Abstract
Retinoic acid (RA), the active metabolite of vitamin A, is required for spermatogenesis and many other biological processes. RA formation requires irreversible oxidation of retinal to RA by aldehyde dehydrogenase enzymes of the 1A family (ALDH1A). While ALDH1A1, ALDH1A2, and ALDH1A3 all form RA, the expression pattern and relative contribution of these enzymes to RA formation in the testis is unknown. In this study, novel methods to measure ALDH1A protein levels and intrinsic RA formation were used to accurately predict RA formation velocities in individual human testis samples and an association between RA formation and intratesticular RA concentrations was observed. The distinct localization of ALDH1A in the testis suggests a specific role for each enzyme in controlling RA formation. ALDH1A1 was found in Sertoli cells, while only ALDH1A2 was found in spermatogonia, spermatids, and spermatocytes. In the absence of cellular retinol binding protein (CRBP)1, ALDH1A1 was predicted to be the main contributor to intratesticular RA formation, but when CRBP1 was present, ALDH1A2 was predicted to be equally important in RA formation as ALDH1A1. This study provides a comprehensive novel methodology to evaluate RA homeostasis in human tissues and provides insight to how the individual ALDH1A enzymes mediate RA concentrations in specific cell types.
Keywords: aldehyde dehydrogenase 1A, testes, mass spectrometry
All-trans-retinoic acid (atRA) is essential for a multitude of biological processes, including reproduction, maintenance of the immune system, skin integrity, fetal development, and cell cycle regulation (1–5). Most importantly, in all of these processes, the concentrations of atRA are tightly regulated to obtain time- and location-specific signaling. This requires careful control of the expression of enzymes responsible for synthesizing and eliminating atRA. Yet, the specific enzymology of how atRA concentrations are regulated in a tissue- and cell type-specific manner is not well-established (6–8). In various in vitro systems, three aldehyde dehydrogenases (ALDHs), ALDH1A1, ALDH1A2, and ALDH1A3 have been shown to generate atRA from retinaldehyde (9–11), but the tissue- and cell type-specific localization, ontogeny, and specific activity of these enzymes is poorly understood.
The importance of ALDH1A2 and ALDH1A3 in atRA formation during fetal development has been established by the fact that ALDH1A2−/− mice die during embryonic development, displaying classic malformations associated with atRA deficiency, and ALDH1A3−/− mice die shortly after birth (12, 13). In contrast, ALDH1A1−/− mice are viable, fertile, and protected against obesity (14). The significance of ALDH1A1 in atRA formation in mice is implied from the increased serum retinal concentrations when the ALDH1A1−/− mice are treated with retinol (15, 16). Yet, as the quantitative protein expression and intrinsic RA formation capacity of these enzymes in specific tissues is unknown, the importance of the three ALDH1A enzymes to atRA formation in different atRA target tissues in humans or animals is currently undetermined.
In humans, ALDH1A1 mRNA has been detected in the liver, kidney, testis, brain, lung, red blood cells, and lens of the eye (17–19). In contrast, ALDH1A2 mRNA is found predominantly in the testis, uterus, and skeletal muscle, while ALDH1A3 mRNA is localized in the prostate, trachea, intestine, and testis (18). These mRNA expression patterns strongly suggest that all three ALDH1A enzymes contribute to atRA synthesis during postnatal life. In addition, ALDH1A1 expression has been proposed as a cancer stem cell marker and associated with poor prognosis in certain types of cancers, including clear cell renal, esophageal squamous cell, and papillary thyroid carcinomas (20–22). Conversely, ALDH1A2 has been identified as a candidate tumor suppressor in a transgenic animal model of prostate cancer, and gene variants of ALDH1A2 have been found to be associated with osteoarthritis and increased newborn kidney size (23–26). Taken together these studies point to tissue-specific central roles that ALDH1A enzymes play in human physiology, and the critical importance for obtaining information concerning the expression and activity of these enzymes in human tissues.
One of the major challenges in evaluating vitamin A and atRA homeostasis and the importance of ALDH1A enzymes in these processes has been the fact that serum concentrations do not readily reflect tissue retinoid concentrations, as atRA is believed to be synthesized in the target tissues (27). Because of this, measurements of tissue atRA concentrations, and expression and activity of atRA-synthesizing enzymes in small tissue fractions, such as biopsies from humans, is critically important. To date, methods have not been readily available to determine the specific activity and expression of ALDH1A enzymes in human tissues or for demonstrating that ALDH1A activity influences tissue atRA concentrations and net atRA formation velocity. As such, it has not been possible to predict the pharmacological outcomes within a specific target tissue or globally in the whole body in response to specific ALDH1A enzyme inhibition or induction.
An excellent model organ for the tissue-specific synthesis of atRA is the testis (28), as atRA synthesis within the testis is required for male fertility (29, 30). For example, there is no initiation of spermatogenesis in mice which lack ALDH1A1, ALDH1A2, and ALDH1A3 in the Sertoli cells (30). Similarly, an inhibitor of the ALDH1A enzymes, WIN 18,446, has been shown to affect the ability of multiple mammalian species, including humans, to produce sperm (31–33), likely due to decreased atRA concentrations in the testes (33). However, the specific role of the individual ALDH1A enzymes in atRA formation in the testis has not been evaluated. Hence, the human testis was chosen as a model tissue to establish how atRA synthesis is orchestrated by the three ALDH1A enzymes in a tissue- and cell type-specific manner to regulate atRA concentrations in the organ. A novel LC-MS/MS peptide quantification method was developed that allows accurate quantification of the expression levels of ALDH1As in small biopsy scale samples. This method was coupled with measurements of ALDH1A activity, the formation of atRA in tissue samples from 18 human donors, and measurements of atRA concentrations in paired samples from the same donors. In addition, the effect of cellular retinol binding protein (CRBP) on atRA synthesis in the testis was evaluated. These novel methods allowed the prediction of the relative contribution of each of the ALDH1A enzymes to overall intratesticular RA formation. The validity of the novel methods was confirmed via specific inhibition studies. The importance of individual ALDH1A enzymes to atRA formation in specific cell types in the testis was evaluated via tissue localization studies of ALDH1A enzymes. The approach used can also be applied to measuring atRA formation in any human tissue biopsy by combining quantitative proteomics and sensitive kinetic analyses.
MATERIALS AND METHODS
Human samples used in the study
Samples of testicular tissue used to characterize ALDH1A expression and activity were obtained from 18 anonymous donors at the Center of Reproductive Medicine and Andrology in Munster, Germany. Fifteen of the testes were from men who underwent orchiectomy as an androgen depletion therapy for the treatment of prostate carcinoma and all samples were anonymized following informed consent. All of the men had qualitatively normal spermatogenesis based on histological analysis. The men were 62–87 years old (mean, 70.7 years). In addition, three samples from individuals undergoing male to female sex reassignment surgery after at least two years of estrogen treatment were analyzed. These samples were also anonymized following informed consent and the age range of this group was 49–75 years (mean, 58.3 years). Due to the estrogen treatment, spermatogenesis was severely reduced on histological analysis of the tissue from these individuals. The tissues were collected into ice-cold Leibovitz medium at the surgery site, protected from light, and taken to the laboratory on ice. The testes were decapsulated and fragmented by scalpels and scissors. The fragments were snap-frozen in liquid nitrogen and stored at −80°C until RA measurements or preparation of S10 fractions.
Quantification of ALDH1A1, ALDH1A2, and ALDH1A3 in human testes using LC-MS/MS
Tissue biopsies (33–102 mg) from 18 donors were homogenized on ice using a drill-powered 2 ml Potter-Elvehjem glass homogenizer (Kimbel Glass, Vineland, NJ). Samples were homogenized in 3× volume tissue homogenization buffer (50 mM potassium phosphate and 250 mM sucrose) with an EDTA-free protease inhibitor cocktail (Roche, San Francisco, CA). The samples were transferred to a 1.5 ml microcentrifuge tube, the homogenizer washed with 2× volume of homogenization buffer, and the wash added to the homogenate. The homogenate was centrifuged at 10,000 g for 25 min at 4°C to pellet large organelles and cell membranes. The resulting supernatant (S10 fraction) that contains microsomes and cytosolic proteins was collected, aliquoted, flash-frozen in liquid nitrogen, and stored at −80°C. Total protein concentration in each S10 fraction was measured using a BCA assay (Thermo Fisher, Waltham, MA).
ALDH1A expression in human testicular tissue was measured by peptide quantification using LC-MS/MS. Peptides were chosen based on in silico analysis of their selectivity and analytical performance (34). The FASTA files for each ALDH1A sequence were downloaded from UniProt with the accession numbers P00352, O94788-1, and P47895. The human ALDH1A enzymes were first trypsin digested in silico, and peptides with a predicted m/z greater than 1,100 were excluded due to the m/z range limitations of the mass spectrometer. Next, the predicted peptides were screened against a trypsin-digested human proteome to ensure selectivity. Additionally, sites predicted or reported to have posttranslational modifications or mutations were avoided. Purified recombinant ALDH1A enzymes were trypsin digested in vitro to determine the coverage of ALDH1A digestion. Of the predicted peptides that were within the detectable mass range of the mass spectrometer, 100% were detected for ALDH1A1, 100% for ALDH1A2, and 93% for ALDH1A3.
For the peptides that passed the initial criteria described above, the predicted precursor ion and fragments were incorporated into an LC-MS/MS method. Human testicular S10 protein and purified recombinant ALDH1A were trypsin digested to determine which remaining peptides were detected in human testicular tissue and were specific for ALDH1A. For each ALDH1A, one peptide was selected as a quantification peptide and a second for verification (Table 1). For the quantification peptide, a matching heavy labeled peptide was synthesized as an internal standard (Pierce, Rockford, IL). In order to account for the efficiency of the trypsin digestion, a heavy labeled peptide with extended sequence over the trypsin cleavage site (lagging end) was synthesized for ALDH1A3. This peptide was labeled at the N terminus with a [13C615N2]lysine or [13C615N2]arginine. The lagging end peptide requires two cleavages by trypsin to produce the target peptide. In addition, a separate labeled internal standard for ALDH1A1 and ALDH1A2 quantification peptides was incorporated that did not require trypsin cleavage (Table 1). For each peptide, two fragments from each of the two peptides were used for detection and to confirm the presence of the protein. The stability of the peptides was validated by confirming that there was no significant sample degradation after three freeze-thaw cycles, 24 h at 37°C and 24 h at room temperature.
TABLE 1.
Summary of signature peptides and mass spectrometric parameters used for ALDH1A quantification
| Protein | Peptide (Amino Acid Number) | Precursor Ion (m/z) | Fragments (m/z) | Declustering Potential | Collision Energy |
| ALDH1A1 | ANNTFYGLSAGVFTK* (420-434) | 795.4 | 879.5 | 120 | 20 |
| 1,042.6 | |||||
| VAFTGSTEVGK (241-251) | 548.3 | 778.4 | 71 | 28 | |
| 677.3 | |||||
| ALDH1A2 | ILELIQSGVAEGAK* (370-383) | 714.4 | 846.4 | 120 | 30 |
| 959.5 | |||||
| EEIFGPVQEILR (415-426) | 715.4 | 911.5 | 83 | 35 | |
| 854.5 | |||||
| ALDH1A3 | EEIFGPVQPILK*,# (409-420) | 685.4 | 851.5 | 81 | 34 |
| 794.5 | |||||
| ELGEYALAEYTEVK (487-500) | 807.9 | 839.4 | 90 | 38 | |
| 952.5 |
Two signature peptides were chosen for quantification and confirmation of detection of ALDH1A proteins. The peptides used as an internal standard for quantification are marked with *. All peptides were synthesized with a heavy labeled terminal lysine or arginine. The peptide marked with # was synthesized as EVTDNMRIAKEEIFGPVQPILK and required cleavage by trypsin to be detected. This peptide was used to control for the variability in trypsin digestion across the sample set. For each peptide, values of 10 and 13 were used for the entrance and collision exit potential.
Trypsin digestion of purified ALDH1As and testicular S10 fractions was done in 96-well plates according to the following protocol: First, 15 μl of sample (0–400 nM of ALDH1A or 5.33 mg/ml S10 fraction) were added to each well. Next, 5 μl of 300 nM ALDH1A3 lagging end peptide was added together with 4 μl dithiothreitol (100 mM) and 10 μl ammonium bicarbonate buffer (100 mM, pH 7.8). Next, 5 μl of 10% sodium deoxycholate (Sigma, St. Louis, MO) was added in the well, and the sample was mixed before incubation at 95°C for 5 min. After cooling to room temperature, 4 μl of iodoacetamide (200 mM) was added and the sample was incubated at room temperature in the dark for 20 min. Trypsin was added at a 1:25 trypsin:protein ratio, and the sample was digested for 24 h at 37°C. The incubation was quenched by addition of 20 μl chilled acetonitrile with 8% trifluoroacetic acid containing the non-lagging end heavy labeled ALDH1A1 and ALDH1A2 peptide internal standards. Samples were centrifuged at 3,000 g for 25 min at 4°C and analyzed by LC-MS/MS.
The peptides were quantified by MS using an AB Sciex 5500 qTrap Q-LIT mass spectrometer (AB Sciex, Foster City, CA) equipped with an Agilent 1290 ultra (U)HPLC (Agilent, Santa Clara, CA). The tryptic peptides were separated using an Aeris peptide XB-C18 column (50 × 2.1 mm) with 1.7 μm particle size at 40°C and a SecurityGuard Ultra UHPLC C18-peptide cartridge (Phenomenex, Torrance, CA). The eluting solvents were: A, water + 0.1% formic acid, and B, acetonitrile (ACN) + 0.1% formic acid. For chromatographic separation, the following 18 min linear gradient with a 400 μl/min flow rate was used: 0 → 3.5 min 3% B, then increased by 12.0 min to 40% B, 12.0 → 12.1 min 95% B, and remained at 95% B until 15.0 min, then 15.0 → 15.1 min 3% B, 15.1 → 18 min 3% B.
For quantification of ALDH1As, purified ALDH1A1, ALDH1A2, and ALDH1A3 were used as standards. A nine point standard curve was generated with values of 0.018–7.2 pmol for ALDH1A1, 0.005–1.35 pmol for ALDH1A2, and 0.002–0.72 pmol ALDH1A3. The amount of enzyme detected in each sample was normalized to total S10 protein (0.08 mg).
Mass spectrometric quantification of atRA, 13-cisRA, and retinal
The concentrations of atRA, 13-cisRA, and retinal were measured using an AB Sciex 5500 qTrap Q-LIT mass spectrometer equipped with an Agilent 1290 UHPLC as previously described (35) with several minor modifications. Briefly, atRA and 13-cisRA were monitored using positive ion APCI and MS transitions of m/z 301 → 205 and m/z 301 → 123 with the 205 fragment used for quantification. For quantification, atRA and 13-cisRA peak areas were normalized to the atRA-d5 internal standard area. The concentration of retinal was measured only in the in vitro samples using positive ion APCI and the MS transition m/z 285 → 161 with declustering potential of 66.0, collision energy of 13.0, and collision cell exit potential of 4.0. The at-retinal-d5 was used as an internal standard and was monitored using the m/z 290 → 161 transition.
For in vitro incubation samples, the analytes were separated using a Kinetex C18 column (100 × 2.1 mm, 1.7 μm particle size) heated to 40°C with a SecurityGuard Ultra UHPLC C18 cartridge (Phenomenex) and a linear gradient with A, water + 0.1% formic acid, and B, ACN + 0.1% formic acid. The mobile phase flow was 600 μl/min with the following linear gradient: 0 → 0.25 min 40% B, then increased to 95.0% at 4 min, 4 → 5 min 95.0% B, 5 → 6 min 40% B. A standard curve consisting of RA spiked into buffer containing 0.05 mg/ml S10 protein at concentrations of 0, 1, 5, 10, 25, 50, and 100 nM was prepared simultaneously with all incubations. Analyst software (AB Sciex) was used to quantify peak areas for each analyte.
For testicular tissue analysis, the extraction of RA from tissue was optimized to increase the recovery of RA without causing any isomerization, and chromatographic separation was optimized to avoid endogenous interference from tissue. Briefly, tissue was homogenized with a 2 ml Potter-Elvehjem glass homogenizer (Kimbel Glass, Vineland, NJ) in a 1:1 volume of 0.9% NaCl. After transferring the sample to a 15 ml glass culture tube, a 2:1 volume of ACN with 1% formic acid was added followed by 10 μl of 1 μM atRA-d5, which served as an internal standard. Hexanes (10 ml) were used to extract RA, the organic layer was transferred to a glass tube and dried under nitrogen at 37°C. The sample was reconstituted in 60:40 ACN/water. A standard curve and quality control samples were generated with RA standards spiked into homogenized dog testicular tissue (a kind gift from Dr. Mary Zoulas of the Seattle Animal Shelter) exposed to UV light to destroy endogenous RA. Dog testicular tissue was homogenized in a 1:1 volume with 0.9% NaCl and 198 μl were placed in a 15 ml glass culture tube. atRA and 13-cisRA (2 μl) standards were added to each tube to final concentrations of 0, 1, 2.5, 5, 7.5, and 10 nM for each standard. The quality control samples were prepared at 1.5, 3.75, and 8.75 nM, and duplicate quality control samples were included together with the standard curve in all analytical assays. atRA and 13-cisRA were separated from endogenous interferences using a 150 × 2.1 mm Supelco Ascentis Express reverse phase amide column (Sigma) with 2.7 μm particle size and an Ascentis Express reverse phase amide 2.7 μm guard cartridge. The solvents for the UHPLC analysis were A, water + 0.1% formic acid, and B, ACN/methanol (60/40) + 0.1% formic acid. The solvent flow was 500 μl/min and the following linear gradient was used: 0 → 2 min 40% B, then increased to 95% B at 10 min, 10 → 15 min 95% B, 15 → 17 min 40% B. atRA and 13-cisRA peak areas were normalized to the atRA-d5 internal standard and peak area ratios were used for quantification using Analyst software (AB Sciex). A signal:noise of nine was set as the minimum threshold for quantitation.
To further confirm the identity of retinoids detected in the testes, MS/MS/MS was used. To collect reference spectra, atRA and 13-cisRA standards were spiked into 60:40 ACN:water + 0.1% formic acid. The standards and two separate 75 mg testicular tissue samples were extracted and analyzed as described above with the following modifications on the MS settings. The m/z 205.3+ fragment generated from m/z 301.2+ (RA) was fragmented using the linear ion trap of the API 5500 with a scan rate of 10,000 daltons/second, Q3 entry barrier of 8.0 volts, and a dynamic fill time set for the ion trap. All the fragments generated from m/z 205.3+ between m/z 50 and 200 were collected in the trap and detected. The MS/MS/MS product ion spectrum was collected for each chromatographic peak and the spectra of compounds eluting from the tissue sample were compared with the RA standards. Based on the fact the m/z 159.1+ fragment was the main fragment generated from m/z 205.3+, from RA, the m/z 159.1+ fragment was extracted from the MS/MS/MS total ion chromatograms and monitored as a function of time to generate a chromatogram of the m/z 301.2+ > 205.3+ > 159.1+ transition.
Formation of atRA and 13-cisRA by recombinant ALDH1A enzymes and human testis S10 fractions
Human ALDH1A1 cDNA was purchased from OriGene (SC321535). Full-length human ALDH1A2 cDNA was cloned from human testis RNA using RT-PCR. An open reading frame of ALDH1A1 was subcloned into a pETite expression vector (Lucigen, Middleton, WI) with a 5′ hexahistidine tag and that of ALDH1A2 into a pET28 bacterial expression vector (Novagen, Billerica, MA) containing a 3′ hexahistidine tag. The resulting plasmids were verified through DNA sequencing for correct frame and sequences. Enzymes were expressed in Escherichia coli [BL21(DE3)] grown to an optical density at 600 nm of 0.6–1.0. Expression was induced with 0.5 mM isopropyl-beta-D-l-thio-galactopyranoside and continued for 4 h at 37°C before harvest. ALDH1A1 and ALDH1A2 were purified using the His.Bind resin and buffer kit (catalog # 69755-3, Novagen) as previously described (36). Purified enzyme was dialyzed against 20 mM HEPES buffer (pH 8.5) containing 150 mM KCl and 1 mM EDTA. Protein purity was confirmed by SDS-PAGE followed by Coomassie staining and concentrations were determined using BCA protein assay kit (Thermo Fisher) or protein assay kit II (BioRad, Hercules, CA). Enzyme was stored at 4°C in the dialysis buffer with 1 mM TCEP (Thermo Fisher), a reducing agent. Purified recombinant human ALDH1A3 was purchased from Life Technologies (Grand Island, NY). The lyophilized protein was reconstituted to a final concentration of 0.2 mg/ml with filtered water according to the manufacturer’s directions and stored at −80°C.
To characterize the kinetics of atRA formation by ALDH1A1 and ALDH1A3 enzymes, at-retinal (0–2,000 nM) was incubated with 18 nM ALDH1A in 100 μl of buffer consisting of 750 mM KCl and 50 mM HEPES at a pH of 8.0 with 2 mM NAD+. Following a preincubation of 5 min at 37°C, the incubations were initiated with addition of substrate. The incubations were terminated at 5 min by transferring 75 μl of the incubation into an equal volume of chilled acetonitrile with 100 nM atRA-d5 (internal standard). In order to characterize the atRA formation kinetics by ALDH1A2, the protein concentration was decreased to 2 nM, and the incubations were terminated after 2 min. The formation kinetics of 13-cisRA by ALDH1A1 were characterized by incubating 13-cis-retinal (0–15,000 nM) with 15 nM ALDH1A1 and quenching after 5 min. After termination, all plates were spun at 3,000 g for 25 min, the samples were transferred to a 96-well plate and analyzed by LC-MS/MS for atRA and 13-cisRA concentrations. In order to determine whether 13-cis-retinal was a substrate of ALDH1A2 or ALDH1A3, 5,000 nM 13-cis-retinal was incubated with 50 nM ALDH1A2 and ALDH1A3 for up to 45 min. Over the course of 45 min, no enzyme-dependent 13-cisRA formation greater than 0.0007 pmol/min/pmol (based on assay lower limit of detection) was detected. All incubations were conducted under protein and time linearity and as triplicate measurements. The Michaelis-Menten equation was fitted to the velocity of atRA formation as a function of substrate concentration data to obtain kinetic parameters for each enzyme. When ALDH1A1 was incubated with 13-cis-retinal, 13-cisRA formation demonstrated substrate inhibition kinetics, and the substrate inhibition equation was used to fit the data according to equation 1:
| (Eq. 1) |
where v is the predicted velocity, Vmax is the maximum observed velocity, [S] is the substrate concentration, Km is the affinity constant of 13-cis-retinal with the ALDH1A1, and Ki is the disassociation constant for 13-cis-retinal binding in a way that allows for the enzyme to bind two substrates. All kinetic analyses were done using GraphPad Prism (La Jolla, CA)
atRA and 13-cisRA formation from the corresponding retinals was measured in the individual human testis S10 preparations using a similar method as that described for recombinant ALDH1As. Briefly, at-retinal at nominal concentrations of 100 nM and 1,000 nM or 13-cis-retinal (1,000 nM) were incubated with S10 fractions (5 μg S10 protein/0.1 ml) from each donor. The incubations were initiated with substrate and terminated after 10 min and analyzed as described above. The protein binding of at-retinal and 13-cis-retinal to the nonspecific proteins and membranes in the activity assay was determined by ultracentrifugation, as described before (37). Retinal was added to 0.05 mg/ml pooled S10 protein to final concentrations of 100 and 1,000 nM at-retinal and 1,000 nM 13-cis-retinal. The samples were aliquoted into ultracentrifuge tubes (Beckman 343775) and spun at 435,000 g at 37°C for 90 min using a Sorval Discovery M150 SE ultracentrifuge with a Thermo Scientific S100-AT3 rotor or incubated at 37°C for 90 min in a water bath. The incubated sample or supernatant was added to 100 μl ice-cold acetonitrile containing 100 nM at-retinal-d5. The samples were centrifuged at 3,000 g for 20 min at 4°C. Samples were transferred to a 96-well plate and analyzed using the LC-MS/MS method described. The unbound (free) fraction of retinal in the incubations was calculated as the ratio of retinal concentration in solution with or without ultracentrifugation. This calculation assumes minimal CRBP concentration and binding in the incubations. The insignificant concentration of CRBP was confirmed via direct measurement of CRBP concentrations in the S10 fractions (see below).
ALDH1A inhibition by WIN 18,446
In order to determine reversible inhibition of ALDH1A1, ALDH1A2, and ALDH1A3 by WIN 18,446, recombinant purified enzymes were incubated with 100 nM of at-retinal and increasing concentrations (1–5,000 nM) of WIN 18,446. All activity assays with recombinant enzymes included 18 nM ALDH1A and 2 mM NAD+, and were terminated after 5 min by transferring 75 μl of the incubation into an equal volume of chilled acetonitrile with 100 nM atRA-d5. The samples were prepared and analyzed by LC-MS/MS as described for incubations with recombinant purified ALDH1A. A screen for time-dependent inhibition (TDI) used the dilution method and at-retinal as a substrate (38). Briefly, TDI was determined by incubating 180 nM ALDH1A enzyme with and without 1 μM WIN 18,446 in the presence of 2 mM NAD+ in a final volume of 100 μl of buffer (750 mM KCl and 50 mM HEPES at a pH of 8.0). At time points of 0.25, 3, and 5 min, 10 μl aliquots were diluted 10-fold into 100 μl of buffer (750 mM KCl and 50 mM HEPES at a pH of 8.0) containing NAD+ (2 mM) and a saturating concentration of at-retinal (5,000 nM). All incubations were performed in triplicate with a no NAD+ control for the WIN 18,446 and at-retinal incubations. Because WIN 18,446 was a TDI for ALDH1A2, but not ALDH1A1 or ALDH1A3, the kinetics of ALDH1A2 TDI with WIN 18,446 were characterized using eight concentrations of WIN 18,446 (0–5,000 nM). The dilution method was used with time points of 0.25, 1, 3, and 5 min. The kinact and KI were determined by fitting equation 2 to the data in Graphpad Prism.
| (Eq. 2) |
[I] is the inhibitor concentration, λ is the observed apparent inactivation rate (min−1), kinact is the maximum inactivation rate, and KI is the concentration of inhibitor when the inactivation rate is half of the kinact.
Quantification of CRBP1 expression in human testes and determination of the effect of CRBP1 to RA formation in human testes
Testicular CRBP1 protein expression was quantified in all tissue donors using an ELISA kit obtained from Cloud Clone Corp. (Houston, TX). Standards and samples were prepared according to the manufacturer’s instructions. Testicular S10 protein fractions were diluted 1:20-fold into phosphate buffer (pH 7.4) and CRBP1 quantified based on the standard provided. The amount of CRBP1 in each sample was normalized to milligrams of S10 protein, and the expression was scaled to the concentration of CRBP1 as picomoles per gram testes using the S10 recovery (milligrams S10 protein per gram testes). In addition, the concentration of CRBP1 in the incubations with testicular S10 protein was calculated using the measured concentration in each donor.
The effect of CRBP1 (purchased as purified protein from Origene, Rockville, MD) on atRA formation by ALDH1A was determined using recombinant ALDH1A proteins and testicular S10 protein. CRBP bound with an equal concentration of retinal (holo-CRBP)1 was generated by preincubating 3,000 nM at-retinal for 5 min at room temperature with 3,000 nM CRBP1 in buffer [10% glycerol, 100 mM glycine, 25 mM Tris-HCl (pH 7.3)]. The holo-CRBP1 (300 nM final concentration) or at-retinal (300 nM final concentration) in buffer was used to initiate incubations with 0.05 mg/ml pooled S10 protein from five donors, 18 nM ALDH1A1, 18 nM ALDH1A3, or 2 nM ALDH1A2 in a 100 μl volume. The concentration of 300 nM was chosen for holo-CRBP1 and at-retinal concentrations to mimic expected testicular concentrations as closely as possible. All of the incubations were performed in quadruplicate and contained 2 mM NAD+. Incubations were quenched after 5 min (ALDH1A enzymes) or 10 min (S10 protein) by transferring 75 μl of the incubation into an equal volume of chilled acetonitrile with 100 nM atRA-d5 (internal standard). After termination, the samples were analyzed by LC-MS as described above.
Contribution of individual ALDH1A enzymes to RA formation in human testes
To establish the relative importance of each ALDH1A enzyme in RA formation in human testes, the ALDH1A concentrations for each subject and the in vitro kinetics of RA formation from retinal by the recombinant ALDH1A enzymes were used to predict the overall atRA and 13-cisRA formation velocity in each donor. Due to the different Km and kcat values for ALDH1A enzymes with at-retinal as a substrate, their contribution to atRA formation was predicted at two different substrate concentrations, while 13-cisRA formation was predicted only at one concentration. atRA formation in each testis S10 sample was predicted using equation 3:
| (Eq. 3) |
in which v is the predicted product formation velocity, [ALDH1A] is the measured expression level of each individual ALDH1A enzyme, kcat is the maximum product formation velocity measured with recombinant enzymes, fu is the experimentally determined free (unbound) fraction in S10 preparation, [S] is the total substrate concentration, and Km is the affinity constant of at-retinal with the ALDH1A. Due to the fact 13-cisRA formation by ALDH1A1 from 13-cis-retinal demonstrated substrate inhibition, an equation accounting for substrate inhibition was used (equation 4). Ki is the disassociation constant for 13-cis-retinal binding in a way that allows for the enzyme to bind two substrates.
| (Eq. 4) |
In order to determine the contribution of each ALDH1A enzyme to the total formation of atRA, the predicted velocity of atRA formation by the given ALDH1A was divided by the predicted total velocity at multiple substrate concentrations. The accuracy of the ALDH1A activity predictions was evaluated by comparing the measured velocity to the predicted velocity of atRA and 13-cisRA formation in each subject (S10 fraction) at the different concentrations. The average fold error (afe) of the predictions was calculated for each prediction using equation 5:
| (Eq. 5) |
An afe between 0.5 and 2.0, which indicates a 2-fold prediction error cutoff, was set as the limit for acceptable predictions according to previous studies on clearance predictions (39).
The relative importance of ALDH1A2 and ALDH1As to atRA formation in the testes was confirmed using WIN 18,446. Due to the fact WIN 18,446 inactivates ALDH1A2, but not ALDH1A1 or ALDH1A3, the contribution of ALDH1A2 to atRA formation can be determined using the TDI characteristics of WIN 18,446 and the dilution method described above. Briefly, 625 nM WIN 18,446 and pooled human testes S10 (250 μg S10 protein/0.1 ml) were incubated for 0.25, 15, and 30 min after initiating the reaction with 10 μl of 20 mM NAD+. At the set time points, 2 μl aliquots were diluted 1:50-fold into activity assays containing a final concentration of 2 mM NAD+ and 5,000 nM at-retinal. After 10 min, incubations were terminated by transferring 75 μl of the incubation into an equal volume of chilled acetonitrile with 100 nM atRA-d5. The concentration of WIN 18,446, incubation time, and the dilution were chosen so that 98% of ALDH1A2 was inactivated in 30 min and minimal reversible inhibition of ALDH1A1 and ALDH1A3 would be present. This concentration was determined using equations 2 and 6:
| (Eq. 6) |
A concentration of 625 nM WIN 18,446 results in an inactivation rate of 0.14 min−1 and a decrease of 87% and 98% in ALDH1A2 activity at 15 and 30 min. The 12.5 nM WIN 18,446 remaining after the 1:50 dilution results in minimal reversible inhibition with saturating substrate conditions.
The effect of CRBP1 on ALDH1A activity in the testes was predicted by incorporating the measured in vitro effect of CRBP1 on atRA formation by ALDH1A1 and ALDH1A2 at the physiologically relevant at-retinal concentration of 100 nM. In addition to ALDH1A expression levels and atRA formation kinetics, the observed 52% decrease in atRA formation by ALDH1A1 and 2.7-fold increase of atRA formation by ALDH1A2 in the presence of CRBP1 was incorporated into equation 2 to predict the contribution of each enzyme to atRA formation.
Immunohistochemistry
Testicular tissue from three healthy fertile subjects was used to determine ALDH1A localization. These men volunteered to be part of this research study that was approved in advance by the University of Washington Institutional Review Board. All subjects provided signed written consent prior to any study procedures. Three healthy men aged 28, 52, and 43 years consented to donating testicular biopsies for the study. The tissue slices shown are from a 43 year old subject with left spermatocele, normal sperm counts, and normal hormone levels. Immunohistochemistry was performed using human testis tissue fixed in Bouin’s fixative using commercial rabbit polyclonal antibodies raised against ALDH1A1 (Abcam, ab24343), ALDH1A2 (Proteintech Group, 13951-1-AP), and ALDH1A3 (Abgent, AP7847a) as previously described (40). Antigen retrieval was achieved using citrate buffer (pH 6) at a rolling boil for 5 min. Sections were incubated in primary antibody at a concentration of 0.5 μg/ml (ALDH1A1), 4 μg/ml (ALDH1A2), 2.5 μg/ml (ALDH1A3), or 0.1 μg/ml in 5% normal goat serum/0.1% BSA in phosphate buffered saline at room temperature overnight (∼16 h). Control sections were incubated without primary antibody. Biotinylated goat-anti-rabbit secondary antibody (Invitrogen, 956143b) was applied for 1 h at room temperature, following the manufacturer’s instructions. Streptavidin conjugated horseradish peroxidase (Invitrogen, 956143b) was subsequently applied for 1 h at room temperature. Localization was determined by a brown precipitate formed by horseradish peroxidase activity in the presence of 3,3′-diaminobenzidine tetrahydrochloride (Invitrogen, 002020). Sections were counterstained with a 1:3 dilution Harris hematoxylin (Sigma-Aldrich, HHS32-1L), dehydrated, and mounted under glass coverslips using DPX mounting media (VWR International, 360294H). Cell types were determined using nuclear morphology and location within the testis. Immunohistochemistry was performed on three samples for three individuals to ensure consistent results.
Statistical analysis
Linear regression analysis was used and measured atRA and 13-cisRA formation velocities. Linear regression was also used to test whether the measured and predicted 13-cisRA and atRA formation correlated. The correlation between measured atRA concentrations and atRA and 13-cisRA formation velocities was used to test whether in vitro data could predict in vivo atRA concentrations. Unpaired two-tailed Student’s t-test was used to determine any significant differences between transgender individuals and the rest of the sample set. P values of less than 0.05 were considered significant for all analyses. All statistical tests and kinetic analyses were performed using GraphPad Prism.
RESULTS
ALDH1A protein quantification
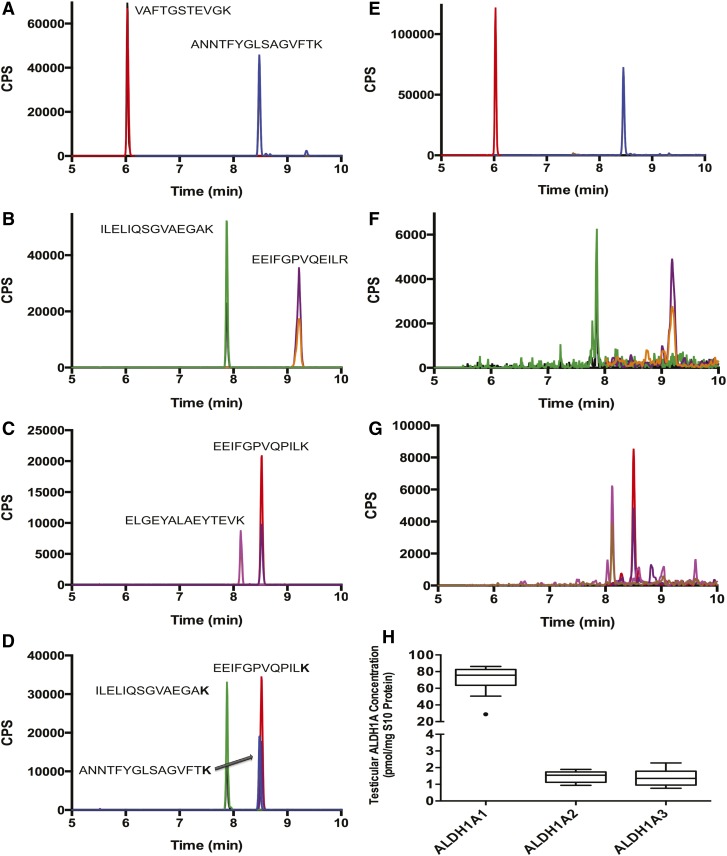
In order to simultaneously quantify all three ALDH1A enzymes in human tissue, a novel LC-MS/MS-based peptide quantification method was developed. Recombinant purified ALDH1A enzymes were used as standards and [13C615N2]lysine- or [13C615N4]arginine-labeled peptides as internal standards. For each ALDH1A enzyme, two trypsin digested peptides were chosen for measurement. One of the peptides was used for quantification and the second for verification of protein identification. The peptides and the MS/MS transitions used for quantification of the ALDH1A enzymes are listed in Table 1. The peptides were chosen based on their predicted specificity and the observed sensitivity. In order to account for the efficiency of the trypsin digestion, also a heavy labeled peptide for ALDH1A3 was synthesized with an extended end over the trypsin cleavage site, and hence its detection required cleavage by trypsin (Table 1). Chromatograms of the digested recombinant proteins and the internal standards showing the specific quantified peptides are depicted in Fig. 1.
Fig. 1.
Quantification of ALDH1A1, ALDH1A2, and ALDH1A3 from testicular tissue S10 fractions. ALDH1A concentrations were measured using a novel LC-MS/MS-based peptide quantification method. Recombinant protein was digested and two peptides (each with two m/z transitions) were chosen for each protein to measure ALDH1A1 (A), ALDH1A2 (B), and ALDH1A3 (C) using LC-MS/MS. The peptide used for quantification was normalized to a corresponding stable isotope-labeled peptide with a [13C615N2]lysine or [13C615N2]arginine (D). Each peptide was observed in all 18 men and representative chromatograms of detection of ALDH1A1 (E), ALDH1A2 (F), and ALDH1A3 (G) in human testis are shown. The expression levels of the three proteins in the human testes of nontransgendered men are shown in (H). All peaks displayed in the chromatograms are scaled to counts per second (CPS).
The expression of ALDH1A1, ALDH1A2, and ALDH1A3 was quantified from 33–102 mg samples of testicular tissue collected from 15 men undergoing orchiectomy for treatment of prostate cancer and from 3 male to female transgendered individuals undergoing orchiectomy as part of their gender reassignment surgery. S10 fractions containing the cytosolic and microsomal proteins were generated from each individual donor sample. The average yield of S10 protein was 48 ± 11 mg S10 protein/gram testis tissue. Eighty micrograms of this S10 protein was used for ALDH1A quantification. Representative chromatograms of the detection of ALDH1A1, ALDH1A2, and ALDH1A3 in the human testis, and the quantification of the three enzymes in the human testis are shown in Fig. 1. Based on the expression levels, ALDH1A1 was the major ALDH1A enzyme present in the human testis. In the nontransgendered individuals, the average expression of ALDH1A1 (70.9 ± 15.1 pmol/mg testis) was approximately 42-fold higher than that of ALDH1A2 (1.7 ± 0.3 pmol/mg testis) and 51-fold higher than ALDH1A3 (1.4 ± 0.5 pmol/mg testis). A positive correlation was observed between the expression levels of ALDH1A1 and ALDH1A2 (R2 = 0.27, P = 0.05) and ALDH1A1 and ALDH1A3 (R2 = 0.30, P = 0.04). However, there was no correlation between the expression levels of ALDH1A2 and ALDH1A3.
Metabolism of at-retinal and 13-cis-retinal by ALDH1As
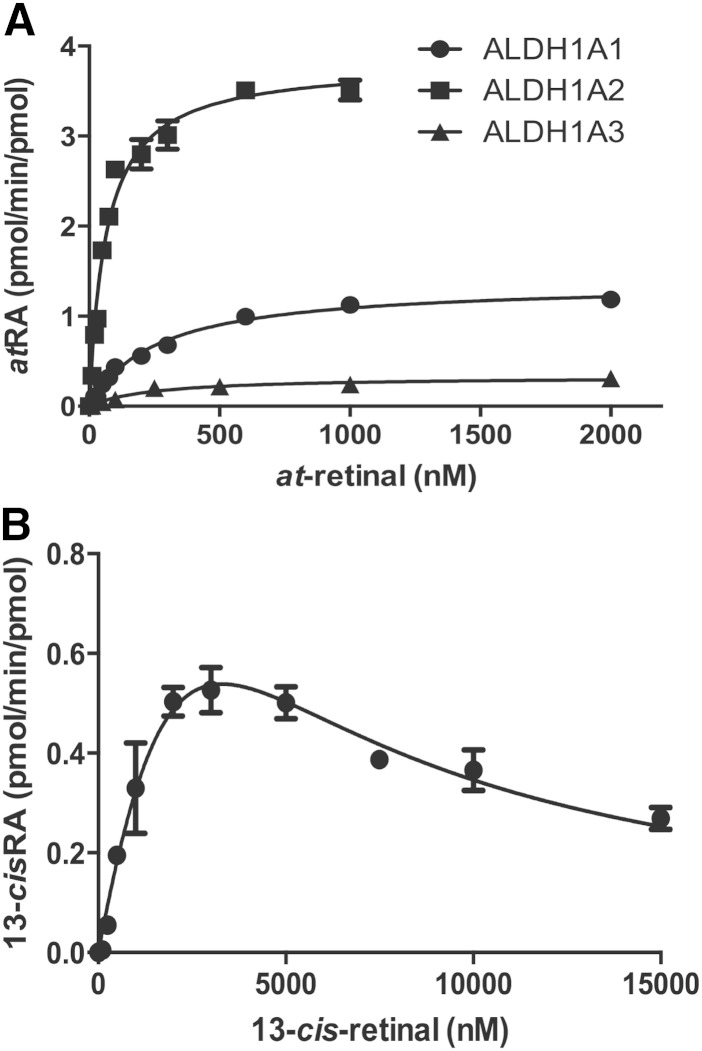
The formation kinetics of atRA from at-retinal and 13-cisRA from 13-cis-retinal by purified ALDH1A1, ALDH1A2, and ALDH1A3 were characterized to determine the specific activity of each of these enzymes in atRA and 13-cisRA formation (Table 2). All three ALDH1A enzymes formed atRA from at-retinal, but only ALDH1A1 showed detectable formation of 13-cisRA from 13-cis-retinal (Fig. 2). Of the ALDH1A enzymes, at-retinal had the highest affinity for ALDH1A2 (Km = 56 nM), and the affinities of at-retinal to ALDH1A1 and ALDH1A3 were both 5-fold lower (Km = 285 and 261 nM). In addition, ALDH1A2 had the greatest efficiency to form atRA. The intrinsic clearance (kcat/Km) of atRA formation by ALDH1A2 was 27-fold greater than that by ALDH1A1 and 60-fold greater than that by ALDH1A3 (Table 2). Interestingly, 13-cisRA formation by ALDH1A1 demonstrated substrate inhibition kinetics (Fig. 2). The kcat, Km, and Ki values relevant for a substrate inhibition model together with kinetic constants obtained for at-retinal with the ALDH1A enzymes are listed in Table 2.
TABLE 2.
Enzyme kinetic parameters for RA formation from retinal by ALDH1A enzymes
| Enzyme | at-Retinal | 13-cis-Retinal | |||||
| kcat (min−1) | Km (nM) | Clint (ml/min/pmol) | kcat (min−1) | Km (nM) | Ki (nM) | Clint (μl/min/pmol) | |
| ALDH1A1 | 1.1 ± 0.1 | 285 ± 24 | 3.9 | 3.0 ± 2.2 | 7,434 ± 6,700 | 1,468 ± 1,320 | 0.4 |
| ALDH1A2 | 3.7 ± 0.1 | 56 ± 5 | 66.1 | N/A | N/A | N/A | N/A |
| ALDH1A3 | 0.3 ± 0.01 | 261 ± 18 | 1.1 | N/A | N/A | N/A | N/A |
Clint, intrinsic clearance calculated as kcat/Km.
Fig. 2.
Kinetics of RA formation by recombinant ALDH1A proteins. Recombinant purified human ALDH1A was used to determine the kinetics of atRA formation by each ALDH1A enzyme (A). Because only ALDH1A1 formed 13-cisRA from 13-cis-retinal, only 13-cisRA formation by ALDH1A1 was characterized (B). The kinetic constants obtained are shown in Table 2.
Characterization of 13-cisRA and atRA formation in human testis samples
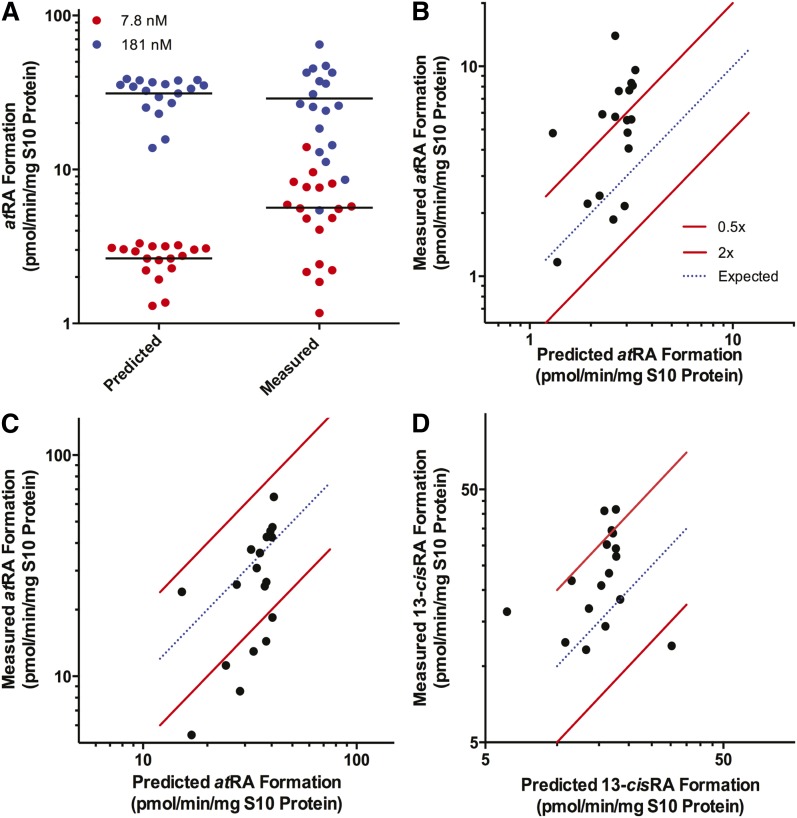
To establish the relative importance of the ALDH1A enzymes in the human testis, and to test whether the measured ALDH1A expression levels and the determined in vitro atRA formation kinetics predict the net atRA formation velocity in the human tissue, atRA formation from at-retinal was measured and predicted at two nominal concentrations (100 and 1,000 nM) of at-retinal for each individual in the study (Fig. 3). The substrate concentrations in the in vitro system were corrected for the protein binding of at-retinal measured at 100 and 1,000 nM at-retinal. The fu of at-retinal was 7.8 ± 0.7% at 100 nM and 18.1 ± 7.8% at 1,000 nM. The measured concentration of CRBP1 in the in vitro incubations was <0.5 nM (see below) and the effect of CRBP binding was considered insignificant for at-retinal in the incubations. Therefore, the free concentrations used in the predictions were 7.8 and 181 nM. Due to the sole contribution of ALDH1A1 to 13-cisRA formation from 13-cis-retinal, the formation of 13-cisRA was measured and predicted only at the nominal concentration of 1,000 nM 13-cis-retinal. The 13-cis-retinal concentration was corrected for the measured unbound fraction in the in vitro system (fu = 60.0 ± 5.1%), and the free concentration of 600 nM 13-cis-retinal was used for all predictions. The average observed and predicted velocities for atRA formation from 7.8 nM free at-retinal in the human testis were 5.6 ± 3.2 pmol/min/mg S10 protein and 2.6 ± 0.6 pmol/min/mg S10 protein, respectively. At 181 nM free at-retinal, the mean observed and predicted velocities were 33.3 ± 7.7 pmol/min/mg S10 protein and 28.9 ± 15.9 pmol/min/mg S10, respectively (Fig. 3). The average observed and predicted 13-cisRA formation velocities at 600 nM 13-cis-retinal were 25.5 ± 9.1 pmol/min/mg S10 and 15.2 ± 3.2 pmol/min/mg S10, respectively. The correlations between the predicted and observed atRA and 13-cisRA formation velocities are shown in Fig. 3. RA formation (ALDH1A activity) was accurately predicted at each retinal concentration with average fold error values of 0.6 (7.8 nM free at-retinal), 1.3 (181 nM free at-retinal), and 0.8 (600 nM free 13-cis-retinal).
Fig. 3.
Comparison of the predicted and measured ALDH1A activity in human testis. atRA formation was measured and predicted for the 18 subjects with at-retinal as the substrate at free concentrations of 8 nM (red circles) and 181 nM (blue circles) (A). Each dot represents an individual in the study. The measured activity was plotted as a function of predicted activity in each subject with 7.8 nM free at-retinal (B), 181 nM free at-retinal (C), and 600 nM free 13-cis-retinal (D). The blue dotted line represents an exact correlation between predicted and measured RA formation (ALDH1A activity). The red lines represent the range between 2-fold overprediction and 0.5-fold underprediction.
Relative importance of individual ALDH1A enzymes to atRA formation in human testis samples
The relative importance of each ALDH1A enzyme in atRA formation [fraction of retinal metabolized by a given enzyme (fm)] was predicted using ALDH1A expression levels and atRA formation kinetics as a function of substrate concentration. The predicted relative importance of each ALDH1A enzyme at increasing at-retinal concentrations is shown in Fig. 4. Overall, ALDH1A1 is predicted to be the main enzyme forming atRA in the testis with >80% contribution to the overall atRA formation. ALDH1A2 was predicted to play a role in atRA formation (∼15%), while the contribution of ALDH1A3 appeared insignificant. Due to the different Km values for at-retinal with ALDH1A2 and ALDH1A1, the predicted contribution of ALDH1A2 increases with decreasing retinal concentrations. The published concentration of retinal in the mouse testis (90.7 pmol/gram) (41) is similar to ALDH1A2 Km. However, the free concentration of retinal in the testis is unknown as well as the fraction of retinal bound to CRBP1 in the testis and hence the contribution of ALDH1A2 could be greater.
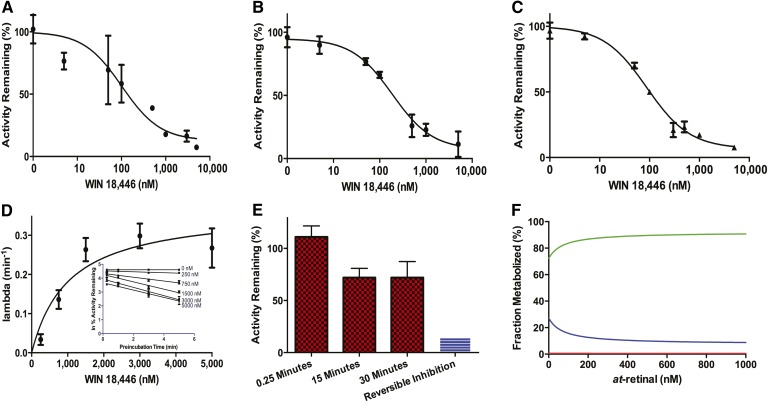
Fig. 4.
Inhibition of ALDH1A and RA formation by WIN 18,446. The inhibition kinetics of WIN 18,446 were characterized and used to determine the fraction of atRA formation by ALDH1A2 in the human testis. The reversible inhibition kinetics of WIN 18,446 with recombinant ALDH1A1 (A) (IC50 = 102 nM) and ALDH1A3 (B) (IC50 = 187 nM) were determined together with the effect of WIN 18,446 on atRA formation in testicular S10 fractions (C) (IC50 = 88 nM). Due to the time-dependent inhibition of ALDH1A2 by WIN 18,446, the rate of inactivation was determined with increasing concentrations of inhibitor (D, inset) and plotted as a function of inhibitor concentration (D) to determine the KI and kinact. In order to measure the fm for ALDH1A2 in the testis, S10 protein was preincubated with inhibitor for 0.25, 10, and 30 min to inactivate any ALDH1A2 protein (E). The preincubation was diluted out to remove reversible inhibition, and the ALDH1A activity was determined. The 25% decrease in ALDH1A activity (E) can be attributed to the contribution by ALDH1A2. The predicted relative contribution of each ALDH1A enzyme (green, ALDH1A1; blue, ALDH1A2; red, ALDH1A3) to atRA formation as a function of at-retinal concentration is shown in (F).
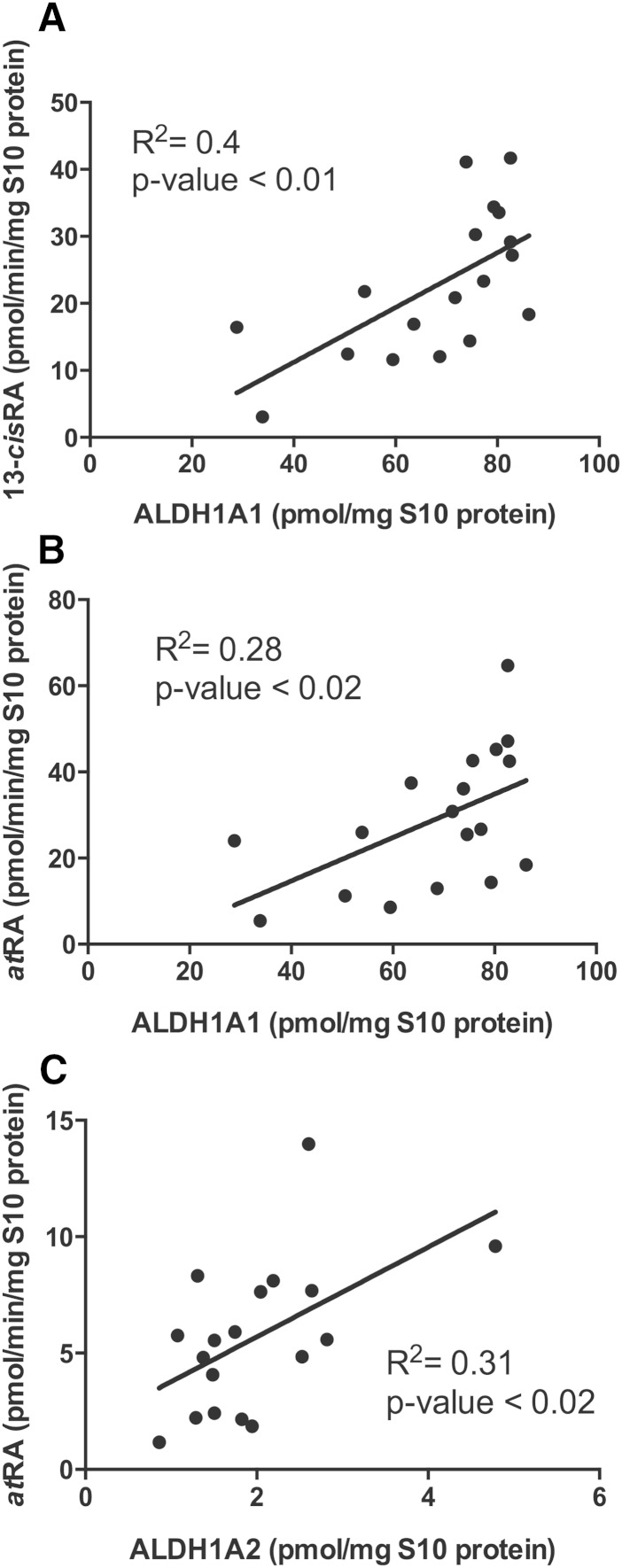
Based on the selective formation of 13-cisRA by ALDH1A1, ALDH1A1 was predicted to be the only contributor to 13-cisRA formation in incubations with at-retinal. In agreement with this, a significant (P < 0.01) correlation between ALDH1A1 protein expression and 13-cisRA formation (R2 = 0.41) was observed (Fig. 5). In addition, ALDH1A1 protein concentration had a significant correlation with atRA formation velocity measured with 181 nM at-retinal (P = 0.02, R2= 0.28) (Fig. 5). However, only ALDH1A2 expression had a positive correlation (P = 0.02, R2 = 0.33) with atRA formation measured at 7.8 nM at-retinal (Fig. 5).
Fig. 5.
The correlation between ALDH1A protein expression and RA formation velocity measured in S10 fraction from testes from each individual in the study. The correlations between 13-cisRA (A) and atRA (B) formation with ALDH1A1 expression are shown with nominal 1,000 nM substrate. The correlation between atRA formation and ALDH1A2 expression at 100 nM nominal substrate concentration is shown in (C).
To further evaluate the relative contributions of ALDH1A enzymes to atRA formation, an inhibitor of ALDH1A enzymes, WIN 18,446, was used as a selective ALDH1A inhibitor. WIN 18,446 was found to be a potent reversible inhibitor of ALDH1A1 and ALDH1A3 (IC50 values 102 ± 2 nM and 187 ± 1 nM), and an efficient time dependent inhibitor (kinact = 22.0 ± 2.4 h−1, KI = 1,026 ± 374 nM) of ALDH1A2 (Fig. 4). Based on the strong inhibition of all three ALDH1A enzymes by WIN 18,446, the effect of WIN 18,446 on atRA formation in human testis S10 fractions was also evaluated. Using a pooled testis S10 fraction of five men, the IC50 for WIN 18,446 against testicular ALDH1A activity (atRA formation) was 88 ± 1 nM. This IC50 value is in excellent agreement with the IC50 with recombinant ALDH1A1 suggesting that this enzyme contributes the majority of atRA formation in the in vitro system (Fig. 4). At concentrations in which WIN 18,446 inhibits recombinant ALDH1A activity >90%, the atRA formation in the testis was inhibited >90% demonstrating that ALDH1A enzymes are the predominant enzymes involved in atRA formation from at-retinal in the testis. To evaluate the specific contribution of ALDH1A2 to atRA formation, the TDI of atRA formation in the testes samples was determined. After a 30 min incubation with WIN 18,446, 97% of the ALDH1A2 is predicted to be inactivated and TDI by WIN 18,446 can be used to determine ALDH1A2 contribution to the atRA formation. The observed decrease in atRA formation after 30 min preincubation with WIN 18,446 was 28 ± 15% (Fig. 4). This observed 28% contribution of ALDH1A2 to testicular atRA formation is consistent with the average 15% contribution predicted from the recombinant enzyme data.
The effect of CRBP1 on RA formation by ALDH1A enzymes and in testes S10 fractions
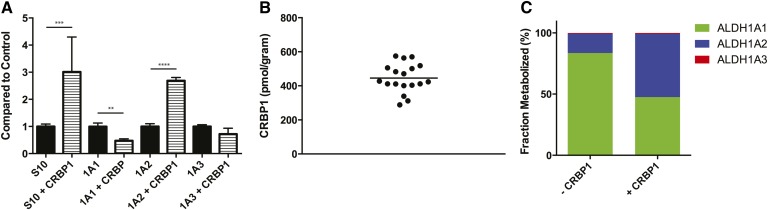
To determine whether CRBPs affect the formation of atRA in human testis, the formation of atRA by recombinant ALDH1A and testes S10 protein was measured with holo-CRBP1. The formation of atRA was increased 3-fold in incubations of testis S10 protein with holo-CRBP1 in comparison to at-retinal (Fig. 6A). In order to determine the enzyme(s) responsible for the increase in atRA formation, the effect of holo-CRBP1 on atRA formation with each ALDH1A enzyme was determined (Fig. 6A). atRA formation by ALDH1A1 was decreased 52% when holo-CRBP1 was used as a substrate compared with at-retinal. In contrast atRA formation by ALDH1A2 was increased 2.7-fold in the presence of CRBP1 when compared with free at-retinal. atRA formation by ALDH1A3 was unchanged in the presence of CRBP1. CRBP1 expression in the human testis was quantified in each study subject and the average concentration of CRBP1 was 445 ± 85 pmol/gram testis (445 nM) (Fig. 6B). The effect of CRBP1 on the contribution of each ALDH1A enzyme to atRA formation in the testis was predicted at 100 nM concentration of at-retinal. In the absence of CRBP1, ALDH1A1 is predicted to be the main contributor to atRA formation (84%). However, if all at-retinal is bound to CRBP1, ALDH1A2 is predicted to contribute to the majority of atRA formation (52%) (Fig. 6C).
Fig. 6.
The effect of CRBP1 on atRA formation and CRBP expression level in the human testis. The effect of CRBP1 on atRA formation by testicular S10 protein and by recombinant ALDH1A1, ALDH1A2, and ALDH1A3 was determined and the formation of atRA from 300 nM holo-CRBP compared with free at-retinal (300 nM) is shown (A). The concentration of 300 nM was chosen to mimic expected in vivo concentrations as closely as possible. CRBP1 expression in the testis S10 fractions was quantified in all subjects by ELISA and the expression levels are shown (B). The relative contribution of each ALDH1A enzyme to atRA formation in the presence or absence of CRBP1 was predicted from a substrate (at-retinal or holo-CRBP) at 100 nM as described in the Materials and Methods and the predicted percent contribution by each ALDH1A is shown in (C).
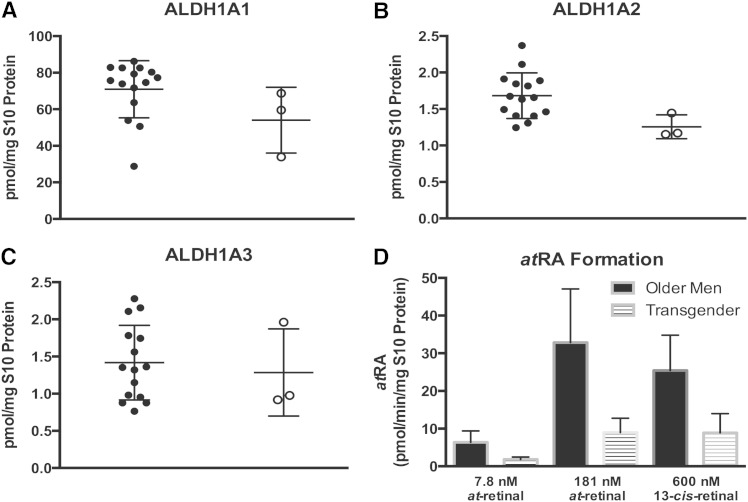
Alterations in the intratesticular ALDH1A activity in transgender individuals
To further evaluate the function of ALDH1A enzymes in the human testis, the ALDH1A activity and protein concentrations were measured in a sample set of three transgender individuals who had undergone hormonal sex reassignment therapy. The average protein concentrations of ALDH1A1, ALDH1A2, and ALDH1A3 were 54.0 ± 14.7 pmol/mg S10 protein, 1.3 ± 0.2 pmol/mg S10 protein, and 1.3 ± 0.5 pmol/mg S10 protein in the transgender donors (Fig. 7). ALDH1A2 expression was significantly (P < 0.05) decreased in the testes of the transgendered individuals compared with the men. Additionally, the ALDH1A1 expression level was 20% lower in these donors than in the other donors, but this difference did not achieve significance (P > 0.05). The ALDH1A mediated atRA formation was significantly (P < 0.02) lower in the testis from transgender donors than in the men undergoing orchiectomy at both concentrations of at-retinal (1.8 ± 0.6 pmol/min/mg S10 protein at 7.8 nM and 9.0 ± 3.4 pmol/min/mg S10 protein at 181 nM) (Fig. 7). Additionally, the formation of 13-cisRA from 13-cis-retinal was significantly (P < 0.01) decreased (8.9 ± 4.1 pmol/min/mg S10 protein) in these individuals when compared with the other donors.
Fig. 7.
Comparison of the ALDH1A protein expression and activity between men and transgender individuals. There was a significant decrease in the ALDH1A2 protein concentration in the three transgender individuals compared with the men (B). However, there was no observed difference in the testicular protein concentration of ALDH1A1 (A) or ALDH1A3 (C). Testicular ALDH1A activity was significantly lower in the transgender individuals at both 7.8 and 181 nM at-retinal (P < 0.02) and 600 nM 13-cis-retinal (P < 0.01) (D).
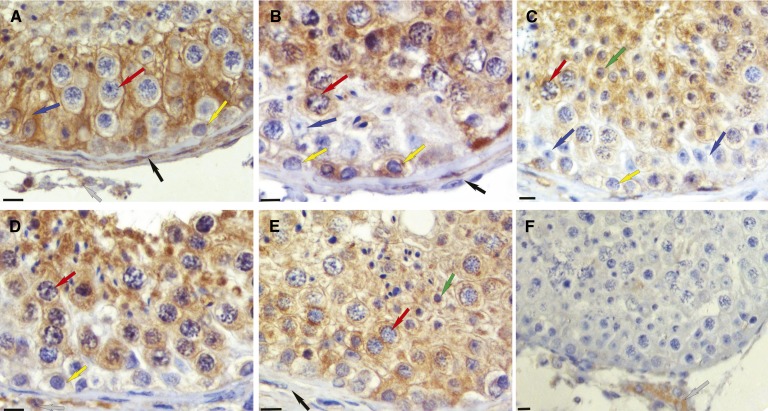
ALDH1A localization in the human testis
The predicted relative contribution of ALDH1A1 to atRA formation in the testis must be reconciled with the fact that male ALDH1A1 knockout mice are fertile (14). Hence, it was hypothesized that the tissue localization of the ALDH1A enzymes within the testis may explain this phenotypic discrepancy. To test this, immunohistochemistry was performed to investigate the cell type-specific expression pattern of the three ALDH1A enzymes within testicular tissue from three healthy fertile men (Fig. 8). The three ALDH1A enzymes were expressed in distinct cell types and the expression patterns were different for each isozyme. Sertoli and peritubular myoid cells were all positive for ALDH1A1, but this enzyme could not be detected within any germ cell type (Fig. 8A). In contrast, ALDH1A2 localized to pachytene spermatocytes and round spermatids with signal also present in some spermatogonia and peritubular myoid cells (Fig. 8B–D). The third isoform, ALDH1A3, displayed a similar expression pattern, with protein detected in pachytene spermatocytes, spermatogonia, and Sertoli cells (Fig. 8E). Under these conditions, cells of the interstitium are positive without primary antibody, and therefore are possibly nonspecifically stained in all. Taken together, the localization patterns of the individual ALDH1A enzymes suggest that the various cell types of the testis can all synthesize RA, but do so utilizing distinct ALDH1A isoforms.
Fig. 8.
Localization of ALDH1A enzymes in the human testis. ALDH1A1, ALDH1A2, and ALDH1A3 have distinct localization patterns in the human testis. Each panel is representative of immunohistochemical results from three different human testes. All were fixed in Bouin’s fluid and embedded in paraffin. Brown stain indicates immunopositive reaction for ALDH1A1 (A), ALDH1A2 (B–D), and ALDH1A3 (E). A negative control (F) used rabbit IgG instead of primary rabbit antibody. The original magnification was 225× for each. The bar at lower left of each panel represents 10 microns. Cell types are indicated by arrows as follows: blue, Sertoli cells; yellow, spermatogonia; red, primary spermatocytes; green, round spermatids; black, peritubular cells; gray, interstitial cells.
Correlation of intratesticular RA concentrations with ALDH1A activity
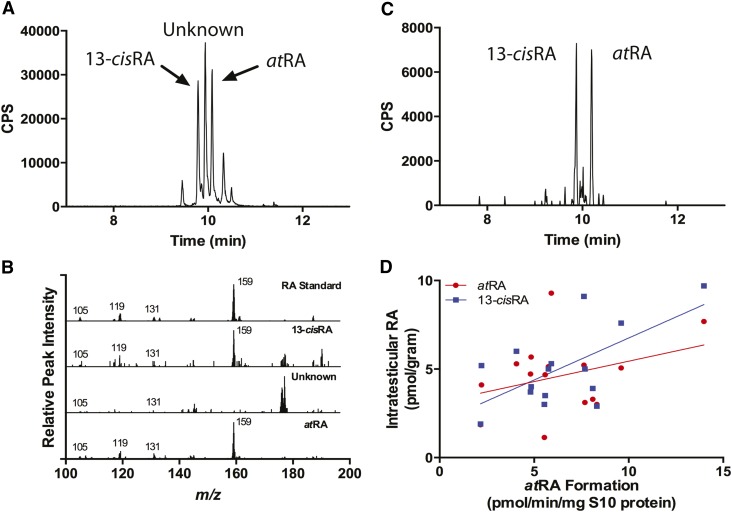
To test whether tissue atRA and 13-cisRA concentrations were affected by ALDH1A activity in the same tissue, intratesticular atRA and 13-cisRA concentrations were measured in the testes samples from the 15 donors with prostate cancer (Fig. 9). The intratesticular RA concentrations were 4.3 ± 0.5 pmol/gram for atRA and 5.1 ± 0.6 pmol/gram for 13-cisRA, and concentrations of atRA and 13-cisRA were positively correlated (R2 = 0.36, P = 0.02). An unknown peak was observed in each sample that eluted between the atRA and 13-cisRA isomers. Based on the fact the 9-cisRA isomer elutes between the atRA and 13-cisRA isomers (35), MS/MS/MS was used to determine whether the unknown peak was a RA isomer. The fragmentation patterns of the atRA and 13-cisRA isomers were identical and generated MS/MS/MS fragments of m/z 159+, 119+, 131+, and 105+ (Fig. 9B). An identical fragmentation pattern was observed for the atRA and 13-cisRA peaks measured from human testicular tissue. However, the fragments generated from the unknown peak were distinctly different and suggest it is not a RA isomer. Based on the abundance of the m/z 159+ fragment formed from RA and not the unknown peak, the 159+ fragment was extracted from the MS/MS/MS data (Fig. 9C). The extraction of the fragment allows for clear identification of RA isomers from matrix components observed using MRM. The ALDH1A activity (atRA formation) measured at the biologically relevant 7.8 nM concentration of at-retinal correlated positively with intratesticular 13-cisRA concentrations (R2 = 0.40, P = 0.01) and total RA concentration (R2 = 0.30, P = 0.03). However, intratesticular atRA concentrations did not correlate significantly with ALDH1A activity at either of the at-retinal concentrations (P = 0.22 at 7.8 nM and 0.94 at 181 nM) (Fig. 9D).
Fig. 9.
Quantification of intratesticular RA and correlation of RA concentration and measured RA formation in human testes. Intratesticular atRA and 13-cisRA were measured using LC-MS/MS and a representative chromatogram is shown (A). To confirm peak identity, a MS/MS/MS experiment was conducted and full MS/MS/MS spectra were collected from ion transition m/z 301.2+ > 205.3+ (B). The spectra shown in (B) are of RA standard and of the three main peaks labeled in (A), 13-cisRA, unknown, and atRA. C: Shows the extracted MS/MS/MS chromatogram of the m/z 301.2+ > 205.3+ > 159.1+ transition. The correlation between formation and atRA or 13-cisRA concentrations in human testis is shown in (D). In addition, total RA had a significant positive correlation with atRA formation.
DISCUSSION
The current study describes the development of novel methods necessary to study the formation of RA by ALDH1A enzymes and the application of these methods to characterization of RA formation in human testis. While the indispensible nature of RA in the testis is well-established, the complement of human enzyme(s) responsible for RA formation has been poorly characterized. The methods developed in this study provide the foundation to determine the tissue-specific importance of each ALDH1A enzyme to RA formation, and establish novel tools to evaluate the cell and tissue-specific mechanisms that allow for generation of RA gradients required for the regulation of biological processes. The developed methods are particularly useful for evaluating the role of altered RA homeostasis in human diseases, as all these methods can be used to study ALDH1A localization, expression level, activity, and RA concentration using a single tissue biopsy of 100 mg.
The testis ALDH1A expression levels reported in this study present, for the first time, the quantitative expression levels of ALDH1A1, ALDH1A2, and ALDH1A3 in any human tissue. Human tissue-specific mRNA expression profiles of each ALDH1A enzyme have been previously shown and based on the mRNA levels, the rank order of ALDH1A expression in the testis is ALDH1A2 (59%) > ALDH1A1 (29%) > ALDH1A3 (12%) (18). This is in contrast to the current study showing ALDH1A1 being the predominant enzyme and emphasizes the need to measure protein expression levels instead of mRNA. Another previous study used in situ hybridization to determine the localization of ALDH1A enzymes in the mouse testis. In that study, ALDH1A1 was found in Leydig cells and Sertoli cells, ALDH1A2 in pachytene spermatocytes and step 1–6 spermatids, and ALDH1A3 in Leydig cells (12). The protein localization reported here is in generally good agreement with the previous mouse mRNA data, although, unlike in mouse mRNA studies, ALDH1A2 protein was found in the spermatogonia of the human testis. The distinct ALDH1A localization in human testicular tissue suggests that each ALDH1A may play a different role in RA formation in different cell types of the testis. Previous work in knockout mice has shown that ALDH1A enzymes in Sertoli cells are responsible for the initial source of RA during spermatogenesis, while ALDH1A in spermatocytes may help maintain RA formation after the first wave of spermatogenesis (30).
This study shows that overall in the human testis, ALDH1A1 and ALDH1A2 contribute significantly to formation of atRA from at-retinal and their relative contributions will depend on the interactions of the ALDH1A enzymes with CRBP1 in the specific cell types. The WIN 18,446 inhibition data unequivocally shows that RA formation in the testis is dependent on ALDH1A enzymes and not other enzymes such as aldehyde oxidases or xanthine oxidases. Together with the correlation between ALDH1A expression and atRA and 13-cisRA formation in vitro, the WIN 18,446 inhibition confirms the validity of the protein quantification and recombinant enzyme kinetic data. The correlation between ALDH1A2 expression and atRA formation together with the predicted importance of ALDH1A2 in the presence of CRBP, suggests that ALDH1A2 makes a significant contribution to atRA formation, and that its expression is at least in part responsible for the interindividual variability in RA formation in the testis. As shown by the expression of CRBP1 in the human testes and the ALDH1A isoform specific effects of CRBP1 on RA formation, prediction of the in vivo contribution of each individual ALDH1A enzyme is complex and likely also affected by the fraction of CRBP1 that is bound by retinol in vivo. The expression of cellular RA binding proteins that may affect product release from ALDH1As may also alter the predicted contributions in vivo. Previously, rat ALDH1A1 has been shown to recognize holo-CRBP1 as a substrate (9), but the effect of CRBP1 on atRA formation by human ALDH1A enzymes has never been reported. Similarly CBRP1 expression in the Sertoli cells of rats (42) has been shown suggesting that CRBPs play an important role in modulating intratesticular RA metabolism. While ALDH1A1 was predicted to be the major contributor to atRA formation in the absence of CRBPs, when the inhibition of ALDH1A1 and activation of ALDH1A2 by CRBP1 was accounted for, ALDH1A2 was predicted to be the predominant enzyme forming atRA in the testis. The observed increase in RA formation in the presence of CRBP1 in the testis S10 fractions is in agreement with the effects observed in the recombinant enzymes and supports the role of ALDH1A2 in RA formation in the testis. The predicted primary contribution of ALDH1A2 to RA formation in the presence of CRBP1 is also in agreement with the fact that ALDH1A1−/− mice are fertile and no obvious defects are detected in these mice at adulthood.
Interestingly, ALDH1A1 was the only ALDH1A enzyme capable of forming 13-cisRA from 13-cis-retinal. The 13-cisRA formation by ALDH1A1 displayed substrate inhibition kinetics suggesting multiple 13-cis-retinal molecules may simultaneously bind ALDH1A1. Despite this, the kinetics determined from the substrate inhibition model could be used to predict 13-cisRA formation velocity in individual human samples in vitro. The fact that ALDH1A1 was the one enzyme able to form 13-cisRA is of interest because in a preliminary study of 24 men, an association between intratesticular 13-cisRA and abnormal semen was revealed, suggesting that altered intratesticular RA homeostasis may be present in infertile men (43). In addition, 13-cisRA treatment has been shown to increase sperm production in humans (44). Based on the results of this study, the decreased intratesticular 13-cisRA concentration may suggest a decreased expression of ALDH1A1 in men with abnormal semen. However, at present the source of intratesticular 13-cisRA is not known and it could be formed either via isomerization from atRA or enzymatically from 13-cis-retinal if it is present in human tissues. Also, it is unknown if 13-cisRA is necessary for spermatogenesis, and the cause of the differences in RA concentrations with different fertility diagnoses has not been identified.
A significant decrease in the RA formation velocity was observed in the testicular tissue of transgender individuals, possibly due to the significant decrease in ALDH1A2 expression. It seems likely that the presurgical hormonal therapy with estradiol commonly given prior to gender reassignment surgery may have affected ALDH1A2 expression. In support of this notion, a mouse model lacking a functional androgen receptor has significantly decreased mRNA for both Aldh1a1 and Aldh1a2 (45). Additionally, ALDH1A2 expression is significantly decreased in infertile men diagnosed with Sertoli cell only syndrome on testicular biopsy (46). The reduced expression of ALDH1A2 in these men is likely related to the reduction in the numbers of developing sperm cells, consistent with our observation that spermatogonia, spermatids, and spermatocytes were the main cell type staining for ALDH1A2 in the histological analysis. Although the donors in this study were men with prostate cancer undergoing orchiectomy, the good agreement with this data and prior clinical findings suggests that these men reflect healthy, fertile men in terms of testicular function. Yet, further studies in different populations of men, including young healthy men and infertile men are needed to fully characterize the role of the different ALDH1A enzymes in male reproduction. Taken together, these data demonstrate the critical importance of understanding the specific enzymology of RA synthesis in the testis.
The correlation discovered between RA formation activity and RA concentration in the human testis described here is, to our knowledge, the first published example of a correlation between human tissue concentrations of an endogenous substance and the activity of the enzyme responsible for its formation. With the evidence suggesting intratesticular RA is formed within the tissue and assuming no interindividual differences in the clearance of intratesticular RA, a correlation between the RA synthesis rate and the measured intratesticular RA is expected. The reason intratesticular 13-cisRA correlated with ALDH1A activity and atRA did not, may be due to the fact 13-cisRA is a poor substrate for CYP26 enzymes compared with atRA (47). Therefore, interindividual differences in CYP26 expression are more likely to affect the intratesticular concentration of atRA.
The results of this study demonstrate the usefulness of novel MS approaches to model RA formation in human tissue, using the testis as an example. RA formation kinetics was accurately characterized using two novel methods to measure ALDH1A concentrations and determine RA formation by ALDH1A in human tissue biopsies. ALDH1A1 is the predominant enzyme in the formation of RA in the Sertoli cells, while the specific localization of ALDH1A2 in the developing sperm indicates a critical role for ALDH1A2 in these cell types. The distinct localization of the ALDH1A enzymes, together with their different interactions with CRBP1, suggests distinct cell type-specific roles of the different ALDH1A enzymes in intratesticular atRA formation. Further studies are needed to establish the full biological relevance of these findings. These results illustrate the need to determine the contribution of each ALDH1A to RA formation and the localization of each enzyme in individual target tissues. Tissue RA concentrations by themselves will not give an accurate representation of how RA formation is controlled within the tissue. Furthermore, given the indispensible role of RA in many other biological functions, the methods described will provide a valuable tool in understanding how RA homeostasis is altered by or potentially leads to disorders associated with changes in ALDH1A expression.
Acknowledgments
The authors wish to thank Dr. Mary Ellen Zoulas, DVM, at the Seattle Animal Shelter for donating the dog testicular tissue, Reinhild Sandhowe for technical support in processing the human tissue samples, and Dr. Maureen Kane for helpful discussion regarding RA quantification.
Footnotes
Abbreviations:
- ACN
- acetonitrile
- afe
- average fold error
- ALDH
- aldehyde dehydrogenase
- atRA
- all-trans-retinoic acid
- CRBP
- cellular retinol binding protein
- fm
- fraction metabolized by a particular enzyme
- fu
- fraction unbound
- holo-CRBP
- cellular retinol binding protein bound with an equal concentration of retinal
- RA
- retinoic acid
- TDI
- time-dependent inhibition
- UHPLC
- ultra HPLC
This study was supported in part by National Institutes of Health Grants R01 GM081569-S1 (to N.I.), R01 GM111772 (to N.I.), U01 HD060408 (to J.K.A.), R01 10808 (to M.D.G.), and National Center for Advancement of Translational Sciences (NCATS) Grant TL1 TR000422 (to S.L.A.). The Eunice Kennedy Shriver National Institute of Child Health and Human Development supported this work through cooperative agreement U54 HD42454 as part of the Cooperative Contraceptive Research Centers Program.
REFERENCES
- 1.Ross A. C. 2007. Vitamin A supplementation and retinoic acid treatment in the regulation of antibody responses in vivo. Vitam. Horm. 75: 197–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chung S. S., Wang X., Wolgemuth D. J. 2009. Expression of retinoic acid receptor alpha in the germline is essential for proper cellular association and spermiogenesis during spermatogenesis. Development. 136: 2091–2100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Noy N. 2010. Between death and survival: retinoic acid in regulation of apoptosis. Annu. Rev. Nutr. 30: 201–217. [DOI] [PubMed] [Google Scholar]
- 4.Clagett-Dame M., Knutson D. 2011. Vitamin A in reproduction and development. Nutrients. 3: 385–428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chapellier B., Mark M., Messaddeq N., Calleja C., Warot X., Brocard J., Gerard C., Li M., Metzger D., Ghyselinck N. B., et al. 2002. Physiological and retinoid-induced proliferations of epidermis basal keratinocytes are differently controlled. EMBO J. 21: 3402–3413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.D’Ambrosio D. N., Clugston R. D., Blaner W. S. 2011. Vitamin A metabolism: an update. Nutrients. 3: 63–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kedishvili N. Y. 2013. Enzymology of retinoic acid biosynthesis and degradation. J. Lipid Res. 54: 1744–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Napoli J. L. 2012. Physiological insights into all-trans-retinoic acid biosynthesis. Biochim. Biophys. Acta. 1821: 152–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Posch K. C., Burns R. D., Napoli J. L. 1992. Biosynthesis of all-trans-retinoic acid from retinal. Recognition of retinal bound to cellular retinol binding protein (type I) as substrate by a purified cytosolic dehydrogenase. J. Biol. Chem. 267: 19676–19682. [PubMed] [Google Scholar]
- 10.Wang X., Penzes P., Napoli J. L. 1996. Cloning of a cDNA encoding an aldehyde dehydrogenase and its expression in Escherichia coli. Recognition of retinal as substrate. J. Biol. Chem. 271: 16288–16293. [DOI] [PubMed] [Google Scholar]
- 11.Graham C. E., Brocklehurst K., Pickersgill R. W., Warren M. J. 2006. Characterization of retinaldehyde dehydrogenase 3. Biochem. J. 394: 67–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vernet N., Dennefeld C., Rochette-Egly C., Oulad-Abdelghani M., Chambon P., Ghyselinck N. B., Mark M. 2006. Retinoic acid metabolism and signaling pathways in the adult and developing mouse testis. Endocrinology. 147: 96–110. [DOI] [PubMed] [Google Scholar]
- 13.Niederreither K., Subbarayan V., Dolle P., Chambon P. 1999. Embryonic retinoic acid synthesis is essential for early mouse post-implantation development. Nat. Genet. 21: 444–448. [DOI] [PubMed] [Google Scholar]
- 14.Ziouzenkova O., Orasanu G., Sharlach M., Akiyama T. E., Berger J. P., Viereck J., Hamilton J. A., Tang G., Dolnikowski G. G., Vogel S., et al. 2007. Retinaldehyde represses adipogenesis and diet-induced obesity. Nat. Med. 13: 695–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fan X., Molotkov A., Manabe S., Donmoyer C. M., Deltour L., Foglio M. H., Cuenca A. E., Blaner W. S., Lipton S. A., Duester G. 2003. Targeted disruption of Aldh1a1 (Raldh1) provides evidence for a complex mechanism of retinoic acid synthesis in the developing retina. Mol. Cell. Biol. 23: 4637–4648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Molotkov A., Duester G. 2003. Genetic evidence that retinaldehyde dehydrogenase Raldh1 (Aldh1a1) functions downstream of alcohol dehydrogenase Adh1 in metabolism of retinol to retinoic acid. J. Biol. Chem. 278: 36085–36090. [DOI] [PubMed] [Google Scholar]
- 17.Sládek N. E., Kollander R., Sreerama L., Kiang D. T. 2002. Cellular levels of aldehyde dehydrogenases (ALDH1A1 and ALDH3A1) as predictors of therapeutic responses to cyclophosphamide-based chemotherapy of breast cancer: a retrospective study. Rational individualization of oxazaphosphorine-based cancer chemotherapeutic regimens. Cancer Chemother. Pharmacol. 49: 309–321. [DOI] [PubMed] [Google Scholar]
- 18.Nishimura M., Naito S. 2006. Tissue-specific mRNA expression profiles of human phase I metabolizing enzymes except for cytochrome P450 and phase II metabolizing enzymes. Drug Metab. Pharmacokinet. 21: 357–374. [DOI] [PubMed] [Google Scholar]
- 19.Zhai Y., Sperkova Z., Napoli J. L. 2001. Cellular expression of retinal dehydrogenase types 1 and 2: effects of vitamin A status on testis mRNA. J. Cell. Physiol. 186: 220–232. [DOI] [PubMed] [Google Scholar]
- 20.Yang L., Ren Y., Yu X., Qian F., Bian B. S., Xiao H. L., Wang W. G., Xu S. L., Yang J., Cui W., et al. 2014. ALDH1A1 defines invasive cancer stem-like cells and predicts poor prognosis in patients with esophageal squamous cell carcinoma. Mod. Pathol. 27: 775–783. [DOI] [PubMed] [Google Scholar]
- 21.Xing Y., Luo D. Y., Long M. Y., Zeng S. L., Li H. H. 2014. High ALDH1A1 expression correlates with poor survival in papillary thyroid carcinoma. World J. Surg. Oncol. 12: 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang K., Chen X., Zhan Y., Jiang W., Liu X., Wang X., Wu B. 2013. Increased expression of ALDH1A1 protein is associated with poor prognosis in clear cell renal cell carcinoma. Med. Oncol. 30: 574. [DOI] [PubMed] [Google Scholar]
- 23.Kim H., Lapointe J., Kaygusuz G., Ong D. E., Li C., van de Rijn M., Brooks J. D., Pollack J. R. 2005. The retinoic acid synthesis gene ALDH1a2 is a candidate tumor suppressor in prostate cancer. Cancer Res. 65: 8118–8124. [DOI] [PubMed] [Google Scholar]
- 24.Touma S. E., Perner S., Rubin M. A., Nanus D. M., Gudas L. J. 2009. Retinoid metabolism and ALDH1A2 (RALDH2) expression are altered in the transgenic adenocarcinoma mouse prostate model. Biochem. Pharmacol. 78: 1127–1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.El Kares R., Manolescu D. C., Lakhal-Chaieb L., Montpetit A., Zhang Z., Bhat P. V., Goodyer P. 2010. A human ALDH1A2 gene variant is associated with increased newborn kidney size and serum retinoic acid. Kidney Int. 78: 96–102. [DOI] [PubMed] [Google Scholar]
- 26.Styrkarsdottir U., Thorleifsson G., Helgadottir H. T., Bomer N., Metrustry S., Bierma-Zeinstra S., Strijbosch A. M., Evangelou E., Hart D., Beekman M., et al. 2014. Severe osteoarthritis of the hand associates with common variants within the ALDH1A2 gene and with rare variants at 1p31. Nat. Genet. 46: 498–502. [DOI] [PubMed] [Google Scholar]
- 27.Napoli J. L. 1996. Retinoic acid biosynthesis and metabolism. FASEB J. 10: 993–1001. [DOI] [PubMed] [Google Scholar]
- 28.Hogarth C. A., Griswold M. D. 2010. The key role of vitamin A in spermatogenesis. J. Clin. Invest. 120: 956–962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tong M. H., Yang Q. E., Davis J. C., Griswold M. D. 2013. Retinol dehydrogenase 10 is indispensible for spermatogenesis in juvenile males. Proc. Natl. Acad. Sci. USA. 110: 543–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Raverdeau M., Gely-Pernot A., Feret B., Dennefeld C., Benoit G., Davidson I., Chambon P., Mark M., Ghyselinck N. B. 2012. Retinoic acid induces Sertoli cell paracrine signals for spermatogonia differentiation but cell autonomously drives spermatocyte meiosis. Proc. Natl. Acad. Sci. USA. 109: 16582–16587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Munson L., Chassy L. M., Asa C. 2004. Efficacy, safety and reversibility of bisdiamine as a male contraceptive in cats. Theriogenology. 62: 81–92. [DOI] [PubMed] [Google Scholar]
- 32.Heller C. G., Moore D. J., Paulsen C. A. 1961. Suppression of spermatogenesis and chronic toxicity in men by a new series of bis(dichloroacetyl) diamines. Toxicol. Appl. Pharmacol. 3: 1–11. [DOI] [PubMed] [Google Scholar]
- 33.Amory J. K., Muller C. H., Shimshoni J. A., Isoherranen N., Paik J., Moreb J. S., Amory D. W., Sr, Evanoff R., Goldstein A. S., Griswold M. D. 2011. Suppression of spermatogenesis by bisdichloroacetyldiamines is mediated by inhibition of testicular retinoic acid biosynthesis. J. Androl. 32: 111–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.MacLean B., Tomazela D. M., Shulman N., Chambers M., Finney G. L., Frewen B., Kern R., Tabb D. L., Liebler D. C., MacCoss M. J. 2010. Skyline: an open source document editor for creating and analyzing targeted proteomics experiments. Bioinformatics. 26: 966–968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Arnold S. L., Amory J. K., Walsh T. J., Isoherranen N. 2012. A sensitive and specific method for measurement of multiple retinoids in human serum with UHPLC-MS/MS. J. Lipid Res. 53: 587–598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fierce Y., de Morais Vieira M., Piantedosi R., Wyss A., Blaner W. S., Paik J. 2008. In vitro and in vivo characterization of retinoid synthesis from beta-carotene. Arch. Biochem. Biophys. 472: 126–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Thatcher J. E., Zelter A., Isoherranen N. 2010. The relative importance of CYP26A1 in hepatic clearance of all-trans retinoic acid. Biochem. Pharmacol. 80: 903–912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Grimm S. W., Einolf H. J., Hall S. D., He K., Lim H. K., Ling K. H., Lu C., Nomeir A. A., Seibert E., Skordos K. W., et al. 2009. The conduct of in vitro studies to address time-dependent inhibition of drug-metabolizing enzymes: a perspective of the pharmaceutical research and manufacturers of America. Drug Metab. Dispos. 37: 1355–1370. [DOI] [PubMed] [Google Scholar]
- 39.Brown H. S., Ito K., Galetin A., Houston J. B. 2005. Prediction of in vivo drug-drug interactions from in vitro data: impact of incorporating parallel pathways of drug elimination and inhibitor absorption rate constant. Br. J. Clin. Pharmacol. 60: 508–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hogarth C. A., Griswold M. D. 2013. Immunohistochemical approaches for the study of spermatogenesis. Methods Mol. Biol. 927: 309–320. [DOI] [PubMed] [Google Scholar]
- 41.Kane M. A., Napoli J. L. 2010. Quantification of endogenous retinoids. Methods Mol. Biol. 652: 1–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kato M., Sung W. K., Kato K., Goodman D. S. 1985. Immunohistochemical studies on the localization of cellular retinol-binding protein in rat testis and epididymis. Biol. Reprod. 32: 173–189. [DOI] [PubMed] [Google Scholar]
- 43.Nya-Ngatchou J. J., Arnold S. L. M., Walsh T. J., Muller C. H., Page S. T., Isoherranen N., Amory J. K. 2013. Intratesticular 13-cis retinoic acid is lower in men with abnormal semen analyses: a pilot study. Andrology 1: 325–331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hoting V. E., Schütte B., Schirren C. 1992. Isotretinoin and acne conglobata: andrological evaluations [article in German]. Fortschr. Med. 110: 427–430. [PubMed] [Google Scholar]
- 45.O’Shaughnessy P. J., Abel M., Charlton H. M., Hu B., Johnston H., Baker P. J. 2007. Altered expression of genes involved in regulation of vitamin A metabolism, solute transportation, and cytoskeletal function in the androgen-insensitive tfm mouse testis. Endocrinology. 148: 2914–2924. [DOI] [PubMed] [Google Scholar]
- 46.Amory J. K., Arnold S., Lardone M. C., Piottante A., Ebensperger M., Isoherranen N., Muller C. H., Walsh T., Castro A. 2014. Levels of the retinoic acid synthesizing enzyme aldehyde dehydrogenase-1A2 are lower in testicular tissue from men with infertility. Fertil. Steril. 101: 960–966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Helvig C., Taimi M., Cameron D., Jones G., Petkovich M. 2011. Functional properties and substrate characterization of human CYP26A1, CYP26B1, and CYP26C1 expressed by recombinant baculovirus in insect cells. J. Pharmacol. Toxicol. Methods. 64: 258–263. [DOI] [PubMed] [Google Scholar]