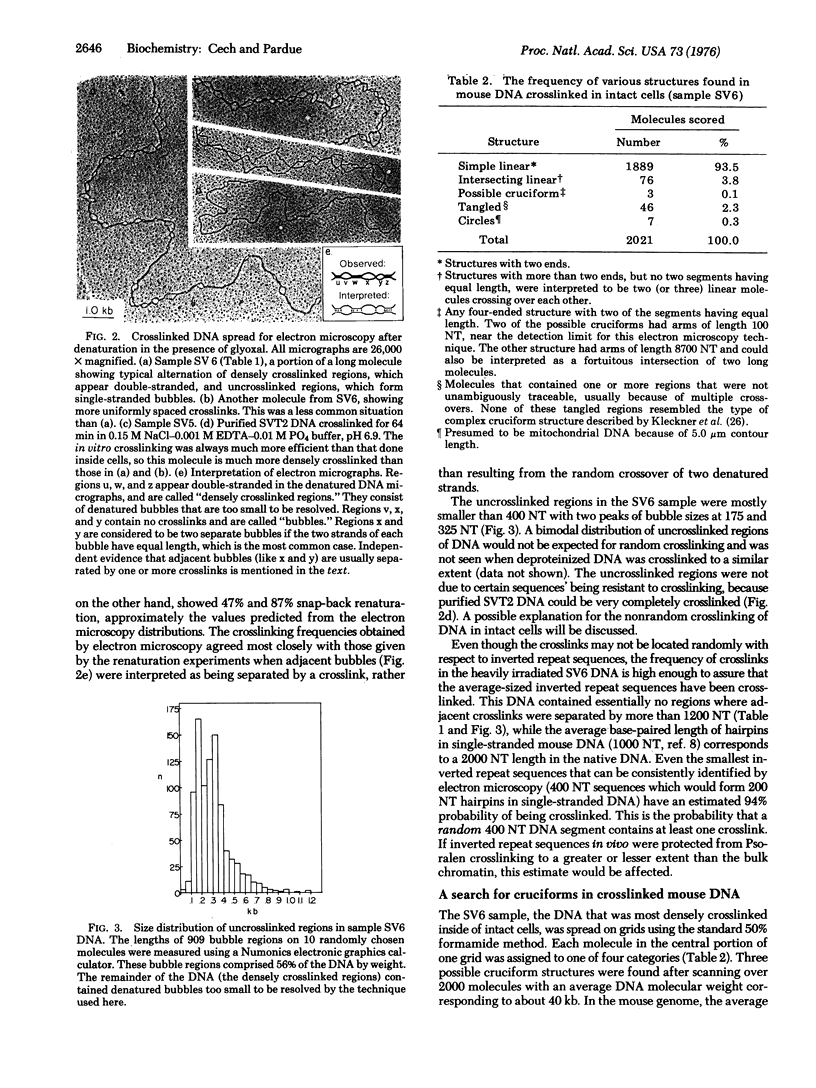
Abstract
It has been suggested that inverted repeat (palindrome) sequences, which are widespread in eukaryotic genomes, exist in two alternate configurations, a linear form and a cruciform. To investigate the relative frequency of these forms, the DNA of intact mouse tissue culture cells was covalently crosslinked with 4,5',8-trimethylpsoralen (me3-psoralen) in order to prevent rearrangement of the DNA secondary structure during DNA isolation. The distribution of me3-psoralen crosslinks was determined by electron microscopy after denaturation of the DNA in the presence of glyoxal. Because of the high frequency and the relatively uniform distribution of the me3-psoralen crosslinks, it could be concluded that almost all of the inverted repeat sequences had been crosslinked. In spite of this, no significant number of cruciforms was detected by electron microscopy. To determine whether the me3-psoralen might itself be disrupting cruciform structures, cruciforms were first produced in isolated Tetrahymena rDNA by heat treatment and then crosslinked in vitro. The crosslinking was found to stabilize rather than disrupt these cruciforms. We conclude that the inverted repeat sequences of the mouse tissue culture cells we tested are predominantly in linear forms rather than in cruciform structures inside the cell.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Broude N. E., Budowsky E. I. The reaction of glyoxal with nucleic acid components. 3. Kinetics of the reaction with monomers. Biochim Biophys Acta. 1971 Dec 30;254(3):380–388. doi: 10.1016/0005-2787(71)90868-9. [DOI] [PubMed] [Google Scholar]
- Cech T. R., Hearst J. E. An electron microscopic study of mouse foldback DNA. Cell. 1975 Aug;5(4):429–446. doi: 10.1016/0092-8674(75)90062-8. [DOI] [PubMed] [Google Scholar]
- Cech T. R., Rosenfeld A., Hearst J. E. Characterization of the most rapidly renaturing sequences in mouse main-band DNA. J Mol Biol. 1973 Dec 15;81(3):299–325. doi: 10.1016/0022-2836(73)90143-5. [DOI] [PubMed] [Google Scholar]
- Cole R. S. Light-induced cross-linking of DNA in the presence of a furocoumarin (psoralen). Studies with phage lambda, Escherichia coli, and mouse leukemia cells. Biochim Biophys Acta. 1970 Sep 17;217(1):30–39. doi: 10.1016/0005-2787(70)90119-x. [DOI] [PubMed] [Google Scholar]
- Cole R. S., Zusman D. Sedimentation properties of phage DNA molecules containing light-induced psoralen cross-links. Biochim Biophys Acta. 1970 Dec 14;224(2):660–662. doi: 10.1016/0005-2787(70)90607-6. [DOI] [PubMed] [Google Scholar]
- Davidson E. H., Graham D. E., Neufeld B. R., Chamberlin M. E., Amenson C. S., Hough B. R., Britten R. J. Arrangement and characterization of repetitive sequence elements in animal DNAs. Cold Spring Harb Symp Quant Biol. 1974;38:295–301. doi: 10.1101/sqb.1974.038.01.033. [DOI] [PubMed] [Google Scholar]
- Engberg J., Andersson P., Leick V., Collins J. Free ribosomal DNA molecules from Tetrahymena pyriformis GL are giant palindromes. J Mol Biol. 1976 Jun 25;104(2):455–470. doi: 10.1016/0022-2836(76)90281-3. [DOI] [PubMed] [Google Scholar]
- Firtel R. A., Kindle K. Structural organization of the genome of the cellular slime mold Dictyostelium discoideum: interspersion of repetitive and single-copy DNA sequences. Cell. 1975 Aug;5(4):401–411. doi: 10.1016/0092-8674(75)90059-8. [DOI] [PubMed] [Google Scholar]
- GEIDUSCHEK E. P. "Reversible" DNA. Proc Natl Acad Sci U S A. 1961 Jul 15;47:950–955. doi: 10.1073/pnas.47.7.950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgiev G. P., Varshavsky A. J., Ryskov A. P., Church R. B. On the structural organization of the transcriptional unit in animal chromosomes. Cold Spring Harb Symp Quant Biol. 1974;38:869–884. doi: 10.1101/sqb.1974.038.01.089. [DOI] [PubMed] [Google Scholar]
- Hanson C. V., Shen C. K., Hearst J. E. Cross-linking of DNA in situ as a probe for chromatin structure. Science. 1976 Jul 2;193(4247):62–64. doi: 10.1126/science.935855. [DOI] [PubMed] [Google Scholar]
- Hsu M. T., Kung H. J., Davidson N. An electron microscope study of Sindbis virus RNA. Cold Spring Harb Symp Quant Biol. 1974;38:943–950. doi: 10.1101/sqb.1974.038.01.096. [DOI] [PubMed] [Google Scholar]
- Johnson D. A new method of DNA denaturation mapping. Nucleic Acids Res. 1975 Nov;2(11):2049–2054. doi: 10.1093/nar/2.11.2049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karrer K. M., Gall J. G. The macronuclear ribosomal DNA of Tetrahymena pyriformis is a palindrome. J Mol Biol. 1976 Jun 25;104(2):421–453. doi: 10.1016/0022-2836(76)90280-1. [DOI] [PubMed] [Google Scholar]
- Kleckner N., Chan R. K., Tye B. K., Botstein D. Mutagenesis by insertion of a drug-resistance element carrying an inverted repetition. J Mol Biol. 1975 Oct 5;97(4):561–575. doi: 10.1016/s0022-2836(75)80059-3. [DOI] [PubMed] [Google Scholar]
- Musajo L., Rodighiero G. Mode of photosensitizing action of furocoumarins. Photophysiology. 1972;(7):115–147. [PubMed] [Google Scholar]
- Musajo L., Rodighiero G. Studies on the photo-C4-cyclo-addition reactions between skin-photosensitizing furocoumarins and nucleic acids. Photochem Photobiol. 1970 Jan;11(1):27–35. doi: 10.1111/j.1751-1097.1970.tb05714.x. [DOI] [PubMed] [Google Scholar]
- Pathak M. A., Krämer D. M. Photosensitization of skin in vivo by furocoumarins (psoralens). Biochim Biophys Acta. 1969 Nov 19;195(1):197–206. doi: 10.1016/0005-2787(69)90616-9. [DOI] [PubMed] [Google Scholar]
- Platt J. R. POSSIBLE SEPARATION OF INTERTWINED NUCLEIC ACID CHAINS BY TRANSFER-TWIST. Proc Natl Acad Sci U S A. 1955 Mar 15;41(3):181–183. doi: 10.1073/pnas.41.3.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SCHWARTZ D. Studies on crossing over in maize and Drosophila. J Cell Physiol Suppl. 1955 May;45(Suppl 2):171–188. doi: 10.1002/jcp.1030450510. [DOI] [PubMed] [Google Scholar]
- Schmid C. W., Deininger P. L. Sequence organization of the human genome. Cell. 1975 Nov;6(3):345–358. doi: 10.1016/0092-8674(75)90184-1. [DOI] [PubMed] [Google Scholar]
- Schmid C. W., Manning J. E., Davidson N. Inverted repeat sequences in the Drosophila genome. Cell. 1975 Jun;5(2):159–172. doi: 10.1016/0092-8674(75)90024-0. [DOI] [PubMed] [Google Scholar]
- Sobell H. M. Molecular mechanism for genetic recombination. Proc Natl Acad Sci U S A. 1972 Sep;69(9):2483–2487. doi: 10.1073/pnas.69.9.2483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tinoco I., Jr, Borer P. N., Dengler B., Levin M. D., Uhlenbeck O. C., Crothers D. M., Bralla J. Improved estimation of secondary structure in ribonucleic acids. Nat New Biol. 1973 Nov 14;246(150):40–41. doi: 10.1038/newbio246040a0. [DOI] [PubMed] [Google Scholar]
- Walker P. M., McLaren A. Binding behaviour on DNA-agar of a naturally occurring cross-linked fraction in rodent DNA. Mol Gen Genet. 1969;104(1):104–106. doi: 10.1007/BF00277366. [DOI] [PubMed] [Google Scholar]
- Wilson D. A., Thomas C. A., Jr Palindromes in chromosomes. J Mol Biol. 1974 Mar 25;84(1):115–138. doi: 10.1016/0022-2836(74)90216-2. [DOI] [PubMed] [Google Scholar]