Abstract
With the advancement of transcriptome profiling by micro-arrays and high-throughput RNA-sequencing, transcriptome complexity and its dynamics are revealed at different levels in cardiovascular development and diseases. In this review, we will highlight the recent progress in our knowledge of cardiovascular transcriptome complexity contributed by RNA splicing, RNA editing and noncoding RNAs. The emerging importance of many of these previously under-explored aspects of gene regulation in cardiovascular development and pathology will be discussed.
Keywords: Cardiovascular diseases, Genes, Molecular biology, Signal transduction
The mature heart develops through complex cellular differentiation involving morphological and functional changes during the embryonic and postnatal periods. Each step of this process, from lineage commitment to morphogenesis, is marked by distinct changes in gene expression profiles in both cardiomyocyte and non-myocyte components.1 Earlier studies have revealed an elaborate transcriptional regulatory network driven by tissue-specific and temporally coordinated expression of transcription factors.2,3 The importance of cardiac gene regulation is underscored by the critical contribution of transcriptional dysfunction to both congenital heart diseases and the pathogenesis of heart failure. In fact, many of the same transcriptional regulators involved in cardiac development also have important roles in cardiac hypertrophy, pathological remodeling and heart failure. Therefore, understanding cardiac transcriptome dynamics and regulation has been a major focus of research in the field of cardiac biology and cardiovascular medicine. However, much of our current knowledge of cardiac gene regulation is based on dynamic changes in the composition and expression level of mRNA. With recent advancement of RNA-sequencing technology, our view of transcriptome complexity has been expanded dramatically. Novel mRNA transcripts derived from alternative splicing and editing, and previously unrecognized species of noncoding RNAs have emerged as dominant features of the mammalian transcriptome, reshaping the fundamental concept of gene regulation, genome structure and genetic/epigenetic contribution to development and diseases. This review will highlight some of the recent progress and the new concepts emerging from studies of cardiac transcriptome regulation, with particular focus on RNA splicing and editing, as well as noncoding RNAs. Considering the fact that microRNA function and regulation have been discussed extensively by several excellent reviews,4–6 we will not cover those issues except when it intercepts with the topics covered by this review.
Alternative RNA Splicing in Transcriptome Regulation
Regulatory Machinery of RNA Splicing
All multi-exon genes require coordinated splicing to generate mature RNA transcripts. Multiple mRNA species can be generated from a single gene through alternative RNA splicing events. Extensive profiling has revealed that alternative splicing is a major contributor to mRNA complexity in the mammalian transcriptome, affecting more than 94% of human transcripts.7–9 At the global transcriptome level, alternative RNA splicing is a highly regulated process associated with physiological and pathological conditions,10 including embryonic stem cell (ESC) differentiation and cancer development.11–14 However, our knowledge of RNA splicing regulation and its role in cardiac development and diseases remains very limited, especially compared with the wealth of information about transcriptional regulation.
The constitutive RNA splicing event excises the intronic sequences according to pre-demarcated exon/intron boundaries. An alternative RNA splicing event, however, can use alternative 5′ or 3′ splice sites, leading to specific exon skipping or inclusion or intron retention7,15,16 Although a basic spliceosome is responsible for constitutive RNA splicing, additional trans-acting factors and cis-acting sequence motifs are responsible for enhancing or repressing the alternative splicing events.9 The RBP superfamily includes serine/arginine-rich (SR) proteins, neuro-oncological ventral antigen (Nova) proteins, heterogeneous nuclear ribonucleoproteins (hnRNPs) and RBFox proteins.17–19 The tissue-specific and signal-regulated expression or activities of the RBPs are the key to coordinated mRNA splicing events20 (Figure 1). More importantly, the RNA splicing machinery is an integral part of gene regulation, and its function has been implicated beyond RNA splicing events to effects on RNA transcription, quality control, transportation and other post-transcriptional processes.21–25 However, very limited information is available on how these splicing factors are regulated in response to developmental or pathological cues to achieve coordinated RNA splicing in the heart.
Figure 1.
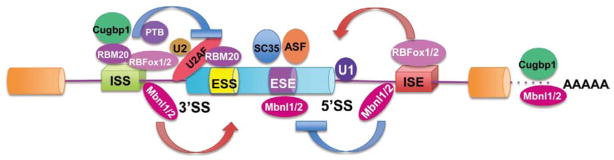
Illustration of key splicing regulators in heart. A representative alternatively spliced exon with ISS (intron splicing silence), ESS (exon splicing silence), ESE (exon splicing enhancer) and ISE (intron splicing enhancer) is shown. Trans-acting splicing regulators that have been implicated in cardiac gene regulation include: CUGBP1/2, MBNL1/2, RBFox-1/2, RBM20, PTB, SC35, ASF. Constitutive RNA splicing factors illustrated include U1/2, U2AF. Arrows indicate enhancing alternative splicing, and block indicates repressing alternative splicing of the neighboring exon.
RNA Splicing Regulation in Cardiac Development and Diseases
Salomonis et al revealed that alternative RNA splicing is important for calcium signaling and cardiomyocyte differentiation from progenitor cells,26,27 In another study, cross-talk between the microRNA regulatory network and alternative splicing is demonstrated to define transcriptome maturation during postnatal cardiac development in the mouse.28 However, a comprehensive analysis of mRNA splicing in the heart during development has not been reported. Therefore, it would be interesting to perform RNA splicing analysis at different stages of cardiac development, a task becoming increasingly feasible with more sensitive high-throughput RNA-sequencing capabilities and more sophisticated bioinformatics tools.29–31
A number of cardiac-enriched RNA splicing regulators, including muscle-blind-like protein 1 (MBNL1), RBFox2, CUG-BPI and CUG-BP2, are highly expressed during early fetal heart development but decreased postnatally. In contrast, the cardiac expression of RBFox 1 is significantly induced only after birth.32–33 On the other hand, the change in the expression of CUGBPI and CUGBP2 is directly regulated by miR-23a/b during cardiac development and this contributes to a significant number of developmentally associated splicing events in the heart.28
In additional to cardiomyocyte differentiation and development, the alternative splicing profiles in the heart are also tightly associated with the pathogenesis of heart failure. Global alternative splicing profiling has been done in the diseased heart, including cardiac hypertrophy and heart failure.34,35 For example, an earlier study compared pressure overload-induced cardiac hypertrophy and heart failure in the mouse heart using deep RNA-sequencing and revealed a global change of alternative splicing in the failing murine heart.35 More recently, Ames et al and other groups also identified a significant number of alternative splicing events during cardiac hypertrophy in rats.34,36 For splicing regulators, it is suggested that the expression of PTB and ASF/SF2 is altered in the pressure overload-induced hypertrophic heart.37 The transcriptome signature and RNA splicing events have also been profiled in human heart disease.38,39 Based on a gene expression profiling analysis, a total of 17 splicing factors were found to be upregulated in the human failing heart, including RBM25. QK1, hnRNPA1 and Tra2a.40
Genetic inactivation of ASF/SF2 in cardiomyocytes causes a hypercontractile phenotype, in part because of aberrant splicing of the Ca2+/calmodulin-dependent kinase II δ(CaMKIIδ) transcript.41 Cardiac-specific ablation of SC35 also causes dilated cardiomyopathy, associated with mis-regulated splicing of cardiac ryanodine receptor 2 (RyR2).42 Disruption of the MBNL1 gene in the mouse also leads to muscle, eye and splicing abnormities mimicking the phenotype of myotonic dystrophy.43 Lastly, as a key regulator for alternative splicing of Titin mRNA, RBM20-deficient rats develop dilated cardiomyopathy.44,45 Therefore, tissue-specific and coordinated RNA splicing events carried out by a well-orchestrated RNA splicing regulatory network are critical to normal cardiac development and physiology.
Stress Signaling in Cardiac RNA Splicing Regulation
Emerging evidence suggests that the RNA splicing machinery is also a common target of pathological signaling. In response to pressure overload, the expression level of dual-specificity tyrosine-phosphorylated and regulated kinase 1A (Dyrk1 A) is significantly upregulated, which represses the expression of the splicing factor ASF and downstream alternative splicing of CaMKIIδ.46 In addition, angiotensin II (AngII) can also affect the expression of another splicing regulator (68-kDa Src) associated during mitosis (SAM68) in the heart.47 SAM68, in turn, regulates the alternative splicing of both mTOR and SMN and plays a key role in spinal muscular atrophy.48,49 In a recent report by el Mabrouk et al,47 AngII was shown to stimulate the binding of SAM68 to a poly-U region through a PI3K-dependent pathway. Lastly, splicing factor hnRNP A1 forms a molecular complex with activated p38 in vivo, which is important for the proper intracellular localization of this splicing factor.50
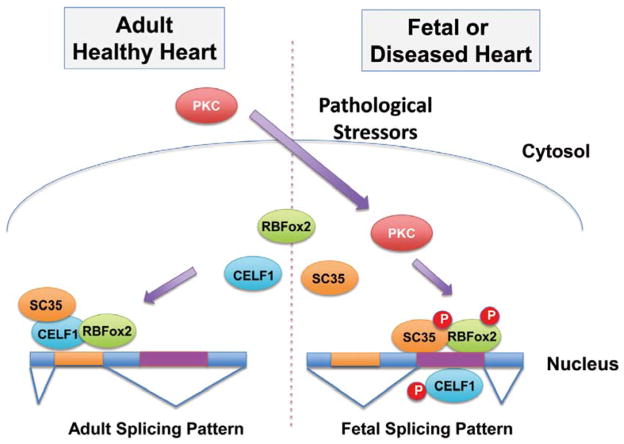
Among the different stress-signaling molecules, the best characterized regulator for RNA splicing is protein kinase C (PKC). Its regulatory role in alternative splicing was first demonstrated for the alternative splicing of Bcl-x pre-mRNA, which generates 3 distinct isoforms, Bcl-xL and Bcl-xs.51 Sarcoplasmic reticulum Ca2+-ATPase1 (SERCA1) splicing is also regulated by PKC.52 A potential underlying mechanism is through PKC-mediated modulation of the activity of an RNA splicing regulator, SC35. In the postnatal rat heart, nucleus-localized PKC can also phosphorylate and activate SC35, which is important for cardiac morphogenesis.53,54 As demonstrated by Cataldi et al, hypoxia in the adult heart can modulate the phosphorylation level of SC35 through a PKC-dependent pathway that contributes to the hypoxia-induced expression of vascular endothelial growth factor (VEGF),55 thus providing a RNA splicing dependent mechanism for hypoxia-induced neoangiogenesis (Figure 2). A recent study has demonstrated that in diabetic cardiomyopathy, a significant number of alternative splicing events switch back to an embryonic pattern. This phenotype is potentially regulated by PKCα/β via phosphorylating and up-regulating of 2 key cardiac splicing regulators: CELF1 and RBFox2.56 All together, it appears that the RNA splicing machinery is an important downstream target of stress signaling in the heart. Considering the molecular complexity of the stress-signaling network and the RNA splicing machinery, there must be more interactions between alternative RNA splicing and stress signaling waiting to be explored.
Figure 2.
PKC-mediated regulation of alternative splicing in cardiac development and diseases. PKC, protein kinase C.
Molecular Targets of Alternative RNA Splicing in the Heart
Alternative RNA splicing affects many genes in the heart. One example is SCN5A, which encodes the Na-channel α-subunit,57,58 whereby its splicing variants are associated with heart failure, arrhythmia and Brugada syndrome in humans.40,59–61 Its splicing is regulated at least in part by RBM25/LUC7L3.40 Another cardiac gene subjected to alternative splicing is KCNQ1 which generates a truncated isoform with a dominant-negative function associated with cardiac arrhythmias in both human and rat hearts.62,63 In addition, the aberrant splicing isoforms of cardiac troponin T (cTnT) has been identified in dilated cardiomyopathy.64 Both in vitro and in vivo studies have revealed that cardiac-enriched splicing regulators, including CUGBP1, CUGBP2 and MBNL,65–67 are responsible for cTnT alternative splicing at different exons. Titin has also been demonstrated to be regulated by alternative splicing during both cardiac development and disease. The inclusion level of the PEVK domain and the alternatively spliced IG domains increases in mature cardiomyocytes, generating the N2B-containing Titin isoform that is critical for cardiomyocytes’ passive tension regulation. During early development or heart failure, however, the dominant isoform of Titin switches to the N2BA isoform.68,69 This alternative splicing switch is a highly conserved splicing event across different species.45,68,70 A recent study further demonstrated that the splicing regulator RBM20 is responsible for regulating Titin alternative splicing, and a mutation of RBM20 is associated with human dilated cardiomyopathy.45,71
The Bnip3L/Nix gene also produces an alternative splicing variant: sNix, which is localized in both the cytoplasm and the nucleus. On stimulation of tumor necrosis factor α (TNFα), sNix rapidly translocates to the nucleus where it activates cardiac gene expression through the nuclear factor κ-B (NF-κB) pathway.72 Finally, transcription co-regulator PGC-1α is also subjected to alternative splicing regulation to generate a splicing variant: PGC-1α4. Instead of targeting classical PGC-1α downstream targets, this splicing variant induces expression of insulin-like growth factor and can further stimulate muscle hypertrophy both in vitro and in vivo.73
In summary, RNA splicing is a prevailing molecular event in cardiac transcriptoine programming and reprogramming during development and pathogenesis. It is an emerging area of research with the advances in high-throughput whole transcriptome profiling. The importance of RNA splicing in cardiac development and diseases is beginning to be recognized. However, our understanding of the molecular nature of the RNA splicing machinery in the heart, the regulatory mechanisms during development and pathogenesis, the molecular targets of RNA splicing as well as their functional impact remains very primitive at this time and need to be further explored in the future.
RNA Editing in Cardiac Development and Disease
RNA Editing Machinery
RNA editing refers to post-transcriptional sequence alterations in mature transcripts different from their genomic sequences.74 The types of RNA editing identified so far include nucleotide insertions, deletions and exchanges. RNA editing can result in amino acid substitutions, alternative splicing and changes in gene expression levels, leading to an expansion of transcriptome complexity.18,75 RNA editing has been suggested to play important roles during multiple biological processes and diseases, including cancer and the immune response.76 By targeting RNA editing, novel therapeutic strategies can be formulated to treat genetic disorders.77,78 With newly developed high-throughput sequencing method and bioinformatics tools, RNA editing is now recognized as a significant phenomena in transcriptome programming during tissue regeneration, including of the heart. Currently, at least 2 major proteins are implicated in RNA editing for the mammalian transcriptome. The ADARs (adenosine deaminases) can convert adenosines to inosines in double-stranded or specific structured regions of RNA.79,80 Another involves deaminating cytidine to uridine, which is carried out by the APOBEC family of cytidine deaminases.81–84 At least 2 members of the APBEC family, APOBEC1 and APOBEC2, have C (cytidine) to U (uridine) editing capability. Although APOBEC1 is ubiquitously expressed among multiple tissues, APOBEC2 is a cardiac- and skeletal muscle-specific RNA editing enzyme.85
RNA Editing of Cardiac Genes
The mRNA expression level of ADAR1 is significantly increased in the developing heart and limbs, correlating with increased proliferation and cellular remodeling.86 In a parallel study, RNA editing events were found to be significantly increased in children with cyanotic congenital heart disease and it is suggested that A-to-I RNA editing in the MED 13 gene was significantly higher among the cyanotic patients and the RNA level of ADAR2 was reduced.87 Furthermore, both the expression level and alternative splicing of ADARs are tightly regulated in the heart. ADAR2 is the key enzyme responsible for Q/R site editing in the GluR-B transcript, and ADAR2 itself is regulated by alternative splicing. Recently, a novel alternative splicing variant including part of the ADAR2 intron 7 was found to be highly expressed in heart and skeletal muscle, but not in the brain. The inclusion of this novel exon generates stop codons that potentially can affect the total ADAR2 expression.88
One of the key substrates for APOBEC 1 is the translational repressor, NAT1. APOBEC1 deaminates specific cytidine bases in NAT1 to uridine, changing a codon for glutamine into a premature stop codon. In the embryonic heart, NAT1 expression level is high in both atrial and ventricular myocytes based on immunohistochemical staining analysis; however, western blotting showed that the different isoforms of NAT1 generated by RNA editing have distinct expression levels during cardiac development.89 Interestingly, during cardiac hypertrophy, the mRNA level of NAT1 is significantly increased without an associated increase at the total protein level. However, western blot analysis confirms the presence of different protein species because of RNA editing, providing a possible link between NAT1 RNA editing regulation and cardiac hypertrophy.90 Another important substrate for APOBEC1 is apolipoprotein B (ApoB). ApoB mRNA editing involves converting a single C into U at the codon of glutamine 2153, resulting in an in-frame stop codon (UAA) and a truncated protein. The RNA editing of ApoB mRNA is tissue-specific and developmentally regulated.91 In the heart, it is suggested that the regulatory role of APOBEC1 in ApoB editing involves another chaperone regulator, Bcl2-associated athanogene-4 (BAG-4). BAG-4 is predominantly expressed in the heart and brain, and by interacting with both Hsp90 and APOBEC1. BAG-4 functions as a negative regulator for APOBEC1-mediated RNA editing by shuttling APOBEC1 from the nucleus to the cytoplasm.92 ApoB RNA editing could also be regulated by PKC.93 Further, it is suggested that, at least in vitro, increasing the extracellular calcium concentration or depleting SR calcium stores is sufficient to change the level of mRNA editing of ApoB.94 Lastly, the cardiac-enriched splicing regulator, CUGBP2, can also regulate ApoB mRNA editing. According to Anant et al,95 CUGBP2 forms a complex with APOBEC1 together with another factor, ACF, CUGBP2 binds to the AU-rich sequence located upstream of the edited cytidine in ApoB RNA and inhibits the RNA editing process.
In summary, RNA editing is a newly recognized phenomena in cardiac transcriptome programming and reprogramming. Much of the data so far offers only a correlative indication of its relevance in cardiac development and pathological process. APOBEC1 knockout mice have an abnormal lipoprotein profile because of abnormal editing of ApoB RNA, and APOBEC2 knockout leads to muscle-type switch and myopathy.96,97 While genetic evidence for other RNA editing enzymes is still lacking, the functional significance of RNA editing in cardiac development and diseases remains to be established.
Long Noncoding RNA in Cardiac Development and Disease
Genesis of Long Noncoding RNA
Although the definition of long noncoding RNA (IncRNA) continues to evolve, the generally accepted hallmarks include: (1) longer than 200 nucleotides, (2) no functional open reading frames, and (3) poor conservation at the sequence level.98 Although the concept of IncRNA was established decades ago, few studies have explored their functional significance beyond X chromosome inactivation and imprinting based on the early discovery of Xist and H19.99,100 With deep RNA-sequencing and recently developed IncRNA-sensitive microarray technologies, much more information is now available about the expression profiles and biochemical properties of the IncRNAs.101,102 The identification, annotation and predication of IncRNAs are further facilitated by recent developments in bioinformatics analysis tools,103 including IncRNAMap and NONCODEv4.104–106 In contrast to the previous notion that low copies of IncRNA may represent “transcriptional noise” because of aberrant transcriptional initiation,107 it is increasingly recognized that IncRNAs actually are expressed in a developmental and cell type-specific manner.108,109 The IncRNAs species are generally classified into 4 categories depending on their relationship to neighboring coding transcripts, including (a) overlapping with annotated gene bodies with transcription initiating from either exons or introns from the sense or (b) antisense strands, (c) lying within the cis-regulatory regions of genes, and (d) lying in the intergenic regions.110,111 Although IncRNAs are widely detected, it remains controversial to what extent their expressions are functional in development and human diseases.
LncRNAs in Cardiac Development
The expression of IncRNAs shows distinct profiles in different tissues (eg, brown adipose tissue vs. skeletal muscle)112 and in different cell types (eg, erythroblasts, megakaryocytes and mega-karyocyte-erythroid precursors).113 Although the sequence conservation of IncRNAs is relatively low compared with mRNAs and miRNAs, multiple evidence suggests that IncRNAs can also be highly conserved at the functional region. In addition to mammals, IncRNAs are also widely expressed and tightly regulated in zebrafish,114,115 pigs and tetrapods,116–118 suggesting a potentially highly conserved function among some IncRNA species. The critical role of IncRNAs in development was reported for neurogenesis, involving a large screening approach demonstrating that inactivation of IncRNAs can block human ESC differentiation into mature neurons.119
In the heart, microarray and deep RNA-sequencing analyses have revealed a large number of IncRNAs with expression profiles associated with both cardiac development and disease120–122 (Table). In a recent study,121 a total of 1,237 IncRNAs were found to have different expression levels during development, with a particular potential effect on developmental processes, metabolic processes and mTOR signaling pathways, according to gene ontology (GO) analysis of neighboring mRNAs. The first IncRNA identified to play a critical role in cardiovascular lineage commitment was Braveheart (Bvht).123–126 This 590-nt transcript was identified during screening of IncRNAs potentially important for ESC cardiac lineage commitment. Another IncRNA that also controls lineage commitment and cell differentiation in cardiomyocytes is Fendrr. 127,128 As more IncRNAs are discovered in the heart, their functional role in cardiac development will be expanded beyond gene expression to morphogenesis and growth regulation.
Table.
List of LncRNAs implicated in Cardiovascular Development and Diseases
| Category | IncRNAs | Reference |
|---|---|---|
| Upregulated IncRNAs during cardiac development | Bvht | Klattenhoff et al124 |
| Kcnq1 | Korostowski et al122 | |
| DT903035 | Zhu et al121 | |
| BC049716 | ||
| AK085135 | ||
| AK013988 | ||
| Uc008hzy.1 | ||
| Uc008mey.1 | ||
| MM9LINCRNAEXON10678 | ||
| NR_029457 | ||
|
| ||
| Downregulated IncRNAs during cardiac development | Fendrr | Grote et al127,128 |
| AK045554 | Zhu et al121 | |
| AK050713 | ||
| Uc007xf.1 | ||
| BC024929 | ||
| AK008015 | ||
| AK019733 | ||
| Gm16133 | ||
|
| ||
| Upregulated IncRNAs in diseased heart | CHRF | Wang et al139 |
| N339730 | Song et al132 | |
| N408065 | Yang et al133 | |
| KCONS_0000467 | Liu et al103 | |
| N406465 | ||
|
| ||
| Downregulated IncRNAs in diseased heart | N383233 | Song et al132 |
| N407211 | Yang et al133 | |
| N339159 | Liu et al103 | |
IncRNA, long noncoding RNA.
LncRNAs in Cardiovascular Diseases
The expression profiles of IncRNAs have been demonstrated to be dynamically regulated under different disease conditions.129–131 In a study by Song et al of human ventricular septal defects (VSD), more than 1,500 IncRNAs showed altered expression associated with the disease state.132 Moreover, changes in IncRNA expression are significantly associated with the expression of their neighboring genes, suggesting a possible cis-regulatory relationship between IncRNAs and coding mRNAs. A more recent study comparing ischemic and dilated human failing heart samples pre- and post-LVAD (left ventricular assisted device) therapy with non-failing human heart samples using deep RNA-sequencing analysis also revealed a significant number of IncRNAs with dysregulated expression in the diseased hearts and some of these IncRNAs were normalized in post-LVAD hearts.133 More interestingly, the IncRNA expression profiles in the human failing hearts were found to be a more significant indicator for the different etiologies than the mRNA profiles, suggesting a potential contribution to the different etiologies of heart failure. In addition, IncRNAs detected in the plasma of a mouse heart failure model were also significantly associated with heart failure, and thus may serve as independent biomarkers of the diseases.120
Taken together, the dynamic expression of IncRNAs during cardiac development and disease suggests a potential link between them and cardiac pathology (Table). Beyond these correlative analyses, however, the direct contribution of IncRNAs to cardiovascular diseases is only beginning to be revealed.
Molecular Mechanisms of LncRNA-Mediated Regulation
As IncRNA species proliferate, the variations in the underlying mechanisms have also expanded to involve almost every aspect of gene regulation. By binding directly to the promoter region of coding genes, or facilitating transcription factor or miR binding, the IncRNAs are able to activate/silence expression in both a cis and trans manner.134–137
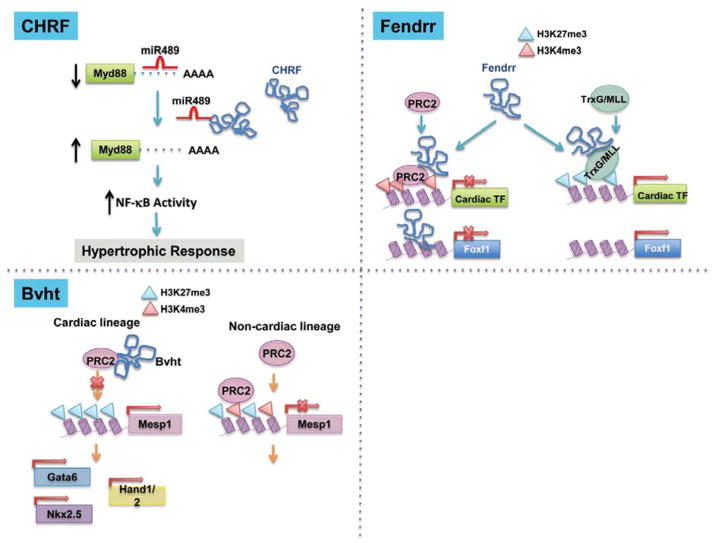
The IncRNAs can function as a functional sponge for miRNAs to interfere global gene expression.138 A recently identified IncRNA, cardiac hypertrophy related factor (CHRF),139 is one of the IncRNAs showing expression changes in AngII-treated NRVM. This 1843-nt IncRNA plays a critical role in the cardiomyocytes’ hypertrophic response by interacting directly with miR-489, which in turn regulates Myd88 expression, leading to the activation of NF-κB pathway.
Finally, IncRNAs can act as a scaffold for the histone modification complex, antagonizing the localization and regulatory function of the SWI/SNF chromatin-modifying complex or directly interacting with the polycomb repressive complex 2 (PRC2) and the TrxG/MII complex in order to modify the status of histone methylation and acetylation.127,140,141 This is one of the most important mechanisms for IncRNA regulation of cell differentiation.140 In the heart, the IncRNA, Bvht, acts as a molecular scaffold140,142 by interacting with SUZ12, a core subunit of PRC2, and together the complex regulates the promoter activity of genes critical for the cardiac lineage, including MesP1, Gata6, Hand1 and Nkx2.5 by changing the histone lysine methylation status. In contrast, Fendrr regulates downstream target expression either in -cis by binding to Foxf1 promoter or in -trans by binding to the Pitx2 promoter.
The IncRNAs are a newly recognized transcriptome component with an emerging significance in development and diseases. 143,144 Studies of IncRNAs in the heart have just begun and many questions about IncRNA function, regulation and underlying mechanisms are yet to be addressed (Figure 3). More investigations are needed to demonstrate the diagnostic and therapeutic value of IncRNAs in heart diseases.
Figure 3.
Molecular mechanisms of known long noncoding RNA functions in the heart.
Small Nucleolar RNAs and Cardiovascular Disease
During RNA metabolism, another noncoding RNA species present in the transcriptome is the small nucleolar RNAs (snoRNAs). These are a family of noncoding small RNAs playing important roles in guiding the modification of other noncoding RNAs, including ribosomal, small nuclear and transfer RNAs.145–147 Although originally considered as a biological byproducts of alternative splicing, the dynamic expression profiles of snoRNAs suggest they could have functional effects in disease, including cancer.148,149
The expression of snoRNAs associated with cardiac diseases has been demonstrated at the whole transcriptome level. By comparing right ventricular tissue samples from 16 infants with nonsyndromic tetralogy of Fallot (TOF) and 8 normal samples, more than 100 snoRNA expression profiles were identified to be significantly changed. Interestingly, it should be noted that the expression profiles of snoRNAs in the infants with TOF resembled the expression profiles observed in the fetal myocardium.150 To date, this is the first evidence of a global change in snoRNA expression associated with cardiovascular disease. As regulation of snoRNA expression in the heart is still poorly studied and the molecular mechanisms for snoRNA functions in cardiovascular diseases remain elusive. snoRNAs in the cardiovascular system should be an interesting area for future investigations.
Conclusions
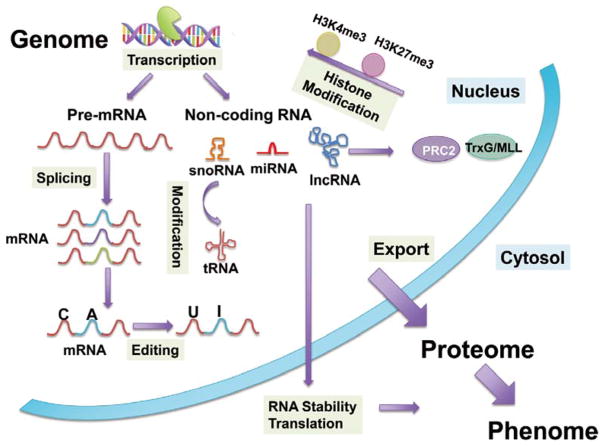
Gene regulation has been a central issue in modern molecular biology, and understanding the underlying mechanisms and functional roles is at the core of cardiac research, both in fundamental understanding for cardiac development and in translational investigation of disease mechanisms. With the rapid expansion of transcriptome complexity, the functional paths from genome to phenome have become even more complicated. The emerging roles of RNA splicing, RNA editing, IncRNAs and snoRNAs in the heart add additional intricacy to the regulatory network of cardiac gene expression, and also reveal more ways of potential perturbation in response to pathological stressors (Figure 4). Although much of the work remains to be accomplished, current progress has demonstrated the potential of these new insights of cardiac transcriptome regulation for diagnostic and therapeutic applications. Revealing and understanding cardiac transcriptome complexity should be an important goal for future efforts.
Figure 4.
Transcriptome programming contributed by RNA splicing, editing and noncoding RNA-mediated gene regulation.
References
- 1.Miquerol L, Kelly RG. Organogenesis of the vertebrate heart. Wiley Interdiscip Rev Dev Biol. 2013;2:17–29. doi: 10.1002/wdev.68. [DOI] [PubMed] [Google Scholar]
- 2.Bruneau BG. Signaling and transcriptional networks in heart development and regeneration. Cold Spring Harb Perspect Biol. 2013;5:a008292. doi: 10.1101/cshperspect.a008292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rana MS, Christoffels VM, Moorman AF. A molecular and genetic outline of cardiac morphogenesis. Acta Physiol (Oxf) 2013;207:588–615. doi: 10.1111/apha.12061. [DOI] [PubMed] [Google Scholar]
- 4.Condorelli G, Latronico MV, Cavarretta E. microRNAs in cardiovascular diseases: Current knowledge and the road ahead. J Am Coll Cardiol. 2014 Feb 26; doi: 10.1016/j.jacc.2014.01,050.. [DOI] [PubMed] [Google Scholar]
- 5.Boettger T, Braun T. A new level of complexity: The role of microRNAs in cardiovascular development. Circ Res. 2012;110:1000–1013. doi: 10.1161/CIRCRESAHA.111.247742. [DOI] [PubMed] [Google Scholar]
- 6.Da Costa Martins PA, De Windt LJ. MicroRNAs in control of cardiac hypertrophy. Cardiovasc Res. 2012;93:563–572. doi: 10.1093/cvr/cvs013. [DOI] [PubMed] [Google Scholar]
- 7.Irimia M, Blencowe BJ. Alternative splicing: Decoding an expansive regulatory layer. Curr Opin Cell Biol. 2012;24:323–332. doi: 10.1016/j.ceb.2012.03.005. [DOI] [PubMed] [Google Scholar]
- 8.Hallegger M, Llorian M, Smith CW. Alternative splicing: Global insights. FEBS J. 2010;277:856–866. doi: 10.1111/j.1742-4658.2009.07521.x. [DOI] [PubMed] [Google Scholar]
- 9.Witten JT, Ule J. Understanding splicing regulation through RNA splicing maps. Trends Genet. 2011;27:89–97. doi: 10.1016/j.tig.2010.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kalsotra A, Cooper TA. Functional consequences of developmentally regulated alternative splicing. Nat Rev Genet. 2011;12:715–729. doi: 10.1038/nrg3052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.David CJ, Manley JL. Alternative pre-mRNA splicing regulation in cancer: pathways and programs unhinged. Genes Dev. 2010;24:2343–2364. doi: 10.1101/gad.1973010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ohta S, Nishida E, Yamanaka S, Yamamoto T. Global splicing pattern reversion during somatic cell reprogramming. Cell Rep. 2013;5:357–366. doi: 10.1016/j.celrep.2013.09.016. [DOI] [PubMed] [Google Scholar]
- 13.Venables JP, Lapasset L, Gadea G, Fort P, Klinck R, Irimia M, et al. MBNL1 and RBFOX2 cooperate to establish a splicing programme involved in pluripotent stem cell differentiation. Nat Commun. 2013;4:2480. doi: 10.1038/ncomms3480. [DOI] [PubMed] [Google Scholar]
- 14.Choudhury R, Roy SG, Tsai YS, Tripathy A, Graves LM, Wang Z. The splicing activator DAZAP1 integrates splicing control into MEK/Erk-regulated cell proliferation and migration. Nat Commun. 2014;5:3078. doi: 10.1038/ncomms4078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McManus CJ, Graveley BR. RNA structure and the mechanisms of alternative splicing. Curr Opin Genet Dev. 2011;21:373–379. doi: 10.1016/j.gde.2011.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen M, Manley JL. Mechanisms of alternative splicing regulation: Insights from molecular and genomics approaches. Nat Rev Mol Cell Biol. 2009;10:741–754. doi: 10.1038/nrm2777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Han SP, Tang YH, Smith R. Functional diversity of the hnRNPs: Past, present and perspectives. Biochem J. 2010;430:379–392. doi: 10.1042/BJ20100396. [DOI] [PubMed] [Google Scholar]
- 18.Licatalosi DD, Darnell RB. Applications of next-generation sequencing RNA processing and its regulation: Global insights into biological networks. Nat Rev Genet. 2010;11:75–87. doi: 10.1038/nrg2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shepard PJ, Hertel KJ. The SR protein family. Genome Biol. 2009;10:242. doi: 10.1186/gb-2009-10-10-242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xiao X, Lee JH. Systems analysis of alternative splicing and its regulation. Wiley Interdiscip Rev Syst Biol Med. 2010;2:550–565. doi: 10.1002/wsbm.84. [DOI] [PubMed] [Google Scholar]
- 21.Mathieu O, Bouche N. Interplay between chromatin and RNA processing. Curr Opin Plant Biol. 2014;18C:60–65. doi: 10.1016/j.pbi.2014.02.006. [DOI] [PubMed] [Google Scholar]
- 22.Zlotorynski E. Transcription: Splicing keeps RNA polymerase II in check. Nat Rev Mol Cell Biol. 2014;15:222. doi: 10.1038/nrm3774. [DOI] [PubMed] [Google Scholar]
- 23.Yap K, Makeyev EV. Regulation of gene expression in mammalian nervous system through alternative pre-mRNA splicing coupled with RNA quality control mechanisms. Mol Cell Neurosci. 2013;56:420–428. doi: 10.1016/j.mcn.2013.01.003. [DOI] [PubMed] [Google Scholar]
- 24.Takemura R, Takeiwa T, Taniguchi I, McCloskey A, Ohno M. Multiple factors in the early splicing complex are involved in the nuclear retention of pre-mRNAs in mammalian cells. Genes Cells. 2011;16:1035–1049. doi: 10.1111/j.1365-2443.2011.01548.x. [DOI] [PubMed] [Google Scholar]
- 25.Escudero-Paunetto L, Li L, Hernandez FP, Sandri-Goldin RM. SR proteins SRp20 and 9G8 contribute to efficient export of herpes simplex virus 1 mRNAs. Virology. 2010;401:155–164. doi: 10.1016/j.virol.2010.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Salomonis N, Nelson B, Vranizan K, Pico AR, Hanspers K, Kuchinsky A, et al. Alternative splicing in the differentiation of human embryonic stem cells into cardiac precursors. PLoS Comput Biol. 2009;5:el000553. doi: 10.1371/journal.pcbi.1000553.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Salomonis N, Schlieve CR, Pereira L, Wahlquist C, Colas A, Zambon AC, et al. Alternative splicing regulates mouse embryonic stem cell pluripotency and differentiation. Proc Natl Acad Sci USA. 2010;107:10514–10519. doi: 10.1073/pnas.0912260107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kalsotra A, Wang K, Li PF, Cooper TA. MicroRNAs coordinate an alternative splicing network during mouse postnatal heart development. Genes Dev. 2010;24:653–658. doi: 10.1101/gad.1894310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vitting-Seerup K, Porse BT, Sandelin A, Waage J. spliceR: An R package for classification of alternative splicing and prediction of coding potential from RNA-seq data. BMC Bioinformatics. 2014;15:81. doi: 10.1186/1471-2105-15-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gatto A, Torroja-Fungairino C, Mazzarotto F, Cook SA, Barton PJ, Sanchez-Cabo F, et al. FineSplice, enhanced splice junction detection and quantification: A novel pipeline based on the assessment of diverse RNA-Seq alignment solutions. Nucleic Acids Res. 2014 Feb 25; doi: 10.1093/nar/gku166.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Boley N, Stoiber MH, Booth BW, Wan KH, Hoskins RA, Bickel PJ, et al. Genome-guided transcript assembly by integrative analysis of RNA sequence data. Nat Biotechnol. 2014;32:341–346. doi: 10.1038/nbt.2850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kalsotra A, Xiao X, Ward AJ, Castle JC, Johnson JM, Burge CB, et al. A postnatal switch of CELF and MBNL proteins reprograms alternative splicing in the developing heart. Proc Natl Acad Sci USA. 2008;105:20333–20338. doi: 10.1073/pnas.0809045105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ladd AN, Stenberg MG, Swanson MS, Cooper TA. Dynamic balance between activation and repression regulates pre-mRNA alternative splicing during heart development. Dev Dyn. 2005;233:783–793. doi: 10.1002/dvdy.20382. [DOI] [PubMed] [Google Scholar]
- 34.Ames EG, Lawson MJ, Mackey AJ, Holmes JW. Sequencing of mRNA identifies re-expression of fetal splice variants in cardiac hypertrophy. J Mol Cell Cardiol. 2013;62:99–107. doi: 10.1016/j.yjmcc.2013.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lee JH, Gao C, Peng G, Greer C, Ren S, Wang Y, et al. Analysis of transcriptome complexity through RNA sequencing in normal and failing murine hearts. Circ Res. 2011;109:1332–1341. doi: 10.1161/CIRCRESAHA.111.249433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Park JY, Li W, Zheng D, Zhai P, Zhao Y, Matsuda T, et al. Comparative analysis of mRNA isoform expression in cardiac hypertrophy and development reveals multiple post-transcriptional regulatory modules. PLoS One. 2011;6:e2239l. doi: 10.1371/journal.pone.0022391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kim T, Kim JO, Oh JG, Hong SE, Kim do H. Pressure-overload cardiac hypertrophy is associated with distinct alternative splicing due to altered expression of splicing factors. Mol Cells. 2014;37:81–87. doi: 10.14348/molcells.2014.2337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Joehanes R, Ying S, Huan T, Johnson AD, Raghavachari N, Wang R, et al. Gene expression signatures of coronary heart disease. Arterioscler Thromb Vasc Biol. 2013;33:1418–1426. doi: 10.1161/ATVBAHA.112.301169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ricci M, Xu Y, Hammond HL, Willoughby DA, Nathanson L, Rodriguez MM, et al. Myocardial alternative RNA splicing and gene expression profiling in early stage hypoplastic left heart syndrome. PLoS One. 2012;7:e29784. doi: 10.1371/journal.pone.0029784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gao G, Xie A, Huang SC, Zhou A, Zhang J, Herman AM, et al. Role of RBM25/LUC7L3 in abnormal cardiac sodium channel splicing regulation in human heart failure. Circulation. 2011;124:1124–1131. doi: 10.1161/CIRCULATIONAHA.111.044495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xu X, Yang D, Ding JH, Wang W, Chu PH, Dalton ND, et al. ASF/SF2-regulated CaMKIIdelta alternative splicing temporally reprograms excitation-contraction coupling in cardiac muscle. Cell. 2005;120:59–72. doi: 10.1016/j.cell.2004.11.036. [DOI] [PubMed] [Google Scholar]
- 42.Ding JH, Xu X, Yang D, Chu PH, Dalton ND, Ye Z, et al. Dilated cardiomyopathy caused by tissue-specific ablation of SC35 in the heart. EMBO J. 2004;23:885–896. doi: 10.1038/sj.emboj.7600054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kanadia RN, Johnstone KA, Mankodi A, Lungu C, Thornton CA, Esson D, et al. A muscleblind knockout model for myotonic dystrophy. Science. 2003;302:1978–1980. doi: 10.1126/science.1088583. [DOI] [PubMed] [Google Scholar]
- 44.Guo W, Pleitner JM, Saupe KW, Greaser ML. Pathophysiological defects and transcriptional profiling in the RBM20−/− rat model. PLoS One. 2013;8:e84281. doi: 10.1371/journal.pone.0084281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Guo W, Schafer S, Greaser ML, Radke MH, Liss M, Govindarajan T, et al. RBM20, a gene for hereditary cardiomyopathy, regulates titin splicing. Nat Med. 2012;18:766–773. doi: 10.1038/nm.2693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yao JSH, Lu XC, Gu QQ, Zhu JH. Metoprolol attenuaes pressure overload-induced myocardial hypertrophy through modulating Dryk1A-ASF-CaMKIId signaling pathways. Zhonghua Xin Xue Guan Bing Za Zhi. 2013;41:1029–1033. [PubMed] [Google Scholar]
- 47.El Mabrouk M, Diep QN, Benkirane K, Touyz RM, Schiffrin EL. SAM68: A downstream target of angiotensin II signaling in vascular smooth muscle cells in genetic hypertension. Am J Physiol Heart Circ Physiol. 2004;286:H1954–H1962. doi: 10.1152/ajpheart.00134.2003. [DOI] [PubMed] [Google Scholar]
- 48.Huot ME, Vogel G, Zabarauskas A, Ngo CT, Coulombe-Huntington J, Majewski J, et al. The Sam68 STAR RNA-binding protein regulates mTOR alternative splicing during adipogenesis. Mol Cell. 2012;46:187–199. doi: 10.1016/j.molcel.2012.02.007. [DOI] [PubMed] [Google Scholar]
- 49.Pedrotti S, Bielli P, Paronetto MP, Ciccosanti F, Fimia GM, Stamm S, et al. The splicing regulator Sam68 binds to a novel exonic splicing silencer and functions in SMN2 alternative splicing in spinal muscular atrophy. EMBO J. 2010;29:1235–1247. doi: 10.1038/emboj.2010.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shimada N, Rios I, Moran H, Sayers B, Hubbard K. p38 MAP kinase-dependent regulation of the expression level and subcellular distribution of heterogeneous nuclear ribonucleoprotein A1 and its involvement in cellular senescence in normal human fibroblasts. RNA Biol. 2009;6:293–304. doi: 10.4161/rna.6.3.8497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Revil T, Toutant J, Shkreta L, Garneau D, Cloutier P, Chabot B. Protein kinase C-dependent control of Bcl-x alternative splicing. Mol Cell Biol. 2007;27:8431–8441. doi: 10.1128/MCB.00565-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhao Y, Koebis M, Suo S, Ohno S, Ishiura S. Regulation of the alternative splicing of sarcoplasmic reticulum Ca(2)(+)-ATPase 1 (SERCA1) by phorbol 12-myristate 13-acetate (PMA) via a PKC pathway. Biochem Biophys Res Commun. 2012;423:212–217. doi: 10.1016/j.bbrc.2012.05.033. [DOI] [PubMed] [Google Scholar]
- 53.Zara S, Bosco D, Di Giulio C, Antonucci A, Cataldi A. Protein kinase Calpha early activates splicing factor SC-35 during postnatal rat heart development. J Biol Regul Homeost Agents. 2009;23:45–54. [PubMed] [Google Scholar]
- 54.Zara SFM, Rapino M, Zago M, Orsini G, Mazzotti G, Cataldi A, Teti G. pPKCd activates SC35 splicing factor during H9c2 myoblastic differentiation. Histol Histopathol. 2011;26:59–69. doi: 10.14670/HH-26.59. [DOI] [PubMed] [Google Scholar]
- 55.Cataldi A, Zingariello M, Rapino M, Zara S, Daniele F, Di Giulio C, et al. Effect of hypoxia and aging on PKC delta-mediated SC-35 phosphorylation in rat myocardial tissue. Anat Rec (Hoboken) 2009;292:1135–1142. doi: 10.1002/ar.20936. [DOI] [PubMed] [Google Scholar]
- 56.Verma SK, Deshmukh V, Liu P, Nutter CA, Espejo R, Hung ML, et al. Reactivation of fetal splicing programs in diabetic hearts is mediated by protein kinase C signaling. J Biol Chem. 2013;288:35372–35386. doi: 10.1074/jbc.M113.507426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.van Stuijvenberg L, Yildrim C, Kok BG, van Veen TA, Varro A, Winckels SK, et al. Alternative promoter usage and splicing of the human SCN5A gene contribute to transcript heterogeneity. DNA Cell Biol. 2010;29:577–587. doi: 10.1089/dna.2009.0999. [DOI] [PubMed] [Google Scholar]
- 58.Murphy LL, Moon-Grady AJ, Cuneo BF, Wakai RT, Yu S, Kunic JD, et al. Developmentally regulated SCN5A splice variant potentiates dysfunction of a novel mutation associated with severe fetal arrhythmia. Heart Rhythm. 2012;9:590–597. doi: 10.1016/j.hrthm.2011.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gao G, Dudley SC., Jr SCN5A splicing variants and the possibility of predicting heart failure-associated arrhythmia. Expert Rev Cardiovasc Ther. 2013;11:117–119. doi: 10.1586/erc.12.180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wahbi K, Algalarrondo V, Becane HM, Fressart V, Beldjord C, Azibi K, et al. Brugada syndrome and abnormal splicing of SCN5A in myotonic dystrophy type 1. Arch Cardiovasc Dis. 2013;106:635–643. doi: 10.1016/j.acvd.2013.08.003. [DOI] [PubMed] [Google Scholar]
- 61.Gao G, Dudley SC., Jr RBM25/LUC7L3 function in cardiac sodium channel splicing regulation of human heart failure. Trends Cardiovasc Med. 2013;23:5–8. doi: 10.1016/j.tcm.2012.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yamada Y, Chen X, Kobayashi T, Kamada Y, Nagashima M, Tsutsuura M, et al. A truncated splice variant of KCNQ1 cloned from rat heart. Biochem Biophys Res Commun. 2002;294:199–204. doi: 10.1016/S0006-291X(02)00459-X. [DOI] [PubMed] [Google Scholar]
- 63.Mohammad-Panah R, Demolombe S, Neyroud N, Guicheney P, Kyndt F, van den Hoff M, et al. Mutations in a dominant-negative isoform correlate with phenotype in inherited cardiac arrhythmias. Am J Hum Genet. 1999;64:1015–1023. doi: 10.1086/302346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Biesiadecki BJ, Elder BD, Yu ZB, Jin JP. Cardiac troponin T variants produced by aberrant splicing of multiple exons in animals with high instances of dilated cardiomyopathy. J Biol Chem. 2002;277:50275–50285. doi: 10.1074/jbc.M206369200. [DOI] [PubMed] [Google Scholar]
- 65.Warf MB, Berglund JA. MBNL binds similar RNA structures in the CUG repeats of myotonic dystrophy and its pre-mRNA substrate cardiac troponin T. RNA. 2007;13:2238–2251. doi: 10.1261/rna.610607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ho TH, Bundman D, Armstrong DL, Cooper TA. Transgenic mice expressing CUG-BP1 reproduce splicing mis-regulation observed in myotonic dystrophy. Hum Mol Genet. 2005;14:1539–1547. doi: 10.1093/hmg/ddi162. [DOI] [PubMed] [Google Scholar]
- 67.Goo YH, Cooper TA. CUGBP2 directly interacts with U217S snRNP components and promotes U2 snRNA binding to cardiac troponin T pre-mRNA. Nucleic Acids Res. 2009;37:4275–4286. doi: 10.1093/nar/gkp346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Warren CM, Krzesinski PR, Campbell KS, Moss RL, Greaser ML. Titin isoform changes in rat myocardium during development. Mech Dev. 2004;121:1301–1312. doi: 10.1016/j.mod.2004.07.003. [DOI] [PubMed] [Google Scholar]
- 69.Guo W, Bharmal SJ, Esbona K, Greaser ML. Titin diversity--alternative splicing gone wild. J Biomed Biotechnol. 2010 Mar 21; doi: 10.1155/2010/753675.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Nelson OL, Robbins CT, Wu Y, Granzier H. Titin isoform switching is a major cardiac adaptive response in hibernating grizzly bears. Am J Physiol Heart Circ Physiol. 2008;295:H366–H371. doi: 10.1152/ajpheart.00234.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Li S, Guo W, Dewey CN, Greaser ML. Rbm20 regulates titin alternative splicing as a splicing repressor. Nucleic Acids Res. 2013;41:2659–2672. doi: 10.1093/nar/gks1362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Chen Y, Decker KF, Zheng D, Matkovich SJ, Jia L, Dorn GW., 2nd A nucleus-targeted alternately spliced Nix/Bnip3L protein isoform modifies nuclear factor kappaB (NFkappaB)-mediated cardiac transcription. J Biol Chem. 2013;288:15455–15465. doi: 10.1074/jbc.M113.452342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ruas JL, White JP, Rao RR, Kleiner S, Brannan KT, Harrison BC, et al. A PGC-la isoform induced by resistance training regulates skeletal muscle hypertrophy. Cell. 2012;151:1319–1331. doi: 10.1016/j.cell.2012.10.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Chateigner-Boutin AL, Small I. Organellar RNA editing. Wiley Interdiscip Rev RNA. 2011;2:493–506. doi: 10.1002/wrna.72. [DOI] [PubMed] [Google Scholar]
- 75.Farajollahi S, Maas S. Molecular diversity through RNA editing: A balancing act. Trends Genet. 2010;26:221–230. doi: 10.1016/j.tig.2010.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Galeano F, Tomaselli S, Locatelli F, Gallo A. A-to-I RNA editing: The “ADAR” side of human cancer. Semin Cell Dev Biol. 2012;23:244–250. doi: 10.1016/j.semcdb.2011.09.003. [DOI] [PubMed] [Google Scholar]
- 77.Decher N, Netter MF, Streit AK. Putative impact of RNA editing on drug discovery. Chem Biol Drug Des. 2013;81:13–21. doi: 10.1111/cbdd.12045. [DOI] [PubMed] [Google Scholar]
- 78.Athanasiadis A. Zalpha-domains: At the intersection between RNA editing and innate immunity. Semin Cell Dev Biol. 2012;23:275–280. doi: 10.1016/j.semcdb.2011.11.001. [DOI] [PubMed] [Google Scholar]
- 79.Pullirsch D, Jantsch MF. Proteome diversification by adenosine to inosine RNA editing. RNA Biol. 2010;7:205–212. doi: 10.4161/rna.7.2.11286. [DOI] [PubMed] [Google Scholar]
- 80.Nishikura K. Functions and regulation of RNA editing by ADAR deaminases. Annu Rev Biochem. 2010;79:321–349. doi: 10.1146/annurev-biochem-060208-105251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Conticello SG. The AID/APOBEC family of nucleic acid mutators. Genome Biol. 2008;9:229. doi: 10.1186/gb-2008-9-6-229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Conticello SG, Thomas CJ, Petersen-Mahrt SK, Neuberger MS. Evolution of the AID/APOBEC family of polynucleotide (deoxy) cytidine deaminases. Mol Biol Evol. 2005;22:367–377. doi: 10.1093/molbev/msi026. [DOI] [PubMed] [Google Scholar]
- 83.Roberts SA, Lawrence MS, Klimczak LJ, Grimm SA, Fargo D, Stojanov P, et al. An APOBEC cytidine deaminase mutagenesis pattern is widespread in human cancers. Nat Genet. 2013;45:970–976. doi: 10.1038/ng.2702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Smith HC, Bennett RP, Kizilyer A, McDougall WM, Prohaska KM. Functions and regulation of the APOBEC family of proteins. Semin Cell Dev Biol. 2012;23:258–268. doi: 10.1016/j.semcdb.2011.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Liao W, Hong SH, Chan BH, Rudolph FB, Clark SC, Chan L. APOBEC-2, a cardiac- and skeletal muscle specific member of the cytidine deaminase supergene family. Biochem Biophys Res Commun. 1999;260:398–404. doi: 10.1006/bbrc.1999.0925. [DOI] [PubMed] [Google Scholar]
- 86.Witman NM, Behm M, Ohman M, Morrison JI. ADAR-related acivation of adenosine-to-inosine RNA editing during regeneration. Stem Cells Dev. 2013;22:2254–2267. doi: 10.1089/scd.2013.0104. [DOI] [PubMed] [Google Scholar]
- 87.Borik S, Simon AJ, Nevo-Caspi Y, Mishali D, Amariglio N, Rechavi G, et al. Increased RNA editing in children with cyanotic congenital heart disease. Intensive Care Med. 2011;37:1664–1671. doi: 10.1007/s00134-011-2296-z. [DOI] [PubMed] [Google Scholar]
- 88.Agranat L, Sperling J, Sperling R. A novel tissue-specific alternatively spliced form of the A-to-I RNA editing enzyme ADAR2. RNA Biol. 2010;7:253–262. doi: 10.4161/rna.7.2.11568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Pak BJ, Pang SC. Developmental regulation of the translational repressor NAT1 during cardiac development. J Mol Cell Cardiol. 1999;31:1717–1724. doi: 10.1006/jmcc.1999.1008. [DOI] [PubMed] [Google Scholar]
- 90.Sangaralingham SJ, Pak BJ, Tse MY, Angelis E, Adams MA, Smallegange C, et al. Expression of the translational repressor NAT1 in experimental models of cardiac hypertrophy. Mol Cell Biochem. 2003;245:183–190. doi: 10.1023/a:1022884515544. [DOI] [PubMed] [Google Scholar]
- 91.Funahashi T, Giannoni F, DePaoli AM, Skarosi SF, Davidson NO. Tissue-specific, developmental and nutritional regulation of the gene encoding the catalytic subunit of the rat apolipoprotein B mRNA editing enzyme: Functional role in the modulation of apoB mRNA editing. J Lipid Res. 1995;36:414–428. [PubMed] [Google Scholar]
- 92.Lau PP, Chan L. Involvement of a chaperone regulator, Bcl2-associated athanogene-4, in apolipoprotein B mRNA editing. J Biol Chem. 2003;278:52988–52996. doi: 10.1074/jbc.M310153200. [DOI] [PubMed] [Google Scholar]
- 93.Chen ZG, Eggerman TL, Patterson AP. Phosphorylation is a regulatory mechanism in apolipoprotein B mRNA editing. Biochem J. 2001;357:661–672. doi: 10.1042/0264-6021:3570661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Chen Z, Eggerman TL, Potosky D, Arborati M, Patterson AP. Calcium increases apolipoprotein B mRNA editing. Biochem Biophys Res Commun. 2000;277:221–227. doi: 10.1006/bbrc.2000.3668. [DOI] [PubMed] [Google Scholar]
- 95.Anant S, Henderson JO, Mukhopadhyay D, Navaratnam N, Kennedy S, Min J, et al. Novel role for RNA-binding protein CUGBP2 in mammalian RNA editing: CUGBP2 modulates C to U editing of apolipoprotein B mRNA by interacting with apobec-1 and ACF, the apobec-1 complementation factor. J Biol Chem. 2001;276:47338–47351. doi: 10.1074/jbc.M104911200. [DOI] [PubMed] [Google Scholar]
- 96.Sato Y, Probst HC, Tatsumi R, Ikeuchi Y, Neuberger MS, Rada C. Deficiency in APOBEC2 leads to a shift in muscle fiber type, diminished body mass, and myopathy. J Biol Chem. 2010;285:7111–7118. doi: 10.1074/jbc.M109.052977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Nakamuta M, Chang BH, Zsigmond E, Kobayashi K, Lei H, Ishida BY, et al. Complete phenotypic characterization of apobec-1 knockout mice with a wild-type genetic background and a human apolipoprotein B transgenic background, and restoration of apolipoprotein B mRNA editing by somatic gene transfer of Apobec-1. J Biol Chem. 1996;271:25981–25988. doi: 10.1074/jbc.271.42.25981. [DOI] [PubMed] [Google Scholar]
- 98.Guttman M, Amit I, Garber M, French C, Lin MF, Feldser D, et al. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature. 2009;458:223–227. doi: 10.1038/nature07672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Penny GD, Kay GF, Sheardown SA, Rastan S, Brockdorff N. Requirement for Xist in X chromosome inactivation. Nature. 1996;379:131–137. doi: 10.1038/379131a0. [DOI] [PubMed] [Google Scholar]
- 100.Leighton PA, Ingram RS, Eggenschwiler J, Efstratiadis A, Tilghman SM. Disruption of imprinting caused by deletion of the H19 gene region in mice. Nature. 1995;375:34–39. doi: 10.1038/375034a0. [DOI] [PubMed] [Google Scholar]
- 101.Huang W, Long N, Khatib H. Genome-wide identification and initial characterization of bovine long non-coding RNAs from EST data. Anim Genet. 2012;43:674–682. doi: 10.1111/j.1365-2052.2012.02325.x. [DOI] [PubMed] [Google Scholar]
- 102.Pang KC, Dinger ME, Mercer TR, Malquori L, Grimmond SM, Chen W, et al. Genome-wide identification of long noncoding RNAs in CD8+ T cells. J Immunol. 2009;182:7738–7748. doi: 10.4049/jimmunol.0900603. [DOI] [PubMed] [Google Scholar]
- 103.Liu MX, Chen X, Chen G, Cui QH, Yan GY. A computational framework to infer human disease-associated long noncoding RNAs. PLoS One. 2014;9:e84408. doi: 10.1371/journal.pone.0084408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Chan WL, Huang HD, Chang JG. IncRNAMap: A map of putative regulatory functions in the long non-coding transcriptome. Comput Biol Chem. 2014 Jan 23; doi: 10.1016/j.compbiolchem.2014.01.003.. [DOI] [PubMed] [Google Scholar]
- 105.Yang X, Gao L, Guo X, Shi X, Wu H, Song F, et al. A network based method for analysis of IncRNA-disease associations and prediction of IncRNAs implicated in diseases. PLoS One. 2014;9:e87797. doi: 10.1371/journal.pone.0087797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Xie C, Yuan J, Li H, Li M, Zhao G, Bu D, et al. NONCODEv4: Exploring the world of long non-coding RNA genes. Nucleic Acids Res. 2014;42:D98–D103. doi: 10.1093/nar/gkt1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Louro R, Smirnova AS, Verjovski-Almeida S. Long intronic non-coding RNA transcription: Expression noise or expression choice? Genomics. 2009;93:291–298. doi: 10.1016/j.ygeno.2008.11.009. [DOI] [PubMed] [Google Scholar]
- 108.Ponting CP, Oliver PL, Reik W. Evolution and functions of long noncoding RNAs. Cell. 2009;136:629–641. doi: 10.1016/j.cell.2009.02.006. [DOI] [PubMed] [Google Scholar]
- 109.Dong R, Jia D, Xue P, Cui X, Li K, Zheng S, et al. Genome-wide analysis of long noncoding RNA (IncRNA) expression in hepatoblastoma tissues. PLoS One. 2014;9:e85599. doi: 10.1371/journal.pone.0085599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Yang L, Froberg JE, Lee JT. Long noncoding RNAs: Fresh perspectives into the RNA world. Trends Biochem Sci. 2014;39:35–43. doi: 10.1016/j.tibs.2013.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Guttman M, Rinn JL. Modular regulatory principles of large non-coding RNAs. Nature. 2012;482:339–346. doi: 10.1038/nature10887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Zhang J, Cui X, Shen Y, Pang L, Zhang A, Fu Z, et al. Distinct expression profiles of LncRNAs between brown adipose tissue and skeletal muscle. Biochem Biophys Res Commun. 2014;443:1028–1034. doi: 10.1016/j.bbrc.2013.12.092. [DOI] [PubMed] [Google Scholar]
- 113.Paralkar VR, Mishra T, Luan J, Yao Y, Kossenkov AV, Anderson SM, et al. Lineage and species-specific long noncoding RNAs during erythro-megakaryocytic development. Blood. 2014;123:1927–1937. doi: 10.1182/blood-2013-12-544494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Pauli A, Valen E, Lin MF, Garber M, Vastenhouw NL, Levin JZ, et al. Systematic identification of long noncoding RNAs expressed during zebrafish embryogenesis. Genome Res. 2012;22:577–591. doi: 10.1101/gr.133009.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Ulitsky I, Shkumatava A, Jan CH, Sive H, Bartel DP. Conserved function of lincRNAs in vertebrate embryonic development despite rapid sequence evolution. Cell. 2011;147:1537–1550. doi: 10.1016/j.cell.2011.11.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Necsulea A, Soumillon M, Wamefors M, Liechti A, Daish T, Zeller U, et al. The evolution of IncRNA repertoires and expression patterns in tetrapods. Nature. 2014;505:635–640. doi: 10.1038/nature12943. [DOI] [PubMed] [Google Scholar]
- 117.Du ZQ, Eisley CJ, Onteru SK, Madsen O, Groenen MA, Ross JW, et al. Identification of species-specific novel transcripts in pig reproductive tissues using RNA-seq. Anim Genet. 2014;45:198–204. doi: 10.1111/age.12124. [DOI] [PubMed] [Google Scholar]
- 118.Kaushik K, Leonard VE, Kv S, Lalwani MK, Jalali S, Patowary A, et al. Dynamic expression of long non-coding RNAs (IncRNAs) in adult zebrafish. PLoS One. 2013;8:e83616. doi: 10.1371/journal.pone.0083616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Ng SY, Johnson R, Stanton LW. Human long non-coding RNAs promote pluripotency and neuronal differentiation by association with chromatin modifiers and transcription factors. EMBO J. 2012;31:522–533. doi: 10.1038/emboj.2011.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Li D, Chen G, Yang J, Fan X, Gong Y, Xu G, et al. Transcriptome analysis reveals distinct patterns of long noncoding RNAs in heart and plasma of mice with heart failure. PLoS One. 2013;8:e77938. doi: 10.l37l/journal.pone.0077938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Zhu JG, Shen YH, Liu HL, Liu M, Shen YQ, Kong XQ, et al. Long noncoding RNAs expression profile of the developing mouse heart. J Cell Biochem. 2014;115:910–918. doi: 10.1002/jcb.24733. [DOI] [PubMed] [Google Scholar]
- 122.Korostowski L, Sedlak N, Engel N. The Kcnq lot 1 long non-coding RNA affects chromatin conformation and expression of Kcnq1, but does not regulate its imprinting in the developing heart. PLoS Genet. 2012;8:e1002956. doi: 10.1371/journal.pgen.1002956.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Hambridge JA, Turner A, Baker AL. BraveHeart begins: Pilot results of group cognitive behaviour therapy for depression and anxiety in cardiac patients. Aust NZ J Psychiatry. 2009;43:1171–1177. doi: 10.3109/00048670903270415. [DOI] [PubMed] [Google Scholar]
- 124.Klattenhoff CA, Scheuermann JC, Surface LE, Bradley RK, Fields PA, Steinhauser ML, et al. Braveheart, a long noncoding RNA required for cardiovascular lineage commitment. Cell. 2013;152:570–583. doi: 10.1016/j.cell.2013.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Srivastava D, Cordes Metzler KR. Fending for a Braveheart. EMBO J. 2013;32:1211–1213. doi: 10.1038/emboj.2013.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Kataoka M, Huang ZP, Wang DZ. Build a braveheart: The missing linc (RNA) Circ Res. 2013;112:1532–1534. doi: 10.1161/CIRCRESAHA.113.301519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Grote P, Herrmann BG. The long non-coding RNA Fendrr links epigenetic control mechanisms to gene regulatory networks in mammalian embryogenesis. RNA Biology. 2013;10:1579–1585. doi: 10.4161/rna.26165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Grote P, Wittler L, Hendrix D, Koch F, Wahrisch S, Beisaw A, et al. The tissue-specific IncRNA Fendrr is an essential regulator of heart and body wall development in the mouse. Dev Cell. 2013;24:206–214. doi: 10.1016/j.devcel.2012.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Prensner JR, Chen W, Iyer MK, Cao Q, Ma T, Han S, et al. PCAT-1, a long noncoding RNA, regulates BRCA2 and controls homologous recombination in cancer. Cancer Res. 2014;74:1651–1660. doi: 10.1158/0008-5472.CAN-13-3159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Yang F, Zhang H, Mei Y, Wu M. Reciprocal regulation of HIF-1 alpha and lincRNA-p21 modulates the Warburg effect. Mol Cell. 2014;53:88–100. doi: 10.1016/j.molcel.2013.11.004. [DOI] [PubMed] [Google Scholar]
- 131.Chen FJ, Sun M, Li SQ, Wu QQ, Ji L, Liu ZL, et al. Upregulation of the long non-coding RNA HOTAIR promotes esophageal squamous cell carcinoma metastasis and poor prognosis. Mol Carcinog. 2013;52:908–915. doi: 10.1002/mc.21944. [DOI] [PubMed] [Google Scholar]
- 132.Song G, Shen Y, Zhu J, Liu H, Liu M, Shen YQ, et al. Integrated analysis of dysregulated IncRNA expression in fetal cardiac tissues with ventricular septal defect. PLoS One. 2013;8:e77492. doi: 10.1371/journal.pone.0077492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Yang KC, Yamada KA, Patel AY, Topkara VK, George I, Cheema FH, et al. Deep RNA sequencing reveals dynamic regulation of myocardial noncoding RNAs in failing human heart and remodeling with mechanical circulatory support. Circulation. 2014;129:1009–1021. doi: 10.1161/CIRCULATIONAHA.113.003863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Vance KW, Sansom SN, Lee S, Chalei V, Kong L, Cooper SE, et al. The long non-coding RNA Paupar regulates the expression of both local and distal genes. EMBO J. 2014;33:296–311. doi: 10.1002/embj.201386225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Ng SY, Bogu GK, Soh BS, Stanton LW. The long noncoding RNA RMST interacts with SOX2 to regulate neurogenesis. Mol Cell. 2013;51:349–359. doi: 10.1016/j.molcel.2013.07.017. [DOI] [PubMed] [Google Scholar]
- 136.Zhao X, Tang Z, Zhang H, Atianjoh FE, Zhao JY, Liang L, et al. A long noncoding RNA contributes to neuropathic pain by silencing Kcna2 in primary afferent neurons. Nat Neurosci. 2013;16:1024–1031. doi: 10.1038/nn.3438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Tripathi V, Shen Z, Chakraborty A, Giri S, Freier SM, Wu X, et al. Long noncoding RNA MALAT1 controls cell cycle progression by regulating the expression of oncogenic transcription factor B-MYB. PLoS Genet. 2013;9:e1003368. doi: 10.1371/journal.pgen.1003368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Kallen AN, Zhou XB, Xu J, Qiao C, Ma J, Yan L, et al. The imprinted H19 IncRNA antagonizes let-7 microRNAs. Mol Cell. 2013;52:101–112. doi: 10.1016/j.molcel.2013.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Wang K, Liu F, Zhou LY, Long B, Yuan SM, Wang Y, et al. A Long noncoding RNA, CHRF regulates cardiac hypertrophy by targeting miR-489. Circ Res. 2014 Feb 20; doi: 10.1161/circre-saha.114.302476.. [DOI] [PubMed] [Google Scholar]
- 140.Tsai MC, Manor O, Wan Y, Mosammaparast N, Wang JK, Lan F, et al. Long noncoding RNA as modular scaffold of histone modification complexes. Science. 2010;329:689–693. doi: 10.1126/science.1192002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Prensner JR, Iyer MK, Sahu A, Asangani IA, Cao Q, Patel L, et al. The long noncoding RNA SChLAP1 promotes aggressive prostate cancer and antagonizes the SWI/SNF complex. Nat Genet. 2013;45:1392–1398. doi: 10.1038/ng.2771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Guttman M, Donaghey J, Carey BW, Garber M, Grenier JK, Munson G, et al. lincRNAs act in the circuitry controlling pluripotency and differentiation. Nature. 2011;477:295–300. doi: 10.1038/nature10398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Maeda N, Kasukawa T, Oyama R, Gough J, Frith M, Engstrom PG, et al. Transcript annotation in FANTOM3: Mouse gene catalog based on physical cDNAs. PLoS Genet. 2006;2:e62. doi: 10.1371/journal.pgen.0020062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Djebali S, Davis CA, Merkel A, Dobin A, Lassmann T, Mortazavi A, et al. Landscape of transcription in human cells. Nature. 2012;489:101–108. doi: 10.1038/nature11233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Decatur WA, Fournier MJ. rRNA modifications and ribosome function. Trends Biochem Sci. 2002;27:344–351. doi: 10.1016/s0968-0004(02)02109-6. [DOI] [PubMed] [Google Scholar]
- 146.Darzacq X, Jady BE, Verheggen C, Kiss AM, Bertrand E, Kiss T. Cajal body-specific small nuclear RNAs: A novel class of 2′-O-methylation and pseudouridylation guide RNAs. EMBO J. 2002;21:2746–2756. doi: 10.1093/emboj/21.11.2746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Clouet d’Orval B, Bortolin ML, Gaspin C, Bachellerie JP. Box C/D RNA guides for the ribose methylation of archaeal tRNAs: The tRNA Trp intron guides the formation of two ribose-methylated nucleosides in the mature tRNA Trp. Nucleic Acids Res. 2001;29:4518–4529. doi: 10.1093/nar/29.22.4518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Ronchetti D, Mosca L, Cutrona G, Tuana G, Gentile M, Fabris S, et al. Small nucleolar RNAs as new biomarkers in chronic lymphocytic leukemia. BMC Med Genomics. 2013;6:27. doi: 10.1186/1755-8794-6-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Su H, Xu T, Ganapathy S, Shadfan M, Long M, Huang THM, et al. Elevated snoRNA biogenesis is essential in breast cancer. Oncogene. 2013;33:1348–1358. doi: 10.1038/onc.2013.89. [DOI] [PubMed] [Google Scholar]
- 150.O’Brien JE, Jr, Kibiryeva N, Zhou XG, Marshall JA, Lofland GK, Artman M, et al. Noncoding RNA expression in myocardium from infants with tetralogy of Fallot. Circ Cardiovasc Genet. 2012;5:279–286. doi: 10.1161/CIRCGENETICS.111.961474. [DOI] [PubMed] [Google Scholar]