Abstract
We investigated tRNA methyltransferase activities in crude cell extracts from the thermoacidophilic archaeon Thermoplasma acidophilum. We analyzed the modified nucleosides in native initiator and elongator tRNAMet, predicted the candidate genes for the tRNA methyltransferases on the basis of the tRNAMet and tRNALeu sequences, and characterized Trm5, Trm1 and Trm56 by purifying recombinant proteins. We found that the Ta0997, Ta0931, and Ta0836 genes of T. acidophilum encode Trm1, Trm56 and Trm5, respectively. Initiator tRNAMet from T. acidophilum strain HO-62 contained G+, m1I, and m22G, which were not reported previously in this tRNA, and the m2G26 and m22G26 were formed by Trm1. In the case of elongator tRNAMet, our analysis showed that the previously unidentified G modification at position 26 was a mixture of m2G and m22G, and that they were also generated by Trm1. Furthermore, purified Trm1 and Trm56 could methylate the precursor of elongator tRNAMet, which has an intron at the canonical position. However, the speed of methyl-transfer by Trm56 to the precursor RNA was considerably slower than that to the mature transcript, which suggests that Trm56 acts mainly on the transcript after the intron has been removed. Moreover, cellular arrangements of the tRNA methyltransferases in T. acidophilum are discussed.
Keywords: RNA modification, tRNA methyltransferase, archaea
1. Introduction
Thermoplasma acidophilum is a thermoacidophilic archaeon that grows optimally at 59 °C and pH 1.9 [1]. The characteristic property of this archaeon is that the cells are very irregular in shape due to the lack of a cell wall [2,3]. Despite this, the cytoplasmic membrane tolerates an acidic environment at high temperatures. Consequently, components of the membrane have been studied in detail [4]. Furthermore, lipoylation of proteins [5], biosynthesis of lipids [6], cell surface glycoproteins [7] and a channel protein [8] have been also studied in T. acidophilum. Given that a prokaryotic histone-like DNA binding protein was discovered first from T. acidophilum [9,10], the bacterium has been used as a model system to investigate DNA replication, DNA repair, and transcriptional initiation in archaea [11,12,13,14,15,16]. Furthermore, the energy metabolism of T. acidophilum has been studied in detail because it can grow under an extreme microaerophilic environment [17,18]. Genome sequencing elucidated that the T. acidophilum genome encodes only approximately 1500 open reading frames [19]. Consequently, large protein complexes such as the proteasome and chaperonin are composed of a relatively limited number of protein subunits, and thus they have been studied and compared with their more complicated counterparts from eukaryotes [20,21,22,23].
Although T. acidophilum has been investigated from various viewpoints as described above, there is little knowledge about tRNA modifications, with the exception of some early studies [24,25,26,27] and our more recent work [28]. In 1981 and 1982, the sequences of the initiator ([25] and Figure 1A) and elongator tRNAMet ([24] and Figure 1B) were determined. A novel modification at position 15 (N in Figure 1B), which was named later as archaeosine at position 15 (G+15) [29], and the typical archaeal tRNA modification of 2'-O-methylcytidine at position 56 (Cm56) [30] were reported. In 1991, Edmonds et al. [27] reported that a mixture of tRNAs from T. acidophilum contains N7-methylguanine (m7G). In general, the m7G modification is found at position 46 in class I tRNAs from eubacteria and eukaryotes [31,32,33,34,35]; class I tRNAs are defined as tRNAs with a variable region of regular size. To verify the location of the m7G modification in the tRNA, we analyzed tRNA modifications in T. acidophilum [28]. Unexpectedly, we found that the m7G modification was present at a novel position, nucleotide 49 in class II tRNALeu (Figure 1C); class II tRNAs have a long variable region. Furthermore, we found several distinct modifications in this tRNALeu (Figure 1C): 4-thiouridine at position 9 (s4U9) [36,37,38,39], G+13 [29,40], and 5-carbamoylmethyluridine at position 34 (ncm5U34) [41,42]. The modifications s4U9 and ncm5U34 have been not found in other archaeal tRNAs and G+13 has not been reported in any other tRNA [31,32]. In the current study, we tested the tRNA methyltransferase activities in crude cell extract from T. acidophilum, analyzed the methylated nucleosides in purified tRNAs and characterized the tRNA methyltransferases by expressing recombinant proteins in Escherichia coli.
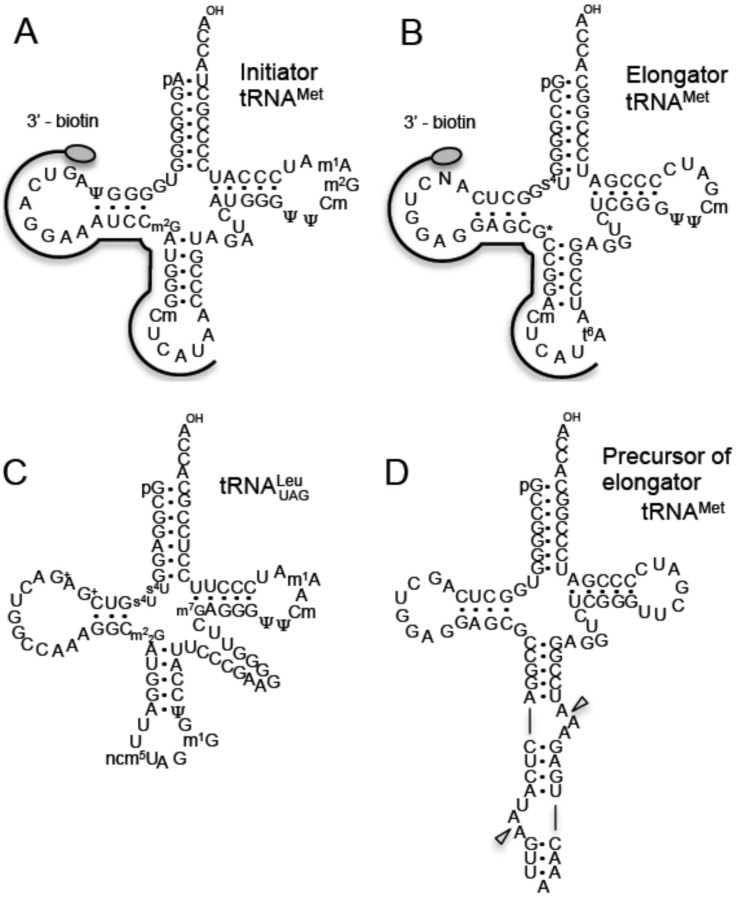
Figure 1.
tRNAs and precursor tRNA from T. acidophilum. The sequences of initiator tRNAMet (A), elongator tRNAMet (B) and tRNALeuUAG (C) are depicted by cloverleaf structures. The regions to which the 3’-biotinylated DNA probes hybridize are illustrated. The abbreviations are as follows: pseudouridine, ψ; N2-methylguanosine, m2G; 2'-O-methylcytidine, Cm; 1-methyladenosine, m1A; 4-thiouridine, s4U; unknown modification, N; unidentified G modification, G*; N6-thereonylcarbamoyladenosine, t6A; archaeosine, G+; N2,N2-dimethylguanosine, m22G; 5-carbamoylmethyluridine, ncm5U; 1-methylguanosine, m1G; 7-methylguanosine, m7G. The precursor of elongator tRNAMet contains an intron at the canonical site (D). Triangles show the cleavage sites of tRNA splicing endonuclease.
2. Results and Discussion
2.1. Methylated Nucleosides in T. acidophilum tRNAs
Figure 1A–C show the cloverleaf structures of initiator tRNAMet, elongator tRNAMet and tRNALeuUAG, respectively. In the current study, we utilized these tRNA sequences as a basis to characterize tRNA methyltransferases in T. acidophilum. As shown in Table 1, various methylated nucleosides are present in these tRNAs. We predicted the candidate genes for the enzymes responsible for the modifications by BLAST searches [28] and analyzed the corresponding recombinant proteins. For example, given that the G26 in elongator tRNAMet was reported to be a modified G (G*) ([24] and Figure 1B) we investigated whether recombinant Trm1 methylated G26 in the elongator tRNAMet transcript. The results of the current study are summarized in Table 1. Furthermore, it should be mentioned that the sequence of the initiator tRNAMet that is encoded in the genome of T. acidophilum strain HO-62 differs from that reported in the earlier study ([25] and Figure 1A): the nucleotide at position 57 is A instead of G in strain HO-62. In archaeal tRNAs, A57 is often modified to 1-methylinosine (m1I57) via 1-methyladenosine (m1A57) by TrmI and deamination [43,44,45]. Some possible explanations for this discrepancy are elaborated in the Discussion section. Moreover, in the case of the precursor of elongator tRNAMet, a standard intron is inserted at the canonical position between nucleotides 38 and 39 (Figure 1D). Therefore, it is possible that the presence of the intron might affect the methylations by tRNA methyltransferases.
Table 1.
Methylated nucleosides in T. acidophilum tRNAs.
| Modification and Position | Candidate Enzyme and Gene | Results from This Study | |
|---|---|---|---|
| Intiator tRNAMet | |||
| m2G26 | Trm1 | Ta0997 | m22G26 and m2G26 formed by Trm1. |
| Cm32 | TrmJ? | Ta1010m? | 1 We did not analyze this gene product. |
| Cm56 | Trm56 | Ta0931 | Cm56 formed by Trm56. |
| m2G57 | ? | 2 This position in the tRNA gene is A57. | |
| m1A58 | TrmI | Ta0852? | 3 We could not detect m1A58 formation activity in the recombinant protein. |
| Elongator tRNAMet | |||
| G*26 | Trm1 | Ta0997 | m22G26 and m2G26 formed by Trm1. |
| Cm32 | TrmJ? | Ta1010m? | 1 We did not analyze this gene product. |
| Cm56 | Trm56 | Ta0931 | Cm56 formed by Trm56. |
| tRNALeuUAG | |||
| m22G26 | Trm1 | Ta0997 | m22G26 formed by Trm1. |
| m1G37 | Trm5 | Ta0836 | m1G37 formed by Trm5. |
| m7G49 | ? | Ta0679? +α? | We could not obtain soluble recombinant protein. |
| Cm56 | Trm56 | Ta0931 | Cm56 formed by Trm56. |
| m1A58 | TrmI | Ta0852? | 3 We could not detect m1A58 formation activity in the recombinant protein. |
1 During the course of the current study, it has been reported that Sulfolobus acidocaldarius TrmJ generates the Cm32 modification in tRNA. The candidate gene in T. acidophilum was predicted by a BLAST search; 2 The sequence of the initiator tRNAMet that is encoded in the genome of the T. acidophilum strain HO-62 differs from that reported in the earlier study [25] (see Figure 1A and Figure 6A): the nucleotide at position 57 is A instead of G in strain HO-62. Furthermore, the tRNA gene in the genome of strain HO-62 contains additional nucleotides, A20b and C22; 3 The Ta0852 gene product was expressed in Escherichia coli as a soluble protein. However, we could not detect any ability to form m1A58; ?, the corresponding enzyme is unknown.
2.2. Transfer RNA Methyltransferase Activities in the Crude Cell Extract
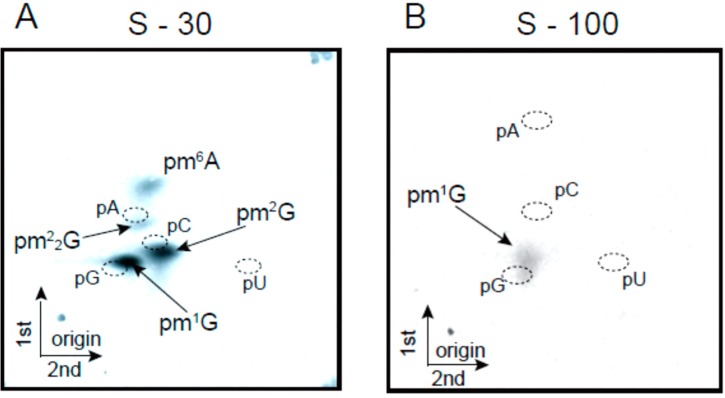
Next, we tested tRNA methyltransferase activities in crude extract from T. acidophilum cells. The supernatant fraction from centrifugation at 30,000× g (S-30) was prepared and then the tRNALeuUAG transcript was subjected to methylation by the S-30 extract with 14C-S-adenosyl-l-methionine (AdoMet) as the methyl group donor. The methylated tRNA was digested completely with nuclease P1 and then the resultant 14C-methylated nucleotides were analyzed by two-dimensional thin-layer chromatography (2D-TLC). As shown in Figure 2A, four 14C-methylated nucleotides (pm1G, pm2G, pm22G and pm6A) could be detected. On the basis of the sequence of tRNALeuUAG (Figure 1C) and the candidate enzymes (Table 1), pm1G, pm2G and pm22G, and pm6A were expected to be derived from the activities of Trm5, Trm1, and TrmI, respectively: pm6A could be converted from pm1A non-enzymatically [46]. However, unexpectedly, pCm and pm7G were not detected. In general, the formation of pCm by Trm56 is one of the most common tRNA methyltransferase activities found in crude extract from archaeal cells. For example, it was reported that Trm56 activity in relation to the formation of pCm56 is clearly detected in cell extract from Pyrococcus furiosus [47]. Analysis of the T. acidophilum proteome revealed that various proteins form several large (more than 300 kDa) protein complexes and that some protein complexes might interact with the membrane [48]. Consequently, Trm56 and an unknown tRNA (m7G49) methyltransferase might be included in protein complexes and precipitated by centrifugation at 30,000× g. We tested several buffer conditions such as variations in pH, components, detergents, and salt concentrations (data not shown). However, to date, we have not detected enzyme activities responsible for the formation of pCm and pm7G in crude extract from T. acidophilum cells, though there are other possibilities; while we used aluminum oxide to prepare the extract in this experiment, other methods for preparation of cell extract should be tested. In addition, tRNA methyltransferases in the crude extract have different affinities for 14C-AdoMet. The concentration of 14C-AdoMet in the experiment was 19.5 µM. In this case, tRNA methyltransferases, which have relatively high affinity for AdoMet, might preferentially consume the 14C-AdoMet. When the supernatant fraction from centrifugation at 100,000× g (S-100) was used as the cell extract instead of the S-30 fraction, the findings were even more marked: only the formation of pm1G was detectable (Figure 2B). Given that tRNA methyltransferases have a general affinity for RNA, the enzymes often bind to ribosomes and are precipitated by centrifugation at 100,000× g. In fact, the majority of TrmI from Thermus thermophilus [49] is precipitated by centrifugation at 100,000× g [50]. However, our findings with the extract from T. acidophilum are unprecedented. In the current study, we characterized tRNA methyltransferases by analyzing purified recombinant proteins. However, these enzymes might interact with other proteins and form large protein complexes in living cells.
Figure 2.
Activities of tRNA methyltransferases in the crude cell extracts. The activities of tRNA methyltransferases in the S-30 (A) and S-100 (B) fractions were analyzed by 2D-TLC. 14C-methylated nucleotides were monitored by autoradiography. The solvent systems were as follows: first dimension, isobutyric acid, ammonia, water, 66/1/33 v/v/v; second dimension, isopropyl alcohol, HCl, water, 70/15/15 v/v/v.
2.3. Modified Nucleosides in Purified tRNAs
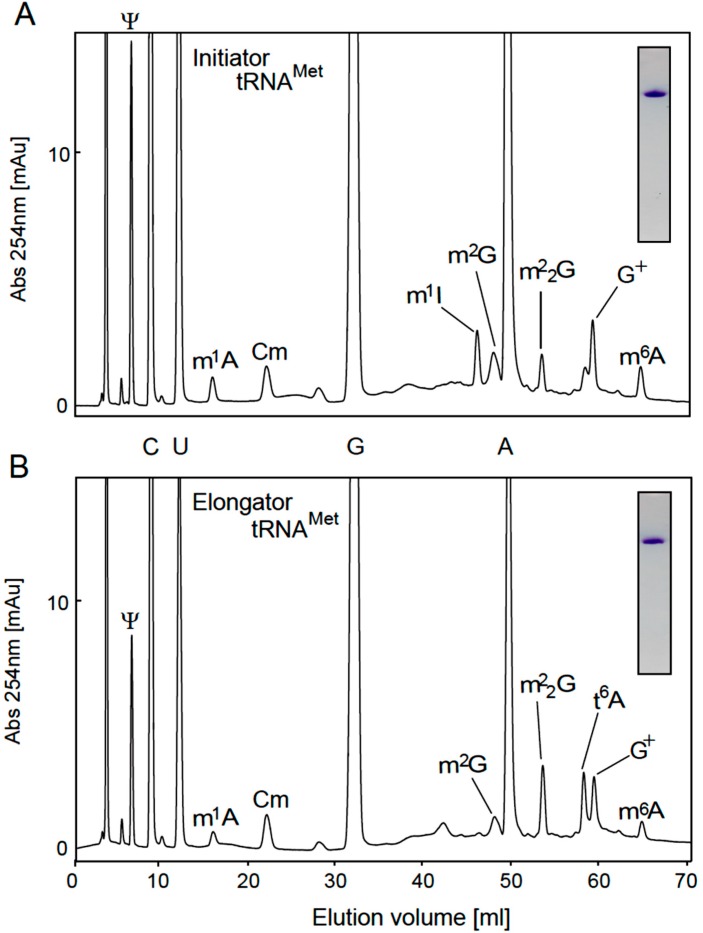
To analyze the modified nucleosides in tRNAs, we purified initiator and elongator tRNAMet by the solid-phase DNA probe method [51], in which tetraalkylammonium salts were used in the hybridization buffer. Tetraalkylammonium salts destabilize the tRNA structure and enhance the formation of DNA-RNA hybrids [51], and thus we were able recently to purify single tRNA species from thermophiles such as Aquifex aeolicus [52], T. thermophilus [50,53], Aerophyrum pernix [54], and T. acidophilum [28] using this approach. In the case of initiator tRNAMet, because the sequence from G15 to U36 was distinct, we designed the DNA probe to this region. In the case of elongator tRNAMet, T. acidophilum two species: the sequence of one was previously determined as shown in Figure 1B [24]. These two tRNAMet species differ in three nucleotides in the D-loop and anticodon arm. Therefore, we designed the DNA probe as shown in Figure 1B. As shown in the insets in Figure 3A,B, initiator and elongator tRNAMet were purified successfully: the 14C-Met-charging activities were checked by the S-100 fraction (data not shown). The purified tRNAs were digested with snake venom phosphodiesterase, RNase A and bacterial alkaline phosphatase, and then the resultant nucleosides were analyzed by HPLC using a C18 column (Figure 3A,B). Snake venom phosphodiesterase can cleave the phosphodiester bond adjacent to 2'-O-methylated nucleotide. As shown in Figure 3A, m1A, Cm, m1I, m2G, m22G and m6A were detected as methylated nucleosides in the initiator tRNAMet sample. The presence of m1A, Cm, m2G and m6A is consistent with the published RNA sequence (Figure 1A). However, the presence of m1I suggests that A57 in this tRNA is modified to m1A57 by TrmI [44,45] and that deamination then generates m1I57 as in the case of Haloferax volcanii [43]. Furthermore, a peak for m22G was detected, which suggests that some proportion of the initiator tRNAMet contains m22G26. Moreover, G+ was clearly detected, which suggests that G15 is modified to G+15 in initiator tRNAMet. In the elongator tRNAMet sample, m1A, Cm, m2G, m22G and m6A were detected. The modifications m2G and m22G have not been reported at any position in elongator tRNAMet, although the uncharacterized G26 modification represents a possible location (G*26 in Figure 1B and Table 1). Consequently, the modified G26 was expected to be a mixture of m2G26 and m22G26. Furthermore, N6-threonylcarbamoyladenosine (t6A) was also detected, which is consistent with the RNA sequence (Figure 1B). After these pilot experiments, we established expression systems in E. coli for the candidate genes shown in Table 1. As mentioned in Table 1, we could not obtain soluble protein from the Ta0679 gene. Furthermore, the Ta0852 gene product did not show TrmI activity. Consequently, these gene products were not analyzed further in the current study.
Figure 3.
Modified nucleosides in native initiator and elongator tRNAMet. Native initiator and elongator tRNAMet were purified by the solid-phase DNA probe method (insets). The hybridization regions of the probes are illustrated in Figure 1A,B. The modified nucleosides in the initiator (A) and elongator (B) tRNAMet were analyzed by reverse phase column chromatography.
2.4. Formation of m1G37 in tRNALeuUAG Transcript by Ta0836 Gene Product
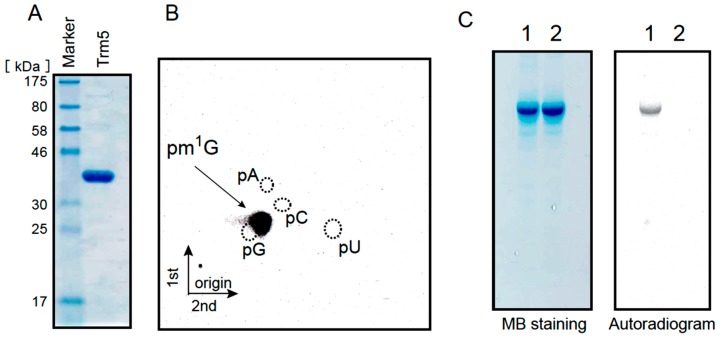
The Ta0836 gene product was expressed in E. coli and purified as shown in Figure 4A. The expected amino acid sequence of the Ta0836 gene product shares a high degree of homology (82%) with that of the identified archaeal Trm5 (Mj0883 of Methanocaldococcus jannaschii) [55,56]. When the purified protein was incubated with the tRNALeuUAG transcript and 14C-AdoMet, the 14C-methyl group was clearly incorporated into the transcript (see Figure 4C lane 1). Analysis of the modified nucleotides by 2D-TLC revealed that the 14C-methylated nucleotide was pm1G (Figure 4B). Furthermore, when the G37 in tRNALeuUAG transcript was replaced by A, no methyl group incorporation was observed (Figure 4C lane 2), which indicates that the methylation site is G37. From these results, we concluded that the Ta0836 gene product is the T. acidophilum Trm5 protein. Given that the m1G modification was previously found only at position 37 in tRNALeuUAG in T. acidophilum ([28] and Figure 1C), the m1G modification activity in the S-100 is probably derived from Trm5.
Figure 4.
The Ta0836 gene product is T. acidophilum Trm5. (A) 10 µg of the purified Ta0836 gene product was analyzed by 15% SDS-PAGE. The gel was stained with Coomassie Brilliant Blue; (B) The tRNALeuUAG transcript was methylated by the Ta0836 gene product and then the generated methylated nucleotide was analyzed by 2D-TLC; (C) The methyl group acceptance activities of the wild-type tRNALeuUAG transcript (lane 1) and the mutant tRNALeuUAG transcript (lane 2), in which G37 was replaced by A, were investigated. The transcripts were individually incubated with the Ta0836 gene product and 14C-AdoMet, and then separated by 10% PAGE (7 M urea). The gel was stained with methylene blue (left panel) and the autoradiogram of the same gel was taken (right panel).
2.5. Ta0997 Gene Product Is a Single Site-Specific Trm1
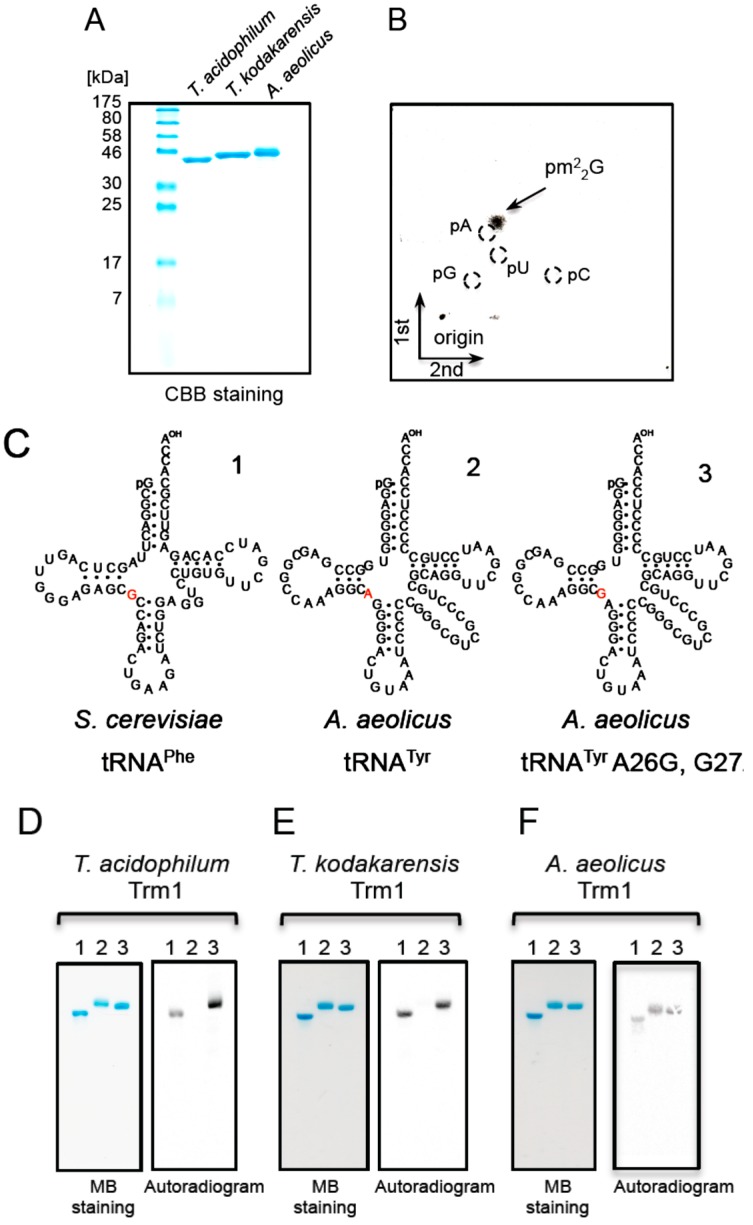
The m22G modification was observed only at position 26 in tRNALeuUAG ([28] and Figure 1C). Trm1 transfers two methyl groups to the 2-amino group in the target guanine and m2G is formed as an intermediate [57,58]. Consequently, the m2G and m22G modifications in tRNALeuUAG transcript by the S-30 (Figure 2A) were expected to be derived from Trm1 activity. Trm1 enzymes can be divided into two types on the basis of their specificity for the target guanosine(s). One is a single-site-specific Trm1, which modifies only G26 and is found in eukaryotes and archaea [57,58,59,60]. The second is a multi-site-specific Trm1, which modifies both G26 and G27 and is found in the hyperthermophilic eubacterium, A. aeolicus [52]. In addition to the Ta0997 gene product (Figure 5A lane 1), we prepared two types of Trm1 enzyme from Thermococcus kodakarensis (Figure 5A lane 2) and A. aeolicus (Figure 5A lane 3) as controls. The Ta0997 gene product methylated the tRNALeuUAG transcript (data not shown) and 14C-nucleotide analysis revealed that the modified nucleotide was pm22G (Figure 5B). These results showed that the Ta0997 gene product is the T. acidophilum Trm1 protein. To distinguish the site specificity, yeast tRNAPhe and A. aeolicus tRNATyr transcripts were prepared (Figure 5C). These tRNA transcripts were used previously to assess the site specificity of A. aeolicus Trm1 [52]. Yeast tRNAPhe contains the sequence G26C27, whereas A. aeolicus tRNATyr contains the sequence A26G27. In addition, the A. aeolicus tRNATyr A26G, G27A mutant transcript has the sequence G26A27. As shown in Figure 5D, T. acidophilum Trm1 methylated yeast tRNAPhe and A. aeolicus tRNATyr A26G, G27A transcripts, which contain G26. In contrast, T. acidophilum Trm1 did not methylate the wild-type tRNATyr transcript (Figure 5D center), which contains A26. Thus, these results demonstrate that T. acidophilum Trm1 is a single-site-specific Trm1, which methylates only G26. Similar to T. acidophilum Trm1, T. kodakarensis Trm1 methylated only the yeast tRNAPhe and A. aeolicus tRNATyr A26G, G27A transcripts (Figure 5E). In contrast, A. aeolicus Trm1 methylated all the transcripts (Figure 5F), which indicates that A. aeolicus Trm1 has multi-site specificity.
Figure 5.
T. acidophilum Trm1 (the Ta0997 gene product) is a single-site-specific enzyme. (A) The T. acidophilum Ta0997 gene product (left), T. kodakaraensis Trm1 (center) and A. aeolicus Trm1 (right) (2 µg each) were analyzed by 15% SDS-PAGE. The gel was stained with Coomassie Brilliant Blue; (B) The tRNALeuUAG transcript was methylated by the Ta0997 gene product and then the generated methylated nucleotide was analyzed by 2D-TLC; (C) The sequences of S. cerevisiae tRNAPhe, A. aeolicus tRNATyr and A. aeolicus tRNATyr A26G, G27A are depicted by cloverleaf structures. These tRNA transcripts were previously used to determine the site specificity of A. aeolicus Trm1, which is a multi-site-specific Trm1. The nucleotides at position 26 in the tRNA transcripts are colored red. The transcript numbers (1, 2, and 3) correspond to the lane numbers in panels (D–F). (D) The tRNA transcripts (0.1 A260 units each) were incubated with the Ta0997 gene product (T. acidophilum Trm1) and 14C-AdoMet at 50 °C for 5 min, separated by 10% PAGE (7 M urea), and then the gel was analyzed by autoradiography. The left and right panels show the gel stained with methylene blue and its autoradiogram, respectively. T. kodakaraensis (E) and A. aeolicus (F) Trm1 proteins were analyzed by the same method.
2.6. T. acidophilum Trm1 Can Modify G26 in Initiator tRNAMet Transcript to m22G26 via m2G26
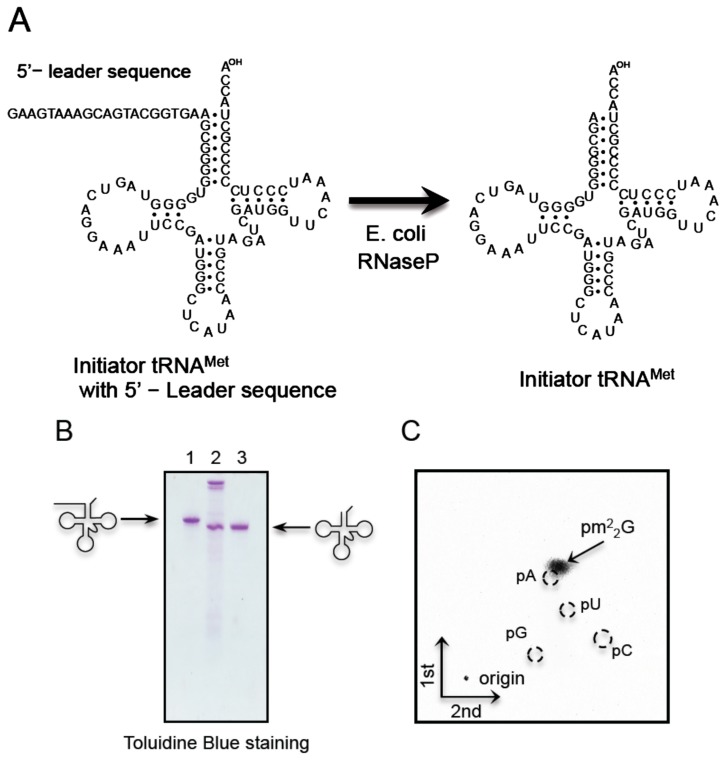
The G26 modification in initiator tRNAMet was reported to be m2G ([25] and Table 1). Consequently, we investigated whether T. acidophilum Trm1 can modify G26 in the initiator tRNAMet transcript to m22G. Given that the 5'-end of initiator tRNAMet is an A, T7 RNA polymerase did not synthesize the transcript efficiently. Consequently, the initiator tRNAMet transcript was synthesized with a 5'-leader sequence (Figure 6A) by T7 RNA polymerase (Figure 6B, lane 1) and then the 5'-leader sequence was removed with E. coli RNase P ([61] and Figure 6B, lane 2). The initiator tRNAMet transcript was then purified by 10% polyacrylamide gel electrophoresis in the presence of 7 M urea (PAGE (7 M urea)), (Figure 6B lane 3). Trm1 from T. acidophilum efficiently methylated the initiator tRNAMet transcript (data not shown), and the methylated nucleotide was pm22G (Figure 6C). Furthermore, the modified nucleoside analysis showed that native initiator tRNAMet contained m22G (Figure 3A). Taking these results together, we conclude that initiator tRNAMet from T. acidophilum contained the m22G26 modification in addition to m2G26 and that TrmI activity was responsible for these modifications.
Figure 6.
T. acidophilum Trm1 methylates the initiator tRNAMet transcript. (A) Given that the 5'-end of the T. acidophilum initiator tRNAMet is A, the transcript is not synthesized efficiently by T7 RNA polymerase. Consequently, the initiator tRNAMet transcript was synthesized with a 5'-leader sequence by T7 RNA polymerase, purified by 10% PAGE (7 M urea), and then the 5'-leader sequence was removed with E. coli RNase P; (B) The cleavage of the 5'-leader sequence was monitored by 10% PAGE (7 M urea). The gel was stained with toluidine blue. The lanes are as follows: left, initiator tRNAMet with the 5'-leader sequence; center, the reaction mixture; right, purified mature initiator tRNAMet transcript. The M1 RNA in RNase P can be observed at the top of the gel in the center lane; (C) The methylated nucleotide was analyzed by 2D-TLC.
2.7. T. acidophilum Trm1 Can Methylate the Precursor of Elongator tRNAMet with an Intron
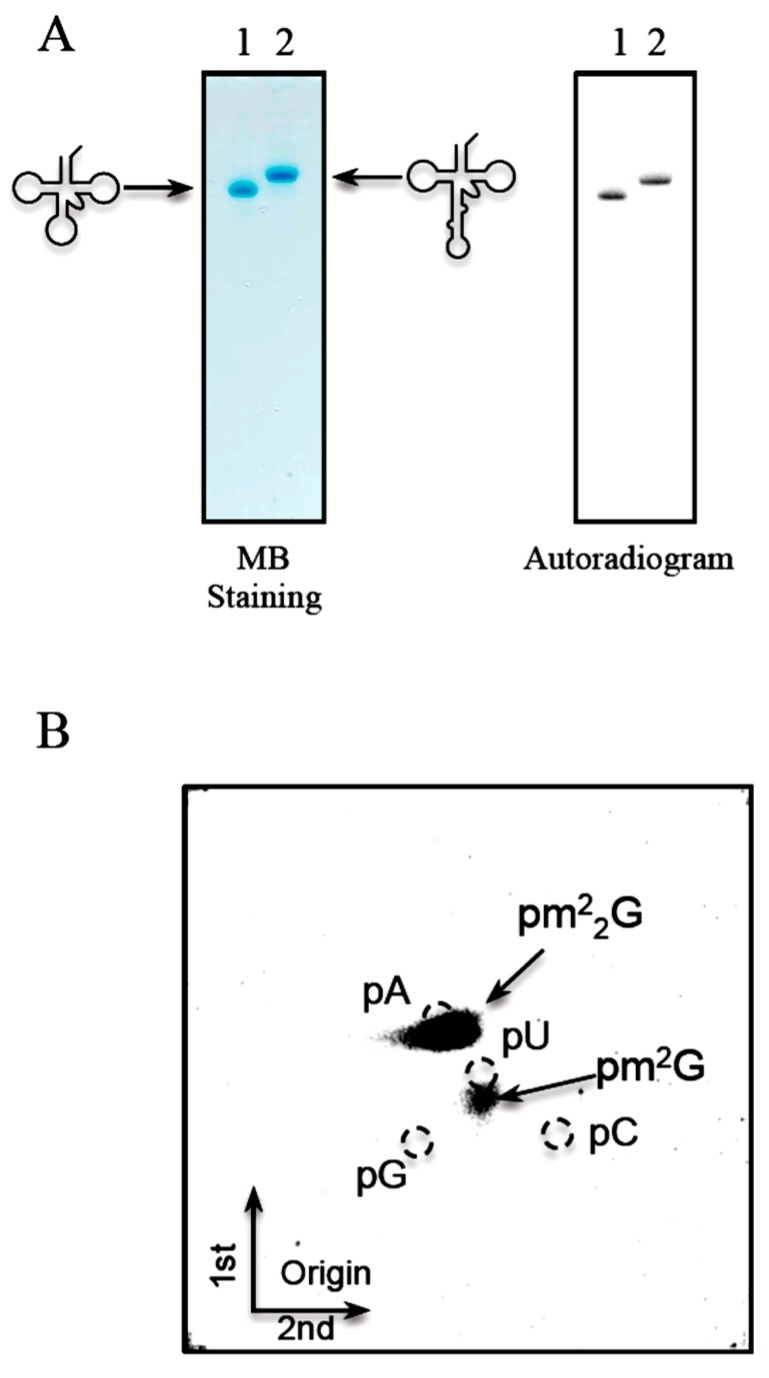
The G26 modification of elongator tRNAMet was uncharacterized ([24], and Table 1). Furthermore, the precursor of elongator tRNAMet contains an intron at the canonical position between nucleotides 38 and 39 (Figure 1D). To verify whether Trm1 could methylate the elongator tRNAMet transcript and its precursor, we analyzed the methyl group acceptance activities of these RNAs (Figure 7A). Both the elongator tRNAMet transcript and its precursor were methylated efficiently by T. acidophilum Trm1. The analysis of modified nucleotides by 2D-TLC revealed that methylated nucleotides were pm2G and pm22G (Figure 7B). The analysis of modified nucleosides revealed that native elongator tRNAMet contained m2G and m22G (Figure 3B). Furthermore, m2G and m22G modifications were not reported in the published sequence of elongator tRNAMet although the modification at G26 was uncharacterized [24]. Taking these results together, we conclude that the uncharacterized G26 modification in the elongator tRNAMet is a mixture of m2G and m22G, which is formed by Trm1.
Figure 7.
T. acidophilum Trm1 can methylate both the elongator tRNAMet transcript and its precursor, which contains an intron. The mature transcript of elongator tRNAMet and its precursor with an intron (Figure 1D) were prepared. (A) These transcripts were incubated with T. acidophilum Trm1 and 14C-AdoMet at 50 °C for 5 min, and then separated by 10% PAGE (7 M urea) (left panel). The gel was stained with methylene blue. The samples are as follows: lane 1, mature transcript of elongator tRNAMet; lane 2, precursor of elongator tRNAMet with an intron. The right panel shows an autoradiogram of the same gel; (B) The 14C-methylated nucleosides in the mature transcript of elongator tRNAMet were analyzed by 2D-TLC.
2.8. Ta0931 Gene Product Is Trm56
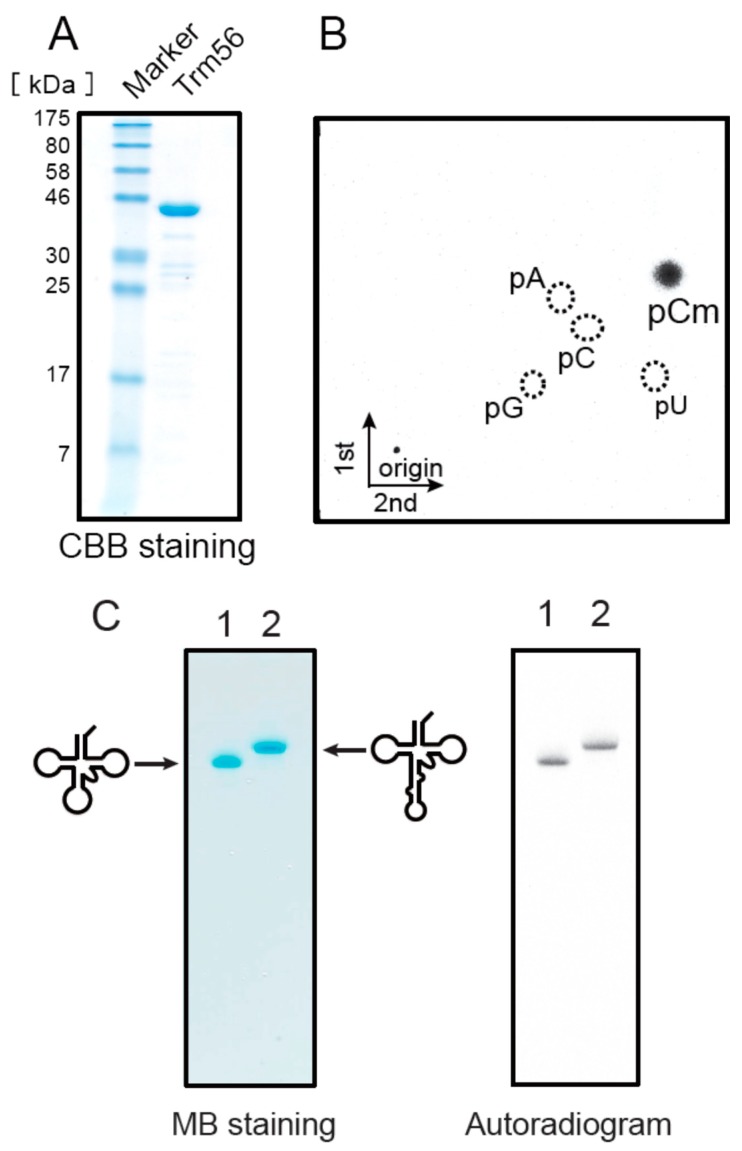
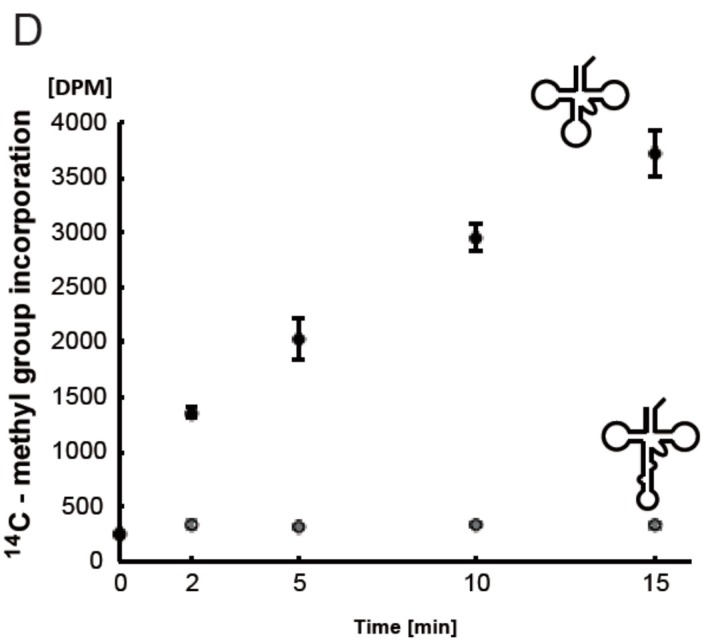
To analyze whether the Ta0931 gene product was Trm56, the recombinant protein was purified as shown in Figure 8A. The purified Ta0931 methylated the tRNALeuUAG transcript (data not shown) and the methylated nucleotide was identified as pCm (Figure 8B). The Cm modification is only found at position 56 in native tRNALeuUAG (Figure 1C). These results showed that the Ta0931 gene product is Trm56. Neither the S-30 nor the S-100 fraction contained activity that was responsible for introducing the Cm modification into the tRNALeuUAG transcript (Figure 2); however, the genome does encode Trm56. This discrepancy is addressed in the Discussion section. Finally, we investigated the influence of the presence of intron on Trm56 activity. Trm56 methylated both the elongator tRNAMet transcript and its precursor (Figure 8C). However, the methyl group acceptance activity of the precursor was considerably lower than that of the mature transcript (Figure 8D). It should be mentioned that the incubation in Figure 8C was performed for 12 h to show the methylation of the precursor tRNA. These results suggest that the methylation by Trm56 occurs mainly after the removal of the intron. Although the mechanism by which Trm56 recognizes tRNA has not been reported thus far, the results of the current study suggest two possibilities. The first is that the presence of the intron results in steric hindrance that prevents Trm56 binding to the substrate tRNA. The second is that Trm56 directly recognizes the anticodon loop in the tRNA. To clarify the mechanism, further study is required.
Figure 8.
The Ta0931 gene product (Trm56) can methylate both the elongator tRNAMet transcript and its precursor with an intron. (A) An aliquot of 4 µg of the Ta0931 gene product was analyzed by 15% SDS-PAGE. The gel was stained with Coomassie Brilliant Blue; (B) The Ta0931 gene product and tRNALeuUAG transcript were incubated at 50 °C for 1 h, and then the 14C-methylated nucleotide was analyzed by 2D-TLC; (C) The incorporation of methyl groups into the mature transcript and the precursor of elongator tRNAMet were investigated. The samples were as follows: lane 1, mature transcript of elongator tRNAMet; lane 2, precursor of elongator tRNAMet. The gel was stained with methylene blue (left panel) and the autoradiogram of the same gel was taken (right panel); (D) The incorporation of methyl groups into the mature transcript and the precursor of elongator tRNAMet were measured by the filter assay. Closed and open circles show the incorporation of 14C-methyl groups into the mature transcript and precursor of elongator tRNAMet, respectively.
3. Discussion
In the current study, we investigated tRNA methyltransferase activities in crude extract from T. acidophilum cells, analyzed the modified nucleosides in native initiator and elongator tRNAMet, and characterized three tRNA methyltransferases (Trm5, Trm1 and Trm56) by purified recombinant proteins. We utilized the sequences of three tRNAs from T. acidophilum (initiator tRNAMet, elongator tRNAMet and tRNALeuUAG), which were reported previously in earlier [24,25] and our recent [28] studies, as a basis to predict the candidate genes for the enzymes responsible for the modifications. As summarized in Table 1, our experiments revealed that the genes Ta0997, Ta0931, and Ta0836 encode Trm1, Trm56 and Trm5, respectively, from T. acidophilum. In archaeal tRNA modifications, there are some reports that different enzymes modify the same modification at the same position in tRNA: for example, Cm56 modification is formed by two systems, Trm56 or C/D sRNP [30]. Therefore, to understand tRNA modification systems precisely, the construction of gene disruptant mutant strains is desirable. However, there is no gene disruption method for T. acidophilum. Therefore, in this study, we could not utilize this approach. Consequently, the other gene product except for Trm1, Trm5 and Trm56 may bring the same modification(s) at the same position(s) in tRNA. Unexpectedly, we could not detect Trm56 activity in the crude cell extracts (S-30 and S-100 fractions). Analysis of the T. acidophilum proteome revealed that various proteins form several large (more than 300 kDa) protein complexes and that some of these protein complexes appear to interact with the membrane [48]. Therefore, T. acidophilum Trm56 might be part of a large complex with other proteins in living cells. Similar to the Trm56 activity, we could not detect tRNA (m7G49) methyltransferase activity in the crude cell extract. To clarify the intracellular localization of these enzymes, further study will be required. Although the recognition of tRNA by Trm56 might be affected by the presence of other proteins in the putative complex, the purified enzyme at least can act on both the mature elongator tRNAMet transcript and its precursor, which contains an intron at the canonical site. The methyl group was transferred to the mature transcript much more rapidly than to the precursor tRNA with the intron. This result suggests that the methylation by Trm56 occurs mainly after the removal of the intron. The activity of Trm5 was clearly detected in the S-100 fraction and the purified recombinant Trm5 methylated the G37 nucleotide in the tRNALeuUAG transcript. Among the tRNA methyltransferases from T. acidophilum, only Trm5 seemed to act as a free enzyme, i.e., was not included in a protein complex. It has been reported that Trm5 recognizes the tertiary interaction between the D- and T-arms [55,62]. Consequently, Trm5 might act mainly during and/or after the three-dimensional core of the tRNA has been reinforced structurally by the introduction of other modifications.
The sequence of the initiator tRNAMet that is encoded in the genome of T. acidophilum strain HO-62 [3] differs from that reported in the earlier study [25]. The sequences of the two initiator tRNAMet differ in the D-arm and at position 57: A20b and C22 are inserted in strain HO-62 and this strain also contains A57 instead of G57. These differences might be derived from the different origins of the strains: the strain HO-62 was isolated from Hakone, Japan [3]. There is another possibility as follows. The m1I 57 modification in archaeal tRNA and the initiator tRNAMet gene from T. acidophilum were not reported in 1982. The authors used the Kuchino’s post-labelling method for tRNA sequencing [25], in which tRNA is partially cleaved by formamide and then the nucleotide at the 5'-end of each fragment is analyzed by 2D-TLC [46,63]. However, the mobility of pm1I on 2D-TLC closely resembles that of pm2G [64]. Furthermore, in general, formamide cleavage of tRNAs from thermophiles is very difficult due to their structural rigidity. Therefore, it might be difficult technically to distinguish the pm1I and pm2G on 2D-TLC. In the current study, we detected the m1A (m6A) formation into the tRNALeuUAG transcript by the S-30 fraction. However we could not identify the trmI gene, which encodes archaeal tRNA (m1A57/m1A58) methyltransferase. Consequently, we could not verify whether T. acidophilum TrmI can methylate the A57 in the initiator tRNAMet, although our analysis of modified nucleosides revealed that m1I is contained in the initiator tRNAMet. To determine the position of m1I modification, the RNA sequence of initiator tRNAMet is required. We are now checking the plasmid vector for TrmI expression and the other gene products. From the results of the current study, we were able to add the following information with respect to initiator tRNAMet from T. acidophilum: (1) initiator tRNAMet from T. acidophilum strain HO-62 contains the modifications G+, m1I, and m22G; (2) the m22G26 modification exists in addition to m2G26; (3) m2G26 and m22G26 are formed by Trm1.
In the current study, we have demonstrated that archaeal Trm1 can methylate the tRNALeuUAG transcript, which has a long variable region. As far as we know, this is the first time that archaeal Trm1 has been shown to act on class II tRNAs. In the case of class I tRNAs, archaeal Trm1 was reported to recognize the D-stem and the size of the variable region [58]. Therefore, archaeal Trm1 might be able to recognize the large variable region in the class II tRNAs. Furthermore, we showed that Trm1 efficiently methylated both the mature elongator tRNAMet transcript and the precursor with an intron. These results agree well with those of a previous study [58], namely, that archaeal Trm1 does not recognize the anticodon loop. Moreover, from the results of the current study, we were able to add the following information in relation to elongator tRNAMet from T. acidophilum: the unidentified G modification at position 26 is a mixture of m2G and m22G, which is formed by Trm1.
During the course of the current study, it has been reported that Sulfolobus acidocaldarius TrmJ is responsible for the Cm32 modification in tRNA [65]. The Cm32 modifications in initiator and elongator tRNAMet are probably formed by this new enzyme as shown in Table 1.
4. Experimental Section
4.1. Materials
[Methyl-14C]-AdoMet (2.14 GBq/mmol) was purchased from PerkinElmer (Tokyo, Japan). Q-Sepharose Fast Flow, HiTrap Q-Sepharose, HiTrap SP-Sepharose, HiTrap Heparin-Sepharose and HiLoad 16/600 Superdex 200 prep grade were bought from GE Healthcare Japan (Tokyo, Japan). DNA oligomers were synthesized by Invitrogen Japan (Tokyo, Japan). Other chemical reagents were of analytical grade.
4.2. Strain, Medium, and Culture
The culture source of T. acidophilum HO-62 was a gift from Prof. Akihiko Yamagishi (Tokyo University of Pharmacy and Life Sciences, Hachioji, Japan). The culture was performed at 56 °C under microaerophilic conditions as described previously [3]. The culture medium contained following components (in grams per liter): yeast extract, 1.0; Casamino acid, 1.0; (NH4)2SO4, 1.3; NaCl, 0.2; KH2PO4, 0.3; MgSO4, 0.25; CaCl2, 0.05; pH was adjusted to 1.8 with H2SO4. Cells in late log-phase were used for the experiments.
4.3. Preparation of S-30 and S-100 Fractions, and Detection of tRNA Methyltransferase Activities
Wet cells (0.3 g) were suspended in 2 mL of buffer A (50 mM Tris-HCl (pH 7.6), 5 mM MgCl2, 6 mM 2-mercaptoethanol, and 50 mM KCl). The cells were ground in a mortar with 0.15 g aluminum oxide and then the suspension was centrifuged at 8000× g for 20 min. The supernatant fraction was centrifuged further at 30,000× g for 2 h. The resultant supernatant fraction was used as the S-30 fraction. The S-100 fraction was the supernatant fraction by that was obtained after centrifugation at 100,000× g for 2 h. Transfer RNA methyltransferase activities in the S-30 and S-100 fractions were analyzed as follows: 30 µg of protein from the S-30 or S-100 fraction, 0.2 A260 units tRNALeuUAG transcript and 0.78 nmol [methyl-14C]-AdoMet were incubated in 40 µL of buffer A at 55 °C for 1 h. The RNA was extracted with phenol-chloroform and then recovered by ethanol precipitation. The RNA pellet was dissolved in 3 µL of 50 mM sodium acetate (pH 5.0), and digested with 2.5 units of nuclease P1 (Wako Pure Chemicals, Osaka, Japan). The sample was separated using 2D-TLC as described previously [64]. The 14C-methylated nucleotides were monitored with a BAS 2000 Bio-imaging Analyzer (Fuji Photo Film, Tokyo, Japan).
4.4. Purification of Initiator and Elongator tRNAMet by the Solid-Phase DNA Probe Method
Initiator and elongator tRNAMet were purified by the solid-phase DNA probe method as described in our previous reports [51,52]. The sequences of the 3'-biotinylated DNA oligomers were as follows: for initiator tRNAMet, 5'-ATG AGC CCA TTG GGA TTT CCT GA-biotin 3'; for elongator tRNAMet, 5'-ATG AGT CCG GTG CTC CTC CAG-biotin 3'. The complementary regions are illustrated in Figure 1A,B. The isolated tRNAs were further purified by 10% PAGE (7 M urea).
4.5. Nucleoside Analysis
Nucleoside analysis was performed as described in our previous reports [50,53]. The standard marker of G+ was kindly provided by Prof. Takashi Yokogawa (Gifu University, Gifu, Japan).
4.6. Selection of Candidate Genes
We searched for the candidate genes in the T. acidophilum HO-62 genome by performing a BLAST search using the amino acid sequences of H. volcanii Trm1 and Trm56, and M. jannaschii Trm5. The identification of the other candidate genes was reported in our previous paper [28].
4.7. Cloning of the Candidate Genes, and Expression of Gene Products
4.7.1. Cloning of Ta0836 (trm5)
The Ta0836 gene was amplified by the polymerase chain reaction (PCR) from genomic DNA from T. acidophilum using the following primers: Ta Trm5F, 5'-GAG ATA TAC ATA TGC CTC CAA AGA AGT TCG TTA-3'. Ta Trm5R, 5'-CTC GAA TTC GGA TCC TTA TTA CTA TGC CTT CTT AAG GGT CAT CG-3'.
4.7.2. Cloning of Ta0997 (trm1)
The Ta0997 gene was amplified by PCR from the genomic DNA using the following primers: Ta Trm1F, 5'-GAG ATA TAC ATA TGA TAG TGA GGG AGG GTT CAG-3'; Ta Trm1R, 5'-CTC GAA TTC GGA TCC TTA TTA TGC CGG CGA TCG TCT GTG CA-3'.
4.7.3. Cloning of Ta0931 (trm56)
The Ta0931 gene was amplified by PCR from the genomic DNA using the following primers: Ta Trm56F, 5'-GGA GAT ATA TAC ATA TGA TAA CCG TAC TGC GGA TAA ATC AC-3'; Ta Trm56R, 5'-CTC GAA TTC GGA TCC TTA TTA GCG TAT TTC ATC GAT ATC CAT ACC-3'.
4.7.4. Expression of Gene Products
The underlined regions show restriction enzyme sites (Nde I and Bam HI). The PCR products were individually inserted individually into the multiple cloning linker of expression vector pET-30a (Novagen, Cambridge, MA, USA). The gene products were expressed in the E. coli BL21 (DE3) Rosetta 2 strain (Novagen) in accordance with the manufacturer’s instructions.
4.8. Purification of Recombinant Proteins
4.8.1. Purification of Trm5
Briefly, Trm5 was purified by heat treatment at 50 °C for 30 min, followed by successive rounds of column chromatography through HiTrap Q-Sepharose, HiTrap Heparin-Sepharose, and Toyopearl CM-650M (Tosoh, Tokyo, Japan). The final eluted sample was dialyzed against buffer B (50 mM Tris-HCl (pH 7.6), 50 mM KCl, 6 mM 2-mercaptoethanol and 5% glycerol) and concentrated with a Vivaspin 15R centrifugal filter device (Sartorius Japan, Tokyo, Japan). Glycerol was added to the sample to a final concentration of 50% v/v and the sample stored at −30 °C.
4.8.2. Purification of Trm1
Trm1 was purified as described previously [52].
4.8.3. Purification of Trm56
Briefly, Trm56 was purified by heat treatment at 50 °C for 30 min, followed by successive rounds of column chromatography through HiTrap Q-Sepharose, HiTrap Heparin-Sepharose, and HiLoad 16/600 Superdex 200 pg. The final eluted protein was dialyzed against buffer B, and concentrated with a Vivaspin 15R centrifugal filter device. Glycerol was added to the purified protein to a final concentration of 50% v/v and the samples stored at −30 °C.
4.9. Measurement of tRNA Methyltransferase Activities
The transcripts were prepared by using T7 RNA polymerase and purified by Q-Sepharose column chromatography and 10% PAGE (7 M urea). The standard assay for the purified enzymes was to measure the incorporation of 14C-methyl groups from [methyl-14C]-AdoMet into the appropriate tRNA transcript. For the reaction, 66 nM enzyme, 4.25 µM transcript, and 17.3 µM [methyl-14C]-AdoMet were incubated in 40 µL of buffer A at 50 °C for 5 min. An aliquot (35 µL) of the reaction was then used for the filter assay. To visualize the methyl-transfer reaction, we used 10% PAGE (7 M urea) and autoradiography. Briefly, tRNA (0.1 A260 units) was incubated with 66 nM enzyme and 17.3 µM [methyl-14C]-AdoMet at 50 °C for 5 min in 40 µL of buffer A, and then loaded onto a 10% polyacrylamide gel that contained 7 M urea. The gel was stained with methylene blue or toluidine blue, and then dried. The incorporation of 14C-methyl groups into the tRNA was monitored with a Typhoon FLA 7000 laser scanner (GE Healthcare). The 2D-TLC was performed as follows. An aliquot of 1 µg of purified protein, 0.2 A260 units of tRNA transcript and 0.69 nmol of [methyl-14C]-AdoMet were incubated in 40 µL of buffer A at 55°C for 1 h. The RNA was extracted with phenol-chloroform and then recovered by ethanol precipitation. The RNA pellet was digested with 1.5 units of nuclease P1. The sample was separated using 2D-TLC as described previously [64]. The 14C-methylated nucleotides were monitored with a Typhoon FLA 7000 laser scanner (GE Healthcare).
4.10. Preparation of E. coli RNase P and Removal of 5'-Leader Sequence
The plasmid vectors for the C5 protein and M1 RNA of E. coli RNase P were a gift from Prof. Takashi Yokogawa (Gifu University). The C5 protein was purified as described in the reference [61]. The M1 RNA was synthesized with T7 RNA polymerase. Active RNase P was generated by mixing C5 protein and M1 RNA in accordance with the method described in the reference [61]. The 5'-leader sequence of E. coli tRNAMetf was used. The 5'-leader sequence was cleaved by RNase P in accordance with the reference [61].
Acknowledgments
We thank Takashi Yokogawa of Gifu University for the gifts of plasmid vectors for E. coli RNase P and the standard marker of G+, and Akira Hirata, Takuma Nakayama, Shiho Omori, Ayano Yamauchi, and Miyu Higashiguchi of Ehime University for providing materials (synthesized E. coli RNase P, expression vector for T. kodakaraensis Trm1, and purified A. aeolicus Trm1) and technical support for the culture of T. acidophilum. This work was partly supported by a Research Fellowship for Young Scientists (20-4827 to Chie Tomikawa) and a Grant-in-aid for Science Research (B) (23350081 to Hiroyuki Hori) from the Japan Society for the Promotion of Science.
Author Contributions
Takuya Kawamura, Ryou Anraku, Chie Tomikawa and Hiroyuki Hori conceived and designed the experiments; Takuya Kawamuram, Ryou Anraku, Takahiro Hasegawa, and Chie Tomikawa performed the experiments; all the authors analyzed the data; Takuya Kawamura, Chie Tomikawa and Hiroyuki Hori wrote the manuscript.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Darland G., Brock T.D., Samsonoff W., Conti S.F. A thermophilic, acidophilic mycoplasma isolated from a coal refuse pile. Science. 1970;170:1416–1418. doi: 10.1126/science.170.3965.1416. [DOI] [PubMed] [Google Scholar]
- 2.Mayberry-Carson K.J., Roth I.L., Harris J.L., Smith P.F. Scanning electron microscopy of Thermoplasma acidophilum. J. Bactriol. 1974;120:1472–1475. doi: 10.1128/jb.120.3.1472-1475.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yasuda M., Oyaizu H., Yamagishi A., Oshima T. Morphological variation of new Thermoplasma acidophilum isolates from Japanese hot springs. Appl. Environ. Microbiol. 1995;61:3482–3485. doi: 10.1128/aem.61.9.3482-3485.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shimada H., Nemoto N., Shida Y., Oshima T., Yamagishi A. Complete polar lipid composition of Thermoplasma acidophilum HO-62 determined by high-performance liquid chromatography with evaporative light-scattering detection. J. Bacteriol. 2002;184:556–563. doi: 10.1128/JB.184.2.556-563.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Posner M.G., Upadhyay A., Bagby S., Hough D.W., Danson M.J. A unique lipoylation system in the Archaea lipoylation in Thermoplamsa acidophilum requires two proteins. FEBS J. 2009;276:4012–4022. doi: 10.1111/j.1742-4658.2009.07110.x. [DOI] [PubMed] [Google Scholar]
- 6.Xu Q., Eguchi T., Mathews I.I., Rife C.L., Chiu H.J., Farr C.L., Feuerhelm J., Jaroszewski L., Klock H.E., Knuth M.W., et al. Insights into substrate specificity of geranylgeranyl reductases revealed by the structure of digeranylgeranylglycerophospholipid reductase, an essential enzyme in the biosynthesis of archaeal membrane lipids. J. Mol. Biol. 2010;404:403–417. doi: 10.1016/j.jmb.2010.09.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vinogradov E., Deschatelets L., Lamoureux M., Patel G.B., Tremblay T.L., Robotham A., Goneau M.F., Cummings-Lorbetskie C., Watson D.C., Brisson J.R., et al. Cell surface glycoproteins from Thermoplasma acidophilum are modified with an N-linked glycan containing 6-C-sulfofucose. Glycobiology. 2012;22:1256–1267. doi: 10.1093/glycob/cws094. [DOI] [PubMed] [Google Scholar]
- 8.Kloda A., Martinac B. Mechanosensitive channel of Thermoplasma, the cell wall-less archaea: Cloning and molecular characterization. Cell Biochem. Biophys. 2001;34:321–347. doi: 10.1385/CBB:34:3:321. [DOI] [PubMed] [Google Scholar]
- 9.Stein D.B., Sercy D.G. Physiologically important stabilization of DNA by a prokaryotic histone-like protein. Science. 1978;202:219–221. doi: 10.1126/science.694528. [DOI] [PubMed] [Google Scholar]
- 10.DeLange R.J. A histone-like protein (HTa) from Thermoplasma acidophilum. J. Biol. Chem. 1981;256:900–904. [PubMed] [Google Scholar]
- 11.Haugland G.T., Shin J.H., Birkeland N.K., Kelman Z. Stimulation of MCM helicase activity by a Cdc6 protein in the archaeon Thermoplasma acidophilum. Nucleic Acids Res. 2006;34:6337–6344. doi: 10.1093/nar/gkl864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ogino H., Ishino S., Mayanagi K., Haugland G.T., Birkeland N.K., Yamagishi A., Ishino Y. The GINS complex from the thermophilic archaeon, Thermoplasma acidophilum may function as a homotetramer in DNA replication. Extremophiles. 2011;15:529–539. doi: 10.1007/s00792-011-0383-2. [DOI] [PubMed] [Google Scholar]
- 13.Roth H.M., Tessmer I., van Houten B., Kisker C. Bax1 is a novel endonuclease: Implications for archaeal nucleotide excision repair. J. Biol. Chem. 2009;284:32272–32278. doi: 10.1074/jbc.M109.055913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Moen M.N., Knævelsrud I., Haugland G.T., Grøsvik K., Birkeland N.K., Klungland A., Bjelland S. Uracil-DNA glycosylase of Thermoplasma acidophilum directs long-patch base excision repair, which is promoted by deoxynucleoside triphosphates and ATP/ADP, into short-patch repair. J. Bacteriol. 2011;193:4495–4508. doi: 10.1128/JB.00233-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kuper J., Wolski S.C., Michels G., Kisker C. Functional and structural studies of the nucleotide excision repair helicase XPD suggest a polarity for DNA translocation. EMBO J. 2012;31:494–502. doi: 10.1038/emboj.2011.374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Buechner C.N., Heil K., Michels G., Carell T., Kisker C., Tessmer I. Strand-specific recognition of DNA damages by XPD provides insights into nucleotide excision repair substrate versatility. J. Biol. Chem. 2014;289:3613–3624. doi: 10.1074/jbc.M113.523001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sun N., Pan C., Nickell S., Mann M., Baumeister W., Nagy I. Quantitative proteome and transcriptome analysis of the archaeon Thermoplasma acidophilum cultured under aerobic and anaerobic conditions. J. Proteome Res. 2010;9:4839–4850. doi: 10.1021/pr100567u. [DOI] [PubMed] [Google Scholar]
- 18.Kim S.M., Paek K.H., Lee S.B. Characterization of NADP+-specific L-rhamnose dehydrogenase from the thermoacidophilic Archaeon Thermoplasma acidophilum. Extremophiles. 2012;16:447–454. doi: 10.1007/s00792-012-0444-1. [DOI] [PubMed] [Google Scholar]
- 19.Ruepp A., Graml W., Santos-Martinez M.L., Koretke K.K., Volker C., Mewes H.W., Frishman D., Stocker S., Lupas A.N., Baumeister W. The genome sequence of the thermoacidophilic scavenger Thermoplasma acidophilum. Nature. 2000;407:508–813. doi: 10.1038/35035069. [DOI] [PubMed] [Google Scholar]
- 20.Brandstetter H., Kim J.-S., Groll M., Huber R. Crystal structure of the tricorn protease reveals a protein disassembly line. Nature. 2001;414:466–470. doi: 10.1038/35106609. [DOI] [PubMed] [Google Scholar]
- 21.Steinbacher S., Ditzel L. Review: Nucleotide binding to the Thermoplasma thermosome: Implications for the functional cycle of group II chaperonins. J. Struct. Biol. 2001;135:147–156. doi: 10.1006/jsbi.2001.4382. [DOI] [PubMed] [Google Scholar]
- 22.Noi K., Hirai H., Hongo K., Mizobata T., Kawata Y. Implications for the functional cycle of group II chaperonins from Thermoplasma acidophilum. Biochemistry. 2009;48:9405–9415. doi: 10.1021/bi900959c. [DOI] [PubMed] [Google Scholar]
- 23.Ruschak A.M., Religa T.L., Breuer S., Witt S., Kay L.E. The proteasome antechamber maintains substrates in an unfolded state. Nature. 2010;467:868–873. doi: 10.1038/nature09444. [DOI] [PubMed] [Google Scholar]
- 24.Kilpatrick M.W., Walker R.T. The nucleotide sequence of the tRNAMMet from the archaebacterium Thermoplasma acidophilum. Nucleic Acids Res. 1981;9:4387–4390. doi: 10.1093/nar/9.17.4387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kuchino Y., Ihara M., Yabusaki Y., Nishimura S. Initiator tRNAs from archaebacteria show common unique sequence characteristics. Nature. 1982;298:684–685. doi: 10.1038/298684a0. [DOI] [PubMed] [Google Scholar]
- 26.Walker R.T. Mycoplasma evolution: A review of the use of ribosomal and transfer RNA nucleotide sequences in the determination of phylogenetic relationships. Yale J. Biol. Med. 1983;56:367–372. [PMC free article] [PubMed] [Google Scholar]
- 27.Edmonds C.G., Crain P.F., Gupta R., Hashizume T., Hocart C.H., Kowalak J.A., Pomerantz S.C., Stetter K.O., McCloskey J.A. Posttranscriptional modification of tRNA in thermophilic archaea (Archaebacteria) J. Bacteriol. 1991;173:3138–3148. doi: 10.1128/jb.173.10.3138-3148.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tomikawa C., Ohira T., Inoue Y., Kawamura T., Yamagishi A., Suzuki T., Hori H. Distinct tRNA modifications in the thermo-acidophilic archaeon, Thermoplasma acidophilum. FEBS Lett. 2013;587:3575–3580. doi: 10.1016/j.febslet.2013.09.021. [DOI] [PubMed] [Google Scholar]
- 29.Gregson J.M., Crain P.F., Edmonds C.G., Gupta R., Hashizume T., Phillipson D.W., McCloskey J.A. Structure of the archaeal transfer RNA nucleoside G*-15 (2-amino-4,7-dihydro-4-oxo-7-beta-d-ribofuranosyl-1H-pyrrolo[2,3-d]pyrimidine-5-carboximidamide (archaeosine)) J. Biol. Chem. 1993;268:10076–10086. [PubMed] [Google Scholar]
- 30.Renalier M.H., Joseph N., Gaspin C., Thebault P., Mougin A. The Cm56 tRNA modification in archaea is catalyzed either by a specific 2'-O-methylase, or a C/D sRNP. RNA. 2005;11:1051–1063. doi: 10.1261/rna.2110805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jühling F., Mörl M., Hartmann R.K., Sprinzl M., Stadler P.F., Pütz J. tRNAdb 2009: Compilation of tRNA sequences and tRNA genes. Nucleic Acids Res. 2009;37:D159–D162. doi: 10.1093/nar/gkn772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Machnicka M.A., Milanowska K., Osman Oglou O., Purta E., Kurkowska M., Olchowik A., Januszewski W., Kalinowski S., Dunin-Horkawicz S., Rother K.M., et al. MODOMICS: A database of RNA modification pathways—2013 update. Nucleic Acids Res. 2013;41:D262–D267. doi: 10.1093/nar/gks1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.De Bie L.G., Roovers M., Oudjama Y., Wattiez R., Tricot C., Stalon V., Droogmans L., Bujnicki J.M. The yggH gene of Escherichia coli encodes a tRNA (m7G46) methyltransferase. J. Bacteriol. 2003;185:3238–3243. doi: 10.1128/JB.185.10.3238-3243.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Okamoto H., Watanabe K., Ikeuchi Y., Suzuki T., Endo Y., Hori H. Substrate tRNA recognition mechanism of tRNA (m7G46) methyltransferase from Aquifex aeolicus. J. Biol. Chem. 2004;279:49151–49159. doi: 10.1074/jbc.M408209200. [DOI] [PubMed] [Google Scholar]
- 35.Alexandrov A., Martzen M.R., Phizicky E.M. Two proteins that form a complex are required for 7-methylguanosine modification of yeast tRNA. RNA. 2002;8:1253–1266. doi: 10.1017/S1355838202024019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kambampati R., Lauhon C.T. Evidence for the transfer of sulfane sulfur from IscS to ThiI during the in vitro biosynthesis of 4-thiouridine in Escherichia coli tRNA. J. Biol. Chem. 2000;275:10727–10730. doi: 10.1074/jbc.275.15.10727. [DOI] [PubMed] [Google Scholar]
- 37.Lauhon C.T., Kambampati R. The iscS gene in Escherichia coli is required for the biosynthesis of 4-thiouridine, thiamin, and NAD. J. Biol. Chem. 2000;275:20096–20103. doi: 10.1074/jbc.M002680200. [DOI] [PubMed] [Google Scholar]
- 38.Liu Y., Zhu X., Nakamura A., Orlando R., Söll D., Whitman W.B. Biosynthesis of 4-thiouridine in tRNA in the methanogenic archaeon Methanococcus maripaludis. J. Biol. Chem. 2012;287:36683–36692. doi: 10.1074/jbc.M112.405688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Miranda H.V., Nembhard N., Su D., Hepowit N., Krause D.J., Pritz J.R., Phillips C., Söll D., Maupin-Furlow J.A. E1- and ubiquitin-like proteins provide a direct link between protein conjugation and sulfur transfer in archaea. Proc. Natl. Acad. Sci. USA. 2011;108:4417–4422. doi: 10.1073/pnas.1018151108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Phillips G., Swairjo M.A., Gaston K.W., Bailly M., Limbach P.A., Iwata-Reuyl D., de Crécy-Lagard V. Diversity of archaeosine synthesis in Crenarchaeota. ACS Chem. Biol. 2012;7:300–305. doi: 10.1021/cb200361w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Keith G., Desgrès J., Pochart P., Heyman T., Kuo K.C., Gehrke C.W. Eukaryotic tRNAs(Pro): Primary structure of the anticodon loop; presence of 5-carbamoylmethyluridine or inosine as the first nucleoside of the anticodon. Biochim. Biophys. Acta. 1990;1049:255–260. doi: 10.1016/0167-4781(90)90095-J. [DOI] [PubMed] [Google Scholar]
- 42.Chen C., Huang B., Anderson J.T., Byström A.S. Unexpected accumulation of ncm5U and ncm5s2U in a trm9 mutant suggests an additional step in the synthesis of mcm5U and mcm5s2U. PLoS One. 2011;6:e20783. doi: 10.1371/journal.pone.0020783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Grosjean H., Constantinesco F., Foiret D., Benachenhou N. A novel enzymatic pathway leading to 1-methylinosine modification in Haloferax volcanii tRNA. Nucleic Acids Res. 1995;23:4312–4319. doi: 10.1093/nar/23.21.4312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Roovers M., Wouters J., Bujnicki J.M., Tricot C., Stalon V., Grosjean H., Droogmans L. A primordial RNA modification enzyme: the case of tRNA (m1A) methyltransferase. Nucleic Acids Res. 2004;32:465–476. doi: 10.1093/nar/gkh191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hamdane D., Guelorget A., Guérineau V., Golinelli-Pimpaneau B. Dynamics of RNA modification by a multi-site-specific tRNA methyltransferase. Nucleic Acids Res. 2015;42:11697–11706. doi: 10.1093/nar/gku820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kuchino Y., Kato M., Sugisaki H., Nishimura S. Nucleotide sequence of starfish initiator tRNA. Nucleic Acids Res. 1979;6:3459–3469. doi: 10.1093/nar/6.11.3459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Constantinesco F., Motorin Y., Grosjean H. Transfer RNA modification enzymes from Pyrococcus furiosus: detection of the enzymatic activities in vitro. Nucleic Acids Res. 1999;27:1308–1315. doi: 10.1093/nar/27.5.1308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sun N., Tamura N., Tamura T., Knispel R.W., Hrabe T., Kofler C., Nickell S., Nagy I. Size distribution of native cytosolic proteins of Thermoplasma acidophilum. Proteomics. 2009;9:3783–3786. doi: 10.1002/pmic.200800892. [DOI] [PubMed] [Google Scholar]
- 49.Droogmans L., Roovers M., Bujnicki J.M., Tricot C., Hartsch T., Stalon V., Grosjean H. Cloning and characterization of tRNA (m1A58) methyltransferase (TrmI) from Thermus thermophilus HB27, a protein required for cell growth at extreme temperatures. Nucleic Acids Res. 2003;31:2148–2156. doi: 10.1093/nar/gkg314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tomikawa C., Yokogawa T., Kanai T., Hori H. N7-Methylguanine at position 46 (m7G46) in tRNA from Thermus thermophilus is required for cell viability through a tRNA modification network. Nucleic Acids Res. 2010;38:942–957. doi: 10.1093/nar/gkp1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yokogawa T., Kitamura Y., Nakamura D., Ohno S., Nishikawa K. Optimization of the hybridization-based method for purification of thermostable tRNAs in the presence of tetraalkylammonium salts. Nucleic Acids Res. 2010;38 doi: 10.1093/nar/gkp1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Awai T., Kimura S., Tomikawa C., Ochi A., Ihsanawati, Bessho Y., Yokoyama S., Ohno S., Nishikawa K., Yokogawa T., Suzuki T., Hori H. Aquifex aeolicus tRNA (N2,N2-guanine)-dimethyltransferase (Trm1) catalyzes transfer of methyl groups not only to guanine 26 but also to guanine 27 in tRNA. J. Biol. Chem. 2009;284:20467–20478. doi: 10.1074/jbc.M109.020024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ishida K., Kunibayashi T., Tomikawa C., Ochi A., Kanai T., Hirata A., Iwashita C., Hori H. Pseudouridine at position 55 in tRNA controls the contents of other modified nucleotides for low-temperature adaptation in the extreme-thermophilic eubacterium Thermus thermophilus. Nucleic Acids Res. 2011;39:2304–2318. doi: 10.1093/nar/gkq1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hirata A., Kitajima T., Hori H. Cleavage of intron from the standard or non-standard position of the precursor tRNA by the splicing endonuclease of Aeropyrum pernix, a hyper-thermophilic Crenarchaeon, involves a novel RNA recognition site in the Crenarchaea specific loop. Nucleic Acids Res. 2011;39:9376–9389. doi: 10.1093/nar/gkr615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Christian T., Evilia C., Williams S., Hou Y.M. Distinct origins of tRNA(m1G37) methyltransferase. J. Mol. Biol. 2004;339:707–719. doi: 10.1016/j.jmb.2004.04.025. [DOI] [PubMed] [Google Scholar]
- 56.Goto-Ito S., Ito T., Ishii R., Muto Y., Bessho Y., Yokoyama S. Crystal structure of archaeal tRNA(m(1)G37)methyltransferase aTrm5. Proteins. 2008;72:1274–1289. doi: 10.1002/prot.22019. [DOI] [PubMed] [Google Scholar]
- 57.Reinhart M.P., Lewis J.M., Leboy P.S. A single tRNA (guanine)-methyltransferase from Tetrahymena with both mono- and di-methylating activity. Nucleic Acids Res. 1986;14:1131–1148. doi: 10.1093/nar/14.3.1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Constantinesco F., Motorin Y., Grosjean H. Characterisation and enzymatic properties of tRNA(guanine 26, N2,N2)-dimethyltransferase (Trm1p) from Pyrococcus furiosus. J. Mol. Biol. 1999;291:375–392. doi: 10.1006/jmbi.1999.2976. [DOI] [PubMed] [Google Scholar]
- 59.Phillips J.H., Kjellin-Straby K. Studies on microbial ribonucleic acid. IV. Two mutants of Saccharomyces cerevisiae lacking N-2-dimethylguanine in soluble ribonucleic acid. J. Mol. Biol. 1967;26:509–518. doi: 10.1016/0022-2836(67)90318-X. [DOI] [PubMed] [Google Scholar]
- 60.Ellis S.R., Morales M.J., Li J.M., Hopper A.K., Martin N.C. Isolation and characterization of the TRM1 locus, a gene essential for the N2,N2-dimethylguanosine modification of both mitochondrial and cytoplasmic tRNA in Saccharomyces cerevisiae. J. Biol. Chem. 1986;261:9703–9709. [PubMed] [Google Scholar]
- 61.Fukunaga J., Gouda M., Umeda K., Ohno S., Yokogawa T., Nishikawa K. Use of RNase P for efficient preparation of yeast tRNATyr transcript and its mutants. J. Biochem. 2006;139:123–127. doi: 10.1093/jb/mvj005. [DOI] [PubMed] [Google Scholar]
- 62.Goto-Ito S., Ito T., Kuratani M., Bessho Y., Yokoyama S. Tertiary structure checkpoint at anticodon loop modification in tRNA functional maturation. Nat. Struct. Mol. Biol. 2009;16:1109–1115. doi: 10.1038/nsmb.1653. [DOI] [PubMed] [Google Scholar]
- 63.Stanley J., Vassilenko S. A different approach to RNA sequencing. Nature. 1978;274:87–89. doi: 10.1038/274087a0. [DOI] [PubMed] [Google Scholar]
- 64.Grosjean H., Droogmans L., Roovers M., Keith G. Detection of enzymatic activity of transfer RNA modification enzymes using radiolabeled tRNA substrates. Methods Enzymol. 2007;425:55–101. doi: 10.1016/S0076-6879(07)25003-7. [DOI] [PubMed] [Google Scholar]
- 65.Somme J., van Laer B., Roovers M., Steyaert J., Versées W., Droogmans L. Characterization of two homologous 2'-O-methyltransferases showing different specificities for their tRNA substrates. RNA. 2014;20:1257–1271. doi: 10.1261/rna.044503.114. [DOI] [PMC free article] [PubMed] [Google Scholar]