Abstract
With this study, we explored the identity and chemistry of fungal endophytes from the roots of yerba mansa [Anemopsis californica (Nutt.) Hook. & Arn. (Saururaceae)], a botanical traditionally used to treat infection. We compared the diversity of fungal endophytes isolated from a wild-harvested A. californica population, and those from plants cultivated for one year in a greenhouse environment. The wild-harvested population yielded thirteen fungal strains (eleven unique genotypes). Of the extracts prepared from these fungi, four inhibited growth of Staphylococcus aureus by >25% at 20 µg/mL, and three inhibited growth of Pseudomonas aeruginosa by ≥20% at 200 µg/mL. By comparison, A. californica roots after one year of cultivation in the greenhouse produced only two unique genotypes, neither of which displayed significant antimicrobial activity. The fungus Chaetomium cupreum isolated from wild-harvested A. californica yielded a new antimicrobial spirolactone, chaetocuprum (1). An additional fourteen known compounds were identified using LC-MS dereplication of the various fungal endophytes. This study provides new insights into the identity and chemistry of A. californica fungal endophytes, and demonstrates the importance of considering growing conditions when pursuing natural product drug discovery from endophytic fungi.
Keywords: Anemopsis californica, antimicrobial, fungal endophyte, Chaetomium cupreum, spirolactone, Cylindrocarpon sp
1. Introduction
Historically, it has been assumed that the biologically active principles of botanical medicines are plant secondary metabolites. However, plants are teeming with microbial symbionts, including endophytes, microbes that live asymptomatically within plant tissue. Endophytes can produce an array of biologically active secondary metabolites, and the potential influence of these compounds on the biological activity of botanicals has been a topic of recent interest (Tan and Zaou, 2001; Strobel and Daisy, 2003; Strobel et al., 2004). Fungal endophytes can, in some cases, produce biologically active compounds. For example, endophytic fungi play a role in the production of hallucinogenic ergot alkaloids in morning glories (genera Ipomoea) (Ahimsa-Müller et al., 2007) and ergot and insecticidal loline alkaloids in tall fescue grass (genera Festuca and Lolium) (Siegel et al., 1990). To further complicate matters, gene transfer can occur between plants and endophytes, such that microbes may acquire the ability to produce the same compounds originally produced by the host plant, or vice versa (El-Elimat et al., 2014b; Kusari et al., 2009; Taghavi et al., 2005). Additionally, the presence of particular microbes may alter the growth and/or secondary constituent profile of the host plant (Eaton et al., 2010; Naveed et al., 2014). In light of this, it is becoming increasingly apparent that endophytes are a potentially important topic of consideration when investigating the biological activity of botanicals.
Endophytes can be transmitted either vertically, from parent to progeny through seeds, or horizontally, entering plant tissue from the environment (Rodriguez et al., 2009). Vertically transmitted endophytes often engage in mutualistic relationships with their hosts, and can be very closely associated with particular plant species (Saikkonen et al., 2004). Horizontally transmitted endophytes, on the other hand, may be more representative of the environment surrounding the plant than of the particular plant species. Because of horizontal endophyte transmission, it is likely that the same genus and species of a plant grown in different environments can have different endophyte profiles (Brem and Leuchtmann, 2002; Saikkonen et al., 2004; Schardl, 1996).
With this study, we focused on fungal endophytes from the botanical medicine Anemopsis californica (Nutt.) Hook. & Arn. (Saururaceae), which is commonly known as yerba mansa. Anemopsis californica was used by the Shoshoni, Pima, Mahuna, Chumash, Paiute, and Costanoan tribes of North America to treat inflammation and infection in wounds and to control pain (Bocek, 1984; Curtin, 1984; Timbrook, 1987; Romero, 1954; Train et al., 1978). This plant is still used today for the treatment of infections, and although it has been sparsely studied, there are several reports of antimicrobial (Bussey et al., 2014; Medina et al., 2005) or cytotoxic (Daniels et al., 2006; Kaminski et al., 2010) activities associated with A. californica extracts or constituents. To date, however, there have been no investigations of the endophyte profile of A. californica plants. Thus, we sought to isolate endophytes from the roots of this botanical and evaluate their antimicrobial activity and chemical composition. As part of this study, we also compared the diversity of fungal endophytes from a wild population of A. californica immediately after harvest and also after one year of cultivation in a greenhouse environment.
2. Results and Discussion
2.1. Influence of environment on fungal diversity
The first question we sought to explore was whether changes in growing conditions would alter the endophyte profile of A. californica plants. Given the commonness of horizontal transmission, (Rodriguez et al., 2009) we expected that this would be the case. To test this experimentally, we isolated endophytes from two different batches of A. californica roots (Table 1). One batch of roots was harvested directly from a wild population and the other came from the same wild population but was allowed to grow in a greenhouse for one year prior to harvest. The difference in diversity of the fungal collections from the wild population and greenhouse samples is striking. The former yielded a diverse array of at least seven distinct fungal endophytes (Table 1). In stark contrast, the roots that had grown for one year in the greenhouse yielded only two fungal species, Phomopsis columnaris and Ilyonectria robusta. Both of these fungal species have been known to infect and kill plants by either causing root rot (I. robusta) or stem death (P. columnaris) (Cabral et al., 2012; Farr et al., 2002; Roy and Mulder, 2014). Interestingly, P. columnaris was the only fungus found to be present in both the field samples and the greenhouse samples. This fungus was isolated only once from the field samples, but repeatedly (10 times) from the greenhouse samples. Our data suggest that cultivation in the greenhouse for one year caused a loss in fungal richness in the A. californica root samples. Thus, the diversity of fungal endophytes obtained from botanical samples can vary greatly depending on environment/method of cultivation. This is an important point for consideration in natural product drug discovery efforts from fungal endophytes.
Table 1.
Endophytic fungi isolated from Anemopsis californica roots, their constituents, and the antimicrobial activities of their extracts. Extracts were tested against Staphylococcus aureus at a concentration of 20 µg/mL and against Pseudomonas aeruginosa at a concentration of 200 µg/mL. The positive controls for the antimicrobial assays were berberine for S. aureus (MIC 150 µg/mL) and ciprofloxacin for P. aeruginosa (MIC 0.125 µg/mL).
| OTU identificationa | Origin/Abundanceb | Compounds identifiedc |
S. aureus inhibition (%)d |
P. aeruginosa inhibition (%) |
|
|---|---|---|---|---|---|
| Field | Greenhouse | ||||
| Colletotrichum coccodes | 2 | 34 ± 1.6 | 11 ± 1.1 | ||
| Penicillium sp. | 1 | 21 ± 1.7 | 29 ± 1.7 | ||
| Hypocreales sp. | 1 | verticillin A, 11’-deoxyverticillin A | 16 ± 1.0 | 12 ± 1.0 | |
| Cylindrocarpon sp. | 5 | equisetin*, 5’-epiequisetin* | 38 ± 0.6 | 24 ± 3.1 | |
| Chaetomium cupreum | 1 | chaetocuprum*, cochliodone A* | 26 ± 1.5 | 79 ± 3.0 | |
| Aspergillus sp. | 1 | 37 ± 1.7 | 15 ± 1.3 | ||
| Fusarium sp. | 2 | apicidin*, apicidin A*, apicidin D2* | 9 ± 1.0 | 15 ± 2.6 | |
| Penicillium sp. | 2 | 21 ± 1.0 | 19 ± 3.7 | ||
| Herpotrichiellaceae sp. | 1 | 0 ± 3.3 | 0 ± 4.4 | ||
| Sordariales sp. | 1 | 10 ± 4.6 | 0 ± 2.1 | ||
| Penicillium sp. | 2 | 17 ± 1.2 | 20 ± 1.5 | ||
| Hypocreales sp. | 1 | 6 ± 2.6 | 3 ± 2.7 | ||
| Nemania serpens | 2 | 7 ± 2.7 | 0 ± 5.2 | ||
| Phomopsis columnaris | 1 | 10 | acremonidin C, trichothecinol B, AGI-7, (E)-8-(3-(oct-2-enoyl)oxiran-2-yl)octanoic acid, 5,8-epidioxyergosta-6,9(11),22-trien-3-ol | 8 ± 1.5 | 14 ± 1.3 |
| Ilyonectria robusta | 1 | chermesinone A | 0 ± 1.0 | 0 ± 1.0 | |
Operational taxonomic unit (OTU). GenBank accession numbers for the isolated fungi are included in Table S1. Isolates were grouped based on 98% ITS rDNA sequence similarity and identified using BLAST search implemented via web platform PlutoF hosted on the UNITE database.
The number of isolates represents the number of times each fungus was isolated from the A. californica roots in the relevant batch.
All of the fifteen compounds listed were identified in at least one of the isolates from each fungal species. Compounds indicated with an asterisk (*) were isolated and verified by NMR. The remaining compounds were identified by matching accurate mass, MS-MS, and retention time with standards in a dereplication database (El-Elimat et al., 2013a), and are shown in Figure S9. Some fungi did not produce any of the compounds in the database at detectable levels, but are likely to produce other compounds. Experiments to identify these via isolation are ongoing.
Growth inhibition is expressed as the mean decrease in absorbance at 600nm for triplicate cultures (± standard deviation, SD). In cases where multiple isolations were obtained for the same fungus, the reported inhibition values are means of those for all strains.
2.2. Antimicrobial activity of A. californica endophytes
A number of the endophyte extracts from batch 1 (wild-harvested roots) displayed pronounced antimicrobial activity against Staphylococcus aureus. Extracts of Colletotrichum coccodes, Cylindrocarpon sp., Chaetomium cupreum, and Aspergillus sp. all inhibited S. aureus growth by >25% at a concentration of 20 µg/mL. By comparison, extracts of the two fungi isolated from the greenhouse cultivated A. californica exhibited only weak antimicrobial activity (8% for Phomopsis columnaris, no activity for Ilyonectria robusta).
Most of the endophytes were either weakly active or completely inactive against Pseudomonas aeruginosa, even though the concentration tested was 10-fold higher (200 µg/mL) than that used for the Staphylococcus aureus growth inhibition assays. It is well known that the Gram-negative bacterium P. aeruginosa is less susceptible to antimicrobial agents than are Gram-positive bacteria (Balode et al., 2013; Henwood et al., 2001; Rodríguez-Rojas et al., 2012), so this result was not surprising. However, two of the endophytes from batch 1, Cylindrocarpon sp. and Chaetomium cupreum, displayed activity against both S. aureus (≥26%) and P. aeruginosa (≥22%). Thus, these fungi were chosen as starting material for isolation of antimicrobial compounds (Section 2.3).
2.3. Isolation and activity of compounds from Chaetomium cuprum and Cylindrocarpon sp
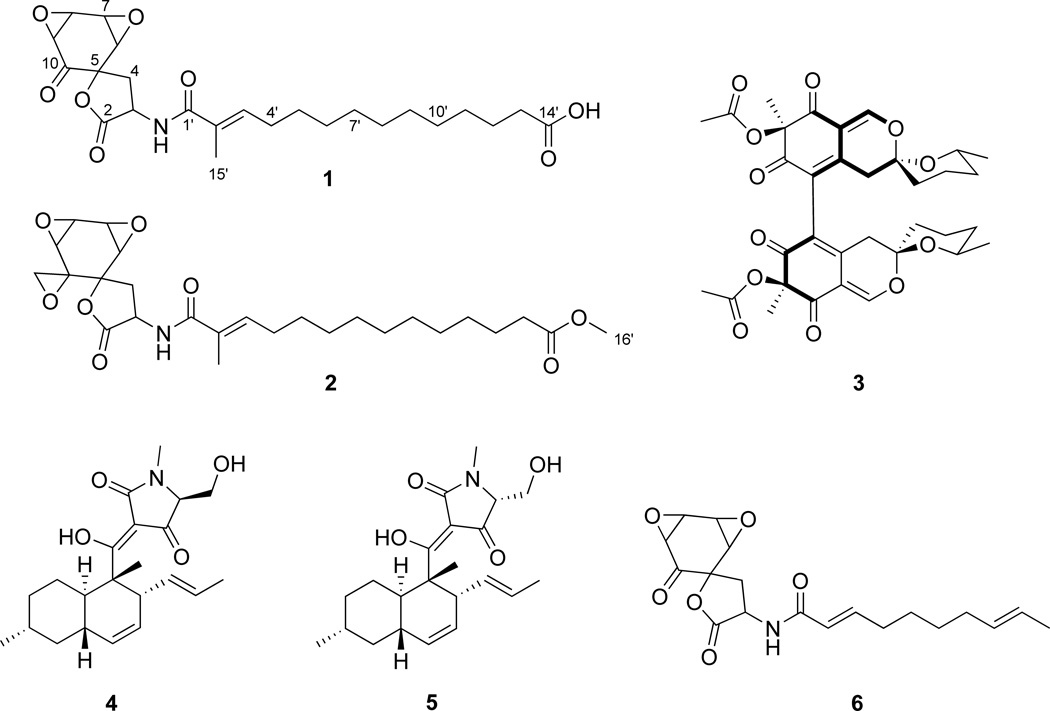
Bioassay-guided isolation from a scaled up extract of Chaetomium cuprum yielded one new compound (1), which we named chaetocuprum. The structure of this compound was confirmed by preparation of its methyl ester derivative (2), as described in Section 3.7. In addition, the known compound cochliodone A (3) was also isolated from C. cuprum. NMR and accurate mass data for this compound matched those reported previously (Phonkerd et al., 2008). Finally, two known compounds, equisetin (4) and 5'-epiequisetin (5), were isolated from Cylindrocarpon sp. Spectroscopic data from these compounds matched literature reports (Phillips et al., 1989).
Chaetocuprum (1) is a spirolactone, which demonstrates some structural similarities to a fungal metabolite (6) that has been reported from Pseudoarachniotus roseus (Garrity et al., 1991). The chemistry of the Chaetomium genus has been investigated previously, (Asai et al., 2013; Panthama et al., 2014) but only two spirolactones have been isolated from this genus. There are a few other examples of natural products containing spirolactone ring systems, including spiromamakone A, aranorosinols, and melettinins (Angawi et al., 2005; Fuse et al., 2013; Roy et al., 1993).
2.3.1 Structure elucidation of chaetocuprum (1)
The molecular formula of chaetocuprum (1) was determined to be C24H33NO8 (9 unsaturations) on the basis of NMR and HRESIMS data. Inspection of the 1H and 13C NMR data (Table 2, Table S2, and Figures S1–S4) in CDCl3 and CD3OD revealed the presence of a methyl singlet, five methine signals, including four oxymethines corresponding to two epoxide units, one olefinic proton, twenty-two methylene protons, and one exchangeable proton. Additionally, 13C NMR data indicated the presence of an oxygenated quaternary carbon, a non-protonated olefinic carbon, and four carbonyl carbons.
Table 2.
NMR spectroscopic data (500 MHz for 1H NMR and 125 MHz for 13C NMR, CDCl3) for chaetocuprum (1).a
| Position | δH (mult., J in Hz) | δC (mult.) | HMBC (1H# → 13C#) |
|---|---|---|---|
| 2 | 173.6 s | ||
| 3 | 4.25 (m) | 48.9 d | C-2, C-4, C-5, C-1' |
| 4α | 2.94 (dd, 10.2, 14.0) | C-2, C-3, C-5, C-6, C-10 | |
| 4β | 2.49 (dd, 10.2, 14.0) | 34.5 t | C-2, C-3, C-5, C-6, C-10 |
| 5 | 83.5 s | ||
| 6 | 3.65 (d, 4.0) | 56.3 d | C-4, C-5, C-7, C-8, C-10 |
| 7 | 3.89 (dd, 2.1, 4.0) | 52.6 d | C-5, C-6, C-8, C-9 |
| 8 | 4.03 (dd, 2.1, 4.0) | 57.9 d | C-6, C-7, C-9, C-10 |
| 9 | 3.58 (d, 4.0) | 55.5 d | C-5, C-7, C-8, C-10 |
| 10 | 196.7 s | ||
| 1' | 169.7 s | ||
| 2' | 129.4 s | ||
| 3' | 6.43 (t, 7.0) | 139.3 d | C-1', C-2', C-4', C-5', C-15' |
| 4' | 2.13 (m) | 28.7 t | C-2', C-3', C-5', C-6' |
| 5' | 1.40 (t, 7.0) | 29.0 t | C-3', C-4', C-7' |
| 6' | 1.30 (m) | 29.9 tb | |
| 7' | 1.30 (m) | 29.4 tb | |
| 8' | 1.30 (m) | 29.3 tb | |
| 9' | 1.30 (m) | 28.6 tb | |
| 10' | 1.30 (m) | 29.4 tb | |
| 11' | 1.30 (m) | 29.1 t | |
| 12' | 1.60 (m) | 24.8 t | C-10', C-14' |
| 13' | 2.32 (t, 7.0) | 33.7 t | C-11', C-14' |
| 14' | 177.0 s | ||
| N-H | 6.34 (d, 4.0)c | C-2, C-3, C-4, C-1', C-2' | |
| 15' | 1.82 (s) | 12.7 q | C-1', C-2', C-3' |
NMR data obtained in CD3OD are provided in Table S2.
13C NMR assignments for C-6' – C-10' can be interchanged.
The chemical shift is variable; this value was observed in CDCl3 at room temperature.
The 1H and 13C NMR signals (Table 2) for H-6 (δH 3.65; J = 4.0 Hz, δC 56.3), H-7 (δH 3.89; J = 4.0, 2.1 Hz, δC 52.6), H-8 (δH 4.03; J = 4.0, 2.1 Hz, δC 57.9), and H-9 (δH 3.58; J = 4.0 Hz, δC 55.5) were indicative of a pair of epoxide groups. Analysis of HSQC, HMBC, and COSY NMR data established the adjacent location of two epoxide units.
HMBC correlations from H-6, H-8, and H-9 to C-10 (δC 196.7) supported the placement of C-8–C-9 epoxide unit at a position alpha to the ketone carbonyl carbon. HMBC correlations from H-6 to an oxygenated quaternary carbon (C-5; δC 83.5) and C-10 suggested the presence of a six-membered ring. Key HMBC correlations from methylene protons H2-4 (δH 2.94 and δH 2.49) to C-5, C-6, and C-10 were consistent with the linkage of this group to C-5. Additional correlations from H2-4 to an ester carbonyl carbon (C-2; δC 173.6) and the adjoining methine carbon (C-3; δC 48.9) in conjunction with the chemical shift of C-5, supported the presence of a lactone, thereby forming a spirocyclic ring system.
HMBC correlations from an exchangeable proton NH (δH 6.34) to C-3, C-1' (δC 169.7), and C-2' (δC 129.4) were also observed when the NMR spectra were collected in CDCl3, establishing the linkage of the amide group to C-3. Key correlations from methyl group protons H3-15' (δH 1.82) to C-1', C-2' (δC 129.4), and C-3' (δC 139.3) extended the side chain to include an α,β-unsaturated olefin. A single spin system including protons H-3' to H-13' was identified primarily by analysis of the COSY NMR data. The remaining NMR data were consistent with the presence of a ten-carbon aliphatic chain. A terminal carboxylic acid group (C-14'; δC 177.0) accounted for the remaining unsaturation and carbon count, thereby completing the assignment of the gross structure of 1.
Compound 1 was treated with excess trimethylsilyldiazomethane (TMSCHN2) and the 1H NMR spectrum for the product (2) showed a methyl singlet at δH 3.65 for the newly formed methoxy group (H3-16'), confirming that 1 contained a carboxylic acid group (Figures S-6 – S-8). However, two new doublets (δH 3.05 and δH 2.85; H2-11) with coupling constants of 4.4 Hz were also observed. Additionally, the 13C NMR signal for the ketone carbon in 1 was replaced by a signal at δc 49.7 (C-11) in 2. Analysis of HSQC and HMBC data were consistent with the assignment of these doublets to the methylene protons resulting from epoxidation of the ketone carbonyl group. HMBC correlations from H2-11 to C-5 (δc 81.2), C-9 (δc 56.6), and C-10 (δc 59.8) confirmed the formation of a geminal epoxide in 2. HRESIMS data [m/z 492.2578 (M+H)+] were consistent with the molecular formula (C26H37NO8) of 2.
NOESY correlations between H3-15' (δH 1.81) and H-4' (δH 2.17) allowed the assignment of E-configuration for the C-2'–C-3' double bond in 1. No correlations were observed between H3-15' and H-3'. NOESY correlations of H-6 with H-7, as well as H-8 with H-9 were consistent with the presence of syn epoxide units. NOESY correlations between H-7 and H-8 were also observed. However, the relative orientation of the two epoxide groups in the ring system and conclusive assignment of the overall relative configuration of chaetocuprum could not be made solely on the basis of NOESY data. Unfortunately, crystallization attempts were also unsuccessful.
2.3.2. Antimicrobial activity of pure compounds
Antimicrobial activity was evaluated for the compounds (1, 4, and 5) that were isolated in sufficient quantity (Table 3). None of these were active against P. aeruginosa (MIC >200 µg/mL). Compound 1 inhibited growth of S. aureus (IC50 of 50 µg/mL), but complete growth inhibition was not achieved against this organism (MIC > 50 µg/mL, the highest concentration tested). The activity of equisetin (4) agreed with literature (MIC of 1 µg/mL against S. aureus and inactive against P. aeruginosa) (Burmeister et al., 1974; Hellwig et al., 2002). 5'-epiequisetin (5) has not been previously evaluated for antimicrobial effects, and it demonstrated an MIC of 1 µg/mL against S. aureus. The positive controls for the antimicrobial assays were berberine (for S. aureus) and ciprofloxacin (for P. aeruginosa), which demonstrated MICs of 150 µg/mL and 0.125 µg/mL, respectively, consistent with previous reports (Chalkley and Koornhof, 1985; Ettefagh et al., 2011).
Table 3.
Minimum inhibitory concentration (MIC) and IC50 of select fungal metabolites against Staphylococcus aureus and Pseudomonas aeruginosa.
| Compound | Staphylococcus aureus | Pseudomonas aeruginosa | ||
|---|---|---|---|---|
| MIC (µg/mL) | IC50 (µg/mL) | MIC (µg/mL) | IC50 (µg/mL) | |
| Chaetocuprum (1) | >50 | 50 | >50 | >50 |
| Equisetin (4) | 1.0 | 0.5 | >200 | >200 |
| 5’-Epiequisetin (5) | 1.0 | 0.5 | >200 | >200 |
| Ciprofloxacin (+ control) | – | – | 0.125 | 0.0625 |
| Berberine (+ control) | 150 | 300 | – | – |
2.4. Additional compounds from A. californica endophytes
As a complementary approach to isolation for identifying chemical constituents of endophyte extracts, all extracts were subjected to LC-MS-MS analysis, and the data were compared to a library of high-resolution mass spectrometry data from fungal compounds, as described previously (El-Elimat et al., 2013a). Using this approach, eleven additional known compounds (Table 1, see structures in Figure S9) were identified. These data suggest that the endophytes not subjected to isolation efforts also biosynthesize structurally diverse secondary metabolites.
3. Experimental
3.1. General experimental procedures
UV spectra were measured by using a Varian Cary 100 Bio UV–vis spectrophotometer. Optical rotation was measured on a Rudolph Research Autopol III polarimeter. ECD data were collected on an Olis DSM 17 CD spectrophotometer. The NMR spectra were recorded in both CDCl3 and CD3OD with references peaks (δH 7.24/ δC 77.2 for CDCl3 and δH 3.31/ δC 49.2 for CD3OD). NMR experiments were conducted using an Agilent-700, JEOL ECA-500, and/or ECS-400 spectrometers (700, 500 or 400 MHz for 1H and 175, 125 or 100 MHz for 13C; Agilent Technologies, Santa Clara, CA, USA; JEOL Ltd., Tokyo, Japan). The HRESIMS data was collected on a Thermo LTQ Orbitrap XL mass spectrometer. Flash chromatography was conducted using a Teledyne Isco CombiFlash Rf system with a RediSep Rf Si-gel Gold column (4 g silica 40 µm). A Varian ProStar HPLC system equipped with ProStar 210 pumps and a ProStar 335 photodiode array detector was used for reversed-phase preparative separations, with a Phenomenex Gemini-NX C18 column (5 µm, 120Å; 250 × 21.2 mm) and the Galaxie Chromatography Workstation Software (version 1.9.3.2). Analytical separations were performed with a Gemini-NX C18 column (5 µm, 120Å; 250 × 4.6 mm) from Phenomenex. For antimicrobial assays, the optical density at 600 nm was read using a POLARstar Optima microplate reader. Müeller-Hinton broth, ciprofloxacin (purity >98% by HPLC), and berberine (purity >98% by HPLC) were purchased from Sigma Aldrich. Other reagents were purchased from Fisher Scientific.
3.2. Plant material
Anemopsis californica (Nutt.) Hook. & Arn. (Saururaceae) plants were collected with permission by Amy Brown of Apache Creek Ranch in Santa Fe, NM (35°35' 56.40"N, 105°50' 27.22”W). A voucher specimen (NCU602027) was deposited in the University of North Carolina Herbarium, and was authenticated by Amy Brown. Fungi were isolated from surface sterilized fresh root samples.
3.3. Endophyte isolation and identification
Isolation of fungal endophytes was performed using methods outlined previously (El-Elimat et al., 2014b; Figueroa et al., 2014). For molecular identification of fungal endophytes isolated from yerba mansa, the internal transcribed spacer region of the ribosomal RNA gene (ITS) was sequenced using methods described previously (El-Elimat et al., 2013b; El-Elimat et al., 2013c; El-Elimat et al., 2014a; El-Elimat et al., 2014b; Figueroa et al., 2014; Figueroa et al., 2013). The ITS sequences from all strains were deposited in GenBank and are listed in Table S1.
3.4. Endophyte culture and extraction
A solid, grain-based medium was used to grow small-scale cultures of fungi in 250 mL Erlenmeyer flasks as previously described (Ayers et al., 2011; Figueroa et al., 2012). Each fungal culture was chopped and shaken overnight (16 h at 100 rpm) in a 1:1 MeOH:CHCl3 solution, subjected to vacuum filtration, and washed with small volumes of MeOH. The filtrate was stirred in a 1:4:5 ratio of MeOH:CHCl3:H2O for 2 h. After separating the organic and aqueous layers, both layers were evaporated to dryness under vacuum. The organic layer was then resuspended in a 1:1:2 mixture of MeOH:CH3CN:hexane. The MeOH:CH3CN and hexane layers were separately dried under vacuum. The residue from the MeOH:CH3CN layer was used for bioassays, dereplication, isolation, and chemical profile comparison between the endophytes and the A. californica extract.
3.5. Isolation
The first stage separations of extracts from Chaetomium cupreum and Cylindrocarpon sp. were conducted with normal-phase flash chromatography (4 g silica gel column) at an 18 mL/min flow rate with a 35 min hexane:CHCl3:MeOH gradient. In the first stage of separation for Chaetomium cupreum (GenBank accession no. KM816761, Figure S10), four fractions were-obtained using normal phase chromatography. Fraction 2 (144 mg) was subjected to a second stage of purification using a reversed phase preparative HPLC with a Gemini-NX C18 column at a 21 mL/min flow rate. A linear CH3CN:H2O gradient starting from 30:70 to 90:10 over 20 min yielded cochliodone A (3), which eluted at 17 min (1 mg, 98 % purity, 0.0004% yield). Fraction 3 was also subjected to the same gradient, and produced sub-fraction 2 (75 mg). This fraction was then purified with an isocratic solvent composition of 50:50 CH3CN: H2O on a preparative HPLC with a Gemini-NX C18 column at a 21 mL/min flow rate over 20 min. Chaetocuprum (1), eluted at 16 min (13 mg, 98.5 % purity, 0.0052% yield). In the first stage of separation for Cylindrocarpon sp. (GenBank accession no. KM816763, Figure S11), four fractions were obtained with normal phase chromatography. Fraction 3 was subjected to further purification using a preparative RP-HPLC with a Gemini-NX C18 column at a 21 mL/min flow rate. A linear CH3CN:H2O gradient starting from 30:70 to 90:10 over 20 min yielded equisetin (4) at 12 min (45 mg, 98 % purity, 0.018% yield) and 5’-epiequisetin (5) at 12.7 min (25 mg, 98 % purity, 0.010% yield).
3.6. Chaetocuprum (1)
Oil; , (c 0.09, CH3OH); ECD (72 µM, CH3OH) λmax (Δε) 214 (+29), 230 (−8), and at 297 (−8), UV/Vis (MeOH) λmax (log ε) 224 (3.5), 1H and 13C NMR data: See Tables 2 and S2; Key NOESY correlations (H-# → H-#): H-4α ↔ H-3, 4β; H-4β↔ H-3, 4α, 6; H-6 ↔ H-4β, 7; H-7 ↔ H-6, 8; H-8 ↔ H-7, 9; H-9 ↔ H-8; H-3' ↔ H2-4', 5'; H3-15' ↔ H2-4'; HRESIMS obsd. m/z 464.2268 [M+H]+, calcd for C24H34NO8, 464.2284.
3.7. Preparation of 10,11-epoxychaetocuprum methyl ester (2)
A sample of 1 (3 mg) was dissolved in 200–300 µL of methanol and 2 M solution of TMSCHN2 in diethyl ether was added dropwise until the yellow color of the TMSCHN2 solution persisted. After stirring for 4 hours at RT, the sample was dried under air. The reaction mixture was analyzed by 1H NMR and then purified by semi-preparative RP-HPLC [CH3CN/H2O (with 0.1% formic acid): 60–100% CH3CN over 15 min] to yield 2 (1.0 mg; tR 14 min).
3.8. 10,11-Epoxychaetocuprum methyl ester (2)
Oil; , (c 0.05, CH3OH); UV/Vis (CH3OH) λmax (log ε) 221 (3.5), NMR data (CD3OD; 700 MHz) δ 6.41 (dt, 1.2, 7.4, H-3'), 4.42 (t, 10.2, H-3), 3.82 (dd, 2.2, 4.4, H-8), 3.78 (dd, 2.2, 4.2, H-7), 3.65 (s, H3-16'), 3.54 (d, 4.2, H-6), 3.05 (d, 4.4, Hα-11), 2.98 (d, 4.2, H-9), 2.91 (dd, 10.2, 13.4, Hα-4), 2.85 (d, 4.4, Hβ-11), 2.36 (dd, 9.9, 13.4, Hβ-4), 2.31 (t, 7.5, H2-13'), 2.19 (m, H2-4'), 1.83 (s, H3-15'), 1.60 (m, H2-12'), 1.46 (m, H2-5'), 1.32 (m; H2-6' – H2-11'); 13C NMR (CD3OD; 175 MHz) δ 176.1 (C-14'), 175.7 (C-2), 171.7 (C-1'), 139.3 (C-3'), 130.9 (C-2'), 81.2 (C-5), 59.8 (C-10), 57.5 (C-6), 56.6 (C-9), 53.9 (C-8), 53.4 (C-7), 52.0 (C-16'), 50.4 (C-3), 49.7 (C-11), 35.6 (C-4), 34.8 (C-13'), 30.2 (C-11'), 29.8 (C-5'), 29.3 (C-4'), 26.0 (C-12'), 12.5 (C-15'), Chemical shifts for five carbons (C-6'–C-10') could not be assigned with confidence but are listed here: δ 30.6, 30.54, 30.53, 30.38, and 30.35; Key HMBC correlations (H-# → C-#): H-3 →C-2, 4, 1'; Hα-4 → C-2, 3, 5, 6, 10; Hα-4→ C-3, 5, 6, 10; H-6 → 5, 8, 10; H-7 → C-6, 8; H-8 → C-7, 9; H-9 → C-5, 7, 10, 11 (wk); H2-11 → C-5, 9, 10; H-3' → C-1', 2' (wk), 4', 5', 15'; H-4' → C-2', 3', 5'; H-5' → C-3', 4'; H-12' → C-13', 14'; H-13' → C-11', 12', 14'; H-15' → C-1', 2', 3'; H-16' → C-14'; HRESIMS obsd. m/z 492.2578 [M+H]+, calcd for C26H38NO8, 492.2592.
3.9. LC-MS dereplication
Each fungal endophyte extract was analyzed with LC-MS-MS in the positive and negative ion modes, using a dereplication method described in detail previously (El-Elimat et al., 2013a).
3.10. Antimicrobial assays
Broth microdilution assays to evaluate antimicrobial susceptibility were performed according to Clinical Laboratory Standards Institute (CLSI) guidelines (2012). S. aureus (strain NCTC 8325-4), (Novick, 1967) and P. aeruginosa (strain NCTC 12903) were used for biological testing. In separate experiments, single colony inocula of S. aureus or P. aeruginosa were grown to log phase in Müeller-Hinton broth and were adjusted to a final assay dilution of 1.0 × 105 CFU/mL based on OD600 of 0.11 for both bacteria. The negative control consisted of 2% DMSO in broth (vehicle), ciprofloxacin served as the positive control with P. aeruginosa, and berberine was used as the positive control with the S. aureus. All treatments and controls were prepared in triplicate wells. For background subtraction, additional wells were included containing the samples without bacteria. OD600 was measured after incubation for 18 h at 37 °C. MIC was defined as the concentration at which no statistically significant difference was observed between the negative control and treated samples. IC50 was defined as the concentration at which there is a 50% decrease in growth observed between the negative control and the treated samples
Supplementary Material
Figure 1.
Chaetocuprum (1), 10,11-epoxychaetocuprum methyl ester (2), cochliodone A (3), equisetin (4), 5’-epiequisetin (5), and a fungal metabolite (6) isolated from Pseudoarachniotus roseus by Merck & Co., Inc. Compound 2 is not a natural product and was prepared by chemical reaction.
HIGHLIGHTS.
Fourteen fungal endophytes were isolated from yerba mansa (Anemopsis californica).
Chaetomium cupreum yielded a new antimicrobial spirolactone, chaetocuprum.
Endophyte diversity was higher from wild-harvested than cultivated roots.
Results indicate the importance of source material for endophyte drug discovery.
Acknowledgments
This research was supported in part by Grant Number 1 R01 AT006860 from the National Center for Complementary and Alternative Medicine (NCCAM), a component of the National Institutes of Health (NIH) and by a Biotechnology Research Grant (2011-BRG-1206) from the North Carolina Biotechnology Center. We thank B. Ehrmann for technical assistance with mass spectrometry data collection, A. Horswill, G. Kaatz, J. Falkinham, I. Dempsey, and R. Cech for helpful advice, A. Brown for supplying the A. californica samples, C. McCormick for preserving botanical vouchers, and X. Tan for translating a reference from Chinese. Mass spectrometry data were collected in the Triad Mass Spectrometry facility.
Appendix A. Supplementary data
Fractionation schemes, 1H and 13C NMR spectra for chaetocuprum (1) and 10,11-epoxychaetocuprum methyl ester (2), and structures of compounds identified with LC-MS-MS dereplication are provided as supplementary data.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ahimsa-Müller MA, Markert A, Hellwig S, Knoop V, Steiner U, Drewke C, Leistner E. Clavicipitaceous Fungi Associated with Ergoline Alkaloid-Containing Convolvulaceae. J. Nat. Prod. 2007;70:1955–1960. doi: 10.1021/np070315t. [DOI] [PubMed] [Google Scholar]
- Angawi RF, Swenson DC, Gloer JB, Wicklow DT. Malettinins B-D: new polyketide metabolites from an unidentified fungal colonist of Hypoxylon Stromata (NRRL 29110) J. Nat. Prod. 2005;68:212–216. doi: 10.1021/np049625r. [DOI] [PubMed] [Google Scholar]
- Asai T, Taniguchi T, Yamamoto T, Monde K, Oshima Y. Structures of Spiroindicumides A and B, Unprecedented Carbon Skeletal Spirolactones, and Determination of the Absolute Configuration by Vibrational Circular Dichroism Exciton Approach. Org. Lett. 2013;15:4320–4323. doi: 10.1021/ol401741z. [DOI] [PubMed] [Google Scholar]
- Ayers S, Graf TN, Adcock AF, Kroll DJ, Matthew S, Carcache de Blanco EJ, Shen Q, Swanson SM, Wani MC, Pearce CJ, Oberlies NH. Resorcylic acid lactones with cytotoxic and NF-kappaB inhibitory activities and their structure-activity relationships. J. Nat. Prod. 2011;74:1126–1131. doi: 10.1021/np200062x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balode A, Punda-Polic V, Dowzicky MJ. Antimicrobial susceptibility of gramnegative and gram-positive bacteria collected from countries in Eastern Europe: results from the Tigecycline Evaluation and Surveillance Trial (T.E.S.T.) 2004–2010. Int. J. Antimicrob. Agents. 2013;41:527–535. doi: 10.1016/j.ijantimicag.2013.02.022. [DOI] [PubMed] [Google Scholar]
- Boeck BR. Ethnobotany of Costanoan Indians, California, based on collections by John P. Harrington. Econ. Bot. 1984;38:240–255. [Google Scholar]
- Brem D, Leuchtmann A. Intraspecific competition of endophyte infected vs uninfected plants of two woodland grass species. Oikos. 2002;96:281–290. [Google Scholar]
- Burmeister HR, Bennett GA, Vesonder RF, Hesseltine CW. Antibiotic Produced by Fusarium equiseti NRRL. Antimicrob. Agents Ch. 1974;5:634–639. doi: 10.1128/aac.5.6.634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bussey RO, 3rd, Sy-Cordero AA, Figueroa M, Carter FS, Falkinham JO, 3rd, Oberlies NH, Cech NB. Antimycobacterial furofuran lignans from the roots of Anemopsis californica. Planta Med. 2014;80:498–501. doi: 10.1055/s-0034-1368352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabral A, Groenewald J, Rego C, Oliveira H, Crous P. Cylindrocarpon root rot: multi-gene analysis reveals novel species within the Ilyonectria radicicola species complex. Mycol. Prog. 2012;11:655–688. [Google Scholar]
- Chalkley LJ, Koornhof HJ. Antimicrobial activity of ciprofloxacin against Pseudomonas aeruginosa, Escherichia coli, and Staphylococcus aureus determined by the killing curve method: antibiotic comparisons and synergistic interactions. Antimicrob. Agents Ch. 1985;28:331–342. doi: 10.1128/aac.28.2.331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clinical and Laboratory Standards Institute (CLSI) M07-A9 – Methods for dilution Antimicrobial Susceptibility Tests for Bacteria that Grow Aerobically, Approved Standard. 9th edition. Wayne, PA: 2012. [Google Scholar]
- Curtin LSM. By the Prophet of the Earth: Ethnobotany of the Pima. Tucson, Ariz: University of Arizona Press; 1984. [Google Scholar]
- Daniels AL, Van Slambrouck S, Lee RK, Arguello TS, Browning J, Pullin MJ, Kornienko A, Steelant WF. Effects of extracts from two Native American plants on proliferation of human breast and colon cancer cell lines in vitro. Oncol. Rep. 2006;15:1327–1331. [PubMed] [Google Scholar]
- Eaton CJ, Cox MP, Ambrose B, Becker M, Hesse U, Schardl CL, Scott B. Disruption of Signaling in a Fungal-Grass Symbiosis Leads to Pathogenesis. Plant. Physiol. 2010;153:1780–1794. doi: 10.1104/pp.110.158451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Elimat T, Figueroa M, Ehrmann BM, Cech NB, Pearce CJ, Oberlies NH. High-resolution MS, MS/MS, and UV database of fungal secondary metabolites as a dereplication protocol for bioactive natural products. J. Nat. Prod. 2013a;76:1709–1716. doi: 10.1021/np4004307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Elimat T, Figueroa M, Raja HA, Adcock AF, Kroll DJ, Swanson SM, Wani MC, Pearce CJ, Oberlies NH. Waol A, trans-dihydrowaol A, and cis-dihydrowaol A: polyketide-derived γ-lactones from a Volutella species. Tetrahedron Lett. 2013b;54:4300–4302. doi: 10.1016/j.tetlet.2013.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Elimat T, Figueroa M, Raja HA, Graf TN, Adcock AF, Kroll DJ, Day CS, Wani MC, Pearce CJ, Oberlies NH. Benzoquinones and terphenyl compounds as phosphodiesterase-4B inhibitors from a fungus of the order Chaetothyriales (MSX 47445) J. Nat. Prod. 2013c;76:382–387. doi: 10.1021/np300749w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Elimat T, Raja HA, Figueroa M, Falkinham JO, 3rd, Oberlies NH. Isochromenones, isobenzofuranone, and tetrahydronaphthalenes produced by Paraphoma radicina, a fungus isolated from a freshwater habitat. Phytochemistry. 2014a;104:114–120. doi: 10.1016/j.phytochem.2014.04.006. [DOI] [PubMed] [Google Scholar]
- El-Elimat T, Raja HA, Graf TN, Faeth SH, Cech NB, Oberlies NH. Flavonolignans from Aspergillus iizukae, a Fungal Endophyte of Milk Thistle (Silybum marianum) J. Nat. Prod. 2014b;77:193–199. doi: 10.1021/np400955q. [DOI] [PubMed] [Google Scholar]
- Ettefagh KA, Burns JT, Junio HA, Kaatz GW, Cech NB. Goldenseal (Hydrastis canadensis L.) Extracts Synergistically Enhance the Antibacterial Activity of Berberine via Efflux Pump Inhibition. Planta Med. 2011;77:835–840. doi: 10.1055/s-0030-1250606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farr DF, Castlebury LA, Rossman AY, Putnam ML. A new species of Phomopsis causing twig dieback of Vaccinium vitis-idaea (lingonberry) Mycol. Res. 2002;106:745–752. [Google Scholar]
- Figueroa M, Graf TN, Ayers S, Adcock AF, Kroll DJ, Yang J, Swanson SM, Munoz-Acuna U, Carcache de Blanco EJ, Agrawal R, Wani MC, Darveaux BA, Pearce CJ, Oberlies NH. Cytotoxic epipolythiodioxopiperazine alkaloids from filamentous fungi of the Bionectriaceae. J. Antibiot. 2012;65:559–564. doi: 10.1038/ja.2012.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figueroa M, Jarmusch AK, Raja HA, El-Elimat T, Kavanaugh JS, Horswill AR, Cooks RG, Cech NB, Oberlies NH. Polyhydroxyanthraquinones as Quorum Sensing Inhibitors from the Guttates of Penicillium restrictum and Their Analysis by Desorption Electrospray Ionization Mass Spectrometry. J. Nat. Prod. 2014. 2014;77:1351–1358. doi: 10.1021/np5000704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figueroa M, Raja H, Falkinham JO, 3rd, Adcock AF, Kroll DJ, Wani MC, Pearce CJ, Oberlies NH. Peptaibols, tetramic acid derivatives, isocoumarins, and sesquiterpenes from a Bionectria sp. (MSX 47401) J. Nat. Prod. 2013;76:1007–1015. doi: 10.1021/np3008842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuse S, Inaba K, Takagi M, Tanaka M, Hirokawa T, Johmoto K, Uekusa H, Shin-ya K, Takahashi T, Doi T. Design and synthesis of 2-phenyl-1,4-dioxaspiro[4.5]deca-6,9-dien-8-ones as potential anticancer agents starting from cytotoxic spiromamakone A. Eur. J. Med. Chem. 2013;66:180–184. doi: 10.1016/j.ejmech.2013.05.030. [DOI] [PubMed] [Google Scholar]
- Garrity GM, Giacobbe R, Greenspan MD, Hensens OD, Joshua H, Matas MTD, Martin I, Milligan JA, Val SMD, Rozdilsky W. Novel hmg-coa synthase inhibitors. Google Patents. 1991 [Google Scholar]
- Hellwig V, Grothe T, Mayer-Bartschmid A, Endermann R, Geschke FU, Henkel T, Stadler M. Altersetin, a new antibiotic from cultures of endophytic Alternaria spp. Taxonomy, fermentation, isolation, structure elucidation and biological activities. J. Antibiot. 2002;55:881–892. doi: 10.7164/antibiotics.55.881. [DOI] [PubMed] [Google Scholar]
- Henwood CJ, Livermore DM, James D, Warner M Pseudomonas Study Group, t. Antimicrobial susceptibility of Pseudomonas aeruginosa: results of a UK survey and evaluation of the British Society for Antimicrobial Chemotherapy disc susceptibility test. J. Antimicrob. Chemoth. 2001;47:789–799. doi: 10.1093/jac/47.6.789. [DOI] [PubMed] [Google Scholar]
- Kaminski CN, Ferrey SL, Lowrey T, Guerra L, Van Slambrouck S, Steelant WF. In vitro anticancer activity of Anemopsis californica. Oncol. Lett. 2010;1:711–715. doi: 10.3892/ol_00000124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusari S, Lamshöft M, Spiteller M. Aspergillus fumigatus Fresenius, an endophytic fungus from Juniperus communis L. Horstmann as a novel source of the anticancer pro-drug deoxypodophyllotoxin. J. Appl. Microb. 2009;107:1019–1030. doi: 10.1111/j.1365-2672.2009.04285.x. [DOI] [PubMed] [Google Scholar]
- Medina AL, Lucero ME, Holguin FO, Estell RE, Posakony JJ, Simon J, O'Connell MA. Composition and antimicrobial activity of Anemopsis californica leaf oil. J. Agr. Food. Chem. 2005;53:8694–8698. doi: 10.1021/jf0511244. [DOI] [PubMed] [Google Scholar]
- Naveed M, Mitter B, Reichenauer TG, Wieczorek K, Sessitsch A. Increased drought stress resilience of maize through endophytic colonization by Burkholderia phytofirmans PsJN and Enterobacter sp. FD17. Environ. Exp. Bot. 2014;97:30–39. [Google Scholar]
- Novick R. Properties of a cryptic high-frequency transducing phage in Staphylococcus aureus. Virology. 1967;33:155–166. doi: 10.1016/0042-6822(67)90105-5. [DOI] [PubMed] [Google Scholar]
- Panthama N, Kanokmedhakul S, Kanokmedhakul K, Soytong K. Chemical constituents from the fungus Chaetomium cupreum RY202. Arch. Pharm. Res. 2014:1–6. doi: 10.1007/s12272-014-0418-1. [DOI] [PubMed] [Google Scholar]
- Phillips NJ, Goodwin JT, Fraiman A, Cole RJ, Lynn DG. Characterization of the Fusarium toxin equisetin: the use of phenylboronates in structure assignment. J Am Chem Soc. 1989;111:8223–8231. [Google Scholar]
- Phonkerd N, Kanokmedhakul S, Kanokmedhakul K, Soytong K, Prabpai S, Kongsearee P. Bis-spiro-azaphilones and azaphilones from the fungi Chaetomium cochliodes VTh01 and C. cochliodes CTh05. Tetrahedron. 2008;64:9636–9645. [Google Scholar]
- Rodriguez RJ, White JF, Arnold AE, Redman RS. Fungal Endophytes: Diversity and Functional Roles. New. Phytol. 2009;182:314–330. doi: 10.1111/j.1469-8137.2009.02773.x. [DOI] [PubMed] [Google Scholar]
- Rodríguez-Rojas A, Oliver A, Blázquez J. Intrinsic and environmental mutagenesis drive diversification and persistence of Pseudomonas aeruginosa in chronic lung infections. J. Infect. Dis. 2012;205:121–127. doi: 10.1093/infdis/jir690. [DOI] [PubMed] [Google Scholar]
- Romero JB. The Botanical Lore of the California Indians. New York: Vintage Press, Inc.; 1954. [Google Scholar]
- Roy BA, Mulder CPH. Pathogens, herbivores, and phenotypic plasticity of boreal Vaccinium vitis-idaea experiencing climate change. Ecosphere. 2014;5:art30. [Google Scholar]
- Roy K, Vijayakumar EKS, Bhat RG, Mukhopadhyay T, Ganguli BN. Aranorosinol A and aranorosinol B, two new metabolites from Pseudoarachniotus roseus: Production, isolation, structure elucidation and biological properties. J. Antibiot. 1992;45:1592–1598. doi: 10.7164/antibiotics.45.1592. [DOI] [PubMed] [Google Scholar]
- Saikkonen K, Wäli P, Helander M, Faeth SH. Evolution of endophyte–plant symbioses. Trends Plant Sci. 2004;9:275–280. doi: 10.1016/j.tplants.2004.04.005. [DOI] [PubMed] [Google Scholar]
- Schardl CL. Epichloe species: Fungal symbionts of grasses. Annu. Rev. Phytopathol. 1996;34:109–130. doi: 10.1146/annurev.phyto.34.1.109. [DOI] [PubMed] [Google Scholar]
- Siegel MR, Latch GC, Bush LP, Fannin FF, Rowan DD, Tapper BA, Bacon CW, Johnson MC. Fungal endophyte-infected grasses: Alkaloid accumulation and aphid response. J. Chem. Ecol. 1990;16:3301–3315. doi: 10.1007/BF00982100. [DOI] [PubMed] [Google Scholar]
- Strobel G, Daisy B. Bioprospecting for microbial endophytes and their natural products. Microbiol. Mol. Biol. R. 2003;67:491–502. doi: 10.1128/MMBR.67.4.491-502.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strobel G, Daisy B, Castillo U, Harper J. Natural products from endophytic microorganisms. J. Nat. Prod. 2004;67:257–268. doi: 10.1021/np030397v. [DOI] [PubMed] [Google Scholar]
- Taghavi S, Barac T, Greenberg B, Borremans B, Vangronsveld J, van der Lelie D. Horizontal gene transfer to endogenous endophytic bacteria from poplar improves phytoremediation of toluene. Appl. Environ. Microb. 2005;71:8500–8505. doi: 10.1128/AEM.71.12.8500-8505.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan RX, Zoua WX. Endophytes: a rich source of functional metabolites. Nat. Prod. Rep. 2001;18:448–459. doi: 10.1039/b100918o. [DOI] [PubMed] [Google Scholar]
- Timbrook J. Virtuous Herbs: Plants in Chumash Medicine. J. Ethnobiol. 1987;7:171–180. [Google Scholar]
- Train P, Henrichs JR, Archer WA. Medicinal Uses of Plants by Indian Tribes of Nevada. Lawrence, Mass: Quarterman Publications; 1978. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.