The isolated Nav channel domain IV paddle motif remains susceptible to toxins that inhibit fast inactivation.
Abstract
Animal toxins that inhibit voltage-gated sodium (Nav) channel fast inactivation can do so through an interaction with the S3b–S4 helix-turn-helix region, or paddle motif, located in the domain IV voltage sensor. Here, we used surface plasmon resonance (SPR), an optical approach that uses polarized light to measure the refractive index near a sensor surface to which a molecule of interest is attached, to analyze interactions between the isolated domain IV paddle and Nav channel–selective α-scorpion toxins. Our SPR analyses showed that the domain IV paddle can be removed from the Nav channel and immobilized on sensor chips, and suggest that the isolated motif remains susceptible to animal toxins that target the domain IV voltage sensor. As such, our results uncover the inherent pharmacological sensitivities of the isolated domain IV paddle motif, which may be exploited to develop a label-free SPR approach for discovering ligands that target this region.
INTRODUCTION
Voltage-gated sodium (Nav) channels constitute a welcome target for venomous animals seeking to disrupt the transmission of electrical signals to incapacitate prey or defend against predators (Kalia et al., 2015). To this end, peptide toxins within these venoms have evolved to interact with a specific region within each of the four Nav channel voltage-sensing domains (VSDs), the S3b–S4 helix-turn-helix motif or paddle motif (Gilchrist et al., 2014). The pharmacological importance of this distinct region was first recognized in voltage-gated potassium (Kv) channels where mutations in the S3b–S4 loop reduced channel sensitivity to hanatoxin, a founding member of the Kv channel gating modifier toxin family (Li-Smerin and Swartz, 2000). Later, structural information revealed that the paddle motif makes few contacts with the rest of the channel protein (Long et al., 2007; Swartz, 2008), which prompted experiments in which the S3b–S4 region was swapped between voltage-gated ion channels without disrupting the voltage-sensing process (Alabi et al., 2007). The paddle motif was also identified in each of the four Nav channel voltage sensors, and transferring these regions from mammalian or insect Nav to Kv channels resulted in functional Kv channels that are sensitive to an array of Nav channel toxins (Bosmans et al., 2008, 2011; Bende et al., 2014). One recurring outcome of these studies is that Kv channels containing the VSD IV paddle motif of donor Nav channels possess slower kinetics when compared with constructs containing paddle motifs from the other three VSDs. These observations fit well with the notion that VSD I–III activation in response to changes in membrane voltage is most important for channel opening, whereas VSD IV plays a distinct role in fast inactivating the channel after it has opened (Bezanilla, 2008; Capes et al., 2013). As such, animal toxins that interact with the paddle motif in VSD I–III generally disrupt channel opening, whereas those that primarily target VSD IV commonly inhibit fast inactivation (Bosmans et al., 2008).
Here, our goal was to examine whether the paddle motif is pharmacologically functional when isolated from its channel background. A positive outcome would be particularly exciting for designing assays geared toward discovering novel ligands that target this region. For example, recent developments with antibodies targeting Nav channel paddles suggest that they may serve as drug targets or diagnostic markers (Chioni et al., 2005; Lee et al., 2014). Given its unique role in channel fast inactivation as well as its discerning sensitivity to animal toxins, we chose to focus on the VSD IV paddle motif and provide proof-of-principle experiments for this concept by synthesizing the paddle peptide and fixing it on sensor chips to be used in surface plasmon resonance (SPR) measurements. In brief, this label-free optical approach uses polarized light to measure the refractive index near a sensor surface to which a molecule of interest (“ligand” in SPR terminology) is attached. When a soluble particle (“analyte” in SPR terminology) binds, surface protein accumulation results in a refractive index alteration that can be measured in real time. The results are then plotted as response or resonance units (RUs) versus time in a “sensorgram.” By fitting kinetics from the association and dissociation phase to a particular adsorption model, the corresponding kinetic rate constants can be calculated (Neumann et al., 2007; Schuck and Zhao, 2010). As partnering analytes, we decided upon α-scorpion toxins, which are small cysteine-rich proteins that potently bind to the VSD IV paddle motif to inhibit Nav channel fast inactivation (Rogers et al., 1996; Gilchrist et al., 2014). Ensuing SPR experiments would determine kinetic or affinity constants between the VSD IV paddle motif and α-scorpion toxins without the need for fluorescent or radioactive probe labeling.
MATERIALS AND METHODS
Two-electrode voltage-clamp recordings from Xenopus laevis oocytes
The DNA sequence of rat (r)Nav1.2a (provided by A. Goldin, University of California, Irvine, Irvine, CA), rKv2.1 (provided by K.J. Swartz, National Institutes of Health), and the rNav1.2a/Kv2.1 VSD II or IV chimera was confirmed by automated DNA sequencing, and cRNA was synthesized using T7 polymerase (mMessage mMachine kit; Life Technologies) after linearizing the DNA with appropriate restriction enzymes. Channels were expressed in Xenopus oocytes (obtained from Xenopus 1) and studied after a 1-d incubation after cRNA injection (incubated at 17°C in 96 mM NaCl, 2 mM KCl, 5 mM HEPES, 1 mM MgCl2, 1.8 mM CaCl2, and 50 µg/ml gentamycin, pH 7.6 with NaOH) using two-electrode voltage-clamp recording techniques (OC-725C; Warner Instruments) with a 150-µl recording chamber. Data were filtered at 4 kHz and digitized at 20 kHz using pClamp 10 software (Molecular Devices). Microelectrode resistances were 0.5–1.5 MΩ when filled with 3 M KCl. For Kv chimera channel experiments, the external recording solution contained (mM): 50 KCl, 50 NaCl, 5 HEPES, 1 MgCl2, and 0.3 CaCl2, pH 7.6 with NaOH. For rNav1.2a experiments, the external recording solution contained (mM): 100 NaCl, 5 HEPES, 1 MgCl2, and 1.8 CaCl2, pH 7.6 with NaOH. All experiments were performed at ∼22°C. Leak and background conductances, identified by blocking the channel with agitoxin-2 (provided by K.J. Swartz), have been subtracted for all of the Kv channel currents shown. Tetrodotoxin (Alomone Labs) subtraction was used to isolate Nav channel currents. After the addition of the toxin to the recording chamber, the equilibration between the toxin and the channel was monitored using weak depolarizations elicited at 5- or 10-s (R1629A/L1630A mutant) intervals. Voltage–activation relationships were obtained by measuring tail currents for Kv channels or steady-state currents and calculating conductance for Nav channels, and a single Boltzmann function was fitted to the data according to I/Imax = [1c + cexp(−zF(Vc − cV1/2)/RT)]−1, where I/Imax is the normalized tail-current amplitude, z is the equivalent charge, V1/2 is the half-activation voltage, F is Faraday’s constant, R is the gas constant, and T is temperature in kelvin. For all channels, we recorded voltage–activation relationships in the absence and presence of toxin. The ratio of currents (I/I0) recorded in the presence (I) and absence (I0) of toxin was calculated for voltages typically ranging from −140 to 10 cmV, depending on the construct. The value of I/I0 measured in the plateau phase at voltages where toxin-bound channels do not open was taken as Fu. The apparent equilibrium dissociation constant (apparent KD) for Kv channels was calculated according to KD = ((1/(1c − cFu1/4))c − c1)[toxin], assuming four independent toxin-binding sites per channel (Swartz and MacKinnon, 1997). Results are given as mean ± SEM unless noted otherwise. Offline data analysis was performed using Clampfit 10 (Molecular Devices), Origin 8.0 (OriginLab), and Excel (Microsoft). All chemicals were obtained from Sigma-Aldrich.
Peptide synthesis
VSD II and IV paddle peptides of rNav1.2a were produced using standard Fmoc solid-phase peptide synthesis on preloaded Fmoc–amino acid Wang resin, HPLC purified (>99.5% purity), and tested for the correct mass using mass spectrometry by ProteoGenix. Circular dichroism (CD) measurements were performed in a standard PBS solution containing 0.1% vol/vol lauryldimethylamine N-oxide on a spectropolarimeter (J-810; JASCO).
SPR experiments
SPR experiments were performed at 25°C on a Biacore T200 instrument (GE Healthcare) using 150 mM NaCl, 3 mM EDTA, 0.005% surfactant P20, and 10 mM HEPES-NaOH, pH 7.4, for the coupling protocol or 50 mM phosphate, pH 7.2, as running buffer. Neutravidin (Thermo Fisher Scientific) was immobilized on CM5 sensor chips (GE Healthcare) using an amine coupling chemistry. All chemicals were obtained from Sigma-Aldrich. Biotinylated paddle peptides (ligands) were injected in experimental flow cells, as well as a SNAP-25-Biotin peptide of comparable length (t-SNARE protein from residues 137 to 206; Südhof and Rothman, 2009) in control flow cells to obtain 50 or 500 fmol of immobilized peptide for kinetic or binding analysis, respectively. In all SPR experiments, nonspecific binding obtained in the control flow cell was subtracted from the signal obtained in the experimental flow cell. For qualitative binding analysis, toxins (100 nM in running buffer containing 0.1% BSA) were injected at a flow rate of 20 µl/min over 2 min. For kinetic studies, varying toxin concentrations (15–2,000 nM) were injected at a flow rate of 20 µl/min. In between injections, the surface of the sensor chip was regenerated by injecting 1 M NaCl (15 s at 30 µl/min). The bulk signal caused by refractive index differences between the flow buffer and the buffer containing the analyte was systematically excluded from the data-fitting process. It is worth noting that isolating a paddle motif (ligand) can induce conformational changes that can affect toxin (analyte) efficacy. In fact, the CD spectrum in Fig. 3 A suggests the presence of an unstructured VSD IV peptide region or pool (∼25%), which may alter toxin susceptibility. Sensorgrams can reveal such ligand heterogeneity by displaying biphasic dissociation curves that cannot be fitted with the frequently used Langmuir model, which assumes a 1:1 stoichiometry of analyte to ligand. To account for a possible heterogeneity of surface sites, we calculated the rate constants and affinity equilibrium dissociation constants (KD) for AaHII by a fit to a heterogeneous ligand model (Schuck and Zhao, 2010) incorporated into the Biacore T200 Evaluation software (v1.0). This model assumes the presence of two sites on the ligand that can bind analyte and can be described by these equations in which A represents the analyte and B1/B2 represents the heterogeneous ligand:
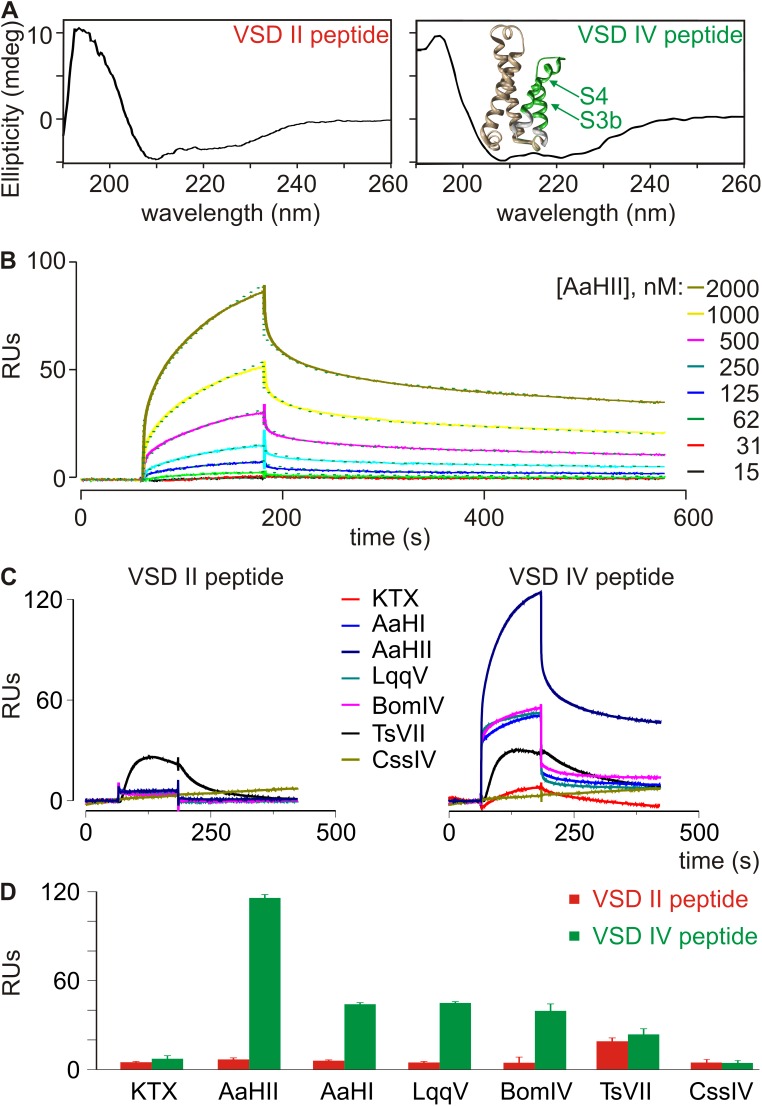
Figure 3.
α-Scorpion toxins interact with the isolated rNav1.2a VSD IV paddle motif. (A) Shown are the CD spectra of the rNav1.2a VSD II and IV paddle peptides used in this paper. Analysis of both spectra revealed the presence of ∼75% structured (α-helix/β-sheet) and ∼25% unstructured peptide. Inset shows the crystal structure of the NavAb voltage sensor (3RVY) (Payandeh et al., 2011) in which the paddle motif is indicated in green; wheat shows S1 and S2 helices, and white indicates the portion of S3 and S4 helices outside the paddle. (B) Representative association and dissociation kinetic curves obtained using SPR after the application of varying concentrations of AaHII (15–2,000 nM) over a sensor chip to which 50 fmol of the rNav1.2a VSD IV paddle peptide was linked. Toxin was applied after obtaining a steady baseline. Colored traces represent toxin binding obtained after subtraction of the signal from the control flow cell. Green dotted lines depict a fit of the data to a heterogeneous surface ligand model, a typical SPR analysis method (Schuck and Zhao, 2010), which yielded a high affinity KD1 of 479 ± 241 nM (RUmax1 = 135) and a lower affinity KD2 of 747 ± 203 nM (RUmax2 = 23; n = 3; all results presented as mean ± SD). The respective contributions of RUmax1 (∼85%) and RUmax2 (∼15%) to the overall RUmax (100%) are reminiscent of the percent structured (∼75%) versus unstructured (∼25%) paddle peptide as observed in the CD spectrum. Toxin concentrations are indicated on the right in a shade of gray corresponding to the sensorgram. (C) Representative SPR traces after the application of 100 nM KTX, AaHI, AaHII, LqqV, BomIV, TsVII, and CssIV over a sensor chip to which the rNav1.2a VSD II (left) or IV (right) paddle peptide (500 fmol) was linked. Toxin was applied after obtaining a steady baseline. (D) Binding capacities of 100 nM α-scorpion toxins AaHI, AaHII, LqqV, the α-like scorpion toxin BomIV, and the β-scorpion toxins TsVII and CssIV to the VSD II and IV paddle motifs using SPR (error bars represent ± SEM). Y-axis represents the maximum RUs obtained after toxin application. Note that 500 fmol paddle peptide was used in C and D as opposed to 50 fmol in B. As a result, RUs differ by a factor of ∼10.
Calculating the KD values involves these components: dB1/dt = −(ka1*A*B1 − kd1*AB1); dB2/dt = −(ka2*A*B2 − kd2*AB2); dAB1/dt = (ka1*A*B1 − kd1*AB1); dAB2/dt = (ka2*A*B2 − kd2*AB2), with A = analyte concentration, B1 = RUmax1 of a particular binding site, B2 = RUmax2, ka and kd = association and dissociation rate constant, and AB1 and AB2 = 0 at the start of each experiment (no complex has been formed). Note that two separate sets of ka and kd constants as well as two KD values describe each binding event.
RESULTS
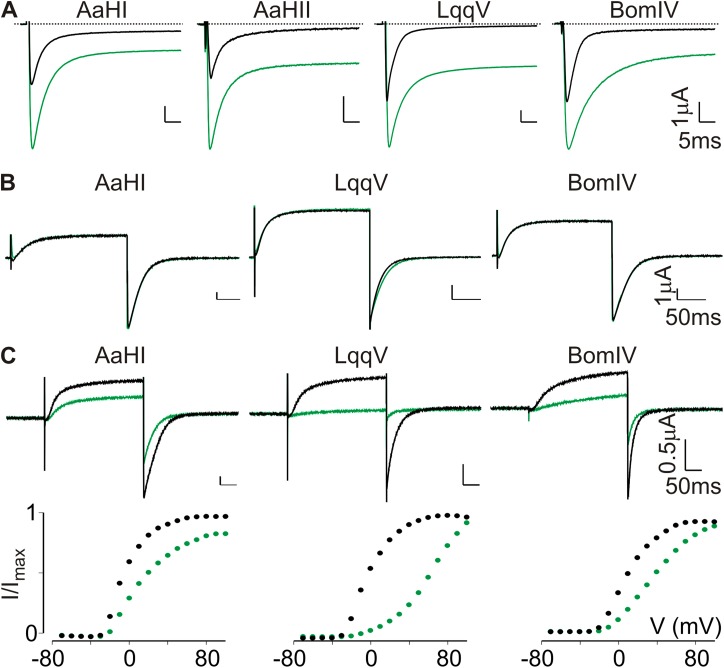
The four Nav channel–selective α-scorpion toxins we selected for our experiments were AaHI and AaHII from Androctonus australis Hector, LqqV from Leiurus quinquestriatus hebraeus, and BomIV from Buthus occitanus mardochei (Martin-Eauclaire and Rochat, 2000; Bende et al., 2014). As negative control, we applied kaliotoxin (KTX) from Androctonus mauretanicus, which blocks the Kv1.1 and Kv1.3 pore but does not influence Nav channel function (Crest et al., 1992). All toxins were purified to homogeneity as reported previously (Crest et al., 1992; Martin-Eauclaire and Rochat, 2000) and tested for functionality on the rNav1.2a isoform expressed in Xenopus oocytes. In all cases, the application of 100 nM AaHI, AaHII, LqqV, or BomIV inhibits rNav1.2a fast inactivation, resulting in the appearance of a large persistent current at the end of a 50-ms test pulse (Fig. 1 A). Because AaHII has already been shown to bind to the VSD IV paddle motif in rNav1.2a (Bosmans et al., 2008), we verified if this was also the case with AaHI, LqqV, and BomIV. To this end, we tested whether 100 nM of each toxin influenced the function of a previously constructed chimera in which the S3b–S4 region of the homotetrameric Kv2.1 channel was swapped for the corresponding WT region in VSD IV from rNav1.2a. As a result, we observed a robust voltage-dependent K+ current inhibition, whereas WT Kv2.1 is insensitive, suggesting that AaHI, LqqV, and BomIV indeed interact with the transferred VSD IV paddle motif (Fig. 1, B and C).
Figure 1.
α-Scorpion toxins interact with the rNav1.2a VSD IV paddle motif. (A) Shown is the effect of 100 nM AaHI, AaHII, LqqV, and BomIV on rNav1.2a channel function. Representative sodium currents were elicited by a 50-ms depolarization to a suitable membrane voltage (−20 to −15 mV) before (black) and after toxin addition (green) from a holding voltage of −90 mV. Clearly, toxin application results in a large persistent current component at the end of the test pulse. Fitting the current decay with a single-exponential function before and after toxin application yields fast inactivation time constants (τ) of 3.2 ± 0.1 and 4.7 ± 0.1 (AaHI); 3.6 ± 0.2 and 4.9 ± 0.1 (AaHII); 2.5 ± 0.1 and 4.6 ± 0.1 (LqqV); and 2.9 ± 0.1 and 8.5 ± 0.1 (BomIV), with n = 3 for each value (mean ± SEM). (B) Shown is the effect of 1 µM AaHI, LqqV, and BomIV on WT rKv2.1. For each toxin, K+ currents were elicited by a 300-ms depolarization to 0 mV from a holding voltage of −90 mV (tail voltage was −60 mV). Currents are shown before (black) and in the presence of toxin (green). (C) Effect of 100 nM AaHI, LqqV, and BomIV on the rNav1.2a/Kv2.1 VSD IV paddle chimera. For each toxin, K+ currents (top) were elicited by a 300-ms depolarization near the foot of the voltage–activation curve (bottom) from a holding voltage of −90 mV. Currents are shown before (black) and in the presence of toxin (green). Representative normalized tail current voltage–activation relationships are shown (bottom), where tail current amplitude (I/Imax) is plotted against test voltage (V) before (black) and in the presence of toxins (green). A Boltzmann fit of the obtained data (n = 3; mean ± SEM) reveals a depolarizing shift in midpoint (V1/2) of ∼15 mV for AaHI, >50 mV for LqqV, and ∼26 mV for BomIV. Holding voltage was −90 mV, and the tail voltage was −60 mV.
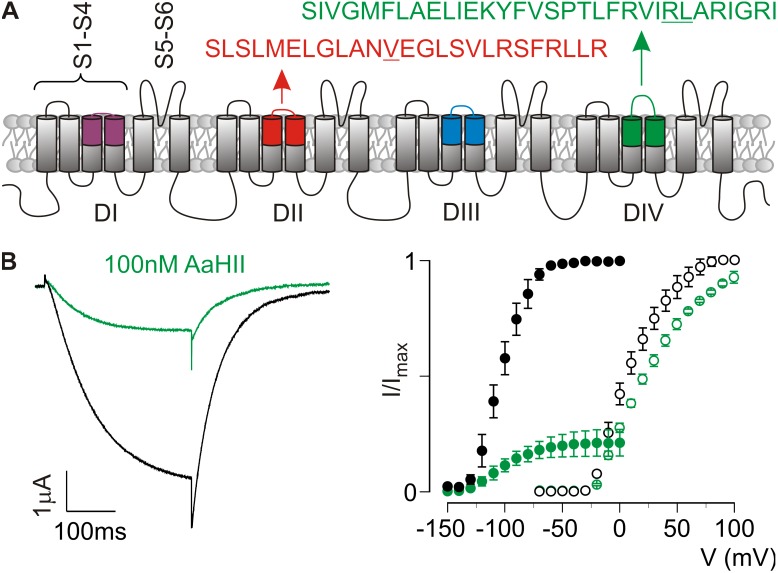
To design the best possible environment for detecting toxin–paddle interactions in our SPR experiments, the isolated VSD IV paddle peptide of rNav1.2a as defined previously (Bosmans et al., 2008) was synthesized with two amino acid substitutions (R1629A/L1630A) that each were shown to increase susceptibility to our model toxin AaHII (Fig. 2 A). Specifically, the affinity of AaHII for the WT rNav1.2a/Kv2.1 VSD IV chimera (apparent KD = 1,902 ± 102 nM) increased to 235 ± 24 nM (R1629A) and 205 ± 23 nM (L1630A). To verify the affinity of AaHII for the double mutant, we introduced R1629A/L1630A into the rNav1.2a/Kv2.1 VSD IV chimera and expressed the construct in Xenopus oocytes. Compared with the tail current voltage–activation relationship of the WT chimera, the R1629A/L1630A substitutions cause a striking hyperpolarizing shift in midpoint (ΔV1/2 of approximately −112 mV; Fig. 2 B). However, it is also clear that 100 nM AaHII strongly inhibits the double mutant, whereas the WT rNav1.2a/Kv2.1 VSD IV chimera is influenced to a much lesser extent (Fig. 2 B).
Figure 2.
AaHII interacts with the rNav1.2a VSD IV paddle motif. (A) Nav channel cartoon embedded in a lipid membrane. Each domain (DI–IV) consists of six transmembrane segments (S1–S6) of which S1–S4 form the VSD and the S5–S6 segments of each domain form the pore. Paddle motif amino acid sequences are shown for VSD II (red) and IV (green). Underlined residues were mutated as reported in Results. (B) Effect of 100 nM AaHII on the rNav1.2a/Kv2.1 VSD IV chimera containing the R1629A/L1630A substitutions. K+ current (left) was elicited by a 300-ms depolarization to −100 mV (tail voltage was −150 mV) after a 500-ms step to −150 mV from a holding voltage of −10 mV (near the Nernst potential for K+). The data show a clear toxin-induced inhibition of the double mutant channel (black, control; green, 100 nM AaHII). Right panel displays normalized tail current voltage–activation relationships of the rNav1.2a/Kv2.1 VSD IV chimera without (open circles) and with the R1629A/L1630A substitutions (closed circles) where tail current amplitude (I/Imax) is plotted against test voltage before (black) and in the presence of 100 nM AaHII (green). Holding voltage for the mutant was −10 mV, followed by a 500-ms hyperpolarizing step to −150 mV to close all channels. Next, 10-mV step depolarizations of 300 ms (V) were trailed by a 300-ms tail voltage step to −150 mV (I). A Boltzmann fit of the obtained data (n = 3; error bars represent mean ± SEM) reveals a shift in midpoint (V1/2) for the double mutant (−105 ± 1 mV; slope, 17.1 ± 1.0) compared with the rNav1.2a/Kv2.1 VSD IV chimera (7 ± 2 mV; slope, 17.3 ± 1.6). Moreover, 100 nM AaHII strongly inhibits the double mutant (apparent KD = 193 ± 42 nM), whereas the rNav1.2a/Kv2.1 VSD IV chimera is influenced less (apparent KD = 1,008 ± 92 nM), with n = 3 for each value (error bars represent mean ± SEM).
Next, we fixed the VSD IV peptide on SPR sensor chips by adding a K-linked biotin group to the C terminus, resulting in a 32–amino acid peptide altogether (Fig. 2 A). This biotin moiety may improve toxin access to its binding site and avoid steric hindrance when the paddle peptide is linked to the sensor chips. Also, the biotin group was attached to the C terminus, as amino acids crucial for AaHII binding are mostly located at the N-terminal region of the rNav1.2a VSD IV paddle (Bosmans et al., 2008). CD measurements reveal that the peptide folds and mainly adopts an α-helical conformation (∼53%; Fig. 3 A), an observation that fits well with an earlier report about the NMR solution structure of an isolated paddle motif from the HsapBK Kv channel (Unnerståle et al., 2009, 2012). To serve as a control for toxins interacting exclusively with the VSD IV paddle motif, we also synthesized the previously defined VSD II paddle motif of rNav1.2a, including a V843A gain-of-function mutation and a C-terminal K-linked biotin group (Bosmans et al., 2008; Zhang et al., 2011). Altogether, this peptide comprises 26 amino acids (Fig. 2 A), and similar to VSD IV, CD measurements of the VSD II paddle motif reveal a folded peptide that mainly adopts an α-helical conformation (Fig. 3 A). For SPR experiments, biotinylated paddle peptides were injected in experimental flow cells to obtain ∼50 or 500 fmol of immobilized peptides for kinetic or binding analysis, respectively.
We started by examining the interaction of AaHII with the VSD IV paddle motif using toxin concentrations ranging from 15 to 2,000 nM. As shown in Fig. 3 B, concentration-dependent association and biphasic dissociation curves ranging from 0 to 100 RUs were obtained, thereby suggesting a possible heterogeneity of biosensor surface sites that may be related to toxin interaction with an unstructured paddle motif region (see Fig. 3 A, Materials and methods, and Schuck and Zhao, 2010). Fits of these sensorgrams using a heterogeneous ligand model yielded two sets of association (ka) and dissociation (kd) rates, i.e., ka1 = 2.33 × 103 ± 0.91 × 103 M−1 s−1 and ka2 = 3.82 × 104 ± 0.47 × 104 M−1 s−1, and kd1 = 9.79 × 10−4 ± 1.40 × 10−4 s−1 and kd2 = 2.81 × 10−2 ± 0.58 × 10−2 s−1, resulting in a high affinity KD1 (kd1/ka1) of 479 ± 241 nM (RUmax1 = 135) and a lower affinity KD2 (kd2/ka2) of 747 ± 203 nM (RUmax2 = 23), which may represent a partially active binding site (n = 3; all results presented as mean ± SD). Although comparing diverse affinity measurements obtained from different systems is not straightforward, KD1 resembles that observed when determining the susceptibility of the rNav1.2a/Kv2.1 VSD IV R1629A/L1630A paddle chimera for AaHII (apparent KD is 193 ± 42 nM; Fig. 2 B). The affinity of the toxin for WT rNav1.2a is ∼5 nM, suggesting that regions outside of the VSD IV paddle motif may enhance toxin efficacy (Wang et al., 2011). However, such secondary interactions seem more crucial for toxins interacting with VSD II (Cestèle et al., 1998, 2006; Leipold et al., 2006; Karbat et al., 2010; Zhang et al., 2012; Bende et al., 2014), a notion that we will explore in the next section. Overall, the biological activity of AaHII in our SPR assay suggests an intrinsic pharmacological sensitivity of the isolated rNav1.2a VSD IV paddle motif and demonstrates the effectiveness of the SPR technique in determining ligand interactions with this region. In contrast to the VSD IV peptide, we did not notice an effect of AaHII when applying up to 20 µM toxin to sensor chips coated with the VSD II paddle motif (Fig. 3, C and D). In concordance, no interaction was observed when assaying AaHII on the rNav1.2a/Kv2.1 VSD II paddle chimera (Bosmans et al., 2008). To further evaluate the robustness of the SPR approach reported here, we examined whether the α-scorpion toxins AaHI, LqqV, and BomIV also interact with the isolated VSD IV paddle peptide at a concentration of 100 nM. We indeed found that all four toxins bind this motif with RUs of ∼40–60, whereas we were unable to obtain a response when testing the VSD II peptide (Fig. 3, C and D).
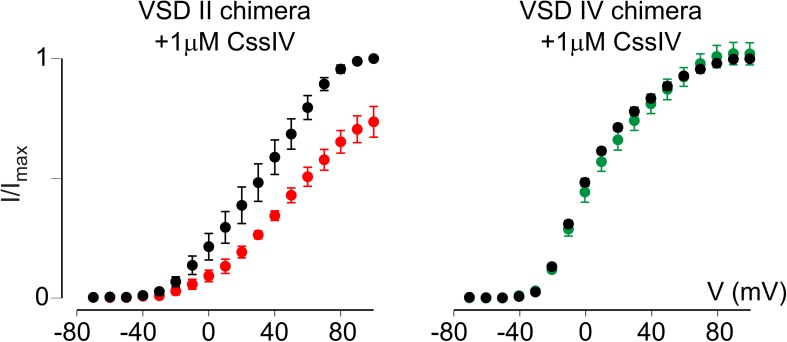
To verify the proper function of the VSD II paddle motif peptide in this SPR assay, we first applied 100 nM TsVII, a β-scorpion toxin from the Brazilian scorpion Tityus serrulatus, which promotes opening of rNav1.2a (apparent KD ranges from 25 to 121 nM) by primarily interacting with the paddle motif in the domain II voltage sensor with an apparent KD of 112 ± 12 nM (Bosmans et al., 2008). However, electrophysiological data from the rNav1.2a/Kv2.1 chimeras revealed that TsVII can also bind to the VSD IV paddle (Bosmans et al., 2008). In concert, both paddle motifs responded to the presence of TsVII in SPR experiments (Fig. 3, C and D), thereby indicating that sensor chips containing the VSD II or IV paddle motifs are capable of binding appropriate ligands. Also, the Kv channel toxin KTX serving as a negative control did not bind to either of the two paddle motif peptides (Fig. 3, C and D). In contrast to α-scorpion toxins for which an interaction with the VSD IV paddle motif seems sufficient to exert their effect (Rogers et al., 1996; Benzinger et al., 1998; Bosmans et al., 2008; Gur et al., 2011; Wang et al., 2011), it is important to mention that residues outside of the VSD II paddle motif are required for proper binding of β-scorpion toxins such as CssIV from Centruroides suffusus suffusus (Cestèle et al., 1998, 2006; Leipold et al., 2006; Karbat et al., 2010; Zhang et al., 2012). Because these interactions are lost when isolating the VSD II paddle motif on an SPR chip, it is reasonable to assume that our assay may not detect binding of particular β-scorpion toxins. To investigate this possibility, we tested whether CssIV (Martin et al., 1987) elicited an SPR response when applied to the VSD II or IV paddle motif. Indeed, no RUs could be detected upon the addition of 100 nM CssIV (Fig. 3, C and D), a concentration that clearly influences WT rNav1.2a gating (Cestèle et al., 1998, 2006). Correspondingly, 1 µM CssIV is required to begin inhibiting the rNav1.2a/Kv2.1 VSD II paddle chimera (Fig. 4), thereby supporting the notion that regions outside of the paddle motif may indeed be crucial for β-scorpion toxin binding. Subsequently, the SPR approach described here is limited to detecting ligand interactions that do not require Nav channel regions outside of the paddle region.
Figure 4.
CssIV interacts with the rNav1.2a VSD II paddle motif. Shown is the effect of 1 µM CssIV on the rNav1.2a/Kv2.1 VSD II and IV paddle chimera. Normalized tail current voltage–activation relationships in which the tail current amplitude (I/Imax) is plotted against test voltage (V) before (black) and in the presence of toxin (red/green) are displayed. A Boltzmann fit of the obtained data (n = 3; error bars represent mean ± SEM) reveals a depolarizing shift in V1/2 of ∼14 mV (from 33 ± 2 mV to 47 ± 1 mV) for the VSD II chimera, whereas 1 µM CssIV does not influence the VSD IV chimera. Holding voltage was −90 mV, and the tail voltage was −60 mV.
DISCUSSION
Collectively, our results suggest that the VSD IV paddle motif as described in this work is a self-contained unit that retains its pharmacological sensitivities toward α-scorpion toxins when isolated from the rest of the Nav channel (Fig. 3). As such, we propose that a label-free SPR method may be used to detect interactions between ligands and Nav channel paddle motifs without the need to express the full-length channel in a heterologous expression system. Because a full measurement cycle—including chip-regeneration steps—takes <10 min, this biosensor approach may constitute a first step toward designing a screening method to uncover interactions between pharmacological ligands and paddle-containing ion channels. One limitation that emerged from our experiments is the inability of this method to detect ligand interactions that require regions outside of the paddle region (e.g., CssIV; Fig. 3). However, in conjunction with a report showing that paddle motif–targeting antibodies may be therapeutically beneficial (Lee et al., 2014) or could serve as diagnostic markers (Chioni et al., 2005), the results reported here may open up a valuable pathway for discovering novel molecules that influence Nav channel function by interacting with the VSDs. Moreover, our methodology can be modified to conduct competition experiments in which the displacement of toxin bound to the VSD IV paddle motif by paddle-targeting drugs can be monitored. The identification of such compounds may be useful in reshaping Nav channel activity in disease conditions associated with an abnormal fast inactivation process (Bennett et al., 1995; Wan et al., 2001; Ulbricht, 2005). In a broader context, this SPR approach could help identify ligands for VSDs not associated with a pore region (Murata et al., 2005; Ramsey et al., 2006; Sasaki et al., 2006).
Acknowledgments
We thank S. Blangy and C. Lévêque for their assistance in performing CD and SPR experiments, respectively.
Parts of this work received funding from the Centre National de la Recherche Scientifique (PEPS-2009/PAGAIE), the NIH under award number R00NS073797 (to F. Bosmans), and support from Marseille Protéomique.
The authors declare no competing financial interests.
Author contributions: M.-F. Martin-Eauclaire, F. Bosmans, and P.E. Bougis conceived the study. M.-F. Martin-Eauclaire, F. Bosmans, and G. Ferracci designed and performed the experiments. All authors were involved in writing the manuscript.
Merritt C. Maduke served as editor.
Footnotes
Abbreviations used in this paper:
- CD
- circular dichroism
- KTX
- kaliotoxin
- Kv
- voltage-gated potassium
- Nav
- voltage-gated sodium
- RU
- resonance unit
- SPR
- surface plasmon resonance
- VSD
- voltage-sensing domain
References
- Alabi A.A., Bahamonde M.I., Jung H.J., Kim J.I., and Swartz K.J.. 2007. Portability of paddle motif function and pharmacology in voltage sensors. Nature. 450:370–375 10.1038/nature06266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bende N.S., Dziemborowicz S., Mobli M., Herzig V., Gilchrist J., Wagner J., Nicholson G.M., King G.F., and Bosmans F.. 2014. A distinct sodium channel voltage-sensor locus determines insect selectivity of the spider toxin Dc1a. Nat. Commun. 5:4350 10.1038/ncomms5350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett P.B., Yazawa K., Makita N., and George A.L. Jr. 1995. Molecular mechanism for an inherited cardiac arrhythmia. Nature. 376:683–685 10.1038/376683a0 [DOI] [PubMed] [Google Scholar]
- Benzinger G.R., Kyle J.W., Blumenthal K.M., and Hanck D.A.. 1998. A specific interaction between the cardiac sodium channel and site-3 toxin anthopleurin B. J. Biol. Chem. 273:80–84 10.1074/jbc.273.1.80 [DOI] [PubMed] [Google Scholar]
- Bezanilla F.2008. How membrane proteins sense voltage. Nat. Rev. Mol. Cell Biol. 9:323–332 10.1038/nrm2376 [DOI] [PubMed] [Google Scholar]
- Bosmans F., Martin-Eauclaire M.F., and Swartz K.J.. 2008. Deconstructing voltage sensor function and pharmacology in sodium channels. Nature. 456:202–208 10.1038/nature07473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosmans F., Puopolo M., Martin-Eauclaire M.F., Bean B.P., and Swartz K.J.. 2011. Functional properties and toxin pharmacology of a dorsal root ganglion sodium channel viewed through its voltage sensors. J. Gen. Physiol. 138:59–72 10.1085/jgp.201110614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capes D.L., Goldschen-Ohm M.P., Arcisio-Miranda M., Bezanilla F., and Chanda B.. 2013. Domain IV voltage-sensor movement is both sufficient and rate limiting for fast inactivation in sodium channels. J. Gen. Physiol. 142:101–112 10.1085/jgp.201310998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cestèle S., Qu Y., Rogers J.C., Rochat H., Scheuer T., and Catterall W.A.. 1998. Voltage sensor-trapping: Enhanced activation of sodium channels by β-scorpion toxin bound to the S3–S4 loop in domain II. Neuron. 21:919–931 10.1016/S0896-6273(00)80606-6 [DOI] [PubMed] [Google Scholar]
- Cestèle S., Yarov-Yarovoy V., Qu Y., Sampieri F., Scheuer T., and Catterall W.A.. 2006. Structure and function of the voltage sensor of sodium channels probed by a beta-scorpion toxin. J. Biol. Chem. 281:21332–21344 10.1074/jbc.M603814200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chioni A.M., Fraser S.P., Pani F., Foran P., Wilkin G.P., Diss J.K.J., and Djamgoz M.B.A.. 2005. A novel polyclonal antibody specific for the Nav1.5 voltage-gated Na+ channel ‘neonatal’ splice form. J. Neurosci. Methods. 147:88–98 10.1016/j.jneumeth.2005.03.010 [DOI] [PubMed] [Google Scholar]
- Crest M., Jacquet G., Gola M., Zerrouk H., Benslimane A., Rochat H., Mansuelle P., and Martin-Eauclaire M.F.. 1992. Kaliotoxin, a novel peptidyl inhibitor of neuronal BK-type Ca2+-activated K+ channels characterized from Androctonus mauretanicus mauretanicus venom. J. Biol. Chem. 267:1640–1647. [PubMed] [Google Scholar]
- Gilchrist J., Olivera B.M., and Bosmans F.. 2014. Animal toxins influence voltage-gated sodium channel function. Handbook Exp. Pharmacol. 221:203–229 10.1007/978-3-642-41588-3_10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gur M., Kahn R., Karbat I., Regev N., Wang J., Catterall W.A., Gordon D., and Gurevitz M.. 2011. Elucidation of the molecular basis of selective recognition uncovers the interaction site for the core domain of scorpion alpha-toxins on sodium channels. J. Biol. Chem. 286:35209–35217 10.1074/jbc.M111.259507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalia J., Milescu M., Salvatierra J., Wagner J., Klint J.K., King G.F., Olivera B.M., and Bosmans F.. 2015. From foe to friend: Using animal toxins to investigate ion channel function. J. Mol. Biol. 427:158–175 10.1016/j.jmb.2014.07.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karbat I., Ilan N., Zhang J.Z., Cohen L., Kahn R., Benveniste M., Scheuer T., Catterall W.A., Gordon D., and Gurevitz M.. 2010. Partial agonist and antagonist activities of a mutant scorpion beta-toxin on sodium channels. J. Biol. Chem. 285:30531–30538 10.1074/jbc.M110.150888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J.H., Park C.K., Chen G., Han Q., Xie R.G., Liu T., Ji R.R., and Lee S.Y.. 2014. A monoclonal antibody that targets a NaV1.7 channel voltage sensor for pain and itch relief. Cell. 157:1393–1404 10.1016/j.cell.2014.03.064 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Leipold E., Hansel A., Borges A., and Heinemann S.H.. 2006. Subtype specificity of scorpion beta-toxin Tz1 interaction with voltage-gated sodium channels is determined by the pore loop of domain 3. Mol. Pharmacol. 70:340–347. [DOI] [PubMed] [Google Scholar]
- Li-Smerin Y., and Swartz K.J.. 2000. Localization and molecular determinants of the Hanatoxin receptors on the voltage-sensing domains of a K+ channel. J. Gen. Physiol. 115:673–684 10.1085/jgp.115.6.673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long S.B., Tao X., Campbell E.B., and MacKinnon R.. 2007. Atomic structure of a voltage-dependent K+ channel in a lipid membrane-like environment. Nature. 450:376–382 10.1038/nature06265 [DOI] [PubMed] [Google Scholar]
- Martin M.F., Garcia y Perez L.G., el Ayeb M., Kopeyan C., Bechis G., Jover E., and Rochat H.. 1987. Purification and chemical and biological characterizations of seven toxins from the Mexican scorpion, Centruroides suffusus suffusus. J. Biol. Chem. 262:4452–4459. [PubMed] [Google Scholar]
- Martin-Eauclaire M.F., and Rochat H.. 2000. Purification and characterization of scorpion toxins acting on voltage-sensitive Na+ channels. Methods and Tools in Biosciences and Medicine. Springer, Basel: 152–168 10.1007/978-3-0348-8466-2_10 [DOI] [Google Scholar]
- Murata Y., Iwasaki H., Sasaki M., Inaba K., and Okamura Y.. 2005. Phosphoinositide phosphatase activity coupled to an intrinsic voltage sensor. Nature. 435:1239–1243 10.1038/nature03650 [DOI] [PubMed] [Google Scholar]
- Neumann T., Junker H.D., Schmidt K., and Sekul R.. 2007. SPR-based fragment screening: Advantages and applications. Curr. Top. Med. Chem. 7:1630–1642 10.2174/156802607782341073 [DOI] [PubMed] [Google Scholar]
- Payandeh J., Scheuer T., Zheng N., and Catterall W.A.. 2011. The crystal structure of a voltage-gated sodium channel. Nature. 475:353–358 10.1038/nature10238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramsey I.S., Moran M.M., Chong J.A., and Clapham D.E.. 2006. A voltage-gated proton-selective channel lacking the pore domain. Nature. 440:1213–1216 10.1038/nature04700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers J.C., Qu Y., Tanada T.N., Scheuer T., and Catterall W.A.. 1996. Molecular determinants of high affinity binding of alpha-scorpion toxin and sea anemone toxin in the S3-S4 extracellular loop in domain IV of the Na+ channel alpha subunit. J. Biol. Chem. 271:15950–15962 10.1074/jbc.271.27.15950 [DOI] [PubMed] [Google Scholar]
- Sasaki M., Takagi M., and Okamura Y.. 2006. A voltage sensor-domain protein is a voltage-gated proton channel. Science. 312:589–592 10.1126/science.1122352 [DOI] [PubMed] [Google Scholar]
- Schuck P., and Zhao H.. 2010. The role of mass transport limitation and surface heterogeneity in the biophysical characterization of macromolecular binding processes by SPR biosensing. Methods Mol. Biol. 627:15–54 10.1007/978-1-60761-670-2_2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Südhof T.C., and Rothman J.E.. 2009. Membrane fusion: Grappling with SNARE and SM proteins. Science. 323:474–477 10.1126/science.1161748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swartz K.J.2008. Sensing voltage across lipid membranes. Nature. 456:891–897 10.1038/nature07620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swartz K.J., and MacKinnon R.. 1997. Hanatoxin modifies the gating of a voltage-dependent K+ channel through multiple binding sites. Neuron. 18:665–673 10.1016/S0896-6273(00)80306-2 [DOI] [PubMed] [Google Scholar]
- Ulbricht W.2005. Sodium channel inactivation: Molecular determinants and modulation. Physiol. Rev. 85:1271–1301 10.1152/physrev.00024.2004 [DOI] [PubMed] [Google Scholar]
- Unnerståle S., Lind J., Papadopoulos E., and Mäler L.. 2009. Solution structure of the HsapBK K+ channel voltage-sensor paddle sequence. Biochemistry. 48:5813–5821 10.1021/bi9004599 [DOI] [PubMed] [Google Scholar]
- Unnerståle S., Madani F., Gräslund A., and Mäler L.. 2012. Membrane-perturbing properties of two Arg-rich paddle domains from voltage-gated sensors in the KvAP and HsapBK K+ channels. Biochemistry. 51:3982–3992 10.1021/bi300188t [DOI] [PubMed] [Google Scholar]
- Wan X., Chen S., Sadeghpour A., Wang Q., and Kirsch G.E.. 2001. Accelerated inactivation in a mutant Na+ channel associated with idiopathic ventricular fibrillation. Am. J. Physiol. Heart Circ. Physiol. 280:H354–H360. [DOI] [PubMed] [Google Scholar]
- Wang J., Yarov-Yarovoy V., Kahn R., Gordon D., Gurevitz M., Scheuer T., and Catterall W.A.. 2011. Mapping the receptor site for α-scorpion toxins on a Na+ channel voltage sensor. Proc. Natl. Acad. Sci. USA. 108:15426–15431 10.1073/pnas.1112320108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J.Z., Yarov-Yarovoy V., Scheuer T., Karbat I., Cohen L., Gordon D., Gurevitz M., and Catterall W.A.. 2011. Structure-function map of the receptor site for β-scorpion toxins in domain II of voltage-gated sodium channels. J. Biol. Chem. 286:33641–33651 10.1074/jbc.M111.282509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J.Z., Yarov-Yarovoy V., Scheuer T., Karbat I., Cohen L., Gordon D., Gurevitz M., and Catterall W.A.. 2012. Mapping the interaction site for a β-scorpion toxin in the pore module of domain III of voltage-gated Na+ channels. J. Biol. Chem. 287:30719–30728 10.1074/jbc.M112.370742 [DOI] [PMC free article] [PubMed] [Google Scholar]