Abstract
Background: Circulating trans fatty acids (TFAs), which cannot be synthesized by humans, are linked to adverse health outcomes. Although TFAs are obtained from diet, little is known about subsequent influences (e.g., relating to incorporation, metabolism, or intercompetition with other fatty acids) that could alter circulating concentrations and possibly modulate or mediate impacts on health.
Objective: The objective was to elucidate novel biologic pathways that may influence circulating TFAs by evaluating associations between common genetic variation and TFA biomarkers.
Design: We performed meta-analyses using 7 cohorts of European-ancestry participants (n = 8013) having measured genome-wide variation in single-nucleotide polymorphisms (SNPs) and circulating TFA biomarkers (erythrocyte or plasma phospholipids), including trans-16:1n–7, total trans-18:1, trans/cis-18:2, cis/trans-18:2, and trans/trans-18:2. We further evaluated SNPs with genome-wide significant associations among African Americans (n = 1082), Chinese Americans (n = 669), and Hispanic Americans (n = 657) from 2 of these cohorts.
Results: Among European-ancestry participants, 31 SNPs in or near the fatty acid desaturase (FADS) 1 and 2 cluster were associated with cis/trans-18:2; a top hit was rs174548 (β = 0.0035, P = 4.90 × 10−15), an SNP previously associated with circulating n–3 and n–6 polyunsaturated fatty acid concentrations. No significant association was identified for other TFAs. rs174548 in FADS1/2 was also associated with cis/trans-18:2 in Hispanic Americans (β = 0.0053, P = 1.05 × 10−6) and Chinese Americans (β = 0.0028, P = 0.002) but not African Americans (β = 0.0009, P = 0.34); however, in African Americans, fine mapping identified a top hit in FADS2 associated with cis/trans-18:2 (rs174579: β = 0.0118, P = 4.05 × 10−5). The association between rs174548 and cis/trans-18:2 remained significant after further adjustment for individual circulating n–3 and n–6 fatty acids, except arachidonic acid. After adjustment for arachidonic acid concentrations, the association between rs174548 and cis/trans-18:2 was nearly eliminated in European-ancestry participants (β-coefficient reduced by 86%), with similar reductions in Hispanic Americans and Chinese Americans.
Conclusions: Our findings provide novel evidence for genetic regulation of cis/trans-18:2 by the FADS1/2 cluster and suggest that this regulation may be influenced/mediated by concentrations of arachidonic acid, an n–6 polyunsaturated fat.
Keywords: arachidonic acid, genome-wide association, meta-analysis, phospholipid, trans fatty acids
INTRODUCTION
trans Fatty acids (TFAs),8 unsaturated fatty acids with one or more double bonds in the trans configuration, have adverse health effects (1, 2). Concentrations of TFAs in circulating phospholipids are linked to greater systematic inflammation (3, 4), sudden cardiac death, and fatal coronary artery disease (5, 6). Because trans bonds cannot be synthesized by humans, exposure to TFAs is only from dietary consumption, especially from packaged or prepared foods containing partially hydrogenated vegetable oils but also from meats or milk products from ruminants (e.g., cattle, sheep, and goats) that contain small amounts of TFAs (7). However, circulating TFA concentrations are only partly correlated with dietary intakes of these foods (7), suggesting potential nondietary influences. Yet, following consumption, little is known about the metabolic fate of TFA or its determinants. For instance, endogenous mechanisms of incorporation, metabolism, and/or competition with other fatty acids could influence circulating TFA concentrations (8) and modulate or mediate their impact on health.
Phospholipid fatty acids, the major components of cellular membranes, influence the function of membrane-bound receptors and channels that are involved in various signal transductions and act as precursors to a myriad of active lipid metabolites (9). They also serve as biomarkers for the potential effects of individual fatty acids on health and disease. For example, phospholipid concentrations of some TFA subtypes, such as those with 16 carbons and 1 double bond at the seventh N-terminal carbon (i.e., trans-16:1n–7), are not associated with coronary artery disease and have been linked to lower risk of type 2 diabetes mellitus (7, 10). In contrast, phospholipid concentrations of other TFA subtypes, especially those with 18 carbons and 2 double bonds (i.e., trans-18:2 fatty acids), are strongly associated with inflammation and cardiac death, even more so than concentrations of trans-18:1 fatty acids, the most common TFA in the diet (3–6). Trans-18:2 fatty acids are particularly enigmatic because their circulating concentrations are associated with adverse clinical outcomes at very low concentrations, and these circulating concentrations are also not strongly correlated with dietary intakes of foods high in partially hydrogenated vegetable oils (7). The latter finding suggests that 18:2 TFAs may be influenced by metabolism or come from other industrial sources, such as vegetable oil deodorization or high-temperature frying (8).
To elucidate the biologic processes that might affect circulating phospholipid TFAs, including potentially novel metabolic pathways, we performed a collaborative investigation that pooled data from 7 prospective genome-wide association studies (GWASs) of phospholipid TFA concentrations across more than 8000 participants of European descent, as part of the Cohorts for Heart and Aging Research in Genomic Epidemiology (CHARGE) Consortium (11). To our knowledge, this is the first report of this genetic association. In addition, we tested whether the top associations we observed from our GWAS meta-analyses in European populations replicated among African Americans, Chinese, and Hispanic populations in the discovery cohorts. We also evaluated the extent to which the associations of single-nucleotide polymorphisms (SNPs) with TFA concentrations that did replicate might be affected by differences in circulating concentrations of cis-PUFAs, which may compete with TFA in their metabolism (8).
METHODS
Populations
We performed a prospectively designed, prespecified pooled GWAS analyses including 8013 participants of European descent across 7 cohort studies participating in the CHARGE Consortium Fatty Acids Working Group. The cohorts were the Coronary Artery Risk Development in Young Adults (CARDIA), Cardiovascular Health Study (CHS), Genetics of Lipid-Lowering Drugs and Diet Network (GOLDN), Health Professionals Follow-Up Study (HPFS), Multi-Ethnic Study of Atherosclerosis (MESA), Nurses’ Health Study (NHS), and Women's Genome Health Study (WGHS) (Table 1). All participants provided informed written consent, including consent to participate in genetic studies and all studies received approval from local ethical oversight committees. Details of participating cohorts are presented in the Supplemental Methods.
TABLE 1.
Characteristics of participants of European ancestry in the CHARGE Consortium Fatty Acids Working Group1
| Circulating phospholipid fatty acid concentration, % of total fatty acids2 |
||||||||
| Cohort | n | Age, y | Women, % | trans-16:1n–7 | total trans-18:1 | cis/trans-18:2 | trans/cis-18:2 | trans/trans-18:2 |
| CARDIA | 1507 | 45.8 ± 3.4 | 53.3 | 0.056 ± 0.028 | 1.489 ± 0.632 | 0.048 ± 0.021 | 0.133 ± 0.053 | 0.030 ± 0.021 |
| CHS | 2404 | 75.0 ± 5.1 | 61.6 | 0.190 ± 0.05 | 2.010 ± 0.71 | 0.080 ± 0.02 | 0.130 ± 0.05 | 0.050 ± 0.04 |
| GOLDN | 793 | 48.3 ± 15.9 | 50.4 | 0.073 ± 0.029 | 1.442 ± 0.424 | 0.086 ± 0.023 | 0.097 ± 0.032 | 0.043 ± 0.024 |
| HPFS | 1295 | 64.3 ± 8.6 | 0 | 0.137 ± 0.045 | 1.498 ± 0.639 | 0.116 ± 0.049 | 0.085 ± 0.043 | 0.014 ± 0.018 |
| MESA | 707 | 61.6 ± 10.4 | 61.6 | 0.064 ± 0.027 | 1.493 ± 0.675 | 0.062 ± 0.021 | 0.135 ± 0.059 | 0.038 ± 0.023 |
| NHS | 655 | 59.9 ± 6.5 | 100 | 0.152 ± 0.045 | 1.619 ± 0.711 | 0.176 ± 0.064 | 0.135 ± 0.055 | 0.087 ± 0.059 |
| WGHS | 652 | 54.4 ± 6.5 | 100 | 0.055 ± 0.026 | 1.791 ± 0.527 | 0.089 ± 0.023 | 0.098 ± 0.048 | 0.033 ± 0.012 |
Values are means ± SDs unless otherwise indicated. CARDIA, Coronary Artery Risk Development in Young Adults Study; CHARGE, Cohorts for Heart and Aging Research in Genomic Epidemiology; CHS, Cardiovascular Health Study; GOLDN, Genetics of Lipid-Lowering Drugs and Diet Network; HPFS, Health Professionals Follow-Up Study; MESA, Multi-Ethnic Study of Atherosclerosis; NHS, Nurses’ Health Study; WGHS, Women's Genome Health Study.
Fatty acids were measured in erythrocyte membrane phospholipids (GOLDN, HPFS, and NHS) or plasma phospholipids (CARDIA, CHS, MESA, and WGHS).
Measurements of phospholipid fatty acids
Detailed methods of phospholipid fatty acid measurements are provided in the Supplemental Methods. Four cohorts measured erythrocyte phospholipids, and 3 measured plasma phospholipids; phospholipid fatty acid concentrations in these 2 compartments interchange with each other and are reasonably correlated (12, 13), with correlations of 0.68 for trans-16:1n–9, 0.71–0.84 for trans-18:1 isomers, and 0.56 for trans-18:2 (I King, University of New Mexico, personal communication, October 7, 2014). Concentrations of each TFA were expressed as weight percentage of total fatty acids. We separately evaluated trans-16:1n–7, total trans 18:1 (all available isomers), and trans/cis, cis/trans, and trans/trans-18:2. Trans-18:1 isomers were summed given their very high intercorrelations (in each cohort, the correlation between each individual trans-18:1 isomer and total trans-18:1 was >0.90). In contrast, concentrations of trans-16:1n–7, total trans-18:1, and the 3 trans-18:2 isomers were only modestly intercorrelated (Supplemental Tables 1–7), consistent with heterogeneous dietary sources and/or metabolic determinants.
Genotyping and genome-wide analysis
Genotyping was done in each cohort separately by using high-density SNP marker platforms (CARDIA, GOLDN, and MESA: Affymetrix 6.0; HPFS and NHS: Illumina 550k, 610Q, 660Q, Affymetrix 6.0; CHS Caucasians: Illumina 370; CHS African Americans: Illumina Omni 1M; WGHS: Illumina HumanHap300 Duo+). Samples with call rates <95%–98% (depending on the cohort) at genotyped markers were excluded. Genotypes were imputed to approximately 2.5 million HapMap SNPs by using Beagle (14) (CARDIA), Bimbam (15) (for Caucasians) and Beagle (14) (for African Americans) (CHS), Impute (16) (MESA), or Mach (17) (GOLDN, HPFS, NHS, and WGHS). SNPs for which Hardy-Weinberg equilibrium testing resulted in P < 10−4 to < 10−6 (cohort-specific) were excluded from imputation. Additional details on genotyping and imputation in each cohort are provided in the Supplemental Methods.
Association analysis between genotype and each fatty acid was performed separately within each cohort according to a prespecified analysis plan. All studies conducted linear regression analysis with robust variance estimators by using an additive genetic model (i.e., regression of phenotype on the number of reference alleles) or equivalently the imputed dosage for imputed genotypes. All analyses adjusted for age, sex, site of recruitment where appropriate, and population substructure by using principal component analysis [derived in each cohort from subsets (e.g., 100,000–200,000) of the genotyped SNPs] to account for possible population genetic substructure. We did not consider potential effects of pharmacotherapy on gene-TFA associations, given no known effects of common medications on TFA concentrations.
Meta-analysis
For each SNP and fatty acid, study-specific GWAS results were combined by using inverse-variance weighted meta-analysis (Metal, www.sph.umich.edu/csg/abecasis/metal). SNPs with minor allele frequency (MAF) ≤1% and imputation quality <0.30 were excluded from the meta-analyses, with visual evaluation of Q-Q plots to assess the potential for systematic bias. P values <5 × 10−8 were considered statistically significant in the meta-analysis of GWASs among Caucasians. The proportion of variance in fatty acid concentrations explained by a particular variant allele was calculated from the formula [β2 × 2 × MAF(1 − MAF)]/Var(Y)], where β was the regression coefficient for one copy of the allele; MAF, the minor allele frequency; and Var(Y), the variance of the fatty acid.
Analyses of top SNPs in cohorts of African, Chinese, and Hispanic descent
We investigated whether genome-wide significant SNPs identified in the meta-analysis were associated with phospholipid TFA concentrations in African American (CHS and MESA; n = 1082), Chinese American (MESA; n = 669), and Hispanic American (MESA; n = 657) populations. The SNPs were directly genotyped as part of candidate gene studies in MESA or were available from genome-wide scans in the CHS. We used linear regression and additive models as described above. We used Bonferroni correction to adjust for the number of genome-wide significant SNPs evaluated in these minority populations (α = 0.05/number of SNPs), with the association in each minority population considered a separate hypothesis. In addition, because our top hit with 18:2ct replicated in Chinese and Hispanics but not in African Americans, we explored the full loci of 2 genes in this region [fatty acid desaturase (FADS) 1/2 region: 61566604–61635303, ∼70 kb, in Human Genome build 37; 61323180–61391879 in build 36] in African Americans by using fine mapping (1000 Genomes imputation data) to assess whether absence of top hit association might be due to differences in genetic architecture between races. We used Bonferroni correction to adjust for the 123 independent SNPs in this fine mapping region, estimated by using an eigendecomposition approach (18, 19) (α = 0.05/123 = 0.004).
Influence of other fatty acids
Because one of the identified associations was in a gene strongly associated with circulating concentrations of cis-PUFA (20), which may compete with the metabolism of TFA by means of substrate inhibition, product inhibition, or competitive inhibition (8), we also evaluated the extent to which the observed significant associations between the top SNP in this region and TFA concentrations might change with differences in circulating concentrations of cis-PUFA. We also evaluated potential mediation of the association by total trans-18:1. Each significant SNP-TFA association, previously adjusted for age, sex, study site, and population substructure, was further evaluated in each cohort with and without further adjustment for specific individual circulating fatty acids, including total trans-18:1, individual cis n–3 PUFA (18:3n–3, α-linolenic acid; 20:5n–3, eicosapentaenoic acid; 22:5n–3, docosapentaenoic acid; 22:6n–3, docosahexaenoic acid), and individual cis n–6 PUFA (18:2n–6, linoleic acid; 20:4n–6, arachidonic acid; 22:4n–6, adrenic acid). We qualitatively compared the proportional difference (ratio of β-coefficients) as well as the 95% CI in the pooled SNP-TFA association with and without adjustment for each cis fatty acid.
RESULTS
The characteristics of participants in the cohorts included in the meta-analyses are described in Table 1 as well as in prior reports (20, 21). Briefly, the mean (±SD) age in the participating cohorts ranged from 45.8 ± 3.4 to 75.0 ± 5.1 y. Five TFAs measured in the phospholipid fraction were available for analysis: trans-16:1n–7, total trans-18:1, cis/trans-18:2, trans/cis-18:2, and trans/trans-18:2. As expected, total trans-18:1 was the most abundant TFA, with mean concentration as percentage of total fatty acids ranging from 1.44 ± 0.42 in GOLDN to 1.79 ± 0.53 in WGHS.
Genome-wide associations with circulating TFAs
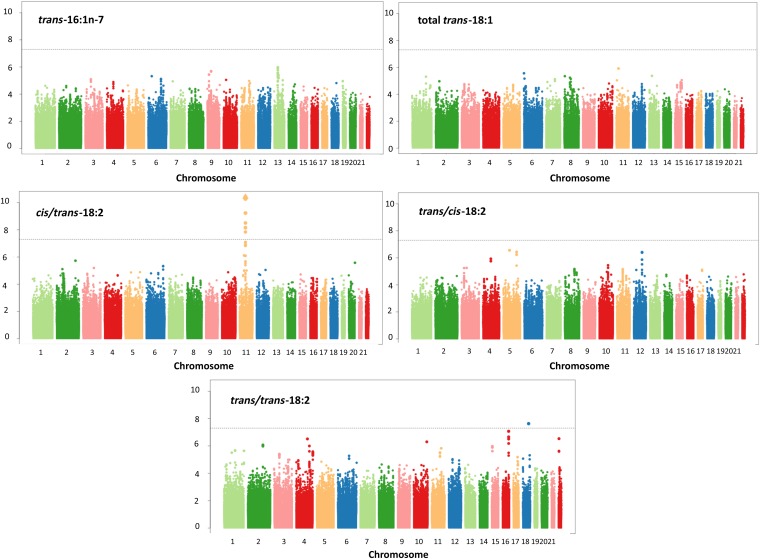
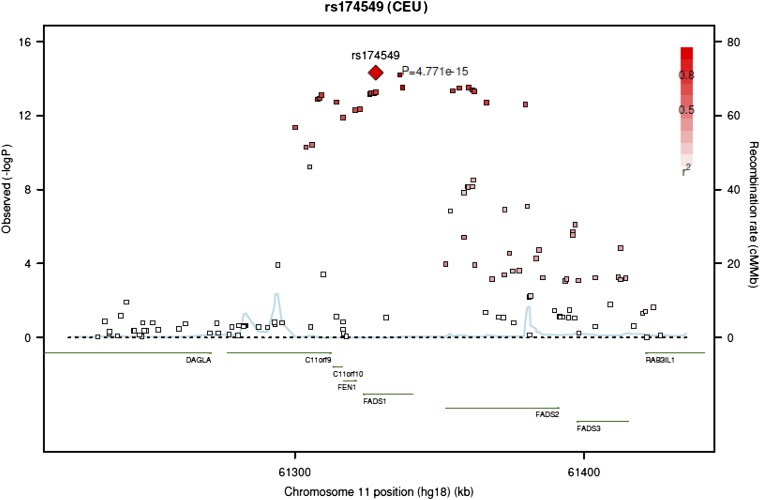
In meta-analyses of the 7 European populations, no significant associations were identified for trans-16:1n–7, trans-18:1, or trans/cis-18:2 (Figure 1, Supplemental Table 8). A total of 31 SNPs on chromosome 11 attained genome-wide significance for cis/trans-18:2. These SNPs were in moderate to high linkage disequilibrium with each other and located in or near the FADS1/2 gene cluster; a top SNP was rs174548 (P = 4.90 × 10−15) (Figure 2, Table 2). Each copy of the G allele of rs174548 was associated with 0.0035 unit (percentage of total fatty acids) higher concentrations of cis/trans-18:2, with consistent direction of association across all 7 cohorts. Across cohorts, the single variant explained an average of 1.1% of the variation in circulating cis/trans-18:2.
FIGURE 1.
Meta-analyses of genome-wide associations for circulating concentrations of trans fatty acids.
FIGURE 2.
Regional association plot for SNP rs174549 and cis/trans-18:2, showing high linkage disequilibrium with 30 other significantly associated SNPs in an area of genes centered on the FADS1/2 cluster. rs174549 was in strong linkage disequilibrium (r2 = 0.95) with rs174548 (P = 4.90 × 10−15), on which we focus in this article for comparability with our prior analysis of circulating n–3 fatty acid concentrations (20). FADS, fatty acid desaturase; SNP, single-nucleotide polymorphism.
TABLE 2.
Top SNPs significantly associated with phospholipid cis/trans-18:2 concentrations1
| Population | n | Chromosome | Genes of interest | SNP | Coded allele (frequency) | P value | β-coefficient ± SE |
| Overall GWAS meta-analysis | |||||||
| European ancestry | 8103 | 11 | FADS1/2 | rs1745492 | A/G (0.29) | 4.77 × 10−15* | 0.0035 ± 0.0004 |
| rs1745482 | G/C (0.29) | 4.90 × 10−15* | 0.0035 ± 0.0004 | ||||
| Replication in non-European ancestry populations | |||||||
| Hispanic Americans | 657 | 11 | FADS1/2 | rs174548 | G/C (0.53) | 1.05 × 10−6** | 0.0053 ± 0.0011 |
| Chinese Americans | 669 | G/C (0.58) | 0.002** | 0.0028 ± 0.0009 | |||
| African Americans | 1082 | G/C (0.21) | 0.34** | 0.0009 ± 0.0010 | |||
| Fine mapping of FADS1/2 region in African Americans | |||||||
| African Americans | 1082 | 11 | FADS1/2 | rs174579 | T/C (0.05) | 4.05 × 10−5*** | 0.0118 ± 0.0029 |
*Evaluated at genome-wide significance (α = 5.0 × 10−8); **a single SNP (rs174548) was evaluated at α = 0.05 for each race-ethnicity; ***evaluated with Bonferroni correction for the number of independent SNPs in this region (α = 0.05/123 = 0.004). FADS, fatty acid desaturase; GWAS, genome-wide association study; SNP, single-nucleotide polymorphism.
A total of 29 other SNPs in this region, in moderate to high linkage disequilibrium with rs174549 and rs174548, were also significantly associated with cis/trans-18:2 concentrations in the meta-analysis, with P values ranging from 6.34 × 10−15 to 1.45 × 10−8 (see Supplemental Table 3). rs174549, which showed the strongest association with cis/trans-18:2 concentrations, was in strong linkage disequilibrium (r2 = 0.95) with rs174548. We focus on rs174548 in this article for comparability with our prior analysis of circulating n–3 fatty acid concentrations (20).
The T allele in SNP rs10469266 on chromosome 18 was nominally associated with higher concentrations of trans/trans-18:2 (P = 2.34 × 10−8). However, this SNP had a very low MAF (1.6%) and was not located in any known gene, and its nominal association was largely driven by association within a single cohort (CARDIA), with missing data in 2 cohorts (Supplemental Table 2). We therefore did not further evaluate this likely spurious finding.
Associations in participants of African, Chinese, and Hispanic descent
We next evaluated the associations of rs174548 with cis/trans-18:2 concentrations in non-European participants from these cohorts (Bonferroni-corrected α = 0.05). This association replicated in Hispanic Americans (P = 1.05 × 10−6) and Chinese Americans (P = 0.002) but not in African Americans (P = 0.34) (Table 2). Because of differences in MAFs for rs174548 on chromosome 11 between races, we used fine mapping to further evaluate this FADS1 and FADS2 region for association with cis/trans-18:2 in African Americans. The meta-analysis demonstrated a top hit of rs174579 in the FADS2 region (P = 4.05 × 10−5), meeting Bonferroni-corrected statistical significance (α = 0.004) (Supplemental Figure 1).
Among European-ancestry participants, 8 other SNPs outside the FADS1/2 region approached genome-wide significance—namely, rs16958148 (P = 8.49 × 10−8), rs16894446 (P = 2.74 × 10−7), rs17099388 (P = 3.63 × 10−7), rs11104877 (P = 3.83 × 10−7), rs7566684 (P = 8.39 × 10−7), rs1399212 (P = 3.07 × 10−7), rs11248534 (P = 4.99 × 10−7), and rs5752209 (P = 2.94 × 10−7) (Supplemental Table 8). In exploratory analyses, none of these 8 SNPs was significantly associated with TFA concentrations in any of the non-European populations (P > 0.05 ÷ 8 = 0.00625 each) (Supplemental Table 9).
Influence of other fatty acids
The association of rs174548 in FADS1/2 with cis/trans-18:2 remained statistically significant after adjusting for circulating concentrations of trans-18:1 and various individual n–3 and n–6 PUFAs except for cis-20:4n–6 (arachidonic acid) (Table 3). After adjustment for 20:4n–6, the association between rs174548 and cis/trans-18:2 was nearly completely attenuated and no longer statistically significant (β-coefficient: 0.0005; 95% CI: −0.00048, 0.00148; P = 0.33). Similar findings were seen among Chinese American and Hispanic American populations in MESA, in whom the association between rs174548 and cis/trans-18:2 was no longer evident after further adjustment for phospholipid concentrations of 20:4n–6 (data not shown). Interestingly, the correlation between phospholipid 20:4n–6 and cis/trans-18:2 concentrations was not high (r = −0.19). In analyses within the CHS cohort in which 20:4n–6 was the dependent variable, rs174548 remained significantly inversely associated with 20:4n–6 concentrations after adjustment for cis/trans 18:2 concentrations (β-coefficient: −1.74; P = 5 × 10−253).
TABLE 3.
Meta-analyses for the relation between rs174548 in the FADS1 gene and cis/trans-18:2 concentrations before and after adjusting for other individual circulating fatty acids also measured in erythrocyte membrane or plasma phospholipids1
| Further adjustment for phospholipid fatty acid concentrations | Association between rs174548 and cis/trans-18:22 | P value |
| None | 0.0035 ± 0.0004 | 4.9 × 10−15 |
| Total trans-18:1 | 0.0033 ± 0.0004 | 1.12 × 10−19 |
| 18:3n–3 (α-linolenic acid) | 0.0031 ± 0.0005 | 4.29 × 10−12 |
| 20:5n–3 (eicosapentaenoic acid) | 0.0027 ± 0.0004 | 5.89 × 10−10 |
| 22:5n–3 (docosapentaenoic acid) | 0.0029 ± 0.0004 | 9.02 × 10−11 |
| 22:6n–3 (docosahexaenoic acid) | 0.0031 ± 0.0004 | 1.48 × 10−12 |
| 18:2n–6 (linoleic acid) | 0.0022 ± 0.0005 | 1.28 × 10−6 |
| 20:4n–6 (arachidonic acid) | 0.0005 ± 0.0005 | 0.33 |
| 22:4n–6 (adrenic acid) | 0.0049 ± 0.0008 | 2.23 × 10−10 |
Findings presented were among Caucasians. Results were similar in the non-European populations: the association among Chinese Americans was no longer significant after adjusting for either linoleic acid or arachidonic acid, the association among Hispanic Americans was no longer significant after adjusting for arachidonic acid, and further adjustment for the other fatty acids in this table did not appreciably alter the association between rs174548 and cis/trans-18:2 among Chinese or Hispanic Americans. FADS, fatty acid desaturase.
Values are β-coefficients ± SEs.
DISCUSSION
This meta-analysis of 7 separate cohorts provides novel evidence that circulating concentrations of certain phospholipid TFAs are under genetic control. Specifically, associations of multiple SNPs on chromosome 11 suggested that concentrations of cis/trans-18:2 are influenced by common genetic variation located in or near the FADS1/2 gene cluster. Findings were consistent in populations of European, Chinese, and Hispanic ancestry, and hits in the FADS2 region were also identified by using fine mapping among African Americans, suggesting that this genetic control is conserved across several races. In contrast, no evidence for significant genetic control was identified for other TFAs, including trans-16:1n–7, trans-18:1, trans/cis-18:2, and trans/trans-18:2.
The FADS1/2 cluster encodes fatty acid desaturase enzymes that regulate endogenous metabolism of cis n–3 and n–6 PUFA (20, 22). The association of this important regulatory region with circulating concentrations of cis/trans-18:2 implies, for the first time to our knowledge, that this region also plays a role in the metabolism of certain TFAs. Our findings further suggest that this association was mediated by circulating concentrations of arachidonic acid, the archetypical n–6 cis-PUFA product of linoleic acid, in that adjustment for circulating concentrations of arachidonic acid largely abolished the association of the FADS1/2 region with cis/trans-18:2. Arachidonic acid, a key modulator of inflammatory pathways and cellular signaling, is a precursor to eicosanoids that regulate production of leukotrienes, prostaglandins, prostacyclin, and thromboxanes and also a precursor to active resolvers of inflammation such as lipoxins and epoxyeicosatrienoic acids (23, 24). Our demonstration of a relation between variation in FADS1/2, cis/trans-18:2, and arachidonic acid suggests novel pathways of shared influences by cis- and trans-PUFA. For example, the effect of genetic variation in the FADS1/2 gene cluster on cis/trans-18:2 could be mediated by effects of the variant on arachidonic acid. We have previously shown that genetic variation in the FADS1/2 cluster explains more than 20% of variation in circulating arachidonic concentrations, with suggestions that the G allele of rs174548 may reflect lower FADS1/2 activity, resulting in lower conversion of linoleic to arachidonic acid (25). Subsequent differences in arachidonic acid concentrations could then alter activities of FADS2 and stearoyl-CoA desaturase, enzymes that could be involved in metabolism (or, less likely, synthesis) of cis/trans-18:2. Arachidonic acid could also alter the rate of incorporation or removal of cis/trans-18:2 from phospholipids, for instance, by influencing concentrations or activities of lysophosphatidyl acyltransferase or phospholipase A2, respectively. The FADS1/2 cluster could also be directly involved in the metabolism of cis/trans-18:2, for example, converting this fatty acid to (unmeasured) trans-arachidonic acid isomers. In this case, the attenuation of this association following adjustment for arachidonic acid could reflect the correlation of this same genetic variant with arachidonic acid, the concentrations of which may be a sensitive indicator of the flow into the pathway and the genetic variation in FADS1/2. Our novel findings support the need for further investigation of the molecular pathways underlying the relationships between these iconic fatty acids and genetic regions, providing insights into novel regulatory pathways of cis and trans fatty acids.
Genetic variation in the FADS1/2 gene cluster is linked to other traits, including blood lipids (total cholesterol, LDL, HDL, and triglycerides), glycemic responses (glucose and β-cell function), and even resting heart rate (26–31). Relationships of the FADS1/2 region with clinical coronary events have been conflicting, with some but not all studies demonstrating associations (32–38). The specific functional variants and mechanistic pathways underlying these associations remain unclear. Our newly identified association with circulating cis/trans-18:2, as well as its interrelationship with arachidonic acid, provides additional impetus to elucidate the functional significance of the identified genetic variants and their role in modulating lipid and metabolic traits.
Although we found variation in the FADS1/2 locus to be related to cis/trans-18:2 concentrations in whites, Chinese, Hispanics, and blacks, the identified variants were not the same in blacks. Recent analyses suggest that whites and blacks have functional differences in FADS1/2 genetic variants related to PUFA metabolism, perhaps driven by evolutionary adaption to facilitate migration from coastal to inland regions on the African continent (39–41). Nonetheless, although the specific evolutionary history and overall functional activity of this gene cluster may differ between whites and blacks, our findings suggest the presence of a conserved general influence of the FADS1/2 locus on cis/trans-18:2 concentrations.
We did not observe significant genome-wide associations with circulating concentrations of other TFAs. Prior investigations from this CHARGE fatty acid consortium demonstrate ample statistical power to detect associations of genetic variants with other fatty acids, including cis n–3 PUFA and saturated and monounsaturated fatty acid products of de novo lipogenesis (20, 21). These findings together suggest that circulating concentrations of most specific TFAs are not subject to appreciable genetic control; this is consistent with the dietary source of these TFAs and the inability of mammals to synthesize trans double bonds. These findings also imply that prior observations of relationships between circulating concentrations of these TFAs with risk factors and disease outcomes (3–6) might not be substantially influenced by endogenous metabolic processes and support the use of these measurements as biomarkers of dietary exposure.
Our investigation has several strengths. Phospholipid fatty acid measurements represent objective circulating biomarkers that also reflect the biologically relevant pool of membrane and tissue phospholipids. By evaluating genetic predictors of circulating TFAs across 7 cohorts and integrating these findings through meta-analysis, we increased statistical power as well as generalizability. Findings observed in participants of European ancestry were evaluated in Chinese, Hispanic, and African American populations, including using fine mapping in the African Americans. We evaluated potential interrelationships of circulating cis-PUFAs with the observed genetic associations, providing novel evidence for a possible biologic interaction between arachidonic acid, cis/trans-18:2, and the FADS1 locus.
Potential limitations should be considered. We investigated genetic associations, and the biological effects of the identified variants on circulating concentrations of these fatty acids are unknown. The top SNPs that were identified from the GWAS of cis/trans-18:2 are in high linkage disequilibrium with many SNPs in the region, and resequencing of the region will be needed to identify the causal variant(s). As with all genetic association studies of complex traits, the proportion of variance explained was small. The TFAs were measured in plasma phospholipids in some cohorts and erythrocyte phospholipids in others. Yet, although mean concentrations of specific fatty acids can differ in these 2 compartments, plasma and membrane phospholipids freely interexchange, their concentrations are well correlated (12, 42), and the direction of association for the top SNPs was consistent in all 7 cohorts. The study included relatively small numbers of non-Caucasians; nonetheless, we confirmed association of the top SNP or another in the same region in each non-Caucasian ethnic group.
In summary, we identified novel evidence for genetic control of circulating phospholipid concentrations of cis/trans-18:2 by the FADS1/2 cluster, and that such regulation may be mediated by concentrations of arachidonic acid, an n–6 cis-PUFA.
Supplementary Material
Acknowledgments
The authors acknowledge the essential role of the Cohorts for Heart and Aging Research in Genomic Epidemiology (CHARGE) Consortium in the development and support of this research. See http://web.chargeconsortium.com for more details. The authors thank the other investigators, the staff, and the participants of each cohort in this study for their invaluable contributions. A full list of principal CHS investigators and institutions can be found at CHS-NHLBI.org. A full list of participating MESA investigators and institutions can be found at http://www.mesa-nhlbi.org.
The authors’ responsibilities were as follows—DM, EKK, DKA, SSR, EBR, FBH, DIC, and M Fornage: designed the research (project conception, development of overall research plan, study oversight); DM, EKK, MRI, DKA, MYT, SSR, QS, MKJ, EBR, FBH, LW, PMR, DIC, M Fornage, LS, IBK, BMP, DSS, and IY-DC: conducted the research (hands-on conduct of the experiments and data collection); DM, DKA, MYT, SSR, QS, MKJ, LW, PMR, DIC, M Fornage, and LS: provided essential reagents or provided essential materials; EKK, COJ, RNL, H Wiener, MRI, AM, QS, H Wu, LW, DIC, AYC, M Foy, and BM: analyzed data or performed statistical analysis; DM, EKK, RNL, LD, and JHYW: wrote the manuscript; and DM, EKK, and QS: had primary responsibility for the final content. DM reports ad hoc honoraria from Bunge and membership on the Unilever North America Scientific Advisory Board. Harvard University has filed a provisional patent application, which has been assigned to Harvard University, listing DM as a coinventor to the US Patent and Trademark Office for use of trans–palmitoleic acid to prevent and treat insulin resistance, type 2 diabetes, and related conditions. BMP serves on the Data Safety Monitoring Board for a clinical trial funded by the device manufacturer (Zoll LifeCor) and on the Steering Committee of the Yale Open Data Access Project funded by Johnson & Johnson. The other authors reported no conflicts of interest.
Footnotes
Abbreviations used: CARDIA, Coronary Artery Risk Development in Young Adults; CHARGE, Cohorts for Heart and Aging Research in Genomic Epidemiology; CHS, Cardiovascular Health Study; FADS, fatty acid desaturase; GOLDN, Genetics of Lipid-Lowering Drugs and Diet Network; GWAS, genome-wide association study; HPFS, Health Professionals Follow-Up Study; MAF, minor allele frequency; MESA, Multi-Ethnic Study of Atherosclerosis; NHS, Nurses’ Health Study; SNP, single-nucleotide polymorphism; TFA, trans fatty acid; WGHS, Women's Genome Health Study.
REFERENCES
- 1.Mozaffarian D, Katan MB, Ascherio A, Stampfer MJ, Willett WC. Trans fatty acids and cardiovascular disease. N Engl J Med 2006;354:1601–13. [DOI] [PubMed]
- 2.Micha R, Mozaffarian D. Trans fatty acids: effects on metabolic syndrome, heart disease and diabetes. Nat Rev Endocrinol 2009;5:335–44. [DOI] [PubMed] [Google Scholar]
- 3.Mozaffarian D, Pischon T, Hankinson SE, Rifai N, Joshipura K, Willett WC, Rimm EB. Dietary intake of trans fatty acids and systemic inflammation in women. Am J Clin Nutr 2004;79:606–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mozaffarian D, Rimm EB, King IB, Lawler RL, McDonald GB, Levy WC. trans Fatty acids and systemic inflammation in heart failure. Am J Clin Nutr 2004;80:1521–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lemaitre RN, King IB, Raghunathan TE, Pearce RM, Weinmann S, Knopp RH, Copass MK, Cobb LA, Siscovick DS. Cell membrane trans-fatty acids and the risk of primary cardiac arrest. Circulation 2002;105:697–701. [DOI] [PubMed] [Google Scholar]
- 6.Lemaitre RN, King IB, Mozaffarian D, Sotoodehnia N, Rea TD, Kuller LH, Tracy RP, Siscovick DS. Plasma phospholipid trans fatty acids, fatal ischemic heart disease, and sudden cardiac death in older adults: the Cardiovascular Health Study. Circulation 2006;114:209–15. [DOI] [PubMed] [Google Scholar]
- 7.Micha R, King IB, Lemaitre RN, Rimm EB, Sacks F, Song X, Siscovick DS, Mozaffarian D. Food sources of individual plasma phospholipid trans fatty acid isomers: the Cardiovascular Health Study. Am J Clin Nutr 2010;91:883–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sebedio JL, Christie WW. The AOCS Lipid Library: metabolism of trans polyunsaturated fatty acids formed during frying [Internet]. 2009 [updated 2009 May 12; cited 2014 Oct 10]. http://lipidlibrary.aocs.org/frying/n-tpufa/index.htm.
- 9.Quehenberger O, Dennis EA. The human plasma lipidome. N Engl J Med 2011;365:1812–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mozaffarian D, Cao H, King IB, Lemaitre RN, Song X, Siscovick DS, Hotamisligil GS. Trans-palmitoleic acid, metabolic risk factors, and new-onset diabetes in U.S. adults: a cohort study. Ann Intern Med 2010;153:790–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Psaty BM, O'Donnell CJ, Gudnason V, Lunetta KL, Folsom AR, Rotter JI, Uitterlinden AG, Harris TB, Witteman JC, Boerwinkle E. Cohorts for Heart and Aging Research in Genomic Epidemiology (CHARGE) Consortium: design of prospective meta-analyses of genome-wide association studies from 5 cohorts. Circ Cardiovasc Genet 2009;2:73–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Haest CWM. Distribution and movement of membrane lipids: red cell membrane transport in health and disease. Berlin: Springer; 2003.
- 13.Patel PS, Sharp SJ, Jansen E, Luben RN, Khaw KT, Wareham NJ, Forouhi NG. Fatty acids measured in plasma and erythrocyte-membrane phospholipids and derived by food-frequency questionnaire and the risk of new-onset type 2 diabetes: a pilot study in the European Prospective Investigation into Cancer and Nutrition (EPIC)-Norfolk cohort. Am J Clin Nutr 2010;92:1214–22. [DOI] [PubMed] [Google Scholar]
- 14.Browning BL, Browning SR. A unified approach to genotype imputation and haplotype-phase inference for large data sets of trios and unrelated individuals. Am J Hum Genet 2009;84:210–23. [DOI] [PMC free article] [PubMed]
- 15.Servin B, Stephens M. Imputation-based analysis of association studies: candidate regions and quantitative traits. PLoS Genet 2007;3:e114. [DOI] [PMC free article] [PubMed]
- 16.Marchini J, Howie B, Myers S, McVean G, Donnelly P. A new multipoint method for genome-wide association studies by imputation of genotypes. Nat Genet 2007;39:906–13. [DOI] [PubMed]
- 17.Aslibekyan S, Kabagambe EK, Irvin MR, Straka RJ, Borecki IB, Tiwari HK, Tsai MY, Hopkins PN, Shen J, Lai CQ, et al. A genome-wide association study of inflammatory biomarker changes in response to fenofibrate treatment in the Genetics of Lipid Lowering Drug and Diet Network. Pharmacogenet Genomics 2012;22:191–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gao X, Becker LC, Becker DM, Starmer JD, Province MA. Avoiding the high Bonferroni penalty in genome-wide association studies. Genet Epidemiol 2010;34:100–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gao X. Multiple testing corrections for imputed SNPs. Genet Epidemiol 2011;35:154–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lemaitre RN, Tanaka T, Tang W, Manichaikul A, Foy M, Kabagambe EK, Nettleton JA, King IB, Weng LC, Bhattacharya S, et al. Genetic loci associated with plasma phospholipid n–3 fatty acids: a meta-analysis of genome-wide association studies from the CHARGE Consortium. PLoS Genet 2011;7:e1002193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wu JH, Lemaitre RN, Manichaikul A, Guan W, Tanaka T, Foy M, Kabagambe EK, Djousse L, Siscovick D, Fretts AM, et al. Genome-wide association study identifies novel loci associated with concentrations of four plasma phospholipid fatty acids in the de novo lipogenesis pathway: results from the CHARGE consortium. Circ Cardiovasc Genet 2013;6:171–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tanaka T, Shen J, Abecasis GR, Kisialiou A, Ordovas JM, Guralnik JM, Singleton A, Bandinelli S, Cherubini A, Arnett D, et al. Genome-wide association study of plasma polyunsaturated fatty acids in the InCHIANTI Study. PLoS Genet 2009;5:e1000338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Serhan CN, Krishnamoorthy S, Recchiuti A, Chiang N. Novel anti-inflammatory–pro-resolving mediators and their receptors. Curr Top Med Chem 2011;11:629–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang G, Kodani S, Hammock BD. Stabilized epoxygenated fatty acids regulate inflammation, pain, angiogenesis and cancer. Prog Lipid Res 2014;53:108–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Guan W, Steffen BT, Lemaitre RN, Wu JH, Tanaka T, Manichaikul A, Foy M, Rich SS, Wang L, Nettleton JA, et al. Genome-wide association study of plasma N6 polyunsaturated fatty acids within the cohorts for heart and aging research in genomic epidemiology consortium. Circ Cardiovasc Genet 2014;7:321–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Aulchenko YS, Ripatti S, Lindqvist I, Boomsma D, Heid IM, Pramstaller PP, Penninx BW, Janssens AC, Wilson JF, Spector T, et al. Loci influencing lipid levels and coronary heart disease risk in 16 European population cohorts. Nat Genet 2009;41:47–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dupuis J, Langenberg C, Prokopenko I, Saxena R, Soranzo N, Jackson AU, Wheeler E, Glazer NL, Bouatia-Naji N, Gloyn AL, et al. New genetic loci implicated in fasting glucose homeostasis and their impact on type 2 diabetes risk. Nat Genet 2010;42:105–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kathiresan S, Willer CJ, Peloso GM, Demissie S, Musunuru K, Schadt EE, Kaplan L, Bennett D, Li Y, Tanaka T, et al. Common variants at 30 loci contribute to polygenic dyslipidemia. Nat Genet 2009;41:56–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Eijgelsheim M, Newton-Cheh C, Sotoodehnia N, de Bakker PI, Muller M, Morrison AC, Smith AV, Isaacs A, Sanna S, Dorr M, et al. Genome-wide association analysis identifies multiple loci related to resting heart rate. Hum Mol Genet 2010;19:3885–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hellstrand S, Sonestedt E, Ericson U, Gullberg B, Wirfalt E, Hedblad B, Orho-Melander M. Intake levels of dietary long-chain PUFAs modify the association between genetic variation in FADS and LDL-C. J Lipid Res 2012;53:1183–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lu Y, Feskens EJ, Dolle ME, Imholz S, Verschuren WM, Muller M, Boer JM. Dietary n–3 and n–6 polyunsaturated fatty acid intake interacts with FADS1 genetic variation to affect total and HDL-cholesterol concentrations in the Doetinchem Cohort Study. Am J Clin Nutr 2010;92:258–65. [DOI] [PubMed] [Google Scholar]
- 32.Martinelli N, Girelli D, Malerba G, Guarini P, Illig T, Trabetti E, Sandri M, Friso S, Pizzolo F, Schaeffer L, et al. FADS genotypes and desaturase activity estimated by the ratio of arachidonic acid to linoleic acid are associated with inflammation and coronary artery disease. Am J Clin Nutr 2008;88:941–9. [DOI] [PubMed] [Google Scholar]
- 33.Teslovich TM, Musunuru K, Smith AV, Edmondson AC, Stylianou IM, Koseki M, Pirruccello JP, Ripatti S, Chasman DI, Willer CJ, et al. Biological, clinical and population relevance of 95 loci for blood lipids. Nature 2010;466:707–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schunkert H, Konig IR, Kathiresan S, Reilly MP, Assimes TL, Holm H, Preuss M, Stewart AF, Barbalic M, Gieger C, et al. Large-scale association analysis identifies 13 new susceptibility loci for coronary artery disease. Nat Genet 2011;43:333–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kwak JH, Paik JK, Kim OY, Jang Y, Lee SH, Ordovas JM, Lee JH. FADS gene polymorphisms in Koreans: association with omega6 polyunsaturated fatty acids in serum phospholipids, lipid peroxides, and coronary artery disease. Atherosclerosis 2011;214:94–100. [DOI] [PubMed] [Google Scholar]
- 36.Aslibekyan S, Jensen MK, Campos H, Linkletter CD, Loucks EB, Ordovas JM, Deka R, Rimm EB, Baylin A. Fatty acid desaturase gene variants, cardiovascular risk factors, and myocardial infarction in the Costa Rica study. Front Genet 2012;3:72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lu Y, Vaarhorst A, Merry AH, Dolle ME, Hovenier R, Imholz S, Schouten LJ, Heijmans BT, Muller M, Slagboom PE, et al. Markers of endogenous desaturase activity and risk of coronary heart disease in the CAREMA cohort study. PLoS ONE 2012;7:e41681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li SW, Lin K, Ma P, Zhang ZL, Zhou YD, Lu SY, Zhou X, Liu SM. FADS gene polymorphisms confer the risk of coronary artery disease in a Chinese Han population through the altered desaturase activities: based on high-resolution melting analysis. PLoS ONE 2013;8:e55869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mathias RA, Fu W, Akey JM, Ainsworth HC, Torgerson DG, Ruczinski I, Sergeant S, Barnes KC, Chilton FH. Adaptive evolution of the FADS gene cluster within Africa. PLoS ONE 2012;7:e44926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sergeant S, Hugenschmidt CE, Rudock ME, Ziegler JT, Ivester P, Ainsworth HC, Vaidya D, Case LD, Langefeld CD, Freedman BI, et al. Differences in arachidonic acid levels and fatty acid desaturase (FADS) gene variants in African Americans and European Americans with diabetes or the metabolic syndrome. Br J Nutr 2012;107:547–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mathias RA, Sergeant S, Ruczinski I, Torgerson DG, Hugenschmidt CE, Kubala M, Vaidya D, Suktitipat B, Ziegler JT, Ivester P, et al. The impact of FADS genetic variants on omega6 polyunsaturated fatty acid metabolism in African Americans. BMC Genet 2011;12:50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.King IB, Lemaitre RN, Kestin M. Effect of a low-fat diet on fatty acid composition in red cells, plasma phospholipids, and cholesterol esters: investigation of a biomarker of total fat intake. Am J Clin Nutr 2006;83:227–36. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.