicted sequences are authentic binding sites for miR-34b and miR-34c.
Fig. 3. miR-34b and miR-34c target α-syn 3’UTR.
SH-SY5Y cells were co-transfected with pre-miR-34b (A) or pre-miR-34c (B) along with reporter constructs containing control vector, α-syn 3’UTR or its mutants (34b-M1, 34b-M2, 34b-M1/M2 and 34c-M) and pSV-β-galactosidase. Luciferase activity was normalized against β-galactosidase activity. Schematic representations of mutant α-syn 3’-UTR reporter constructs are shown below the respective graphs. Data are shown as mean ± SD. ***p<0.001.
Inhibition of miR-34b or miR-34c increase α-syn aggregation
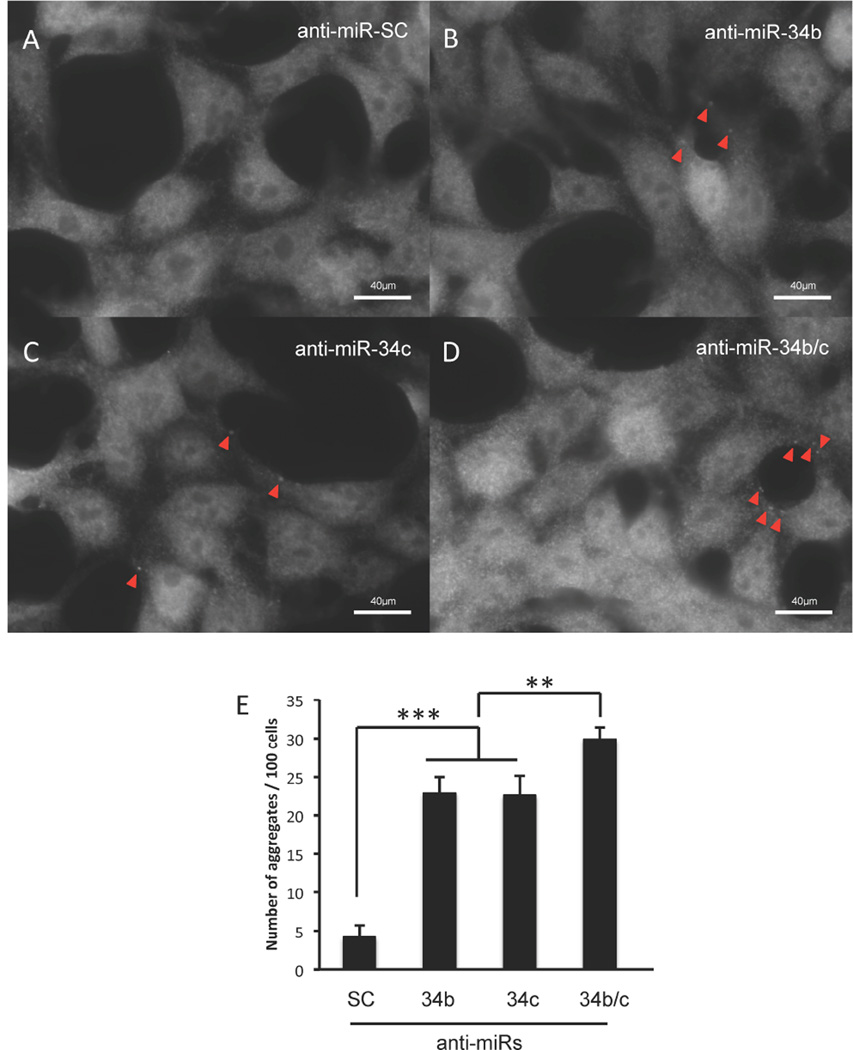
As inhibition of miR-34b or miR-34c led to increased α-syn expression (Fig. 1), we investigated whether transfection of anti-miR-34b or anti-miR-34c could also lead to the formation of α-syn-containing aggregates. To this end, SH-SY5Y cells were transfected with anti-miR-34b, anti-miR-34c or both, and the formation of aggregates were assessed by immunocytochemistry using anti-α-syn antibody. As shown in Fig. 4, inhibiting either miR-34b or miR-34c led to an increase in the formation of α-syn-containing aggregates, and inhibition of both miRs led to further increase in aggregate formation. Therefore, we suggest that reduced levels of miR-34b and miR-34c found in PD brains could significantly increase α-syn expression and the formation of α-syn-containing aggregates in dopaminergic neurons.
Fig. 4. Inhibition of miR-34b or miR-34c increases α-syn aggregation.
Representative pictures showing α-syn-containing aggregates upon transfection of SHSY5Y cells with anti-miR-SC (A), anti-miR-34b (B), anti-miR-34c (C) and combination of anti-miR-34b and anti-miR-34c (D) for 48 hours. Aggregates are shown with red arrowheads. (D) Quantification of average number of α-syn aggregates for every 100 cells. Data are shown as mean ± SEM. **p<0.01 ***p<0.001.
A SNP in α-syn 3’UTR makes resistant to miR-34b
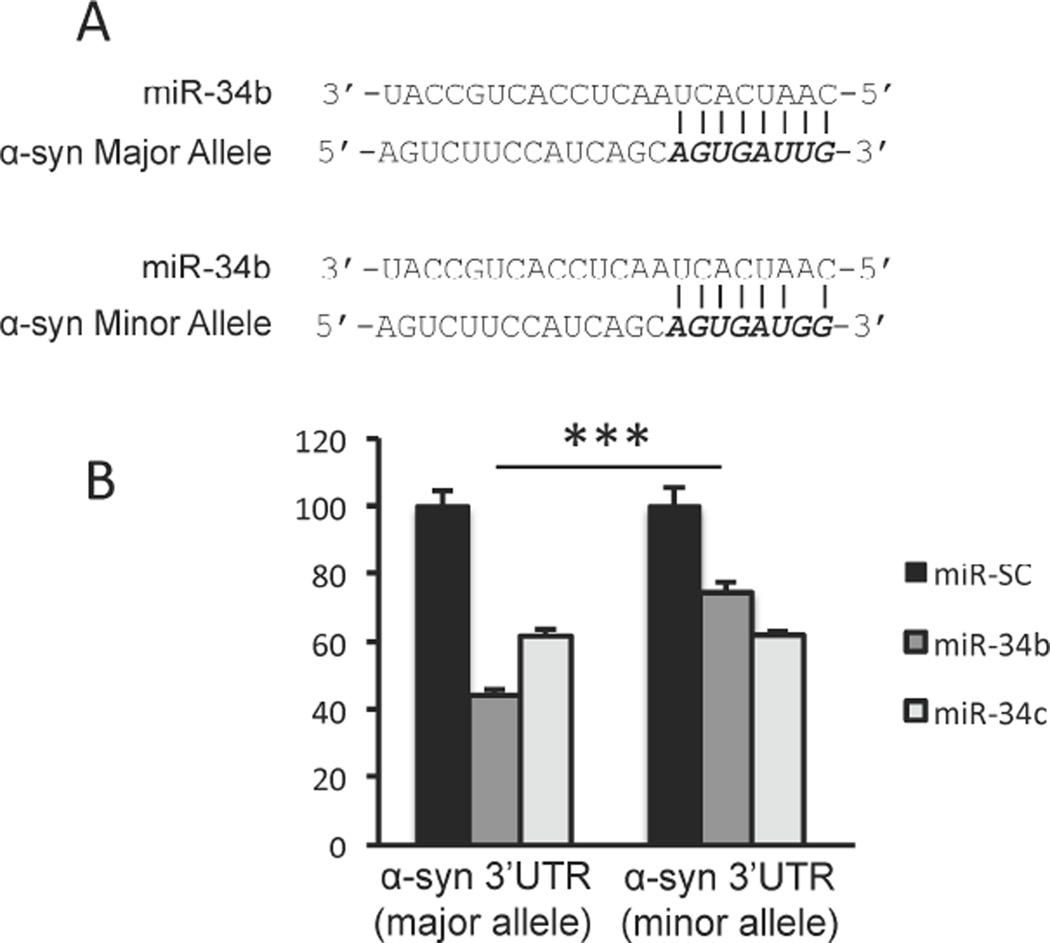
We identified a polymorphic variation (rs10024743) in the 3’-UTR of human α-syn from the GenBank SNP database, although its association with PD is not known. This particular variation lies within the target site 1 of miR-34b. To examine the functional impact of this SNP on the α-syn 3’-UTR, base substitution at position 548 was introduced from thymine (major allele) to guanine (minor allele) (Fig. 5A) in the α-syn 3’-UTR luciferase reporter construct. Co-transfection of the construct containing the major α-syn 3’-UTR allele with pre-miR-34b decreased luciferase activity by 57% as expected, whereas this repression was only 25% for the construct containing the minor allele of α-syn 3’UTR (Fig. 5B). In contrast, miR-34c repressed major and minor allele to a similar extent (Fig. 5B). Therefore, we suggest that individuals harboring the rs10024743 SNP could be resistant to miR-34b-mediated repression of α-syn expression, and this could result in increased α-syn expression and subsequent aggregate formation.
Fig. 5. SNP rs10024743 reduces miR-34b-mediated α-syn repression.
(A) A schematic representation of the major and minor alleles of SNP rs10024743 present in α-syn 3’-UTR. (B) SH-SY5Y cells were co-transfected with reporter constructs containing α-syn 3’-UTR or SNP rs10024743 along with miR-SC or miR-34b or miR-34c for 48h. Luciferase activity was normalized against β-galactosidase activity. Data are shown as mean ± SD. ***p<0.001.
Discussion
Accumulating evidence suggests that alterations in miR expression in the brain is involved in the pathogenesis of PD14, 15, 17–20. Minones-Moyano et al. initially reported the diminished expression of miR-34b and miR-34c in the substantia nigra14, which was confirmed by a recent report15. Inhibition of miR-34b and miR-34c in differentiated SH-SY5Y neuroblastoma cells led to loss of mitochondrial membrane potential and oxidative stress, which are known biochemical abnormalities associated with PD. In addition, both parkin and DJ-1 expression levels were shown to be marginally decreased upon inhibiting the activity of miR-34b and miR-34c. Considering that expression of target genes is expected to increase upon inhibiting their respective miRs, it is likely that miR-34b and miR-34c do not directly target parkin and DJ-1. Consequently, identifying the physiological targets of miR-34b and miR-34c is necessary to better understand the role of these miRNAs in the pathogenesis of PD.
The present investigation provides a mechanistic correlation between the decrease in miR-34b and miR-34c found in the brains of PD patients and α-syn, a key protein in PD. In addition to accumulation of α-syn as fibrillar aggregates in Lewy bodies and Lewy neurites in affected brain regions, the level of α-syn appears to be critical in PD pathogenesis since multiplication of its gene locus leads to autosomal dominant PD. Two miRs, miR-7 and miR-153, have been shown to repress α-syn levels post-transcriptionally by binding to the 3’-UTR of the α-syn mRNA12, 13. However, alterations in the expression of these miRs have yet to be demonstrated in PD. In this report, we show that miR-34b and miR-34c target the 3’-UTR of α-syn mRNA, thus decreasing α-syn protein levels to the similar extent as miR-7 and miR-153. Specifically, miR-34b targets α-syn mRNA at two distinct sites, while miR-34c targets at least one site, as ascertained by utilizing luciferase construct with α-syn 3’-UTR. The repression mediated by miR-34b was completely rescued upon mutating its two binding site in the 3’UTR of α-syn mRNA, confirming the mechanism of action. However, lack of complete derepression after mutation of miR-34c binding site in the α-syn 3’-UTR suggests a possibility of weaker binding of miR-34c to mutated site or presence of other target sites. Further, inhibition of miR-34b and miR-34c leads to increase in α-syn expression and the formation of α-syn-containing aggregates. Combined with the observation that the level of miR-34b and miR-34c are decreased in PD brains, we suggest that elevated expression of α-syn due to loss of miR-34b and miR-34c might contribute to the pathogenesis of PD.
In addition to the multiplication of α-syn locus as a cause of PD, SNPs in the α-syn gene have also been associated with increased risk of the disease21–23. Also, SNPs in miRNA target sites in the 3’-UTR of mRNA affect the interactions between miRNAs and their targets, altering protein expression and potentially impacting disease risk. For example, a significant association has been reported in PD with SNP (rs12720208 [C/T]) located in the 3’-UTR of FGF20 mRNA. This genetic variation lies within a predicted binding region for miR-43324. The risk allele impedes the binding of the mRNA with miR-433 and augments FGF20 expression. Further, this variation seems to enhance the vulnerability to PD through increased α-syn expression, which is mediated by FGF20. However, other reports have failed to replicate an association between SNPs of FGF20 and PD risk, or the connection between the variation rs12720208 and α-syn protein levels25, 26. In the present study, we identified a polymorphic variation (rs10024743) in the 3’-UTR of human α-syn from the GenBank SNP database, although its association with PD is not known. This particular variation lies in the potential target site of miR-34b. We show that this polymorphism is associated with resistance to miR-34b action and subsequently leads to increased α-syn protein levels. It will be interesting to investigate if this genetic variation (rs10024743) is associated with increased risk of PD.
In conclusion, our data suggest that the reduction in miR-34b and miR-34c levels in PD patients’ brains could result in increased α-syn levels and the formation of α-syn-containing aggregates. We also demonstrate that a polymorphism in the miR-34b target sequence of the α-syn 3’-UTR that diminishes the repressive activity of this miR can lead to increased α-syn levels. Thus, dysregulation of these miRs as well as sequence variations in their target sequence might modulate disease risk.
Supplementary Material
SH-SY5Y cells were transfected with pre-miR-34b and pre-miR-34c for 48 hours and total RNA was extracted. β-synuclein mRNA was quantified using Quantitative RT-PCR and values are normalized to 18s rRNA. Data are shown as mean ± SD. ***p<0.001.
After transfection of respective miRs with 48 hours, SH-SY5Y cells were lysed and Western Blot analysis was performed using anti-α-syn antibody. Band intensities were quantified from triplicated samples, and a representative blot is shown. Data are shown as mean ± SD. ***p<0.001.
Highlights.
miR-34b and miR-34c repress α-syn expression by targeting the 3’-UTR of its mRNA.
Inhibition of miR-34b or miR-34c enhances α-syn expression and aggregate formation.
SNP in the 3’-UTR reduces miR-34b-mediated repression of α-syn.
Acknowledgements
This work was supported by NIH grant NS070898 (E.J). M.M.M. is supported by NIH grants NS073994 and AT006868.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Spillantini MG, Crowther RA, Jakes R, Hasegawa M, Goedert M. alpha-Synuclein in filamentous inclusions of Lewy bodies from Parkinson's disease and dementia with lewy bodies. Proceedings of the National Academy of Sciences of the United States of America. 1998;95:6469–6473. doi: 10.1073/pnas.95.11.6469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Narhi L, Wood SJ, Steavenson S, Jiang Y, Wu GM, Anafi D, Kaufman SA, Martin F, Sitney K, Denis P, Louis JC, Wypych J, Biere AL, Citron M. Both familial Parkinson's disease mutations accelerate alpha-synuclein aggregation. The Journal of biological chemistry. 1999;274:9843–9846. doi: 10.1074/jbc.274.14.9843. [DOI] [PubMed] [Google Scholar]
- 3.Singleton AB, Farrer M, Johnson J, Singleton A, Hague S, Kachergus J, Hulihan M, Peuralinna T, Dutra A, Nussbaum R, Lincoln S, Crawley A, Hanson M, Maraganore D, Adler C, Cookson MR, Muenter M, Baptista M, Miller D, Blancato J, Hardy J, Gwinn-Hardy K. alpha-Synuclein locus triplication causes Parkinson's disease. Science. 2003;302:841. doi: 10.1126/science.1090278. [DOI] [PubMed] [Google Scholar]
- 4.Farrer M, Kachergus J, Forno L, Lincoln S, Wang DS, Hulihan M, Maraganore D, Gwinn-Hardy K, Wszolek Z, Dickson D, Langston JW. Comparison of kindreds with parkinsonism and alpha-synuclein genomic multiplications. Annals of neurology. 2004;55:174–179. doi: 10.1002/ana.10846. [DOI] [PubMed] [Google Scholar]
- 5.Maraganore DM, de Andrade M, Elbaz A, Farrer MJ, Ioannidis JP, Kruger R, Rocca WA, Schneider NK, Lesnick TG, Lincoln SJ, Hulihan MM, Aasly JO, Ashizawa T, Chartier-Harlin MC, Checkoway H, Ferrarese C, Hadjigeorgiou G, Hattori N, Kawakami H, Lambert JC, Lynch T, Mellick GD, Papapetropoulos S, Parsian A, Quattrone A, Riess O, Tan EK, Van Broeckhoven C and Genetic Epidemiology of Parkinson's Disease, C. Collaborative analysis of alpha-synuclein gene promoter variability and Parkinson disease. Jama. 2006;296:661–670. doi: 10.1001/jama.296.6.661. [DOI] [PubMed] [Google Scholar]
- 6.Schlaudraff F, Grundemann J, Fauler M, Dragicevic E, Hardy J, Liss B. Orchestrated increase of dopamine and PARK mRNAs but not miR-133b in dopamine neurons in Parkinson's disease. Neurobiology of aging. 2014;35:2302–2315. doi: 10.1016/j.neurobiolaging.2014.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Grundemann J, Schlaudraff F, Haeckel O, Liss B. Elevated alpha-synuclein mRNA levels in individual UV-laser-microdissected dopaminergic substantia nigra neurons in idiopathic Parkinson's disease. Nucleic acids research. 2008;36:e38. doi: 10.1093/nar/gkn084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yelamanchili SV, Fox HS. Defining larger roles for "tiny" RNA molecules: role of miRNAs in neurodegeneration research. Journal of neuroimmune pharmacology. 2010;5:63–69. doi: 10.1007/s11481-009-9172-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Junn E, Mouradian MM. MicroRNAs in neurodegenerative diseases and their therapeutic potential. Pharmacology & therapeutics. 2012;133:142–150. doi: 10.1016/j.pharmthera.2011.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Harraz MM, Dawson TM, Dawson VL. MicroRNAs in Parkinson's disease. Journal of chemical neuroanatomy. 2011;42:127–130. doi: 10.1016/j.jchemneu.2011.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Qiu L, Zhang W, Tan EK, Zeng L. Deciphering the Function and Regulation of microRNAs in Alzheimer's Disease and Parkinson's Disease. ACS chemical neuroscience. 2014 doi: 10.1021/cn500149w. [DOI] [PubMed] [Google Scholar]
- 12.Junn E, Lee KW, Jeong BS, Chan TW, Im JY, Mouradian MM. Repression of alpha-synuclein expression and toxicity by microRNA-7. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:13052–13057. doi: 10.1073/pnas.0906277106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Doxakis E. Post-transcriptional regulation of alpha-synuclein expression by mir-7 and mir-153. The Journal of biological chemistry. 2010;285:12726–12734. doi: 10.1074/jbc.M109.086827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Minones-Moyano E, Porta S, Escaramis G, Rabionet R, Iraola S, Kagerbauer B, Espinosa-Parrilla Y, Ferrer I, Estivill X, Marti E. MicroRNA profiling of Parkinson's disease brains identifies early downregulation of miR-34b/c which modulate mitochondrial function. Human molecular genetics. 2011;20:3067–3078. doi: 10.1093/hmg/ddr210. [DOI] [PubMed] [Google Scholar]
- 15.Villar-Menendez I, Porta S, Buira SP, Pereira-Veiga T, Diaz-Sanchez S, Albasanz JL, Ferrer I, Martin M, Barrachina M. Increased striatal adenosine A2A receptor levels is an early event in Parkinson's disease-related pathology and it is potentially regulated by miR-34b. Neurobiology of disease. 2014;69:206–214. doi: 10.1016/j.nbd.2014.05.030. [DOI] [PubMed] [Google Scholar]
- 16.Rehmsmeier M, Steffen P, Hochsmann M, Giegerich R. Fast and effective prediction of microRNA/target duplexes. Rna. 2004;10:1507–1517. doi: 10.1261/rna.5248604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cardo LF, Coto E, de Mena L, Ribacoba R, Moris G, Menendez M, Alvarez V. Profile of microRNAs in the plasma of Parkinson's disease patients and healthy controls. Journal of neurology. 2013;260:1420–1422. doi: 10.1007/s00415-013-6900-8. [DOI] [PubMed] [Google Scholar]
- 18.Martins M, Rosa A, Guedes LC, Fonseca BV, Gotovac K, Violante S, Mestre T, Coelho M, Rosa MM, Martin ER, Vance JM, Outeiro TF, Wang L, Borovecki F, Ferreira JJ, Oliveira SA. Convergence of miRNA expression profiling, alpha-synuclein interacton and GWAS in Parkinson's disease. PloS one. 2011;6:e25443. doi: 10.1371/journal.pone.0025443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Asikainen S, Rudgalvyte M, Heikkinen L, Louhiranta K, Lakso M, Wong G, Nass R. Global microRNA expression profiling of Caenorhabditis elegans Parkinson's disease models. Journal of molecular neuroscience : MN. 2010;41:210–218. doi: 10.1007/s12031-009-9325-1. [DOI] [PubMed] [Google Scholar]
- 20.Kim J, Inoue K, Ishii J, Vanti WB, Voronov SV, Murchison E, Hannon G, Abeliovich A. A MicroRNA feedback circuit in midbrain dopamine neurons. Science. 2007;317:1220–1224. doi: 10.1126/science.1140481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mata IF, Shi M, Agarwal P, Chung KA, Edwards KL, Factor SA, Galasko DR, Ginghina C, Griffith A, Higgins DS, Kay DM, Kim H, Leverenz JB, Quinn JF, Roberts JW, Samii A, Snapinn KW, Tsuang DW, Yearout D, Zhang J, Payami H, Zabetian CP. SNCA variant associated with Parkinson disease and plasma alpha-synuclein level. Archives of neurology. 2010;67:1350–1356. doi: 10.1001/archneurol.2010.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yu L, Xu P, He X, Hu F, Lin Z, Zhu M, Liu Z, He L, Xu Y. SNP rs7684318 of the alpha-synuclein gene is associated with Parkinson's disease in the Han Chinese population. Brain research. 2010;1346:262–265. doi: 10.1016/j.brainres.2010.05.069. [DOI] [PubMed] [Google Scholar]
- 23.Pan F, Dong H, Ding H, Ye M, Liu W, Wu Y, Zhang X, Chen Z, Luo Y, Ding X. SNP rs356219 of the alpha-synuclein (SNCA) gene is associated with Parkinson's disease in a Chinese Han population. Parkinsonism & related disorders. 2012;18:632–634. doi: 10.1016/j.parkreldis.2012.01.025. [DOI] [PubMed] [Google Scholar]
- 24.Wang G, van der Walt JM, Mayhew G, Li YJ, Zuchner S, Scott WK, Martin ER, Vance JM. Variation in the miRNA-433 binding site of FGF20 confers risk for Parkinson disease by overexpression of alpha-synuclein. Am J Hum Genet. 2008;82:283–289. doi: 10.1016/j.ajhg.2007.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.de Mena L, Cardo LF, Coto E, Miar A, Diaz M, Corao AI, Alonso B, Ribacoba R, Salvador C, Menendez M, Moris G, Alvarez V. FGF20 rs12720208 SNP and microRNA-433 variation: no association with Parkinson's disease in Spanish patients. Neuroscience letters. 2010;479:22–25. doi: 10.1016/j.neulet.2010.05.019. [DOI] [PubMed] [Google Scholar]
- 26.Wider C, Dachsel JC, Soto AI, Heckman MG, Diehl NN, Yue M, Lincoln S, Aasly JO, Haugarvoll K, Trojanowski JQ, Papapetropoulos S, Mash D, Rajput A, Rajput AH, Gibson JM, Lynch T, Dickson DW, Uitti RJ, Wszolek ZK, Farrer MJ, Ross OA. FGF20 and Parkinson's disease: no evidence of association or pathogenicity via alpha-synuclein expression. Movement disorders. 2009;24:455–459. doi: 10.1002/mds.22442. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
SH-SY5Y cells were transfected with pre-miR-34b and pre-miR-34c for 48 hours and total RNA was extracted. β-synuclein mRNA was quantified using Quantitative RT-PCR and values are normalized to 18s rRNA. Data are shown as mean ± SD. ***p<0.001.
After transfection of respective miRs with 48 hours, SH-SY5Y cells were lysed and Western Blot analysis was performed using anti-α-syn antibody. Band intensities were quantified from triplicated samples, and a representative blot is shown. Data are shown as mean ± SD. ***p<0.001.