Abstract
Background
The dysregulation of transforming growth factor-β (TGF-β) signaling plays a crucial role in ovarian carcinogenesis and in maintaining cancer stem cell properties. Classified as a member of the ATP-binding cassette (ABC) family, ABCA1 was previously identified by methylated DNA immunoprecipitation microarray (mDIP-Chip) to be methylated in ovarian cancer cell lines, A2780 and CP70. By microarray, it was also found to be upregulated in immortalized ovarian surface epithelial (IOSE) cells following TGF-β treatment. Thus, we hypothesized that ABCA1 may be involved in ovarian cancer and its initiation.
Results
We first compared the expression level of ABCA1 in IOSE cells and a panel of ovarian cancer cell lines and found that ABCA1 was expressed in HeyC2, SKOV3, MCP3, and MCP2 ovarian cancer cell lines but downregulated in A2780 and CP70 ovarian cancer cell lines. The reduced expression of ABCA1 in A2780 and CP70 cells was associated with promoter hypermethylation, as demonstrated by bisulfite pyro-sequencing. We also found that knockdown of ABCA1 increased the cholesterol level and promoted cell growth in vitro and in vivo. Further analysis of ABCA1 methylation in 76 ovarian cancer patient samples demonstrated that patients with higher ABCA1 methylation are associated with high stage (P = 0.0131) and grade (P = 0.0137). Kaplan-Meier analysis also found that patients with higher levels of methylation of ABCA1 have shorter overall survival (P = 0.019). Furthermore, tissue microarray using 55 ovarian cancer patient samples revealed that patients with a lower level of ABCA1 expression are associated with shorter progress-free survival (P = 0.038).
Conclusions
ABCA1 may be a tumor suppressor and is hypermethylated in a subset of ovarian cancer patients. Hypermethylation of ABCA1 is associated with poor prognosis in these patients.
Electronic supplementary material
The online version of this article (doi:10.1186/s13148-014-0036-2) contains supplementary material, which is available to authorized users.
Keywords: Ovarian cancer, Epigenetics, ABCA1
Background
Ovarian cancer is the most lethal tumor in women and the second most frequent gynecological malignancy [1]. As ovarian cancer has few symptoms early in its course, patients are generally diagnosed with advanced-stage tumors. Despite advances in chemotherapy, the poor prognosis for patients with late-stage ovarian cancer is reflected by the 5-year survival rate of less than 20% [2]. Current prognostic indicators using clinicopathological variables, including stage and grade, neither accurately predict clinical outcomes nor provide biological insight into the disease. Thus, a better understanding of the molecular changes in ovarian cancer would provide insight for better diagnosis and prognosis of this deadly disease.
The transforming growth factor-β (TGF-β) signaling pathway plays an important role in the regulation of ovarian growth [3]. During ovulation, the ovarian surface epithelial (OSE) cell covering the ovary undergoes rupture, followed by proliferation-mediated repair. This growth is inhibited by TGF-β, which plays a key role in preventing overproliferation of OSE cells [4]. In this regard, the dysregulation of TGF-β signaling may be an early step in the development of epithelial ovarian cancer. This is in agreement with the observation that resistance to TGF-β signaling is commonly seen in ovarian cancer, suggesting that reduced responsiveness to TGF-β is a key event in ovarian cancer [5].
Epigenetic alterations, including DNA methylation and histone modifications, play an important role in controlling gene expression [6,7]. Moreover, in cancer, aberrant epigenetic modifications are frequently found to be responsible for the silencing of tumor suppressor genes. Our previous studies using genome-wide techniques have identified several direct targets of TGF-β in immortalized ovarian surface epithelial (IOSE) cells [8]. These targets are silenced epigenetically in ovarian cancer and are associated with poor prognosis in ovarian cancer patients [9-11]. Restoration of the genes inhibited tumor growth, thus suggesting that they are tumor suppressors.
In order to identify additional genes that are hypermethylated in ovarian cancer, we performed methylated DNA immunoprecipitation microarray (mDIP-Chip) in IOSE cells and a panel of ovarian cancer cell lines. Our result showed that ABCA1, which is upregulated by TGF-β [8] and is a major regulator of cellular cholesterol [12], is hypermethylated in a subset of ovarian cancer cell lines. Further analysis demonstrated that ABCA1 methylation was associated with poor prognosis in ovarian cancer patients. These results suggested that ABCA1 may be a tumor suppressor and can serve as a prognostic indicator in ovarian cancer.
Results
ABCA1 is epigenetically silenced in ovarian cancer cells
Our previous studies demonstrated that novel TGF-β targets, FBXO32 and RunX1T1, are epigenetically silenced by promoter hypermethylation in ovarian cancer [10,11]. To identify additional genes regulated by TGF-β that exhibited promoter hypermethylation in ovarian cancer, we performed mDIP-Chip in IOSE cells and a panel of ovarian cancer cell lines [13]. One of the genes, ABCA1, which exhibited promoter methylation in a subset of ovarian cancer cell lines but not in IOSE cells, was selected for further analysis. This gene was also previously found to be upregulated in IOSE cells after TGF-β treatment [8].
To confirm our microarray result, we first examined the mRNA expression level of ABCA1 in various ovarian cancer cell lines (Figure 1A). The result showed that ABCA1 was repressed in A2780 and CP70 cells. Although treatment of HDAC inhibitor (TSA) or EZH2 inhibitor (GSK343) alone did not result in any significant re-expression of ABCA1, treatment of demethylating agent, 5aza, alone resulted in a robust re-expression of ABCA1 in CP70 ovarian cancers, suggesting that DNA methylation is responsible for the suppression of ABCA1 in CP70 cells (Figure 1B). These results were confirmed by bisulfite pyro-sequencing such that CpG sites around the transcriptional start site (TSS) of ABCA1 were heavily methylated in A2780 and CP70 ovarian cancer cells, while such methylation was either undetectable or low in IOSE cells or other ovarian cancer cells (Figure 1C). Taken together, these results suggested that promoter hypermethylation may be responsible for the transcriptional repression of ABCA1 in ovarian cancer.
Figure 1.
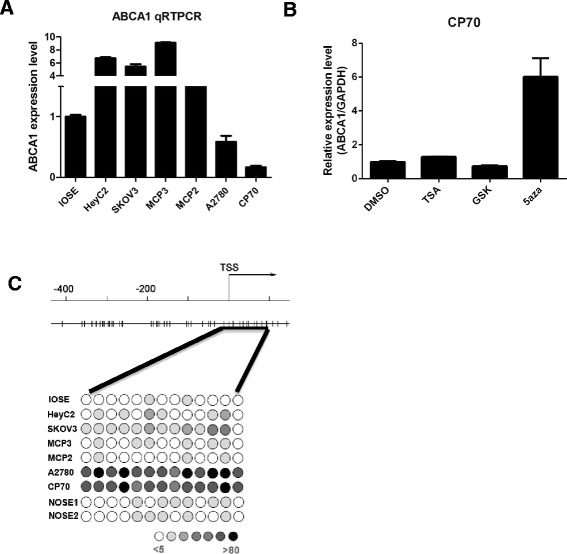
ABCA1 expression and methylation level in IOSE cells and ovarian cancer cell lines. (A) Total RNA was isolated from ovarian cells and converted into cDNA for amplification with specific primers for ABCA1. The relative level of expression after quantitative real-time RT-PCR was compared to IOSE cells (set as one fold). Each bar represents mean ± SD. (B) CP70 cells were treated with TSA (0.5 μM, 12 h), GSK343 (1 μM, 3 days), or 5aza (0.5 μM, 3 days). The expression level of ABCA1 was determined by RT-PCR. Treatment of 5aza, but not TSA or GSK, resulted in robust re-expression of ABCA1 in CP70 cells. Each bar represents mean ± SD. (C) The methylation status of the ABCA1 promoter and TSS region was analyzed by bisulfite pyro-sequencing from −90 to +190 (black line underneath). The upper panel shows the ABCA1 promoter and TSS region and the corresponding CpG sites (vertical bar), and the lower panel illustrates DNA methylation at the interrogated CpG site (circle) in IOSE cells, two NOSE samples, and ovarian cancer cell lines with intensity of gray color indicating methylation level.
Depletion of ABCA1 increases cholesterol level in ovarian cancer cells
As ABCA1 is responsible for cholesterol efflux, we first measured the effect of ABCA1 depletion on the cholesterol level of ovarian cancer cells. Lentiviral knockdown of ABCA1 was performed in MCP3 (Figure 2A) and HeyC2 (Figure 2B) ovarian cancer cells. The result showed that more than 50% repression of ABCA1 was observed in these cells. As expected, depletion of ABCA1 resulted in a significant accumulation of cholesterol in MCP3 (Figure 3A, P < 0.05) and HeyC2 (Figure 3B, P < 0.05) cells.
Figure 2.
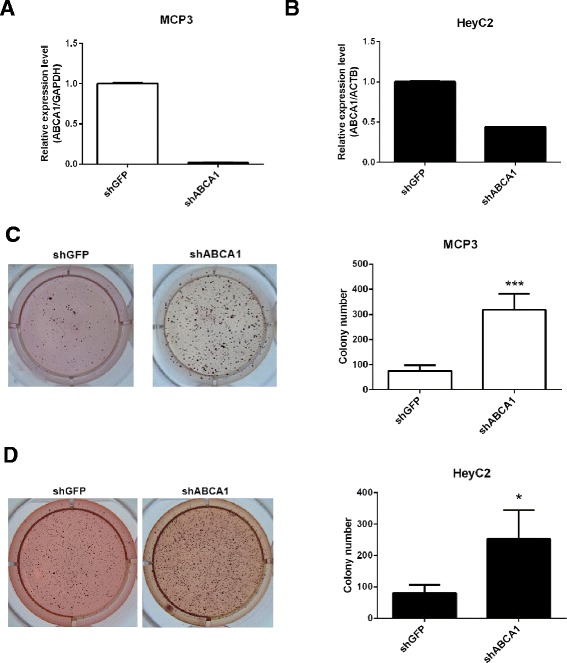
Effects of ABCA1 knockdown on cell growth. Real-time RT-PCR expression of ABCA1 in (A) MCP3 and (B) HeyC2 cells infected by lentivirus against shGFP (control) or shABCA1. Each bar represents mean ± SD. The growth of ABCA1 knockdown (C) MCP3 and (D) HeyC2 cells was examined by soft agar assay. Quantitative analysis of the soft agar assay is also shown. ***P < 0.001; *P < 0.05.
Figure 3.
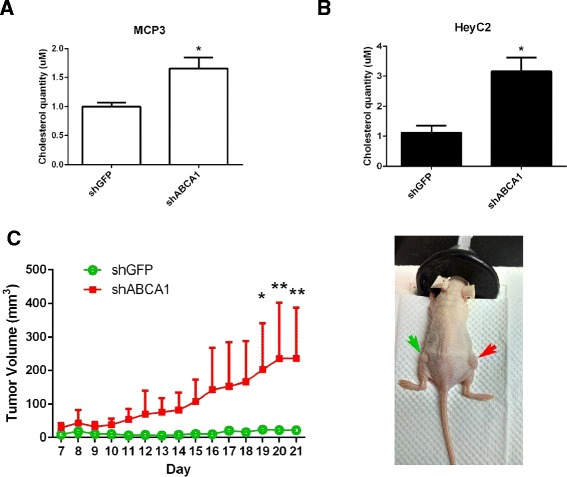
Effects of ABCA1 knockdown on cholesterol level and ovarian cancer growth in vivo . The cholesterol level of ABCA1 knockdown (A) MCP3 and (B) HeyC2 cells was measured by cholesterol quantitation assay. (C) The effect of ABCA1 knockdown on tumor growth in vivo was determined by the nude mice model. HeyC2 cells stably infected with shABCA1 (red arrow) or shGFP (green arrow) were injected subcutaneously into athymic nude mice. One week later, tumor volumes were measured daily. From day 19, the volume of tumors with ABCA1 knockdown was significantly reduced as compared to vector controls (*P < 0.05; **P < 0.005). Data were expressed as mean ± SD (n = 3).
Depletion of ABCA1 promotes cell growth in ovarian cancer cells in vitro and in vivo
Dysregulation of cholesterol homeostasis has been shown to be related to cancer. Therefore, we hypothesized that ABCA1 might act as a tumor suppressor in ovarian cancer. To confirm this hypothesis, we examined the effect of ABCA1 depletion on the growth of ovarian cancer cells. Interestingly, depletion of ABCA1 resulted in an increased cell growth in MCP3 (Figure 2C, P < 0.001) and HeyC2 (Figure 2D, P < 0.05) cells as demonstrated by colony formation assay. Further in vivo experiments also demonstrated that significant increased tumor growth was observed in ABCA1-depleted HeyC2 cells as compared to control (Figure 3C).
Hypermethylation of ABCA1 associates with tumor progression in ovarian cancer patients
Having demonstrated that promoter hypermethylation of ABCA1 can be observed in ovarian cancer cell lines, we then proceeded to examine if such epigenetic event can also be observed in ovarian cancer patient samples (Table 1, Figure 4A, B). Results from bisulphite pyro-sequencing showed that patients with a higher stage (P = 0.026) and grade (P = 0.013) but not age have a significantly higher level of ABCA1 methylation.
Table 1.
Association between methylation of ABCA1 and clinicopathological features of 76 ovarian cancer patients
| Methylation % | P | |
|---|---|---|
| Age | ||
| ≧60 | 5.2 ± 3.01a (53/76)b | 0.379 |
| <60 | 6.71 ± 5.34 (23/76) | |
| Stagec | ||
| Low | 4.36 ± 2.10 (27/76) | |
| High | 6.52 ± 4.44 (49/76) | 0.026 e |
| Graded | ||
| Low | 4.71 ± 3.62 (29/76) | |
| High | 6.26 ± 3.77 (47/76) | 0.013 |
aMean ± SD.
bNumber in parentheses represents the number of cases.
cLow and high stages are defined as FIGO I and II and FIGO III and IV, respectively.
dLow and high grades are defined as grade 1–2 and grade 3, respectively.
eItalicized value indicates P < 0.05.
Figure 4.
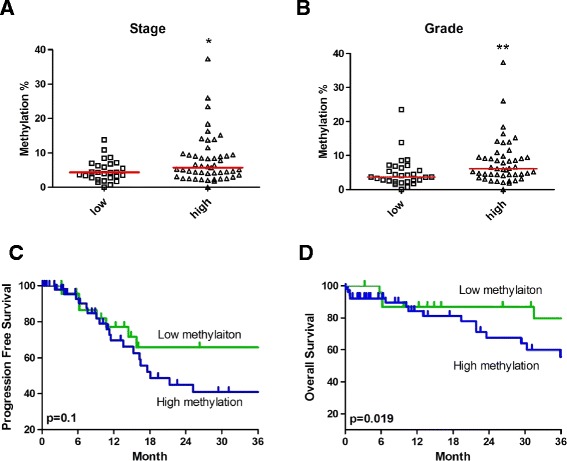
Association between ABCA1 methylation and tumor progression. Dot plot showing the association between ABCA1 methylation in different (A) stages and (B) grades in 76 ovarian cancer patient samples. Methylation of ABCA1 was determined by bisulfite pyro-sequencing. Low stage and low grade represented FIGO I and II and grade 1–2, respectively. While high stage and high grade represented FIGO III and IV and grade 3, respectively. *P < 0.05 by the Mann-Whitney U test. Kaplan-Meier analysis of ABCA1 methylation for (C) progression-free survival and (D) overall survival in 76 ovarian cancer patient samples is shown. Patients were grouped according to methylation of ABCA1 of 3%, which is based on the methylation level of IOSE cells. Patients with “high” ABCA1 methylation (>3% methylation) have significant shorter overall survival (P = 0.019) but not progression-free survival than patients with “low” ABCA1 methylation. Log-rank P values are shown.
Hypermethylation of ABCA1 associates with poor prognosis in ovarian cancer patients
To further investigate if ABCA1 methylation was predictive of survival in ovarian cancer patients, we performed Kaplan-Meier survival analyses. Using the baseline methylation level of ABCA1 in IOSE cells (3%) as a cutoff, a significant association between patients with high ABCA1 methylation and shorter overall survival (OS; P = 0.019) but not progression-free survival (PFS; P = 0.1) was observed (Figure 4C, D).
Because the above approaches based on a cutoff value may have biased the data analysis, the Cox proportional hazards model was used to analyze the predictive value of ABCA1 methylation and other clinicopathological parameters on OS and PFS (Table 2). As expected, a higher stage (OS: hazard ratio: 11.321, P = 0.02; PFS: hazard ratio: 14.278, P < 0.01) and grade (OS: hazard ratio: 22.559, P = 0.03; PFS: hazard ratio: 20.586, P < 0.01) but not age was significantly associated with poor prognosis. Interestingly, methylation of ABCA1 is also associated with poor patient outcome in ovarian cancer (OS: hazard ratio: 1.106, P = 0.033; PFS: hazard ratio: 1.096, P = 0.013). However, this association was not observed in multivariate analysis, thus suggesting that other factors are also involved in the survival of ovarian cancer patients (Table 3). Taken together, methylation of ABCA1 is partially associated with poor prognosis in ovarian cancer patients.
Table 2.
Univariable analysis of survival by the Cox proportional hazards model
| Variable | Overall survival | Progression-free survival | ||
|---|---|---|---|---|
| HR (95% CI) | P | HR (95% CI) | P | |
| ABCA1 methylation | 1.106 (1.008–1.213) | 0.033 a | 1.096 (1.020–1.179) | 0.013 |
| Age | 2.014 (0.712–5.701) | 0.187 | 1.309 (0.579–2.963) | 0.518 |
| Stage | 11.321 (2.48–51.679) | 0.02 | 14.278 (4.233–48.154) | <0.01 |
| Grade | 22.559 (2.849–178.624) | 0.03 | 20.586 (4.817–87.977) | <0.01 |
aItalicized value indicates P < 0.05.
Table 3.
Multivariate analysis of survival by the Cox proportional hazards model
| PFS | OS | |||
|---|---|---|---|---|
| HR (95% CI) | P | HR (95% CI) | P | |
| ABCA1 methylation | 1.013 (0.960–1.069) | 0.636 | 1.016 (0.965–1.069) | 0.554 |
| Age | 0.993 (0.961–1.025) | 0.665 | 1.019 (0.988–1.051) | 0.233 |
| Stage | 1.633 (1.159–2.302) | 0.005 | 2.330 (1.196–4.538) | 0.013 |
| Grade | 1.578 (0.557–4.473) | 0.390 | 1.538 (0.340–6.963) | 0.577 |
Low expression of ABCA1 associates with poor prognosis in ovarian cancer patients
To further investigate the association between expression of ABCA1 and survival of ovarian cancer patients, we performed tissue microarray on a different cohort containing 55 ovarian patient samples (Table 4, Figure 5A). Although expression of ABCA1 is not associated with any clinicopathological parameters of the samples (Table 5), patients with low expression of ABCA1 are associated with shorter progression-free survival (Figure 5B). Similarly, analysis of TCGA ovarian cancer RNA-Seq dataset [14] also found that patients with lower expression of ABCA1 are associated with shorter overall survival (Figure 5C).
Table 4.
Summary of clinicopathological data of 55 ovarian cancer patients in tissue microarray
| Ovarian cancer ( n = 55) | |
|---|---|
| Age at diagnosis (years) | |
| Median | 51 |
| Range | 18–81 |
| Stage | |
| FIGO (I–II) | 22 |
| FIGO (III–VI) | 33 |
| Grade | |
| I | 9 |
| II | 14 |
| III | 32 |
| Pathological type | |
| Clear cell | 4 |
| Serous | 37 |
| Mucinous | 11 |
| Endometroid | 3 |
| Median survival (months) | |
| PFS | 25.3 |
| OS | 36.7 |
PFS progression-free survival, OS overall survival.
Figure 5.
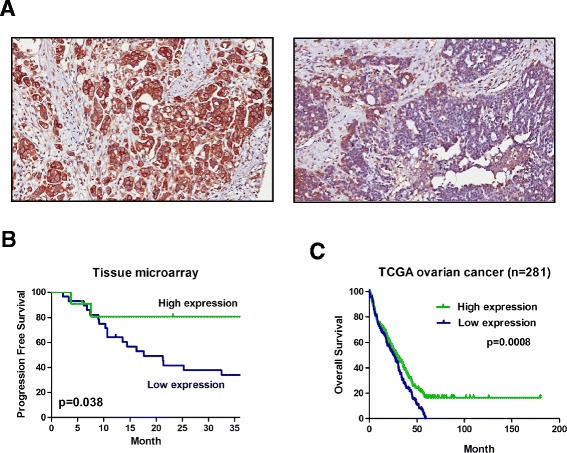
Association between expression of ABCA1 and survival in ovarian cancer patients. Expression of ABCA1 in 55 ovarian cancer patient samples was determined by IHC in tissue microarray. (A) Representative image of ovarian cancer showing high (left panel) and low (right panel) ABCA1 expression on the cell membrane or cytoplasm (×400). (B) Kaplan-Meier analysis found that patients with low ABCA1 expression have shorter progression-free survival than patients with high ABCA1 expression (P = 0.038). (C) Similar results can be observed in TCGA ovarian cancer RNA-Seq dataset that patients with low expression of ABCA1 are associated with shorter overall survival (P = 0.0008). Log-rank P values are shown.
Table 5.
Association between expression of ABCA1 and clinicopathological features of 55 ovarian cancer patients
| Expression score | |||
|---|---|---|---|
| High expression | Low expression | ||
| Age | |||
| ≧60 | 11a (20%) | 26 (47.2%) | |
| <60 | 5 (9.1%) | 13 (23.6%) | 0.572 |
| Stage | |||
| Low | 6 (10.9%) | 16 (29.1%) | |
| High | 10 (18.2%) | 23 (41.8%) | 0.528 |
| Grade | |||
| Low | 3 (5.5%) | 6 (10.9%) | |
| High | 13 (23.6%) | 33 (60%) | 0.522 |
aNumber of cases.
Discussion
In the current study, we demonstrated promoter hypermethylation of ABCA1, a cholesterol transporter in ovarian cancer cell lines and patient samples. This is a cancer-specific event as methylation of ABCA1 was not observed in IOSE and normal ovarian surface epithelial (NOSE) cells. Methylation of ABCA1 was also negatively associated with its expression and increased cell growth in vitro and in vivo. Importantly, ovarian cancer patients with higher ABCA1 methylation were associated with shorter survival. Result from tissue microarray and TCGA ovarian cancer RNA-Seq dataset also demonstrated that ovarian cancer patients with lower ABCA1 expression were associated with shorter survival. Our results were also confirmed by a recent finding that ABCA1 is epigenetically silenced by promoter methylation in prostate cancer cells. Hypermethylation of ABCA1 is associated with high-grade prostate cancer [15]. Taken together, these results suggest that ABCA1 may be a tumor suppressor and are epigenetically silenced in human cancer.
ABCA1, which belongs to the ATP-binding cassette (ABC) protein family, is responsible for cholesterol efflux and metabolism. It has been shown to be essential for the synthesis of high-density lipoprotein (HDL) particles by exporting cellular cholesterol out of the cell. Recent studies suggested that ABCA1 may be a tumor suppressor [16]. For example, Smith and Land demonstrated that overexpression of ABCA1 in colon cancer cells resulted in a decrease of cellular cholesterol and inhibition of tumor growth in vitro and in vivo. This growth inhibition may be due to apoptosis, as overexpression of ABCA1 enhanced cytochrome C release from mitochondria.
Expression of ABCA1 has been previously shown to be activated by the nuclear receptor liver X receptor (LXR) [17]. Interestingly, TGF-β could significantly increase the expression of ABCA1 through activation of LXR signaling [18]. Together with our previous finding that ABCA1 is a TGF-β target in IOSE cells, it is thus suggested that the growth inhibitory effect of TGF-β may be partly due to overexpression of ABCA1 in IOSE cells. Epigenetic alteration of ABCA1 may contribute to the resistance of TGF-β-mediated growth suppression in ovarian cancer cells [9].
Dysregulation of cholesterol homeostasis has been known to be related to cancer [19]. Several studies have demonstrated an elevated level of cholesterol in tumor as compared to normal tissue [20,21]. Recently, Li et al. also demonstrated that ovarian cancer patients with higher serum low-density lipoprotein (LDL) level were significantly associated with shorter progression-free survival and overall survival [22]. It may be due to the fact that lipid serves as an energy source for promoting cell growth and metastasis in ovarian cancer [23]. In this regard, numerous studies have demonstrated that cholesterol-lowering agents, such as statins, exhibit anti-tumor activity against various cancers [24]. For example, statins have been shown to reduce tumor growth in xenograft models [25,26]. In ovarian cancer, a clinical study demonstrated that patients with statins are associated with improved survival [27]. This may be due to the growth inhibition and augmentation of apoptosis by statins in ovarian cancer cells [28-30]. Furthermore, inhibition of HMG-CoA reductase by statins can also lead to a decreased level of mevalonate, which is important for the synthesis of isoprenoids including geranyl pyrophosphate (GPP) and farnesyl pyrophosphate (FPP) [24,31]. Proper activation of several signaling molecules such as small GTP-binding proteins Ras and Rho require prenylation of GPP and FPP. Thus, blocking of cholesterol metabolism would reduce cancer growth by suppression of the proliferation signal.
Conclusions
Promoter methylation of ABCA1 was observed in ovarian cancer cell lines and patient samples. Ovarian cancer patients with higher methylation and lower expression of ABCA1 were associated with shorter survival. The tumor suppressor function of ABCA1 in ovarian cancer warrants further investigation.
Methods
Patient samples
Seventy-six treatment-naive ovarian cancer samples were obtained from Tri-Service General Hospital, Taipei, Taiwan (Table 6). Eight NOSE cell samples were acquired from patients during surgery for benign gynecological disease at either the Tri-Service General Hospital or Indiana University, USA. For this cohort of ovarian cancer patient samples, the median age at the time of diagnosis was 54 years (range, 18–90 years). Forty-nine cases (64.5%) were high stage (International Federation of Gynecology and Obstetrics (FIGO) stages III and IV), and 27 cases (35.5%) were low stage (FIGO stages I and II). Forty-seven cases (61.8%) were grade III, 13 cases (17.1%) were grade I, and 16 cases (21.1%) were grade II. All of the studies involving human ovarian epithelial samples were approved by the institutional review boards of the Tri-Service General Hospital, Taiwan, and Indiana University.
Table 6.
Summary of clinicopathological data of 76 ovarian cancer patients
| Ovarian cancer ( n = 76) | |
|---|---|
| Age at diagnosis (years) | |
| Median | 54 |
| Range | 18–90 |
| Stage | |
| FIGO (I–II) | 27 |
| FIGO (III–VI) | 49 |
| Grade | |
| I | 13 |
| II | 16 |
| III | 47 |
| Pathological type | |
| Serous | 48 |
| Mucinous | 23 |
| Endometroid | 5 |
| Median survival (months) | |
| PFS | 11.51 |
| OS | 17.42 |
PFS progression-free survival, OS overall survival.
Cell culture
IOSE cells were derived by transducing the catalytic subunit of human telomerase and the papilloma virus subunit E7 into primary ovarian epithelial cells, as described previously [32]. The cells were maintained in a 1:1 mixture of Medium 199 (Sigma, St. Louis, MO) and 105 (Sigma) supplemented with 10% fetal bovine serum (FBS) (Invitrogen, Carlsbad, CA), 400 ng/ml hydrocortisone (Sigma), 10 ng/ml EGF, and 50 units/ml of penicillin/streptomycin (Invitrogen). The ovarian cancer cell lines A2780, CP70, MCP2, and MCP3 were propagated in RPMI 1640 (Invitrogen) containing 10% FBS. HeyC2 cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM) containing 5% FBS, 1% NEAA, 1% Gln, and 1% HEPES. SKOV3 cells were cultured in McCoy’s 5A containing 10% FBS, 1% NEAA, 1% Gln, and 1% HEPES.
DNA extraction, RNA extraction, and quantitative reverse transcription-PCR
DNA was extracted with the Tissue & Cell Genomic DNA Purification Kit (GeneMark, Taiwan). It was eluted in 50 μl distilled water and stored at −20°C until use. Total RNA from cell lines was extracted using TRIzol (Invitrogen), as previously described. Briefly, 1 μg of total RNA was treated with DNase I (amplification grade, Invitrogen) before first-strand cDNA synthesis using reverse transcriptase (Superscript II RT, Invitrogen). The real-time PCR reactions were carried out using the ABI StepOne real-time PCR system (Applied Biosystems, Foster City, CA) with specific primers (Additional file 1: Table S1). The relative expression of ABCA1 was calculated using the comparative Ct method.
Bisulfite conversion and pyro-sequencing
Bisulphite pyro-sequencing was performed as described previously [11]. Briefly, 0.5 μg of genomic DNA was bisulfite-modified using the EZ DNA Methylation Kit (Zymo Research, Orange, CA), according to the manufacturer’s protocol. The bisulfite-modified DNA was subjected to PCR amplification using a tailed reverse primer in combination with a biotin-labeled universal primer. The PCR and sequencing primers were designed using PyroMark Assay Design 2.0 (Qiagen GmbH, Hilden, Germany). The ABCA1 TSS (−90 to +190) was PCR-amplified with specific primers (Additional file 1: Table S1) in a 25-μl reaction containing 2× RBC SensiZyme Hotstart Taq premix (RBC Bioscience, Taiwan). Prior to pyro-sequencing, 1.5 μl of each PCR reaction was analyzed on 1% agarose gel. The pyro-sequencing was performed on the PyroMark Q24 instrument (Qiagen) using the Pyro Gold Reagents (Qiagen), according to the manufacturer’s protocol. The methylation level of 11 CpG sites, which are located −13 to +95 with respect to the TSS, was measured. The methylation percentage of each cytosine was determined using the fluorescence intensity of cytosines divided by the sum of the fluorescence intensity of cytosines and thymines at each CpG site. In vitro methylated DNA (Millipore) was included as positive control for pyro-sequencing.
Knockdown of ABCA1 by shRNA
The small hairpin RNA (shRNA) of ABCA1 were acquired from the National RNAi Core Facility Platform at the Institute of Molecular Biology/Genomic Research Center, Academia Sinica, Taiwan. Briefly, 293T cells were transfected with shRNA (TRCN0000029093), pMDG, and pCMVR 8.91 using the ProFection Mammalian Transfection System (Promega) to prepare the shABCA1 lentivirus. Infected ovarian cancer cells were selected by incubating with puromycin (2 μg/ml; Sigma) for at least 2 days.
Soft agar assay for colony formation
Trypsinized cells (1,000 cells) were seeded and mixed in 1.5 ml of 0.35% top layer agar supplemented with DMEM with 10% FBS. This suspension was overlaid on the bottom layer of 0.5% agar in DMEM with 10% FBS in a six-well plate. Plates were allowed to solidify and then incubated at 37°C for around 3 weeks. Colony formation was monitored daily by microscopic observation. At the end of the experiments, the plates were stained with iodonitrotetrazolium (INT) stain (Sigma) for 48 h at 37°C. The number of colonies was counted.
Immunohistochemical analysis on ovarian tissue microarray
The paraffin-embedded ovarian cancer patients’ tissue microarray were prepared and retrieved from the Department of Pathology, Tri-Service General Hospital as described previously [33]. The tissue microarray contained 55 samples of ovarian cancer patients (Table 4). The immunohistochemistry procedure followed a standard protocol, using a rabbit polyclonal anti-human ABCA1 antibody (NB400-105, Novus Biologicals). All tissue microarray slides were examined and scored by two pathologists. Immunohistochemistry (IHC) scoring was determined by distribution of positively stained cell × intensity of the staining. A scoring of 60 was used as a cutoff value.
In vivo tumorigenicity assay
A total of three, 8-week-old, athymic nude mice (BALB/cByJNarl) were obtained from the National Laboratory Animal Center, Taiwan. All mice were kept under specific pathogen-free conditions using a laminar airflow rack with free access to sterilized food and autoclaved water. All experiments were performed under license from Animal Experimentation Ethics Committee of the National Chung Cheng University. HeyC2 cells (1 × 106) stably infected with pLKO.1/shABCA1 or pLKO.1/shGFP were re-suspended in 0.1 ml of medium and Matrigel (BD Biosciences, San Jose, CA) mixture (1:1). The cell suspension was then injected subcutaneously into the flank of each mouse (day 0). Tumor size was measured daily with calipers in length (L) and width (W). Tumor volume was calculated using the formula (L × W2/2). At the end of the experiment, all mice were sacrificed by cervical dislocation.
Analysis of cellular cholesterol content
Ovarian cancer cells (1 × 104) infected with shABCA1 or shGFP were seeded in a six-well dish and cultured for 48 h. After 48 h, the cells were washed with cold PBS twice and scraped. The cells were re-suspended in 10% RIPA buffer (50 mM HEPES-KOH, pKa 7.55, 500 mM LiCl, 1 mM EDTA, 0.1% NP-40, and 0.07% Na-deoxycholate) and kept on ice for 15 min. Cholesterol content was determined by the Amplite™ Fluorimetric Cholesterol Quantitation Kit (AAT Bioquest, Sunnyvale, CA) according to the manufacturer’s protocol. Fluorescence of cholesterol was detected by Ex/Em at 540/590 nm.
Statistical analysis
PFS and OS were assessed by Kaplan-Meier analysis using the log-rank test. Progression-free survival was defined as the duration from the day of diagnosis or chemotherapy to the detection of new lesions or progression of residual lesions. Overall survival was defined as the duration from the day of diagnosis to death. A DNA methylation level at 3%, which is the level of methylation in OSE cells, was used as a cutoff. Fisher’s exact test or the Mann-Whitney U test was also used to compare parameters of different groups. All statistical calculations were performed using the statistical package SPSS version 13.0 for Windows (SPSS, Inc., Chicago, IL). P < 0.05 was considered to be statistically significant. For TCGA ovarian cancer RNA-Seq dataset, a mean RPKM value of 4 was used as a cutoff.
Acknowledgements
This study was supported by research grants from the Ministry of Science and Technology, Taiwan (NSC102-2320-B-194-006; NSC102-2628-B-038-010-MY3, MOST103-2320-B-194-002) and National Chung Cheng University, Taiwan (NCCU Interdisciplinary Science grant 103-03).
Additional file
Primer sequences used in the study. Table S1. lists all the primers for RT-PCR and bisulphite pyro-sequencing.
Footnotes
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
JLC analyzed the methylation microarray, performed the bisulfite pyro-sequencing, conducted the cell line experiments, and drafted the manuscript. RLH performed the IHC on tissue microarray. JS and LYC assisted with the cell line and methylation studies. SJL assisted with the knockdown experiments. PSY co-designed the project and assisted with the methylation microarray. WTC assisted with the cholesterol analyses. YHL performed the in vivo tumorigenicity assay. YLL assisted with the in vivo tumorigenicity assay. TKC analyzed the IHC results. CIL coordinated the knockdown experiments. CKT and SFW coordinated the in vivo tumorigenicity assay. KPN provided the NOSE samples. THH co-designed the project. HCL provided the patient samples and clinicopathological data, assisted with the interpretation of the data, co-designed the project, and secured the funding. MWC performed the methylation microarray, assisted with the interpretation of the data, co-designed the project, secured the funding, and revised the manuscript. All authors read and approved the final manuscript.
Contributor Information
Jian-Liang Chou, Email: alien711021@gmail.com.
Rui-Lan Huang, Email: gyntsgh@gmail.com.
Jacqueline Shay, Email: yjshieh@gmail.com.
Lin-Yu Chen, Email: maple_916@yahoo.com.tw.
Sheng-Jie Lin, Email: gon80592@yahoo.com.tw.
Pearlly S Yan, Email: pearlly.yan@osumc.edu.
Wei-Ting Chao, Email: wtchao@thu.edu.tw.
Yi-Hui Lai, Email: psynicki@gmail.com.
Yen-Ling Lai, Email: bigeyes0007@hotmail.com.
Tai-Kuang Chao, Email: chaotai.kuang@msa.hinet.net.
Cheng-I Lee, Email: chengi.leeccu@gmail.com.
Chien-Kuo Tai, Email: write2ck@gmail.com.
Shu-Fen Wu, Email: 441autoimmune@gmail.com.
Kenneth P Nephew, Email: knephew@indiana.edu.
Tim H-M Huang, Email: HUANGT3@uthscsa.edu.
Hung-Cheng Lai, Email: hclai@s.tmu.edu.tw.
Michael W Y Chan, Email: biowyc@ccu.edu.tw.
References
- 1.Wakabayashi MT, Lin PS, Hakim AA. The role of cytoreductive/debulking surgery in ovarian cancer. J Natl Compr Canc Netw. 2008;6:803–10. doi: 10.6004/jnccn.2008.0060. [DOI] [PubMed] [Google Scholar]
- 2.Nossov V, Amneus M, Su F, Lang J, Janco JM, Reddy ST, et al. The early detection of ovarian cancer: from traditional methods to proteomics. Can we really do better than serum CA-125? Am J Obstet Gynecol. 2008;199:215–23. doi: 10.1016/j.ajog.2008.04.009. [DOI] [PubMed] [Google Scholar]
- 3.Wong AS, Leung PC. Role of endocrine and growth factors on the ovarian surface epithelium. J Obstet Gynaecol Res. 2007;33:3–16. doi: 10.1111/j.1447-0756.2007.00478.x. [DOI] [PubMed] [Google Scholar]
- 4.Nilsson EE, Skinner MK. Role of transforming growth factor beta in ovarian surface epithelium biology and ovarian cancer. Reprod Biomed Online. 2002;5:254–8. doi: 10.1016/S1472-6483(10)61828-7. [DOI] [PubMed] [Google Scholar]
- 5.Hu W, Wu W, Nash MA, Freedman RS, Kavanagh JJ, Verschraegen CF. Anomalies of the TGF-beta postrecep signaling pathway in ovarian cancer cell lines. Anticancer Res. 2000;20:729–33. [PubMed] [Google Scholar]
- 6.Simon JA, Lange CA. Roles of the EZH2 histone methyltransferase in cancer epigenetics. Mutat Res. 2008;647:21–9. doi: 10.1016/j.mrfmmm.2008.07.010. [DOI] [PubMed] [Google Scholar]
- 7.McKenna ES, Roberts CW. Epigenetics and cancer without genomic instability. Cell Cycle. 2009;8:23–6. doi: 10.4161/cc.8.1.7290. [DOI] [PubMed] [Google Scholar]
- 8.Qin H, Chan MW, Liyanarachchi S, Balch C, Potter D, Souriraj IJ, et al. An integrative ChIP-chip and gene expression profiling to model SMAD regulatory modules. BMC Syst Biol. 2009;3:73. doi: 10.1186/1752-0509-3-73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chan MW, Huang YW, Hartman-Frey C, Kuo CT, Deatherage D, Qin H, et al. Aberrant transforming growth factor beta1 signaling and SMAD4 nuclear translocation confer epigenetic repression of ADAM19 in ovarian cancer. Neoplasia. 2008;10:908–19. doi: 10.1593/neo.08540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chou JL, Su HY, Chen LY, Liao YP, Hartman-Frey C, Lai YH, et al. Promoter hypermethylation of FBXO32, a novel TGF-beta/SMAD4 target gene and tumor suppressor, is associated with poor prognosis in human ovarian cancer. Lab Invest. 2010;90:414–25. doi: 10.1038/labinvest.2009.138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yeh KT, Chen TH, Yang HW, Chou JL, Chen LY, Yeh CM, et al. Aberrant TGFbeta/SMAD4 signaling contributes to epigenetic silencing of a putative tumor suppressor, RunX1T1 in ovarian cancer. Epigenetics. 2011;6:727–39. doi: 10.4161/epi.6.6.15856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fielding PE, Nagao K, Hakamata H, Chimini G, Fielding CJ. A two-step mechanism for free cholesterol and phospholipid efflux from human vascular cells to apolipoprotein A-1. Biochemistry. 2000;39:14113–20. doi: 10.1021/bi0004192. [DOI] [PubMed] [Google Scholar]
- 13.Yeh CM, Shay J, Zeng TC, Chou JL, Huang TH, Lai HC, et al. Epigenetic silencing of ARNTL, a circadian gene and potential tumor suppressor in ovarian cancer. Int J Oncol. 2014;45:2101–7. doi: 10.3892/ijo.2014.2627. [DOI] [PubMed] [Google Scholar]
- 14.The Cancer Genome Atlas Research Network Integrated genomic analyses of ovarian carcinoma. Nature. 2011;474:609–15. doi: 10.1038/nature10166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee BH, Taylor MG, Robinet P, Smith JD, Schweitzer J, Sehayek E, et al. Dysregulation of cholesterol homeostasis in human prostate cancer through loss of ABCA1. Cancer Res. 2013;73:1211–8. doi: 10.1158/0008-5472.CAN-12-3128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Smith B, Land H. Anticancer activity of the cholesterol exporter ABCA1 gene. Cell Rep. 2012;2:580–90. doi: 10.1016/j.celrep.2012.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Laffitte BA, Joseph SB, Walczak R, Pei L, Wilpitz DC, Collins JL, et al. Autoregulation of the human liver X receptor alpha promoter. Mol Cell Biol. 2001;21:7558–68. doi: 10.1128/MCB.21.22.7558-7568.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hu YW, Wang Q, Ma X, Li XX, Liu XH, Xiao J, et al. TGF-beta1 up-regulates expression of ABCA1, ABCG1 and SR-BI through liver X receptor alpha signaling pathway in THP-1 macrophage-derived foam cells. J Atheroscler Thromb. 2010;17:493–502. doi: 10.5551/jat.3152. [DOI] [PubMed] [Google Scholar]
- 19.Potter VR. The biochemical approach to the cancer problem. Fed Proc. 1958;17:691–7. [PubMed] [Google Scholar]
- 20.Dessi S, Batetta B, Anchisi C, Pani P, Costelli P, Tessitore L, et al. Cholesterol metabolism during the growth of a rat ascites hepatoma (Yoshida AH-130) Br J Cancer. 1992;66:787–93. doi: 10.1038/bjc.1992.361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kolanjiappan K, Ramachandran CR, Manoharan S. Biochemical changes in tumor tissues of oral cancer patients. Clin Biochem. 2003;36:61–5. doi: 10.1016/S0009-9120(02)00421-6. [DOI] [PubMed] [Google Scholar]
- 22.Li AJ, Elmore RG, Chen IY, Karlan BY. Serum low-density lipoprotein levels correlate with survival in advanced stage epithelial ovarian cancers. Gynecol Oncol. 2010;116:78–81. doi: 10.1016/j.ygyno.2009.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nieman KM, Kenny HA, Penicka CV, Ladanyi A, Buell-Gutbrod R, Zillhardt MR, et al. Adipocytes promote ovarian cancer metastasis and provide energy for rapid tumor growth. Nat Med. 2011;17:1498–503. doi: 10.1038/nm.2492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Osmak M. Statins and cancer: current and future prospects. Cancer Lett. 2012;324:1–12. doi: 10.1016/j.canlet.2012.04.011. [DOI] [PubMed] [Google Scholar]
- 25.Gao J, Jia WD, Li JS, Wang W, Xu GL, Ma JL, et al. Combined inhibitory effects of celecoxib and fluvastatin on the growth of human hepatocellular carcinoma xenografts in nude mice. J Int Med Res. 2010;38:1413–27. doi: 10.1177/147323001003800423. [DOI] [PubMed] [Google Scholar]
- 26.Kochuparambil ST, Al-Husein B, Goc A, Soliman S, Somanath PR. Anticancer efficacy of simvastatin on prostate cancer cells and tumor xenografts is associated with inhibition of Akt and reduced prostate-specific antigen expression. J Pharmacol Exp Ther. 2011;336:496–505. doi: 10.1124/jpet.110.174870. [DOI] [PubMed] [Google Scholar]
- 27.Elmore RG, Ioffe Y, Scoles DR, Karlan BY, Li AJ. Impact of statin therapy on survival in epithelial ovarian cancer. Gynecol Oncol. 2008;111:102–5. doi: 10.1016/j.ygyno.2008.06.007. [DOI] [PubMed] [Google Scholar]
- 28.Scoles DR, Xu X, Wang H, Tran H, Taylor-Harding B, Li A, et al. Liver X receptor agonist inhibits proliferation of ovarian carcinoma cells stimulated by oxidized low density lipoprotein. Gynecol Oncol. 2010;116:109–16. doi: 10.1016/j.ygyno.2009.09.034. [DOI] [PubMed] [Google Scholar]
- 29.Liu H, Liang SL, Kumar S, Weyman CM, Liu W, Zhou A. Statins induce apoptosis in ovarian cancer cells through activation of JNK and enhancement of Bim expression. Cancer Chemother Pharmacol. 2009;63:997–1005. doi: 10.1007/s00280-008-0830-7. [DOI] [PubMed] [Google Scholar]
- 30.Martirosyan A, Clendening JW, Goard CA, Penn LZ. Lovastatin induces apoptosis of ovarian cancer cells and synergizes with doxorubicin: potential therapeutic relevance. BMC Cancer. 2010;10:103. doi: 10.1186/1471-2407-10-103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Goldstein JL, Brown MS. Regulation of the mevalonate pathway. Nature. 1990;343:425–30. doi: 10.1038/343425a0. [DOI] [PubMed] [Google Scholar]
- 32.Gillan L, Matei D, Fishman DA, Gerbin CS, Karlan BY, Chang DD. Periostin secreted by epithelial ovarian carcinoma is a ligand for alpha(V)beta(3) and alpha(V)beta(5) integrins and promotes cell motility. Cancer Res. 2002;62:5358–64. [PubMed] [Google Scholar]
- 33.Liu CY, Chao TK, Su PH, Lee HY, Shih YL, Su HY, et al. Characterization of LMX-1A as a metastasis suppressor in cervical cancer. J Pathol. 2009;219:222–31. doi: 10.1002/path.2589. [DOI] [PubMed] [Google Scholar]


