Why seizures arise from normal-appearing human cortex is unclear. Using a pairwise transcriptional method to compare high and low-spiking tissues from patients with epilepsy, Dachet et al. generate a ‘cellular interactome’ that reveals millimetre-sized ‘microlesions’ in deeper cortical layers of high-spiking regions. These layer-specific abnormalities may underlie many ‘non-lesional’ forms of epilepsy.
Keywords: localization-related epilepsy, refractory epilepsy, epilepsy genetics, transcriptomics, epilepsy surgery
Abstract
Although epilepsy is associated with a variety of abnormalities, exactly why some brain regions produce seizures and others do not is not known. We developed a method to identify cellular changes in human epileptic neocortex using transcriptional clustering. A paired analysis of high and low spiking tissues recorded in vivo from 15 patients predicted 11 cell-specific changes together with their ‘cellular interactome’. These predictions were validated histologically revealing millimetre-sized ‘microlesions’ together with a global increase in vascularity and microglia. Microlesions were easily identified in deeper cortical layers using the neuronal marker NeuN, showed a marked reduction in neuronal processes, and were associated with nearby activation of MAPK/CREB signalling, a marker of epileptic activity, in superficial layers. Microlesions constitute a common, undiscovered layer-specific abnormality of neuronal connectivity in human neocortex that may be responsible for many ‘non-lesional’ forms of epilepsy. The transcriptional clustering approach used here could be applied more broadly to predict cellular differences in other brain and complex tissue disorders.
Introduction
Epilepsy is an often life-long brain disorder causing seizures or convulsions, which affects ∼1% of the world population (Annegers, 1993). One of the most common forms of epilepsy is temporal lobe epilepsy often associated with glial scarring or sclerosis of the hippocampus where seizures begin. However, patients with seizures starting from the neocortex, while associated with many nearby pathological lesions including dysplasia, tumours, infections, and strokes, often have no clear underlying pathology in regions that produce epileptic activities (Palmini et al., 1991, 2004; Lee et al., 2000; Seo et al., 2009). Around 30% of epileptic patients have refractory epilepsy and they fail to respond to medical therapies, but can improve with resective surgery to remove epileptic brain regions (Spencer and Huh, 2008). Epileptic brain regions consist of areas of the brain producing abnormal electrical activities created by synchronous firing of large populations of neurons. As part of a two-stage surgical procedure for epilepsy, these areas are identified using long-term, in vivo intracranial recordings that show both where seizures start together with areas with frequent electrical discharges between seizures (interictal spiking). Although these two electrical abnormalities are often in the same brain regions (Asano et al., 2003), the relationship between interictal spiking and seizures is not known (Gotman, 1991; Staley et al., 2011). However, removing both of these regions is associated with a better surgical outcome for refractory seizures (Kanazawa et al., 1996; Bautista et al., 1999).
We have developed a unique systems biology approach to measure differential gene expression using an internally controlled experimental design where neocortical regions, where seizures begin, are compared to nearby ‘control’ tissue regions from the same patient that show minimal epileptic activity (Rakhade et al., 2005; Beaumont et al., 2012). This required methods to quantify the in vivo electrical properties of each piece of brain tissue before its removal (Loeb, 2010, 2011; Barkmeier et al., 2012a). Recently, this work has revealed a common group of genes and pathways involving MAPK and CREB activation in superficial neocortical layers in regions where seizures first start that was also associated with a marked increase of synapses (Rakhade et al., 2005; Beaumont et al., 2012). Many of these transcriptional changes correlate better with interictal spiking activity than with seizures (Rakhade et al., 2007), can be reproduced in an animal model of interictal spiking, and can be suppressed with a drug that inhibits MAPK signalling (Barkmeier et al., 2012a). This raises the possibility that interictal spiking is a major driving force for transcriptional changes underlying epileptic disorders.
As a means to understand the cellular and molecular networks that underlie neocortical brain regions that produce spontaneous epileptic spiking and seizures, we performed a pairwise analysis of gene expression between low and high-spiking tissues from 15 patients with neocortical epilepsy. Using a statistical clustering algorithm, we found a subgroup of the differentially-expressed genes arranged into 11 tight clusters, each predicting a change in a specific cell type. The interrelationships of these clusters formed a ‘cellular interactome’ leading to histological predictions that were then validated on tissue sections from the same brain regions. In addition to global increases in vascularity and microglia, transcriptional clustering identified a multicellular ‘microlesion’ present in high-spiking regions from almost all patients examined regardless of other associated brain abnormalities. Microlesions may represent a common epileptic lesion by which focal disruptions in deeper cortical layer connectivity promote epileptic hypersynchrony in more superficial cortical layers.
Materials and methods
Isolation of human tissues and RNA preparation
All human brain tissues were removed as part of planned surgery for refractory epilepsy. Informed consent was obtained through an approved Institutional Review Board protocol at Wayne State University. The 15 patients implicated in this research underwent a two-stage surgery with long-term subdural electrocorticography and at least 3 days of continuous in vivo monitoring (Table 1). Tissue samples were mapped precisely to regions of neocortex displaying different levels of interictal spiking (by averaging of 3 × 10 min segments on different days) based on long-term electrocorticography as described (Beaumont et al., 2012; Lipovich et al., 2012). Tissue regions chosen for this study showed no clear pathological changes and were devoid of surgical haemorrhage. Total RNA was isolated from high and low interictal spiking areas from each patient. Alternating strips of full-thickness (layers I–VI) grey matter from the neocortex were pooled to produce total RNA representative of a given recording electrode location. For each tissue region, a portion of the tissue was also fixed in 4% paraformaldehyde for histological studies as previously described.
Table 1.
Patient and tissue information
| Patient | ILAE classification | Age of onset (years) | Age at surgery | Sex | MRI findings | Outcome (Engel 6 months) | Other tissue diagnoses | Spike frequency |
Tissue use: microarray (M) or histology (H) | |
|---|---|---|---|---|---|---|---|---|---|---|
| Low | High | |||||||||
| ep122 | PC, SG | NA | 15 | F | P | NA | P | 0 | 6 | M, H |
| ep132 | NA | NA | 10 | F | PO | NA | DG, AI, NLP | 1 | 116 | M, H |
| ep143 | SG | 9 | 11 | F | BSH | NA | H | 0 | 5 | M, H |
| ep148 | SG | 5 | 7 | F | Normal | NA | MG | 0 | 425 | M, H |
| ep150 | NA | NA | 33 | M | Normal | I | NTL | 157 | 576 | M, H |
| ep159 | SG | NA | 27 | M | LHA | NA | WG | 2 | 27 | M, H |
| ep158 | NA | 0.3 | 1 | M | IT2 | III | MG | 0 | 85 | M, H |
| ep164 | ES, PC | 0.4 | 3 | F | PE | I | WG, SupH | 66 | 141 | M, H |
| ep165 | ES | 0.5 | 3 | F | BGW | I | MG | 56 | 212 | M, H |
| ep167 | SG | NA | 7 | F | P & W | I | CD, MG | 25 | 215 | M, H |
| ep168 | ES | 2 | 6 | F | TC, (CD?) | I | MG | 26 | 124 | M, H |
| ep169 | ES | 0.5 | 8 | M | NA | III | MG | 3 | 172 | M, H |
| ep185 | PC | 49 | 56 | F | SubID | I | HS | 51 | 299 | M, H |
| ep188 | PC, SG | 0 | 16 | M | PO | I | DG | 44 | 176 | M, H |
| ep189 | SG | 6 | 11 | F | Normal | I | MCD | 2 | 66 | M, H |
| EP160 | ES | NA | 1 | F | Normal | III | MG | 140 | H | |
| EP162 | ES | 0.3 | 1 | F | NSH | I | H, P | 74; 202* | H | |
| EP175 | NA | NA | 49 | M | MA | I | MG | 114 | H | |
| EP178 | ES | 0.3 | 1 | F | focal CD | NA | TS | 104 | H | |
| EP179 | SG | 10 | 23 | M | Normal | II | MG | 188 | H | |
| EP180 | PC | 2.5 | 5 | M | TC, (CD?) | I | CD | 67 | H | |
| EP181 | PC | 7 | 13 | M | NSH | NA | SubBH | 95 | H | |
| EP182 | ES | 0.4 | 3 | M | TS | I | TS | 114 | H | |
| EP183 | NA | 0 | 22 | F | PO | I | HS, GC | 134 | H | |
| EP190 | PC | 0.1 | 17 | M | RVI | III | PNLG | 140 | H | |
| EP195 | PC | 3 | 7 | M | ACVD | NA | G, HS | 23 | H | |
| EP196 | NA | NA | 2 | M | NA | NA | E | 72 | H | |
| EP199 | NA | NA | 2 | M | NA | NA | WG, WD | 57; 83* | H | |
| EP200 | NA | NA | 17 | M | NA | NA | WG, WD | 78 | H | |
Clinical demographics, spike frequencies, and pathological diagnoses from patients with neocortical epilepsy used for this study are shown. Note that the pathological diagnosis was from a different brain region and not present in the tissue used for microarrays. Only the tissues with normal-appearing histology were used for microarrays and histological validation.
*For this patient two different brain regions were used while identifying microlesions. Cells left blank indicate data not available.
ACVD = arachnoid cyst ventricular dilatation; AI = acute inflammation; BGW = blurring of grey-white junction; BSH = bifrontal subcortical heterotopias; CD = cortical dysplasia; CD? = probable cortical dysplasia; DG = diffuse gliosis; E = encephalomyelitis; GC = gliosis in cyst; H = heterotopia; HS = hippocampal sclerosis; ES = epileptic spasms; IT2 = some increase in T2 in white matter; LHA = left hippocampal atrophy; MA = mild atrophy; MCD = mild cortical dysplasia; MG = mild gliosis; NLP = normal laminar pattern; NSH = nodular subcortical heterotopias; NTL = normal temporal lobe; P = polymicrogyria; PC = partial complex (focal seizures with impairment of consciousness or awareness); PE = periventricular mild increase in FLAIR; PNLG = patchy neuronal loss and gliosis at site of injury; PO = porencephalic cyst; RVI = remote vascular injury; SG = secondary generalized (focal seizures evolving to a bilateral, convulsive seizure); SupH = superficial heterotopias; SubID = subcortical ischaemic disease; SubBH = subcortical band heterotopias; TC = thickened cortex; TS = tuberous sclerosis; W = increased white matter signal; WD = white matter demyelination; WG = white matter gliosis; NA = data not available.
Microarray analysis and detection of differentially expressed genes
To have sufficient accuracy of differentially expressed genes, a quadruplicate, flip-dye experimental design was used for each pair of samples (Beaumont et al., 2012; Lipovich et al., 2012). Labelled antisense RNAs were co-hybridized to 60-mer Agilent human genome-wide oligonucleotide arrays (catalogue #G411A, Agilent) using a two-colour dye-swap method. Microarrays were scanned with an Agilent Technologies Microarray Scanner and the fluorescence was accessed using the software Feature Extraction software (v10.3.1, Agilent Technologies). The differentially expressed genes were detected with a linear mixed model and validated using quantitative PCR on several epileptic gene markers (Lipovich et al., 2012).
Cellular interactome
Although the fluorescence intensities from microarrays cannot reliably assess the absolute quantity of RNA in a given sample because of variable hybridization efficiency and cross hybridization (Draghici et al., 2006; Zahurak et al., 2007), they can be used to indicate how the quantity of RNA varies across tissue samples, as the fluorescence intensity is proportional to the quantity of each hybridized transcript (Lockhart et al., 1996; Cho et al., 1998; Li and Wong, 2001; Hubbell et al., 2002). For each differentially expressed gene, the normalized fluorescence was averaged across the four replicates using the two dyes. Across all microarrays for the 15 high and 15 low-spiking samples, this resulted in a series of 30 fluorescent intensities for each differentially expressed gene. Pearson correlations were then calculated for each differentially expressed gene across the 30 samples resulting in a correlation matrix. While other clustering methods are available, we compared these to the Pearson correlation and did not find any additional clusters. Linkages between two genes were created when the Pearson correlations were ≥0.95. A ‘Simple Interaction File’ (SIF format) was built using these linked genes and loaded into Cytoscape v2.8 software (Smoot et al., 2011). Statistically distinct clusters were identified using the plugin AllegroMCODE (AllegroViva, Santa Clara, CA, USA) based on the degree of linking between each gene: the more a gene receives links from other genes, the more it is moved in the direction of the kernel of a given cluster (as in Fig. 1B). Once gene clusters were identified, we used a literature mining approach to assign each cluster a putative cell type with a custom Python algorithm (see Supplementary material for script). This approach identified the number of PubMed abstracts (hits) containing both a given gene and the name of a given cell type. These were then plotted (Supplementary Fig. 1) and summarized in Supplementary Table 1. Cell types with the greatest number of hits were given a cell type designation. For each cluster, the fluorescence intensity for each gene within a given cluster was averaged together with other genes in that cluster to generate an ‘eigengene’ intensity for the relative quantity of each predicted cell type. Dendrograms were then built using the absolute value of the Pearson correlations between profiles as a measure of similarity as shown in Fig. 2. For downregulated clusters, the order of the values from low to high were interchanged and their profiles drawn as negative values on the y-axis. To understand the interrelationship between predicted cell clusters in high versus low spiking brain regions, the fluorescence intensity ratios between high low spiking for each cell type was also plotted for each patient in a second dendrogram (Fig. 2B).
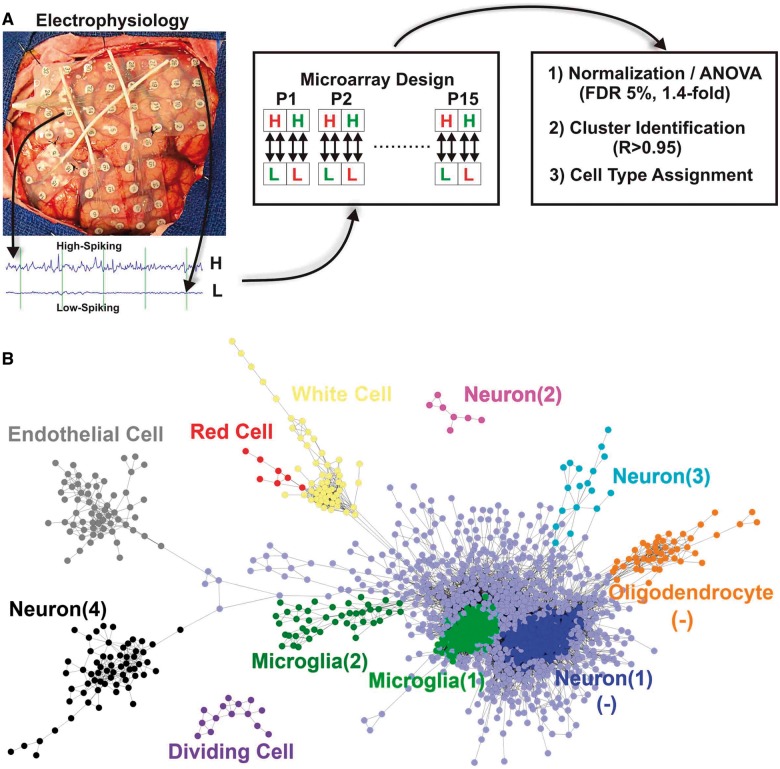
Figure 1.
Paired analysis of low- and high-spiking human neocortex reveals a cellular interactome. (A) Subdural arrays of electrodes placed on the cortex of a human epileptic patient are used to detect low and high interictal spiking areas. Total RNA from each of these areas was used in a quadruplicate dye-swap microarray design to identify differentially expressed genes. (B) The expression of the differentially expressed genes across all patients was used to build a cellular interactome to predict changes in cellular composition in low- versus high-spiking human neocortex. Each cluster of genes represents a different cell type and is displayed with a different colour, where each dot represents a single RNA transcript. The links shown between each gene in a given cluster had a Pearson correlation coefficient ≥0.95. Clusters are themselves clustered based on how closely they are statistically linked to each other. (-) refers to cell clusters that are downregulated.
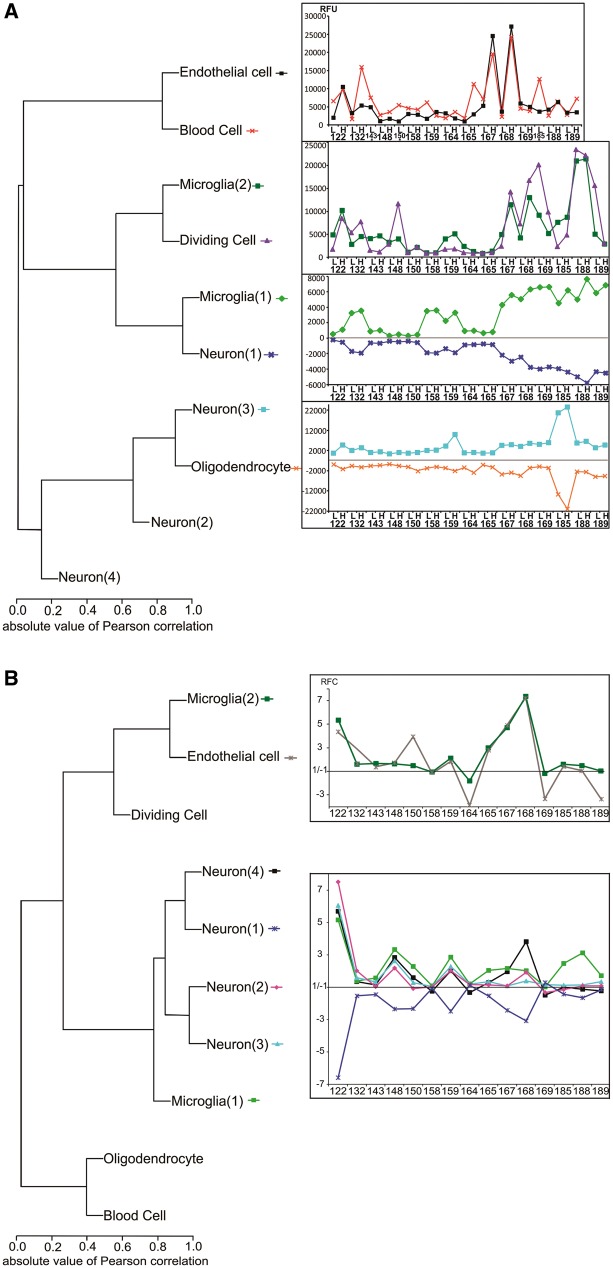
Figure 2.
Hierarchical clustering of assigned cell types predicts both parallel and reciprocal patterns of cellular changes in high-spiking human neocortex. (A) Hierarchical clustering of the predicted cell clusters using Pearson correlations were used to generate their corresponding profile from the average expression of genes within a given cluster. Dendrograms showing similar as well as reciprocal correlations between each cell cluster across patient samples are shown. For each patient sample, ‘L’ indicates low-spiking and ‘H’ high-spiking brain regions. To account for the downregulation of type 1 neurons and oligodendrocytes the values in low-spiking and high-spiking were inverted on the y-axis. (B) Hierarchical clustering of normalized fluorescence ratios between high and low spiking cell clusters segregated into two groups: (i) clustering of neuron types 1, 2, 3 and 4 with type 1 microglia; and (ii) clustering of endothelial cells and type 2 microglia. RFU = relative fluorescence unit; RFC = relative fold change. *This cluster has been computed after removing three samples (high-spiking regions from Patients 143, 150 and 185) because they showed aberrant blood contamination from surgery.
Histology of human neocortex
Brain tissues corresponding to precise electrode locations were prepared as described previously (Beaumont et al., 2012) and 20 µm sections were prepared and stored frozen. Immunostaining was performed after first washing in phosphate-buffered saline (PBS) and incubating for 1 h in a blocking solution (0.05% of Triton™ X-100, 10% goat serum, in PBS). After three more PBS washes, endogenous peroxidases were inactivated using 0.2% sodium azide with 0.3 % H2O2 in TBS (Tris buffered saline) and incubated overnight in the blocking solution with the following antibodies: CD68 (mouse, monoclonal clone KP1, Dako) at a concentration of 1:10; dpERK (mouse, #8159, Sigma) at 1:1000; GFAP (rabbit polyclonal, Dako) at 1:5000; Map (mouse, monoclonal MAB378 anti-MAP2A2B, Millipore) at 1:20 000; NeuN (mouse, monoclonal, cat #MAB377, Millipore) at 1:4000; pCREB (rabbit, #9197, Cell Signaling) at 1:100. The slides were then incubated with a biotinylated secondary antibody anti-mouse or anti-rabbit (Jackson ImmunoResearch) at a concentration of 1:150 for 2 h in blocking solution and amplified with ABC solution (Vectastain, Vector laboratories) followed by 3,3′-diaminobenzidine (DAB, Sigma) as per the manufacturer’s protocol. The slides were dehydrated in three successive baths of ethanol (75%, 90%, 100%) and a final bath of xylene for 5 min each and mounted using Cytoseal™ (Fisher). Antibody specificity for each was confirmed by omitting the primary antibody and through known staining patterns. Blood vessels were revealed by direct DAB labelling (with no antibodies) in the absence of the peroxidase inactivation step. Luxol Fast blue and Periodic acid–Schiff (PAS) stained slides were first washed three times in PBS and then placed overnight in a bath of Luxol Fast blue (Poly Scientific). The next morning, excess reagent was washed away with 95% ethanol, then in water followed by 1 min in 0.005% lithium carbonate (Poly Scientific) and then rinsed with water. The slides were next oxidized using 1% periodic acid (Poly Scientific) for 5 min, rinsed with water before putting them in Schiff reagent (Poly Scientific) for 15 min and then washed with water. The sections were then stained by placing the slides in haematoxylin (Meyer) for 3 min, washed with water and then put in ammonia for 1 min. The sections were then washed with water, dehydrated with 95% ethanol for 3 min, stained in eosin Y (FD NeuroTechnologies) for 8 min, rinsed in 95% ethanol and sealed with Cytoseal™.
Quantitation of blood vessels, microglia and neurons was performed using Metamorph® 6.2 software (Molecular Devices Inc), by counting the cell number per unit surface area. For the blood vessels, the fibre length was used. The significance was assessed using a paired t-test low versus high-spiking.
Quantification of histological staining
MetaMorph® (v7.8, Molecular Devices) was used for all quantitative measures using customized macros. Some blocks of tissue were large and corresponded to multiple recording sites, however, we only used a single slide per block for each quantitative measure. The blood vessel lengths (Fig. 3B) were measured using the function ‘average fibre length’ to quantify the dark brown staining of DAB-labelled vessels in the grey matter. The same function was also used in Fig. 5C to quantity the length of fibres stained by silver inside the layer IV. Identical sized regions of interest (20 × fields) from paired areas high/low spiking at the same layers were used for each quantitation. For silver staining, the areas with reduced NeuN staining versus regions with normal NeuN staining from layer IV regions were selected, blinded from the silver staining pattern on adjacent sections. The average areas of the processes in the layer IV stained by silver (Fig. 5C) were measured with the MetaMorph® function ‘average fibre area’. The processes with a length >20 µm (remove small objects) and a shape between 0 to 0.2 (remove circular objects) were quantified in these two areas for each of five patients. These two filters removed much of the noisy background. The density of microglia (Fig. 3D) and the density of neurons (Fig. 6C) were measured in paired samples using the ‘count’ function, divided by the area used for the quantification. The CD68 antibody used for microglia CD68 is a cellular marker while the NeuN antibody used for neurons is a nuclear marker. The size of microglia (Fig. 3E) and the size of the neuronal nuclei (Fig. 6B) were measured using the function ‘thresolded area’ divided by the number of objects detected and the area for equal areas from high and low spiking regions.
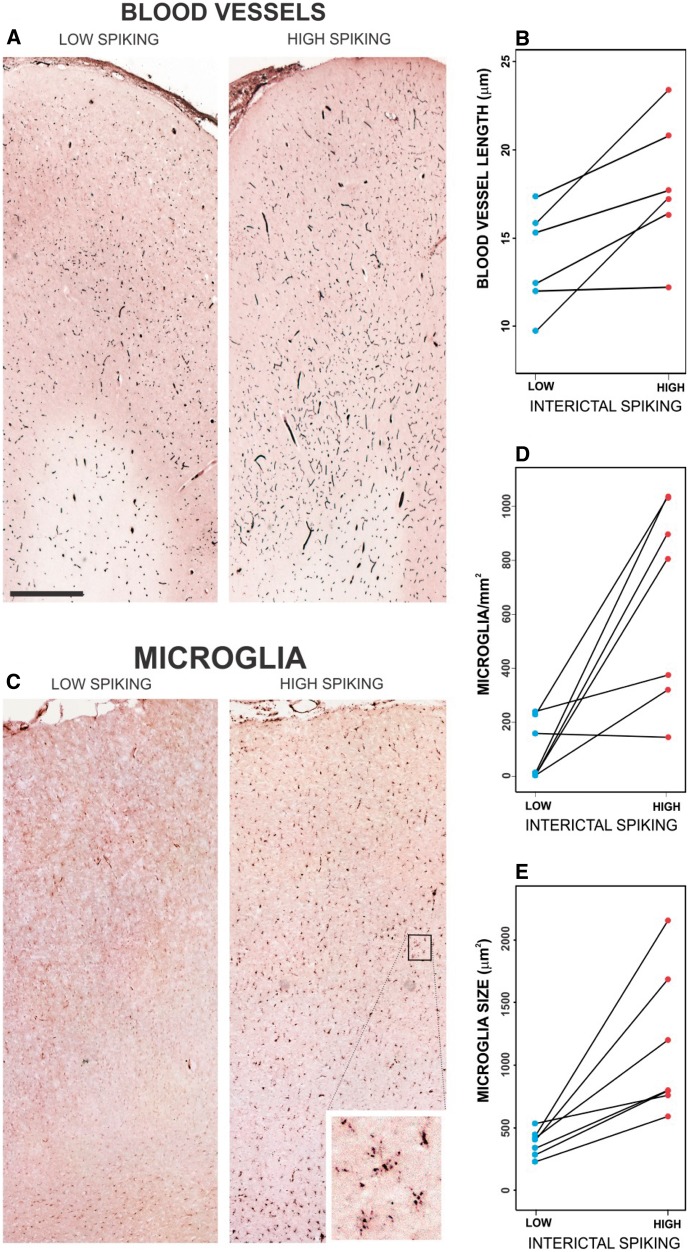
Figure 3.
Validation of increased vascularity and microglial infiltration in high-spiking human neocortex. (A) Staining blood vessels shows an increase in vascularity in high-spiking brain areas. (B) Blood vessel length was significantly increased in the high-spiking brain for seven patients (two-tailed, paired P-value < 0.05). (C–E) Microglia number and size were both increased in high-spiking areas versus low-spiking areas using CD68 staining across six patients (both two-tailed, paired P-value < 0.05). The scale bar in A indicates 1 mm.
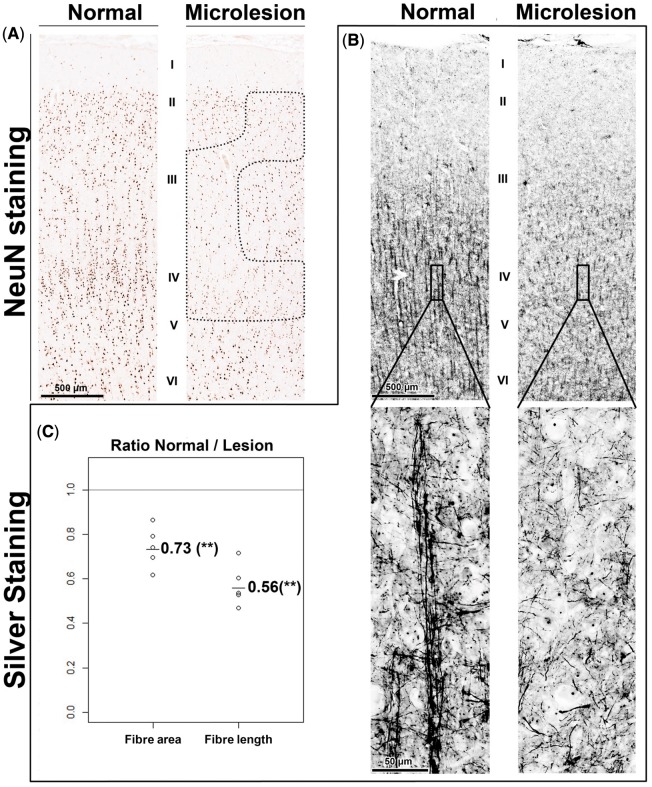
Figure 5.
Silver staining confirms a marked loss of neuronal processes in microlesions. (A) Microlesions identified by NeuN staining show a marked reduction of silver-stained neuronal processes in layer IV (B). (C) Quantitation of fibre density and length from five patients showed a significant reduction of the average fibre density and length in the microlesions. **A paired P-value < 0.01.
Figure 6.
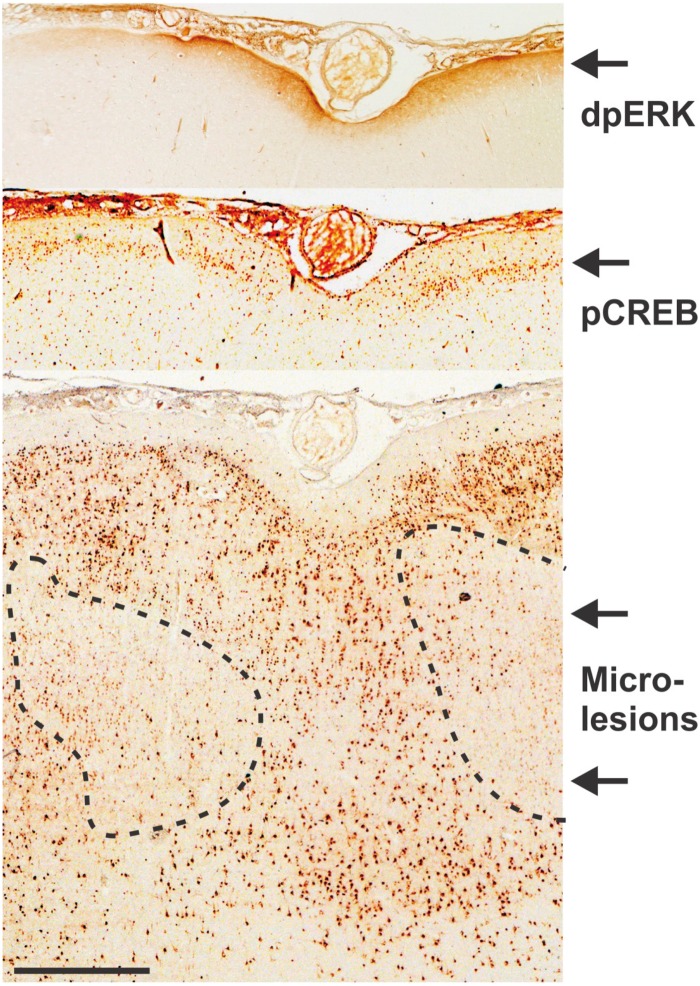
Microlesions were strongly associated with activation of epileptic biomarkers in superficial brain layers. Regions of high interictal spiking showing microlesions with NeuN staining almost always showed nearby activation of epileptic biomarkers including diphosphoERK (dpERK) in layer I and phosphoCREB (pCREB) in layer II. The scale bar indicates 1 mm.
Results
Coordinate modulation of cell-based gene clusters in high-spiking human neocortex
When patients fail to respond to traditional medical treatments, surgical procedures that remove regions of the brain that produce seizures and interictal spikes can be curative. As part of this procedure, abnormal electrical activities produced by increased neuronal synchrony are measured in vivo using subdural arrays of electrodes placed directly on the cortical surface (Fig. 1A). These continuous recordings, lasting from a few days to a week, are used to design a second surgical procedure to remove epileptic regions, with a goal of maximally preserving normal brain functions. For the present study, we have developed an internally controlled experimental design to assess what is different between regions of human neocortex that show high interictal spiking activity compared to nearby low or non-spiking regions from the same patient. Figure 1A shows an example of how low versus high interictal spiking regions are identified from long-term in vivo recordings together with our experimental microarray design to measure differential gene expression between paired regions from each of the 15 patients with neocortical epilepsy (Table 1).
Given that the neocortex is a complex, six-layered network of interconnected neurons and glial cells, understanding the significance of differential gene expression requires consideration of complex brain cytoarchitecture. To make certain that gene expression differences were not due to tissue sampling differences or cortical lesions, two to three strips of full-thickness grey matter were pooled for each RNA sample from histologically normal-appearing low- and high-spiking regions, including patients who had lesions in other brain regions (Table 1). This analysis revealed 2516 highly significant, differentially expressed genes across all 15 patients, 1534 genes were upregulated whereas 982 genes were downregulated in high-spiking brain regions.
One of the major limitations in profiling gene expression in multicellular tissues is the inability to differentiate which gene expression changes occur in which cell types. One approach to solve this has been through physical isolation of individual cell types (Cahoy et al., 2008; Morris et al., 2011); others have used a variety of computational algorithms sometimes combined with high-throughput in situ hybridization data (Shen-Orr et al., 2010; Hawrylycz et al., 2012). The approach used here was to hypothesize that each cell type has a unique transcriptional signature that can be identified through transcriptional clustering. We first used this clustering approach on proteomic data to identify histological changes from the same brain regions (G. Keren-Aviram, in preparation). Cell clusters were predicted using a Pearson correlation measure of expressed genes. Using a correlation cut-off of 0.95 for 15 patients (P-value < 10−3), we identified 725 out of the 2516 genes that fell into 11 clusters of co-expressed genes (Fig. 1B). We then assigned each gene cluster a putative cell type designation using a literature mining Python algorithm we developed that cross-references genes in a given cluster with the names of brain cell types (Supplementary Table 1; see Supplementary material for cell assignation script). As an example, this analysis predicted an increase in endothelial cells in high-spiking brain regions based on known endothelial genes present in the gene cluster (Supplementary Fig. 1). Putative identification of 10 of 11 of these clusters was achieved in this way (Fig. 1B). The 11th cluster (labelled as type 4 neuron) did not produce a statistically significant cell type by this method and was identified manually, gene by gene, using the Allen Brain Atlas database (www.brain-map.org/) and PubMed (NIH) literature mining. Cell types identified as increased in high-spiking cortex include endothelial cells, red and white blood cells, two types of microglia, three types of neurons, and a group of dividing cells, while decreased populations included a group of oligodendrocytes and type 1 neurons. Interestingly, although patients had other known pathologies in other brain regions, we found no clear statistical relationships between the differentially expressed genes and these other pathologies.
The cellular interactome
As a means to examine the interrelationships between these predicted cellular changes, the average expression pattern of each of the 11 predicted cell types for all 30 samples were further organized using dendrograms. Figure 2A shows the relative quantities of each predicted cell type across the 30 human tissue samples. These values were then regrouped by hierarchical clustering to predict statistically significant cellular interactions. Figure 2B shows for each predicted cell type the fold change between paired low and high-spiking tissues for each of the 15 patients.
Interestingly, both highly consistent and exact reciprocal patterns of gene clusters were observed. For example, the upregulated genes forming the cluster labelled as ‘endothelial cell’ correlate best with the cluster formed by upregulated genes from the red and white blood cell clusters, as would be expected for blood vessels. Taken together, these changes predict an increase in vascularity in high-spiking brain regions. The dendrograms also show two microglial clusters, both increased in high-spiking brain regions. However, each of the two microglial clusters segregates to a different group suggesting two distinct microglial roles related to either increased vascularity for type 1 (Fig. 2B) and to changes in neuronal composition for type 2 microglia (Fig. 2B). To assess the subtypes of microglia implicated in high-spiking, we calculated the enrichment for both M1 and M2 microglial subtypes for each of the microglial clusters. We found that type 2 microglial genes were enriched with both M1 and M2 subtypes at around six times higher than expected by chance (Supplementary Table 2). A slight enrichment in type M1 versus M2 was present (M1/M2 = 1.3). The type 1 microglia cluster could not be related to either of the M1 or M2 subtype.
Validation of predictions from the cellular interactome revealed novel ‘microlesions’
The results from the cellular interactome shown in Figs 1 and 2 make predictions about the cellular compositions of high versus low spiking human brain regions. For each block of tissue used for genomic studies, an adjacent piece was analysed histologically to validate cellular predictions (Loeb, 2010, 2011). As predicted in Fig. 2, the density and size of microglia, as well as the density of the microvasculature (composed of endothelial, red and white cells) were significantly increased in high-spiking brain regions relative to low spiking regions within each patient (Fig. 3). This increase in vascularity can be linked directly to a global increase of matrix metallopeptidases 3, 9 and 19 and to the presence in the endothelial cell cluster of the angiogenic factors VEGFC and FGF1.
The bottom panel of Fig. 2B contains a majority of the clustered genes and predicts a complex, reciprocal interplay between four neuronal populations and another microglial subgroup. In fact, of the 11 cell types identified, type 1 neurons (272 genes) and type 1 microglia (179 genes) constitute 62% of all clustered genes and are thus mostly responsible for the cell changes related to interictal spiking. The type 1 neuron cluster is formed by genes downregulated in high-spiking brain and is inversely related to three other neuron types and type 1 microglia (Fig. 2B). This pattern suggests a spatially localized microglial infiltration with a corresponding predicted change in neuronal phenotype in the same high-spiking brain region. To validate these predictions, we identified genes within each group to use for histological validation. One gene in the type 1 neuron cluster was the layer IV-specific neuronal marker RORB (Schaeren-Wiemers et al., 1997), predicting a decrease in the layer IV neurons. Another approach we took was to identify additional genes that correlate with the neuron type 1 cluster even if they were not significantly differentially expressed between high and low spiking tissues at the threshold that we initially chose (fold change ≥ 1.4, FDR ≤ 5%). By lowering this statistical cut-off to 1.3-fold, we identified the well-known histological neuronal marker NeuN (RBFOX3 gene) (Supplementary Fig. 2), which is perhaps the most widely used immunohistochemical marker to label neurons in mammalian cortex (Dent et al., 2010). In fact, NeuN showed a correlation coefficient of 0.92 (P < 0.001) with the type 1 neuron cluster across 15 patients. Taken together, this purely bioinformatic approach predicts a decrease in layer IV neurons showing lower levels of NeuN staining in cortical regions with increased microglia.
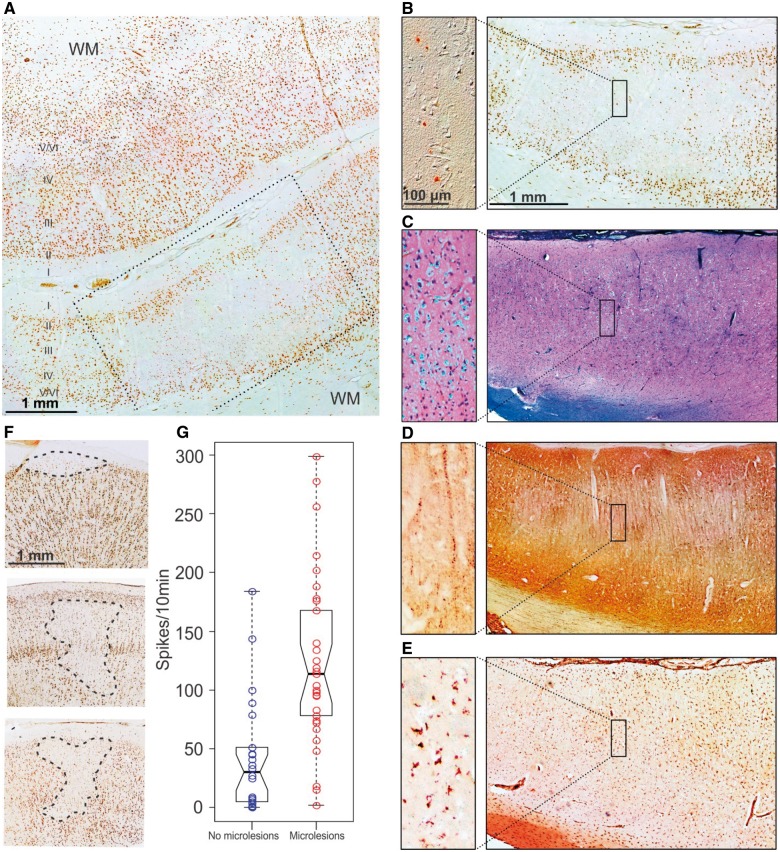
Consistent with the above predictions, an average of 2–3 focal regions of reduced NeuN staining were seen maximally in layer IV high-spiking brain regions in each of 29 high-spiking brain regions from 22 epilepsy patients (Fig. 4 and Table 1). Figure 4A shows two facing regions of human neocortex from a high-spiking region. While a normal six-layered staining pattern is seen on the top half, the facing cortical region below shows a focal loss of NeuN staining. Using serial histological sections from the lower part of this section, the loss of NeuN staining was not due to a loss of neurons, shown by both histological tissue staining methods (Luxol Fast blue-PAS with haematoxylin as shown; cresyl violet data not shown) and DIC (differential interference contrast) microscopy (Fig. 4B and C). However, the neurons in this region were not normal. They showed a significant reduction in MAP2 staining that normally should show rich dendritic arbours (Fig. 4D). To explore this further, we used silver staining to examine neuronal process density and length within microlesions compared to nearby regions with normal NeuN staining (Fig. 5). Consistent with the MAP2 dendritic staining, we found a marked alteration in neuronal process architecture with a significant reduction in both the density and length of neuronal processes in layer IV (Fig. 5B and C). We also quantified the density and nuclear size of the remaining NeuN positive neurons within microlesions, demonstrating a decrease in both neuronal nuclear size and neuronal density compared to nearby regions (Supplementary Fig. 4A–C). Finally, as predicted by the cellular interactome, these regions were filled with microglia (Fig. 4E).
Figure 4.
Evidence for focal microlesions in high-spiking human neocortex. (A) NeuN staining revealed both normal (upper part of section) and regions with reduced NeuN staining (lower part). (B) Regions with decreased NeuN staining still had neurons shown by differential interference contrast microscopy (enlargement) and were centred in layers III and IV, with a normal appearance of layers II, V and VI. Scale bar = 1 mm; inset bar = 100 µm. (C) Traditional histological staining with Luxol Fast blue-PAS and haematoxylin staining shows a normal-appearing laminar pattern of neurons that would not have detected these lesions. (D) NeuN negative regions have markedly reduced staining of neuronal processes using MAP2 antibodies. (E) NeuN negative regions are full of microglia stained with CD68. (F) Three different types of microlesions were seen by NeuN staining involving layers II/III (9%), layers III and IV (34%), or layers II–IV (57%). (G) Microlesions were strongly associated with interictal spike frequency (unpaired t-test, P < 10−5). WM = white matter.
The increases in type 2, 3 and 4 neurons likely represent the ‘missing’ subpopulations of NeuN neurons. Of these, the type 4 neuron best (inversely) correlates with the decrease in type 1 neurons (R = −0.9), suggesting that they represent phenotypically different neurons. Further understanding of these neuronal subtypes is gained from the composition of differentially expressed genes within each cluster. For example, the type 2 neuron gene cluster is well represented by seven genes (ACRC, WNT1, SV2C, NPTX2, GPR68, GABRQ and IGSF22), some of which are known to be highly expressed in layer III (using the Allen Atlas at www.brainatlas.org) (Zeng et al., 2012). A similar analysis for the type 3 neuron cluster shows 18 genes which have been localized to layer IV (ABCC3, ACPL2, AK094995, ALDH1A3, BC063385, BC094703, FNDC9, GFOD1, CHST3, DCLK1, DUSP4, PDYN, PLK3, SCG2, SLC22A4, THC2624202, TIPARP and TTC39C).
Surprisingly, unlike the histopathology normally seen in forms of epilepsy associated with hippocampal sclerosis, no astrogliosis was predicted or seen in these microlesions (Supplementary Fig. 3 shows one example; however, many other patients were also examined). Consistently, astroglial markers (GFAP and ALDH1L1) (Cahoy et al., 2008), were not detected in our gene expression profiles and did not display altered expression in high-spiking regions.
Microlesions are a common in high-spiking epileptic brain regions and associated with epileptic biomarkers
While microlesions varied in size and shape, and have an average cross-sectional area in the range of a few millimetres, the majority were centred at layers II–IV, with three distinct microlesion subtypes: (i) layer II only (9%); (ii) layers III and IV (34%); and (iii) layers II, III and IV (57%) (Fig. 4F). Screening additional patients for the presence and size of these NeuN negative microlesions in both low and high-spiking neocortex revealed the presence of microlesions in 80% of the high-spiking brain regions. Combining all tissue samples together, regions showing high spike frequency had statistically more microlesions than low spiking regions (Fig. 4G).
One limitation of human tissue studies is that they cannot prove cause and effect nor reveal any time-dependent processes, given that the tissue is collected at one point in time after epilepsy is well established. A key unanswered question is whether microlesions in high-spiking epileptic tissue cause or result from interictal spiking or seizures. Although 725 of 2516 differentially expressed genes fell into tight cellular clusters, the remaining genes are also associated with high interictal spiking and likely reflect additional changes in brain structure and function related to epileptic spiking. We recently reported that the constitutive activation of the MAPK/CREB signalling pathway is a biomarker for interictal spiking in superficial neocortical layers I–III in both humans and an animal model of interictal spiking (Rakhade et al., 2005; Barkmeier et al., 2012a; Beaumont et al., 2012; Lipovich et al., 2012). We therefore examined the spatial relationship between microlesions and MAPK/CREB signalling (Fig. 6). This analysis revealed that high-spiking cortical regions show a marked activation of both MAPK and CREB signalling in the superficial cortical layers directly above or adjacent to deeper microlesions. From 19 patients (Table 1) we found that 17 of 21 high-spiking regions (80%) showed activation of ppERK and/or pCREB either directly above or adjacent to the microlesion. These findings raise the intriguing possibility that the layer-specific disruption of deeper cortical layers promotes epileptic hyperexcitability in adjacent superficial layers as a putative mechanism for neocortical epilepsy.
Discussion
Epilepsy remains one of the least understood brain disorders that can produce many types of clinical seizures depending on where abnormal brain activity occurs and how it electrically spreads. In many ‘non-lesional’ cases, including many in our patient group, no clear pathological abnormalities are seen within epileptic brain regions. Even for patients with a wide variety of associated brain lesions nearby, the epileptic focus itself is often histologically normal (Annegers, 1993). Using transcriptional clustering of low- versus high-spiking human neocortex from 15 unique epileptic patients we predicted and validated a common interactome of cellular changes in high-spiking neocortex. This cellular interactome predicted an increase in vascularity and microglia and the presence of microlesions centred in deeper cortical layers superimposed with superficial activation of synaptic plasticity genes. Microlesions were easily identified by a loss of NeuN neuronal staining and are associated both with a significant reduction in MAP2 and silver-stained neuronal processes together with a focal infiltration of microglia. Subdural electrodes did not cause these lesions, as both paired tissues examined had the same amount of exposure to the given grids. An attractive hypothesis that emerges is that disrupted connectivity from deeper cortical layers induces hypersynchrony in superficial layers. In addition to providing new insights in human neocortical epilepsy, the transcriptional clustering methods developed here could be useful in predicting the cellular abnormalities in other brain or complex tissue disorders.
High throughput systems biology can be a powerful tool to generate new research directions by identifying genes and pathways linked to human disease. However, for human or other tissue studies, the expanding number of variables together with the cellular complexity makes the output from these studies difficult to interpret. Nowhere is this more difficult than in the brain, which is composed of many different cell types. One approach is to know a priori which cells are of interest and selectively isolate and profile RNA from each of these cell types (Cahoy et al., 2008; Shen-Orr et al., 2010). We have taken an alternative approach using a paired experimental design to measure consistent transcriptional differences between low- and high-spiking human cortex from 15 patients. We used clustering to identify subsets of differentially expressed genes that fell into 11 statistically significant groups. We then developed a literature-mining tool to assign a likely cell type for each. This analysis not only predicted changes in different cell groups, but also predicted their relative interactions with each other producing a global, ‘cellular interactome’ for high-spiking human neocortex.
One of the most intriguing questions from this work is how deep cortical microlesions relate to epileptic spiking. We found that brain areas with microlesions had increased interictal spiking, suggesting a potential cause and effect relationship. In fact, microlesions were strongly associated with sustained MAPK and CREB activation in more superficial cortical layers I–III directly above or adjacent to the microlesions (Beaumont et al., 2012). One possible explanation is that the loss of axo-dendritic neuronal processes seen both with MAP2 and silver staining in deeper cortical layers reduces normal inhibition resulting in aberrant synchrony in superficial brain layers. Once a small region of hypersynchrony forms, it can spread laterally to adjacent cortical regions (Barkmeier et al., 2012b; Beaumont et al., 2012).
Key predictions made by this cellular interactome were validated in human epileptic neocortex. This is made possible because every block of tissue from our bank of electrically mapped tissues is subdivided for RNA, protein, metabolites and histology (Loeb, 2010). A key prediction led to the identification of millimetre-sized microlesions present in almost all high-spiking tissues examined, independent of any other nearby pathologies that ranged from mild subcortical (white matter) gliosis to more severe lesions such as polymicrogyria, heterotopias and dysplasias (Table 1). It is important to reiterate that, although some patients had these lesions nearby, the neocortical tissues used here was deemed ‘normal’ by traditional histological analysis.
Clues to the presence of cortical microlesions in high-spiking cortex came predominantly from the coordinate downregulation of a group of 272 genes we call type 1 neurons. While microlesions are not readily seen with conventional histological staining methods, a more detailed look in these focal brain regions revealed a marked loss of axo-dendritic arborizations and extensive microglial infiltration. Microlesions are readily seen by staining with antibodies against NeuN, a splicing factor called RBFOX3 (previously known as Fox-3; Kim et al., 2009) that has been shown to be downregulated following neuronal injury (Unal-Cevik et al., 2004). Furthermore, the upregulation of type 1 microglia perfectly mirrored the downregulation of type 1 neurons, and, at the same time, three other neuronal types (type 2, 3 and 4) increased in direct proportion to the degree of the loss of type 1 neurons. This suggests the possible replacement or phenotypic change of type 1 neurons with these other neuronal types in high-spiking areas.
Exactly how and when these lesions occur during the development of the epileptic disorder is not clear. The marked reduction in neuronal processes indicates that these small, focal brain areas are not normal and could be due to developmental defects in neuronal migration or lamination (Bentivoglio et al., 2003; Powell et al., 2003). Further studies are needed; however, as many of the neurons within these lesions are smaller with reduced neuronal branching, it is also possible that they could be interpreted as type IA focal cortical dysplasias that lack large, abnormal neurons, but show changes in the laminar organization (Palmini et al., 2004). Another possibility is that they result from a past injury or infection (Choi et al., 2009), which is supported by pronounced microglial infiltration in these same areas. While microlesions could be the ‘cause’ of the epileptic spiking, it is also possible that they instead ‘result’ from spiking. In fact, neurodegenerative changes with reduced neuronal density have been described after prolonged seizures (Holmes, 2002; Sutula et al., 2003), but have not been reported for interictal spiking.
Validated predictions also included a global increase in microglia and blood vessels. Not surprisingly, samples with increased vascularity also showed a consistent upregulation of the angiogenic factors VEGFC, FGF1 and the matrix metalloproteinases 3, 9 and 19 that are known to promote the growth of blood vessels. These changes could have resulted from a higher oxygen requirement of more active epileptic tissues. There have been a number of reports of increased angiogenesis coupled with aberrant vascularization in epileptic models (Pitkanen and Sutula, 2002; Morin-Brureau et al., 2011). In addition to the type 1 microglia associated with microlesions, the second group of microglia (type 2) correlated strongly with increased vascularity, suggesting an independent role for microglia that interact with blood vessels. Both of these gene clusters contained genes from M1 and M2 subtypes. Microglia have long been implicated in epileptic disorders and suggest the presence of a chronic, inflammatory state (Vezzani et al., 2011). Microglia are known to cause hyperexcitability in damaged brain areas by stripping away inhibitory neurons (Trapp et al., 2007) and they have been reported to be associated with cortical dysplasias associated with epileptic brain regions (Aronica et al., 2005; Boer et al., 2006). While microglial transcriptomes have been demonstrated based on their state of activation (Parakalan et al., 2012), the transcriptional clustering approach used here has divided microglia into two clear groups based on their gene expression patterns. In fact MCP1 (now known as CCL2), a potent chemo-attractant for monocytes, was significantly upregulated in 11 of 15 patients in high-spiking brain regions and has previously been linked to epileptic disorders (Fabene et al., 2010).
One of the most highly studied and best-characterized epileptic lesions is hippocampal sclerosis. Hippocampal sclerosis has been observed in children following prolonged febrile seizures and histologically shows massive neuronal loss with extensive ‘scarring’ that is readily seen with antibodies against GFAP that recognizes reactive astrocytes (Thom et al., 2010; Cersosimo et al., 2011). Unlike hippocampal sclerosis, the cellular changes predicted and described here for neocortical spiking did not show a significant drop out of neurons (Unal-Cevik et al., 2004) nor any detectable gliosis in high-spiking grey matter, even within the microlesions. However, many of the neocortical samples we studied did have subcortical gliosis in the white matter, which was not present in RNA samples (Table 1) as only grey matter was used here. In a previous study where white matter was also included, increased GFAP mRNA expression was in fact observed, consistent with the observed histopathology (Beaumont et al., 2012).
In summary, patients who undergo cortical resections for seizures that do not respond to current medications offer unparalleled opportunities to link the in vivo electrical properties of the human brain to their underlying molecular pathways. Here we have focused on the unique transcriptional profile and predicted and validate novel cellular changes associated with interictal epileptic spiking rather than seizures. Epileptic spiking is more frequent than seizures, often occurs in the exact same brain regions that produce seizures, and therefore has been extensively used to localize seizure foci (Staley et al., 2005). Interictal spiking can also be associated with behavioural abnormalities in humans (Pressler et al., 2005; Fonseca et al., 2008) and in animal models (Zhou et al., 2007; Kleen et al., 2010; Barkmeier et al., 2012a), and can be seen in injured brain regions long before clinical seizures begin (Staley et al., 2005, 2011). However, the relationship between interictal spiking and seizures is not well understood. Using a similar experimental design on a smaller group of patients, we recently identified a complex transcriptional regulatory machinery between coding and long non-coding RNAs that strongly implicates a coding–non-coding MAPK/CREB signalling interactome for interictal spiking (Lipovich et al., 2012). The cellular interactome presented here places these earlier findings in a cellular context and offers further clues to understand the underlying abnormal structural networks underlying human neocortical epilepsy.
Acknowledgements
We thank Drs Eishi Asano, Harry Chugani, Sandeep Mittal, Aashit Shah, Sandeep Sood at the Wayne State Comprehensive Epilepsy Program for ongoing fruitful interactions, help with obtaining tissues from their patients, and EEG data. Microarray scanning was facilitated by the Core Facility of the Environmental Health Sciences Centre at Wayne State University.
Funding
This work was funded by NIH/NINDS Grants R01NS045207 and R01NS058802 (to J.A.L.).
Supplementary material
Supplementary material is available at Brain online.
References
- Annegers J. Philadelphia, PA: Lea & Febiger; 1993. The treatment of epilepsy: principles and practices. [Google Scholar]
- Aronica E, Gorter JA, Redeker S, Ramkema M, Spliet WG, van Rijen PC, et al. Distribution, characterization and clinical significance of microglia in glioneuronal tumours from patients with chronic intractable epilepsy. Neuropathol Appl Neurobiol. 2005;31:280–91. doi: 10.1111/j.1365-2990.2004.00636.x. [DOI] [PubMed] [Google Scholar]
- Asano E, Muzik O, Shah A, Juhasz C, Chugani DC, Sood S, et al. Quantitative interictal subdural EEG analyses in children with neocortical epilepsy. Epilepsia. 2003;44:425–34. doi: 10.1046/j.1528-1157.2003.38902.x. [DOI] [PubMed] [Google Scholar]
- Barkmeier DT, Senador D, Leclercq K, Pai D, Hua J, Boutros NN, et al. Electrical, molecular and behavioral effects of interictal spiking in the rat. Neurobiol Dis. 2012a;47:92–101. doi: 10.1016/j.nbd.2012.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barkmeier DT, Shah AK, Flanagan D, Atkinson MD, Agarwal R, Fuerst DR, et al. High inter-reviewer variability of spike detection on intracranial EEG addressed by an automated multi-channel algorithm. Clin Neurophysiol. 2012b;123:1088–95. doi: 10.1016/j.clinph.2011.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bautista RE, Cobbs MA, Spencer DD, Spencer SS. Prediction of surgical outcome by interictal epileptiform abnormalities during intracranial EEG monitoring in patients with extrahippocampal seizures. Epilepsia. 1999;40:880–90. doi: 10.1111/j.1528-1157.1999.tb00794.x. [DOI] [PubMed] [Google Scholar]
- Beaumont TL, Yao B, Shah A, Kapatos G, Loeb JA. Layer-Specific CREB target gene induction in human neocortical epilepsy. J Neurosci. 2012;32:14389–401. doi: 10.1523/JNEUROSCI.3408-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentivoglio M, Tassi L, Pech E, Costa C, Fabene PF, Spreafico R. Cortical development and focal cortical dysplasia. Epileptic Disord. 2003;5(Suppl 2):S27–S34. [PubMed] [Google Scholar]
- Boer K, Spliet WG, van Rijen PC, Redeker S, Troost D, Aronica E. Evidence of activated microglia in focal cortical dysplasia. J Neuroimmunol. 2006;173:188–95. doi: 10.1016/j.jneuroim.2006.01.002. [DOI] [PubMed] [Google Scholar]
- Cahoy JD, Emery B, Kaushal A, Foo LC, Zamanian JL, Christopherson KS, et al. A transcriptome database for astrocytes, neurons, and oligodendrocytes: a new resource for understanding brain development and function. J Neurosci. 2008;28:264–78. doi: 10.1523/JNEUROSCI.4178-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cersosimo R, Flesler S, Bartuluchi M, Soprano AM, Pomata H, Caraballo R. Mesial temporal lobe epilepsy with hippocampal sclerosis: study of 42 children. Seizure. 2011;20:131–7. doi: 10.1016/j.seizure.2010.11.002. [DOI] [PubMed] [Google Scholar]
- Cho RJ, Campbell MJ, Winzeler EA, Steinmetz L, Conway A, Wodicka L, et al. A genome-wide transcriptional analysis of the mitotic cell cycle. Mol Cell. 1998;2:65–73. doi: 10.1016/s1097-2765(00)80114-8. [DOI] [PubMed] [Google Scholar]
- Choi J, Nordli DR, Jr, Alden TD, DiPatri A, Jr, Laux L, Kelley K, et al. Cellular injury and neuroinflammation in children with chronic intractable epilepsy. J Neuroinflammation. 2009;6 doi: 10.1186/1742-2094-6-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dent MA, Segura-Anaya E, Alva-Medina J, Aranda-Anzaldo A. NeuN/Fox-3 is an intrinsic component of the neuronal nuclear matrix. FEBS Lett. 2010;584:2767–71. doi: 10.1016/j.febslet.2010.04.073. [DOI] [PubMed] [Google Scholar]
- Draghici S, Khatri P, Eklund AC, Szallasi Z. Reliability and reproducibility issues in DNA microarray measurements. Trends Genet. 2006;22:101–9. doi: 10.1016/j.tig.2005.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabene PF, Bramanti P, Constantin G. The emerging role for chemokines in epilepsy. J Neuroimmunol. 2010;224:22–7. doi: 10.1016/j.jneuroim.2010.05.016. [DOI] [PubMed] [Google Scholar]
- Fonseca LC, Tedrus GM, Moraes C, Vicente Machado A, Almeida MP, Oliveira DO. Epileptiform abnormalities and quantitative EEG in children with attention-deficit/hyperactivity disorder. Arq Neuropsiquiatr. 2008;66:462–7. doi: 10.1590/s0004-282x2008000400004. [DOI] [PubMed] [Google Scholar]
- Gotman J. Relationships between interictal spiking and seizures: human and experimental evidence. Can J Neurol Sci. 1991;18(Suppl 4):573–6. doi: 10.1017/s031716710003273x. [DOI] [PubMed] [Google Scholar]
- Hawrylycz MJ, Lein ES, Guillozet-Bongaarts AL, Shen EH, Ng L, Miller JA, et al. An anatomically comprehensive atlas of the adult human brain transcriptome. Nature. 2012;489:391–9. doi: 10.1038/nature11405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes GL. Seizure-induced neuronal injury: animal data. Neurology. 2002;59(9 Suppl 5):S3–S6. doi: 10.1212/wnl.59.9_suppl_5.s3. [DOI] [PubMed] [Google Scholar]
- Hubbell E, Liu WM, Mei R. Robust estimators for expression analysis. Bioinformatics. 2002;18:1585–92. doi: 10.1093/bioinformatics/18.12.1585. [DOI] [PubMed] [Google Scholar]
- Kanazawa O, Blume WT, Girvin JP. Significance of spikes at temporal lobe electrocorticography. Epilepsia. 1996;37:50–5. doi: 10.1111/j.1528-1157.1996.tb00511.x. [DOI] [PubMed] [Google Scholar]
- Kim KK, Adelstein RS, Kawamoto S. Identification of neuronal nuclei (NeuN) as Fox-3, a new member of the Fox-1 gene family of splicing factors. J Biol Chem. 2009;284:31052–61. doi: 10.1074/jbc.M109.052969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleen JK, Scott RC, Holmes GL, Lenck-Santini PP. Hippocampal interictal spikes disrupt cognition in rats. Ann Neurol. 2010;67:250–7. doi: 10.1002/ana.21896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SA, Spencer DD, Spencer SS. Intracranial EEG seizure-onset patterns in neocortical epilepsy. Epilepsia. 2000;41:297–307. doi: 10.1111/j.1528-1157.2000.tb00159.x. [DOI] [PubMed] [Google Scholar]
- Li C, Wong WH. Model-based analysis of oligonucleotide arrays: expression index computation and outlier detection. Proc Natl Acad Sci USA. 2001;98:31–6. doi: 10.1073/pnas.011404098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipovich L, Dachet F, Cai J, Bagla S, Balan K, Jia H, et al. Activity-dependent human brain coding/non-coding gene regulatory networks. Genetics. 2012;192:1133–48. doi: 10.1534/genetics.112.145128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lockhart DJ, Dong H, Byrne MC, Follettie MT, Gallo MV, Chee MS, et al. Expression monitoring by hybridization to high-density oligonucleotide arrays. Nat Biotechnol. 1996;14:1675–80. doi: 10.1038/nbt1296-1675. [DOI] [PubMed] [Google Scholar]
- Loeb JA. A human systems biology approach to discover new drug targets in epilepsy. Epilepsia. 2010;51(Suppl 3):171–7. doi: 10.1111/j.1528-1167.2010.02635.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loeb JA. Identifying targets for preventing epilepsy using systems biology. Neurosci Lett. 2011;497:205–12. doi: 10.1016/j.neulet.2011.02.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morin-Brureau M, Lebrun A, Rousset MC, Fagni L, Bockaert J, de Bock F, et al. Epileptiform activity induces vascular remodeling and zonula occludens 1 downregulation in organotypic hippocampal cultures: role of VEGF signaling pathways. J Neurosci. 2011;31:10677–88. doi: 10.1523/JNEUROSCI.5692-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris J, Singh JM, Eberwine JH. Transcriptome analysis of single cells. J Vis Exp. 2011;50:2634. doi: 10.3791/2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmini A, Andermann F, Olivier A, Tampieri D, Robitaille Y. Focal neuronal migration disorders and intractable partial epilepsy: results of surgical treatment. Ann Neurol. 1991;30:750–7. doi: 10.1002/ana.410300603. [DOI] [PubMed] [Google Scholar]
- Palmini A, Najm I, Avanzini G, Babb T, Guerrini R, Foldvary-Schaefer N, et al. Terminology and classification of the cortical dysplasias. Neurology. 2004;62(6 Suppl 3):S2–8. doi: 10.1212/01.wnl.0000114507.30388.7e. [DOI] [PubMed] [Google Scholar]
- Parakalan R, Jiang B, Nimmi B, Janani M, Jayapal M, Lu J, et al. Transcriptome analysis of amoeboid and ramified microglia isolated from the corpus callosum of rat brain. BMC Neurosci. 2012;13:64. doi: 10.1186/1471-2202-13-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitkanen A, Sutula TP. Is epilepsy a progressive disorder? Prospects for new therapeutic approaches in temporal-lobe epilepsy. Lancet Neurol. 2002;1:173–81. doi: 10.1016/s1474-4422(02)00073-x. [DOI] [PubMed] [Google Scholar]
- Powell EM, Campbell DB, Stanwood GD, Davis C, Noebels JL, Levitt P. Genetic disruption of cortical interneuron development causes region- and GABA cell type-specific deficits, epilepsy, and behavioral dysfunction. J Neurosci. 2003;23:622–31. doi: 10.1523/JNEUROSCI.23-02-00622.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pressler RM, Robinson RO, Wilson GA, Binnie CD. Treatment of interictal epileptiform discharges can improve behavior in children with behavioral problems and epilepsy. J Pediatr. 2005;146:112–7. doi: 10.1016/j.jpeds.2004.08.084. [DOI] [PubMed] [Google Scholar]
- Rakhade SN, Shah AK, Agarwal R, Yao B, Asano E, Loeb JA. Activity-dependent gene expression correlates with interictal spikingin human neocortical epilepsy. Epilepsia. 2007;48(Suppl 5):86–95. doi: 10.1111/j.1528-1167.2007.01294.x. [DOI] [PubMed] [Google Scholar]
- Rakhade SN, Yao B, Ahmed S, Asano E, Beaumont TL, Shah AK, et al. A common pattern of persistent gene activation in human neocortical epileptic foci. Ann Neurol. 2005;58:736–47. doi: 10.1002/ana.20633. [DOI] [PubMed] [Google Scholar]
- Schaeren-Wiemers N, Andre E, Kapfhammer JP, Becker-Andre M. The expression pattern of the orphan nuclear receptor RORbeta in the developing and adult rat nervous system suggests a role in the processing of sensory information and in circadian rhythm. Eur J Neurosci. 1997;9:2687–701. doi: 10.1111/j.1460-9568.1997.tb01698.x. [DOI] [PubMed] [Google Scholar]
- Seo JH, Noh BH, Lee JS, Kim DS, Lee SK, Kim TS, et al. Outcome of surgical treatment in non-lesional intractable childhood epilepsy. Seizure. 2009;18:625–9. doi: 10.1016/j.seizure.2009.07.007. [DOI] [PubMed] [Google Scholar]
- Shen-Orr SS, Tibshirani R, Khatri P, Bodian DL, Staedtler F, Perry NM, et al. Cell type-specific gene expression differences in complex tissues. Nat Methods. 2010;7:287–9. doi: 10.1038/nmeth.1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smoot ME, Ono K, Ruscheinski J, Wang PL, Ideker T. Cytoscape 2.8: new features for data integration and network visualization. Bioinformatics. 2011;27:431–2. doi: 10.1093/bioinformatics/btq675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spencer S, Huh L. Outcomes of epilepsy surgery in adults and children. Lancet Neurol. 2008;7:525–37. doi: 10.1016/S1474-4422(08)70109-1. [DOI] [PubMed] [Google Scholar]
- Staley K, Hellier JL, Dudek FE. Do interictal spikes drive epileptogenesis? Neuroscientist. 2005;11:272–6. doi: 10.1177/1073858405278239. [DOI] [PubMed] [Google Scholar]
- Staley KJ, White A, Dudek FE. Interictal spikes: harbingers or causes of epilepsy? Neurosci Lett. 2011;497:247–50. doi: 10.1016/j.neulet.2011.03.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutula TP, Hagen J, Pitkanen A. Do epileptic seizures damage the brain? Curr Opin Neurol. 2003;16:189–95. doi: 10.1097/01.wco.0000063770.15877.bc. [DOI] [PubMed] [Google Scholar]
- Thom M, Mathern GW, Cross JH, Bertram EH. Mesial temporal lobe epilepsy: how do we improve surgical outcome? Ann Neurol. 2010;68:424–34. doi: 10.1002/ana.22142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapp BD, Wujek JR, Criste GA, Jalabi W, Yin X, Kidd GJ, et al. Evidence for synaptic stripping by cortical microglia. Glia. 2007;55:360–8. doi: 10.1002/glia.20462. [DOI] [PubMed] [Google Scholar]
- Unal-Cevik I, Kilinc M, Gursoy-Ozdemir Y, Gurer G, Dalkara T. Loss of NeuN immunoreactivity after cerebral ischemia does not indicate neuronal cell loss: a cautionary note. Brain Res. 2004;1015:169–74. doi: 10.1016/j.brainres.2004.04.032. [DOI] [PubMed] [Google Scholar]
- Vezzani A, French J, Bartfai T, Baram TZ. The role of inflammation in epilepsy. Nat Rev Neurol. 2011;7:31–40. doi: 10.1038/nrneurol.2010.178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zahurak M, Parmigiani G, Yu W, Scharpf RB, Berman D, Schaeffer E, et al. Pre-processing Agilent microarray data. BMC Bioinformatics. 2007;8:142. doi: 10.1186/1471-2105-8-142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng H, Shen EH, Hohmann JG, Oh SW, Bernard A, Royall JJ, et al. Large-scale cellular-resolution gene profiling in human neocortex reveals species-specific molecular signatures. Cell. 2012;149:483–96. doi: 10.1016/j.cell.2012.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou JL, Lenck-Santini PP, Zhao Q, Holmes GL. Effect of interictal spikes on single-cell firing patterns in the hippocampus. Epilepsia. 2007;48:720–31. doi: 10.1111/j.1528-1167.2006.00972.x. [DOI] [PubMed] [Google Scholar]