Abstract
Bioactive peptides play important roles in metabolic regulation and modulation and many are used as therapeutics. These peptides often possess disulfide bonds, which are important for their structure, function and stability. A systematic network of enzymes—a disulfide bond generating enzyme, a disulfide bond donor enzyme and a redox cofactor—that function inside the cell dictates the formation and maintenance of disulfide bonds. The main pathways that catalyze disulfide bond formation in peptides and proteins in prokaryotes and eukaryotes are remarkably similar and share several mechanistic features. This review summarizes the formation of disulfide bonds in peptides and proteins by cellular and recombinant machinery.
Keywords: bioactive peptides, disulfide bonds, peptide and protein folding, oxidative folding, recombinant technology
1. Introduction
Disulfide bonds are common structural motifs in many bioactive peptides and proteins including hormones, neurotransmitters, growth factors, enzyme inhibitors, and antimicrobial peptides [1,2,3]. They play a critical role in maintaining the overall fold of the peptides and proteins and are thereby often important for the function and stability of proteins and peptides. In nature, such bonds are usually formed during the posttranslational modification stage with the assistance of appropriate enzymes and co-factors whereas, in the laboratory, disulfide bonds in native proteins can be formed randomly in basic buffer via air oxidation or by regioselective methods. This review discusses biological means of disulfide bond formation in biological systems.
2. Folding of Peptides and Proteins in Normal Cells
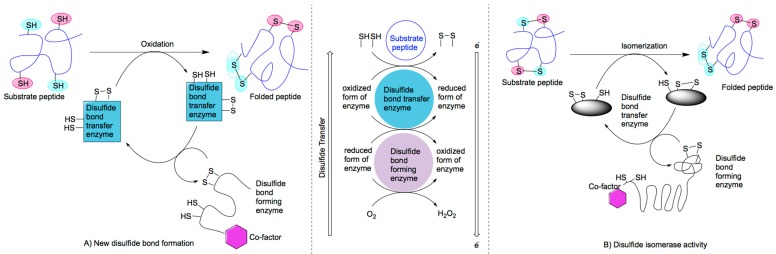
Cysteines, sulfhydryl-containing amino acids, which are located an appropriate distance or next to one another within a polypeptide chain, will form a disulfide bond through their oxidisable thiol groups. This bond will impart a fold in the chain of the protein or bend in its structure. Disulfide bond formation and its effect on protein folding has been a subject of great interest for at least half a century [4]. The first reported study by Anfinsen in 1973 revealed that disulfide bond formation inside the cell is spontaneous and that the amino acid sequence is sufficient to determine correct folding of the peptide or protein [4,5]. It was subsequently shown that there are several active disulfide bond-promoting enzymes and cofactors functioning in the cell [6,7] meaning that disulfide bonds are usually formed by a systematic network of intracellular enzymes [8]. These enzyme systems form a new disulfide bond or reshuffle the existing mispaired disulfide bond in substrate peptides (Figure 1).
Figure 1.
Schematic representation of general mechanisms by which a disulfide bond is formed by an intra-cellular enzyme-cofactor system: (A) Formation of new disulfide bonds; and (B) reshuffling of existing bonds by isomerase activity.
Typically, these systems consist of a disulfide bond generating enzyme, a disulfide bond donor enzyme and a redox cofactor [9]. Interestingly, while these enzymes have very low homology, their functional motifs differ only marginally [10]. A minimum assembly requires C–(X)n–C (where X = amino acid; n = 1–3) motif in the active site associated with a redox co-factor. In some periplasmic enzyme systems, an arginine residue has been observed to stabilize the charge transfer complex between the cysteine and co-factor [11,12,13].
2.1. Mechanisms of Disulfide Bond Formation
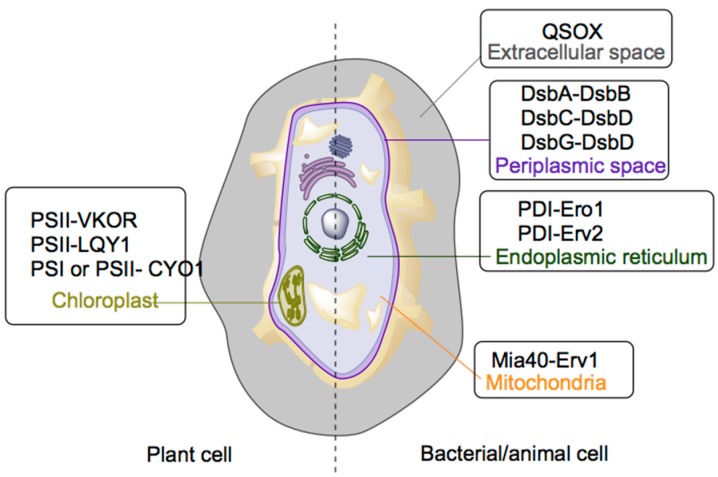
The formation of disulfide bonds in bacterial (prokaryotic) cells is well characterized [14,15]. Generally, bacterial proteins are synthesized by ribosomal mRNA translation and disulfide bonds are subsequently formed as posttranslational modifications catalyzed by various enzymes located in the periplasm [16] or cytoplasm [17,18]. In higher animals the same process is performed in specific cell organelles, such as mitochondria, the endoplasmic reticulum (ER) and chloroplasts (Figure 2, Table 1).
Figure 2.
Cellular representation of enzyme systems and respective organelles.
Table 1.
Cellular compartments and enzyme systems for disulfide bond formation.
| Site | Disulfide Bond Transferring Enzyme | Disulfide Bond Generating Enzyme | Cofactor |
|---|---|---|---|
| Prokaryotic | DsbA | DsbB | Ubiquinone |
| Periplasm | DsbC | DsbD | Ubiquinone |
| DsbG | DsbD | - | |
| Endoplasmic reticulum | PDI | Ero1 | FAD |
| PDI | Erv2 | FAD | |
| Mitochondria | Mia40 | Erv1 | FAD |
| Chloroplast | PSII | LTO1 | Phylloquinone or Hydroquinone |
| PSII | LQY1 | Zn (believed to be a cofactor) | |
| PSI and PSII | CYO1 | Zn (believed to be a cofactor) | |
| Extracellular space | QSOX | QSOX | FAD |
2.1.1. Periplasmic System
In prokaryotic cells, disulfide bond formation predominantly occurs through a network of periplasmic enzymes, the thiol-disulfide oxidoreductase family, called disulfide bond forming enzymes (Dsb) [19,20]. A series of disulfide oxidoreductase enzymes, including DsbA, DsbB, DsbC and DsbD, have been identified over the last 25 years. Elucidation of the crystal structure of E. coli DsbA enabled investigation into the mechanism of disulfide bond formation [21]. This enzyme system introduces a disulfide bond to a newly synthesized protein by means of DsbA-DsbB and ubiquinone (UQ) [22]. DsbA is the primary disulfide bond donor and its active state is the oxidized form with the Cys30–Cys33 disulfide bond. It is kept in the oxidized, active state by membrane bound protein, DsbB, which transmits electrons from DsbA to UQ. DsbB has been predicted to have two periplasmic loops and each of the loops contains one pair of essential cysteines: Cys41–Cys44 and Cys104–Cys130. While the Cys104–Cys130 pair is involved directly in the disulfide exchange with DsbA, the Cys41–Cys44 pair is the target of oxidation by UQ [13,23,24,25,26].
DsbA is known to have no proofreading activity and can form incorrect disulfides in proteins with multiple cysteines. These incorrect disulfide bonds are corrected by a protein disulfide isomerase, DsbC, which is kept in the reduced and active configuration by a membrane-bound protein, DsbD. The DsbC/DsbD isomerization pathway is considered to be isolated from the DsbA/DsbB pathway [27,28,29,30,31].
2.1.2. Endoplasmic Reticulum System
In organisms such as fungi and mammals where protein folding is compartmentalized and complex, disulfide bond formation takes place in specialized organelles such as the ER and mitochondria [32]. The main reasons are: first, the cytosolic environment is reducing due to the high concentration of thioredoxin reductase and glutathione reductase, and second, the availability of supporting systems for appropriate protein folding for disulfide bond formation [33,34]. Ero-1 is the predominant disulfide bond-generating enzyme in ER and Erv2 principally in fungal cells [35]. The most studied transfer enzyme is protein disulfide isomerase (PDI) and the most studied enzyme system in such cells is “PDI-Ero1” [36].
Even though there is very poor sequence homology, the structural features of Ero-1 and Erv2 are similar to DsbB where one “C–(X)n–C” motif generates a disulfide bond together with FAD (flavin adenine dinucleotide) and another “C–(X)n–C” maintains unidirectional propagation of redox equivalents. Moreover, both these enzymes are associated with FAD where an isoalloxazine ring is embedded within the active site [37]. In Ero-1, the first cysteine pair is located in the “C–X–X–C–X–X–C” motif which generates a disulfide bond [38,39]. In the second cysteine pair, “C–(X)n–C”, the residues between two cysteines varies between paralogs of Ero-1 [40]. Similarly, the catalytic core of Erv2 has “C–X–X–C” and “C–(X)n–C” (where n = 1–4) motifs. However, there is no arginine residue as is found in the active site of the DsbB enzyme.
The mammalian ER contains two paralogs of Ero1, three Erv2-like proteins [35] and about twenty PDI family proteins [41]. Transfer of disulfide bonds is carried out by PDI. This oxidoreductase enzyme is found to play a role in isomerization as well as producing a disulfide bond in newly synthesized peptides and proteins. PDI contains two thioredoxin-like domains, a and a', each of which contains an active site within the CXXC motif [42].
2.1.3. Mitochondria
The mitochondrial inter-membrane space (IMS) has a similar environment to that of the cytoplasm of mammalian (eukaryotic) cells. Thus, there is a special enzyme system working in the IMS of mitochondria. The sulfhydryl oxidase Erv1 is a disulfide bond generating enzyme and the redox dependent receptor Mia40 acts as transfer protein [43,44,45]. Disulfide bonds are introduced to the substrate protein via a “C–P–C” motif of Mia40 [46,47]. Mia40, also known as Tim40, was identified as a protein that mediates sorting of Tim proteins [48,49] and the folding of coiled-coil helix coiled-coil helix (CHCH) proteins [43], such as COX [50,51] in mitochondria [52,53]. Thus, it is important to maintain the Mia40 CPC motif in an oxidized form, which is ensured by sulfhydryl oxidase, Erv1 [54,55]. As with Ero1, Erv1 also contains FAD as a cofactor, which generates a disulfide bond between “C–X–X–C” motifs of Erv1 [56,57].
2.1.4. Chloroplasts
Like other eukaryotic organisms, disulfide bond formation in plant cells is carried out in the ER [58,59], and mitochondria [60,61]. However being photosynthetic cells, they possess chloroplasts where there is a huge burden to fold hundreds of enzymes in order to maintain photosynthetic activity [62]. Thus, chloroplasts contain a specialized enzyme system for protein oxidation and folding [63,64].
Vitamin K epoxide reductases (VKOR) are members of a large family of enzymes that exist in a wide range of organisms including bacteria, archaea, vertebrates and plants [65]. Members of this family can mediate disulfide bonding via different mechanisms [66]. However, they all comprise a conserved “C–X–X–C” motif similar to DsbB enzymes in bacteria. This can be oxidized to generate a disulfide bond by transferring electrons to quinone [67]. Arabidopsis VKOR homolog known, as lumen thiol oxidoreductase1 (LTO1) is a plant homologue associated with a quinone moiety in thylakoid. LOT1 reduces quinone to generate a disulfide bond in the “C–X–X–C” motif, which is then transferred to a luminal subunit of Photosystem II (PSII) [68]. There is evidence of quinones such as phylloquinone (vitamin K) [69] and hydroquinone [70] playing roles in the redox cycle. However, the mechanism of electron transfer is not clearly understood.
Other known enzymes include low quantum yield of PSII1 (LQY1), a small Zn-finger protein involved in repair mechanisms by disulfide bond formation [71], and CYO1 (a Japanese word “shi-yo-u” which means cotyledon), an integral membrane protein of thylakoid associated with PSI and PSII [72]. Both of them possess a Zn finger motif [71,72], and are assumed to have PDI activity. However, a lack of structural information limits an understanding of the exact mechanism underlying the electron transfer.
2.1.5. Extracellular Space
Quiescin sulfhydryl oxidase (QSOX) is one of the most important enzymes that carries out disulfide bond formation and protein folding in extracellular and subcellular spaces. QSOX binds a FAD cofactor [73] which enables the generation of disulfide bonds. It also contains thioredoxin [74] motifs which aid in transferring the disulfide to the substrate protein [75]. In mammals, cells secrete QSOXs into the extracellular space after modification by the Golgi complex [76]. Human QSOX1 has two thioredoxin domains, one of which contains a “C–X–X–C” motif similar to prokaryotic DsbA [74] and eukaryotic PDI [77].
3. Folding of Disulfide-Containing Bioactive Peptides and Proteins via Recombinant Technology
Recombinant DNA-mediated polypeptide production remains a popular approach for obtaining properly folded disulfide-containing protein and peptides. Many reviews are available that address the effect of various parameters such as different host organisms [78,79], expression vectors [80], expression rate [81,82], and purification [83,84]. With respect to host systems, many cell lines are available for expressing recombinant peptides and proteins such as E. coli [85], Chinese hamster ovary cells [86], human embryonic kidney cells [87], and S. cerevisiae [88]. The wide range of molecular biology options with E. coli systems provides many options to ensure proper disulfide bond generation [89,90]. E. coli expression is fast and inexpensive to scale-up but not all proteins are amenable to expression in this system [91,92].
A major challenge in obtaining a recombinant protein is correct folding. A protein synthesized in a recombinant cell is often produced in reduced form, which is then oxidized (refolded) using suitable folding conditions [93,94]. The target crude peptide/protein can be obtained in reduced form and then refolded with the aid of various enzymes as disulfide catalyst systems [95,96]. Alternatively, oxidative folding is possible without the use of any enzyme [97,98]. However, such refolding can be a problematic step. Misfolding is a major issue with cysteine-containing polypeptides resulting in low yields. This limitation has necessitated the development of new methodologies to enable the acquisition of correctly folded protein or peptide in its native form in high yields. To assist the refolding, various tactics have been employed including inclusion body formation [99,100], co-expression of supporting enzymes [101,102] and chemical assistance [100].
3.1. Co-Expressing Supporting Enzymes/Peptides
In a recombinant DNA cell, a nascent polypeptide is produced by translation. It is then either folded in the cytoplasm or translocated to a suitable oxidative environment such as the bacterial periplasm. Disrupting reducing pathways in the cytoplasm has been shown to allow disulfide bond formation in the cytoplasm of E. coli [103]. Furthermore, co-expression of Dsb enzymes was found to improve the yield of desired folded protein [89]. These approaches have been extensively used in the expression of disulfide-containing proteins such as chitinase [104], endopolygalcturonase, [105] and anti-freeze proteins [106]. Overexpression of thioredoxin has been shown to act as an oxidant, helping to maintain oxidative conditions in the cytoplasm [107] and resulting in improved yields of folded proteins [108,109].
The most common practice for expressing disulfide-containing proteins in bacterial hosts is to “hijack” the membrane translocation machinery and secrete the polypeptide after translation [110]. A precursor peptide is produced which can be translocated and then folded into its active form. In this case, peptide oxidative folding takes place conveniently but the crucial barrier is the translocation of nascent peptide into the periplasm. A leader peptide sequence is essential for this purpose. The correlation between hydrophobicity of the leader peptide and export mechanism can be a more efficient means of translocating the precursor peptide [111]. A leader peptide could be an endogenous signal peptide sequence [112,113,114], a phage-pIII leader peptide [115] or even a designed synthetic sequence [116]. Another more convenient tactic is the folding of a precursor peptide assisted by co-expression of disulfide bond forming enzymes [117], foldases [118] or even periplasmic chaperones [119].
Although continuous optimization has led to improved yields of expressed and folded protein, there are many more challenges in expressing multiple enzymes/peptides in a recombinant system. These include nonspecific binding of chaperones [120,121], incorrect folding of soluble target protein [122] or direct excretion of translation product into the culture medium [123,124]. Thus at times it can be advantageous to use chemicals to assist the folding of recombinantly-produced peptides.
3.2. Chemical Assistance in Recombinant Folding
Bearing in mind that the main obstacle to the expression of cysteine-containing recombinant proteins is a pronounced tendency to aggregate [125], low molecular weight chaperones have been shown to aid the folding of target proteins either by assisting cellular enzymatic systems or acting as independent chaperones [126,127]. Examples of this approach include the folding of immunotoxins by glycine betaine [128], cytochrome by sucrose [129], and proinsulin by l-arginine [130]. Recent studies have also shown that small molecules can be used to improve yields of recombinant folded peptides [131,132,133]. These include urea and lithium chloride which can minimize the extent of side products generated during oxidative folding [134]. The use of glutathione ester instead of glutathione contributed to improved folding ability of egg lysozyme [135]. Water-soluble reagents such as selenoxides have been successfully used in synthetic as well as recombinant peptides and protein [136].
4. Conclusions
Disulfide bond formation is critical for the proper folding of bioactive peptides and proteins. The increasing understanding of the intricate, complex in vivo disulfide bond forming process is providing important insights into the mechanisms of diseases that are caused by protein misfolding and may contribute to the development of corrective measures with therapeutic applications. This review aims to provide readers with comprehensive details and updated knowledge on how the disulfide network is formed and maintained in biological systems.
Acknowledgments
This research was partly funded by NHMRC (Australia) project grant (1023321) and an ARC Linkage grant (LP120100654). The Victorian Government’s Operational Infrastructure Support Program supported research at the FNI. Nitin A. Patil thanks the University of Melbourne for providing Melbourne International Research Scholarship.
Conflicts of Interest
The authors have no conflict of interest.
References
- 1.Moroder L., Musiol H.J., Gotz M., Renner C. Synthesis of single- and multiple-stranded cystine-rich peptides. Biopolymers. 2005;80:85–97. doi: 10.1002/bip.20174. [DOI] [PubMed] [Google Scholar]
- 2.Góngora-Benítez M., Tulla-Puche J., Albericio F. Multifaceted roles of disulfide bonds. peptides as therapeutics. Chem. Rev. 2013;114:901–926. doi: 10.1021/cr400031z. [DOI] [PubMed] [Google Scholar]
- 3.Marshall E., Costa L.M., Gutierrez-Marcos J. Cysteine-rich peptides (CRPs) mediate diverse aspects of cell-cell communication in plant reproduction and development. J. Exp. Bot. 2011;62:1677–1686. doi: 10.1093/jxb/err002. [DOI] [PubMed] [Google Scholar]
- 4.Haber E., Anfinsen C.B. Regeneration of enzyme activity by air oxidation of reduced subtilisin-modified ribonuclease. J. Biol. Chem. 1961;236:422–424. [PubMed] [Google Scholar]
- 5.Anfinsen C.B. Principles that govern the folding of protein chains. Science. 1973;181:223–230. doi: 10.1126/science.181.4096.223. [DOI] [PubMed] [Google Scholar]
- 6.Heras B., Kurz M., Shouldice S.R., Martin J.L. The name’s bond......disulfide bond. Curr. Opin. Struct. Biol. 2007;17:691–698. doi: 10.1016/j.sbi.2007.08.009. [DOI] [PubMed] [Google Scholar]
- 7.Kadokura H., Beckwith J. Mechanisms of oxidative protein folding in the bacterial cell envelope. Antioxid. Redox Signal. 2010;13:1231–1246. doi: 10.1089/ars.2010.3187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sato Y., Inaba K. Disulfide bond formation network in the three biological kingdoms, bacteria, fungi and mammals. FEBS J. 2012;279:2262–2271. doi: 10.1111/j.1742-4658.2012.08593.x. [DOI] [PubMed] [Google Scholar]
- 9.Inaba K. Structural basis of protein disulfide bond generation in the cell. Genes Cells. 2010;15:935–943. doi: 10.1111/j.1365-2443.2010.01434.x. [DOI] [PubMed] [Google Scholar]
- 10.Hiniker A., Bardwell J.C. Disulfide relay between and within proteins: the Ero1p structure. Trends Biochem. Sci. 2004;29:516–519. doi: 10.1016/j.tibs.2004.08.002. [DOI] [PubMed] [Google Scholar]
- 11.Inaba K., Takahashi Y.H., Fujieda N., Kano K., Miyoshi H., Ito K. DsbB elicits a red-shift of bound ubiquinone during the catalysis of DsbA oxidation. J. Biol. Chem. 2004;279:6761–6768. doi: 10.1074/jbc.M310765200. [DOI] [PubMed] [Google Scholar]
- 12.Kadokura H., Bader M., Tian H., Bardwell J.C., Beckwith J. Roles of a conserved arginine residue of DsbB in linking protein disulfide-bond-formation pathway to the respiratory chain of Escherichia coli. Proc. Natl. Acad. Sci. USA. 2000;97:10884–10849. doi: 10.1073/pnas.97.20.10884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Inaba K., Takahashi Y.-h., Ito K., Hayashi S. Critical role of a thiolate-quinone charge transfer complex and its adduct form in de novo disulfide bond generation by DsbB. Proc. Natl. Acad. Sci. USA. 2006;103:287–292. doi: 10.1073/pnas.0507570103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Messens J., Collet J.-F. Pathways of disulfide bond formation in Escherichia coli. Intl. J. Biochem. Cell Biol. 2006;38:1050–1062. doi: 10.1016/j.biocel.2005.12.011. [DOI] [PubMed] [Google Scholar]
- 15.Hatahet F., Boyd D., Beckwith J. Disulfide bond formation in prokaryotes: History, diversity and design. Biochim. Biophys. Acta. 2014;1844:1402–1414. doi: 10.1016/j.bbapap.2014.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ito K., Inaba K. The disulfide bond formation (Dsb) system. Curr. Opin. Struct. Biol. 2008;18:450–458. doi: 10.1016/j.sbi.2008.02.002. [DOI] [PubMed] [Google Scholar]
- 17.Daniels R., Mellroth P., Bernsel A., Neiers F., Normark S., von Heijne G., Henriques-Normark B. Disulfide bond formation and cysteine exclusion in gram-positive bacteria. J. Biol. Chem. 2010;285:3300–3309. doi: 10.1074/jbc.M109.081398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Inaba K. Disulfide bond formation system in Escherichia coli. J. Biochem. 2009;146:591–597. doi: 10.1093/jb/mvp102. [DOI] [PubMed] [Google Scholar]
- 19.Bardwell J.C.A., McGovern K., Beckwith J. Identification of a protein required for disulfide bond formation in vivo. Cell. 1991;67:581–589. doi: 10.1016/0092-8674(91)90532-4. [DOI] [PubMed] [Google Scholar]
- 20.Grauschopf U., Winther J.R., Korber P., Zander T., Dallinger P., Bardwell J.C.A. Why is DsbA such an oxidizing disulfide catalyst? Cell. 1995;83:947–955. doi: 10.1016/0092-8674(95)90210-4. [DOI] [PubMed] [Google Scholar]
- 21.Martin J.L., Bardwell J.C.A., Kuriyan J. Crystal structure of the DsbA protein required for disulphide bond formation in vivo. Nature. 1993;365:464–468. doi: 10.1038/365464a0. [DOI] [PubMed] [Google Scholar]
- 22.Kobayashi T., Kishigami S., Sone M., Inokuchi H., Mogi T., Ito K. Respiratory chain is required to maintain oxidized states of the DsbA-DsbB disulfide bond formation system in aerobically growing Escherichia coli cells. Proc. Natl. Acad. Sci. USA. 1997;94:11857–11862. doi: 10.1073/pnas.94.22.11857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhou Y., Cierpicki T., Jimenez R.H., Lukasik S.M., Ellena J.F., Cafiso D.S., Kadokura H., Beckwith J., Bushweller J.H. NMR solution structure of the integral membrane enzyme DsbB: Functional insights into DsbB-catalyzed disulfide bond formation. Mol. Cell. 2008;31:896–908. doi: 10.1016/j.molcel.2008.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Inaba K., Murakami S., Suzuki M., Nakagawa A., Yamashita E., Okada K., Ito K. Crystal structure of the DsbB-DsbA complex reveals a mechanism of disulfide bond generation. Cell. 2006;127:789–801. doi: 10.1016/j.cell.2006.10.034. [DOI] [PubMed] [Google Scholar]
- 25.Guddat L.W., Bardwell J.C., Martin J.L. Crystal structures of reduced and oxidized DsbA: Investigation of domain motion and thiolate stabilization. Structure. 1998;6:6757–6767. doi: 10.1016/S0969-2126(98)00077-X. [DOI] [PubMed] [Google Scholar]
- 26.Schirra H.J., Renner C., Czisch M., Huber-Wunderlich M., Holak T.A., Glockshuber R. Structure of reduced DsbA from Escherichia coli in solution. Biochemistry. 1998;37:6263–6276. doi: 10.1021/bi980136y. [DOI] [PubMed] [Google Scholar]
- 27.Katzen F., Beckwith J. Transmembrane electron transfer by the membrane protein DsbD occurs via a disulfide bond cascade. Cell. 2000;103:769–779. doi: 10.1016/S0092-8674(00)00180-X. [DOI] [PubMed] [Google Scholar]
- 28.Cho S.H., Porat A., Ye J., Beckwith J. Redox-active cysteines of a membrane electron transporter DsbD show dual compartment accessibility. EMBO J. 2007;26:3509–3520. doi: 10.1038/sj.emboj.7601799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rozhkova A., Stirnimann C.U., Frei P., Grauschopf U., Brunisholz R., Grutter M.G., Capitani G., Glockshuber R. Structural basis and kinetics of inter- and intramolecular disulfide exchange in the redox catalyst DsbD. EMBO J. 2004;23:1709–1719. doi: 10.1038/sj.emboj.7600178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nagy P. Kinetics and mechanisms of thiol-disulfide exchange covering direct substitution and thiol oxidation-mediated pathways. Antioxid. Redox Signal. 2013;18:1623–1641. doi: 10.1089/ars.2012.4973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Haebel P.W., Goldstone D., Katzen F., Beckwith J., Metcalf P. The disulfide bond isomerase DsbC is activated by an immunoglobulin-fold thiol oxidoreductase: Crystal structure of the DsbC-DsbDalpha complex. EMBO J. 2002;21:4774–4784. doi: 10.1093/emboj/cdf489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bulleid N.J. Disulfide bond formation in the mammalian endoplasmic reticulum. Cold Spring Harb. Perspect. Biol. 2012;4 doi: 10.1101/cshperspect.a013219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Arnér E.S.J. Focus on mammalian thioredoxin reductases—Important selenoproteins with versatile functions. Biochim. Biophys. Acta. 2009;1790:495–526. doi: 10.1016/j.bbagen.2009.01.014. [DOI] [PubMed] [Google Scholar]
- 34.Hebert D.N., Molinari M. In and out of the ER: Protein folding, quality control, degradation, and related human diseases. Physiol. Rev. 2007;87:1377–408. doi: 10.1152/physrev.00050.2006. [DOI] [PubMed] [Google Scholar]
- 35.Sevier C.S., Kaiser C.A. Conservation and diversity of the cellular disulfide bond formation pathways. Antioxid. Redox Signal. 2006;8:797–811. doi: 10.1089/ars.2006.8.797. [DOI] [PubMed] [Google Scholar]
- 36.Sevier C.S., Kaiser C.A. Formation and transfer of disulphide bonds in living cells. Nat. Rev. Mol. Cell Biol. 2002;3:836–847. doi: 10.1038/nrm954. [DOI] [PubMed] [Google Scholar]
- 37.Inaba K., Masui S., Iida H., Vavassori S., Sitia R., Suzuki M. Crystal structures of human Ero1alpha reveal the mechanisms of regulated and targeted oxidation of PDI. EMBO J. 2010;29:3330–3343. doi: 10.1038/emboj.2010.222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Benham A.M., Cabibbo A., Fassio A., Bulleid N., Sitia R., Braakman I. The CXXCXXC motif determines the folding, structure and stability of human Ero1-Lalpha. EMBO J. 2000;19:4493–4502. doi: 10.1093/emboj/19.17.4493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Edman J.C., Ellis L., Blacher R.W., Roth R.A., Rutter W.J. Sequence of protein disulphide isomerase and implications of its relationship to thioredoxin. Nature. 1985;317:267–270. doi: 10.1038/317267a0. [DOI] [PubMed] [Google Scholar]
- 40.Gross E., Kastner D.B., Kaiser C.A., Fass D. Structure of Ero1p, source of disulfide bonds for oxidative protein folding in the cell. Cell. 2004;117:601–610. doi: 10.1016/S0092-8674(04)00418-0. [DOI] [PubMed] [Google Scholar]
- 41.Appenzeller-Herzog C., Riemer J., Christensen B., Sorensen E.S., Ellgaard L. A novel disulphide switch mechanism in Ero1alpha balances ER oxidation in human cells. EMBO J. 2008;27:2977–2987. doi: 10.1038/emboj.2008.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Darby N.J., Freedman R.B., Creighton T.E. Dissecting the mechanism of protein disulfide isomerase: Catalysis of disulfide bond formation in a model peptide. Biochemistry. 1994;33:7937–7947. doi: 10.1021/bi00191a022. [DOI] [PubMed] [Google Scholar]
- 43.Mesecke N., Terziyska N., Kozany C., Baumann F., Neupert W., Hell K., Herrmann J.M. A disulfide relay system in the intermembrane space of mitochondria that mediates protein import. Cell. 2005;121:1059–1069. doi: 10.1016/j.cell.2005.04.011. [DOI] [PubMed] [Google Scholar]
- 44.Allen S., Balabanidou V., Sideris D.P., Lisowsky T., Tokatlidis K. Erv1 mediates the Mia40-dependent protein import pathway and provides a functional link to the respiratory chain by shuttling electrons to cytochrome c. J. Mol. Biol. 2005;353:937–944. doi: 10.1016/j.jmb.2005.08.049. [DOI] [PubMed] [Google Scholar]
- 45.Tokatlidis K. A disulfide relay system in mitochondria. Cell. 2005;121:965–967. doi: 10.1016/j.cell.2005.06.019. [DOI] [PubMed] [Google Scholar]
- 46.Grumbt B., Stroobant V., Terziyska N., Israel L., Hell K. Functional characterization of Mia40p, the central component of the disulfide relay system of the mitochondrial intermembrane space. J. Biol. Chem. 2007;282:37461–37470. doi: 10.1074/jbc.M707439200. [DOI] [PubMed] [Google Scholar]
- 47.Banci L., Bertini I., Cefaro C., Cenacchi L., Ciofi-Baffoni S., Felli I. C., Gallo A., Gonnelli L., Luchinat E., Sideris D., Tokatlidis K. Molecular chaperone function of Mia40 triggers consecutive induced folding steps of the substrate in mitochondrial protein import. Proc. Natl. Acad. Sci. USA. 2010;107:20190–20195. doi: 10.1073/pnas.1010095107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Milenkovic D., Ramming T., Müller J. M., Wenz L.-S., Gebert N., Schulze-Specking A., Stojanovski D., Rospert S., Chacinska A. Identification of the signal directing Tim9 and Tim10 into the intermembrane space of mitochondria. Mol. Biol. Cell. 2009;20:2530–2539. doi: 10.1091/mbc.E08-11-1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sideris D. P., Petrakis N., Katrakili N., Mikropoulou D., Gallo A., Ciofi-Baffoni S., Banci L., Bertini I., Tokatlidis K. A novel intermembrane space-targeting signal docks cysteines onto Mia40 during mitochondrial oxidative folding. J. Cell. Biol. 2009;187:1007–1022. doi: 10.1083/jcb.200905134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Banci L., Bertini I., Cefaro C., Ciofi-Baffoni S., Gallo A., Martinelli M., Sideris D.P., Katrakili N., Tokatlidis K. MIA40 is an oxidoreductase that catalyzes oxidative protein folding in mitochondria. Nat. Struct. Mol. Biol. 2009;16:198–206. doi: 10.1038/nsmb.1553. [DOI] [PubMed] [Google Scholar]
- 51.Bien M., Longen S., Wagener N., Chwalla I., Herrmann J.M., Riemer J. Mitochondrial disulfide bond formation is driven by intersubunit electron transfer in Erv1 and proofread by glutathione. Mol. Cell. 2010;37:516–528. doi: 10.1016/j.molcel.2010.01.017. [DOI] [PubMed] [Google Scholar]
- 52.Naoé M., Ohwa Y., Ishikawa D., Ohshima C., Nishikawa S.-I., Yamamoto H., Endo T. Identification of Tim40 that mediates protein sorting to the mitochondrial intermembrane space. J. Biol. Chem. 2004;279:47815–47821. doi: 10.1074/jbc.M410272200. [DOI] [PubMed] [Google Scholar]
- 53.Chacinska A., Pfannschmidt S., Wiedemann N., Kozjak V., Sanjuan Szklarz L.K., Schulze-Specking A., Truscott K.N., Guiard B., Meisinger C., Pfanner N. Essential role of Mia40 in import and assembly of mitochondrial intermembrane space proteins. EMBO J. 2004;23:3735–3746. doi: 10.1038/sj.emboj.7600389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Coppock D.L., Thorpe C. Multidomain flavin-dependent sulfhydryl oxidases. Antioxid. Redox Signal. 2006;8:300–311. doi: 10.1089/ars.2006.8.300. [DOI] [PubMed] [Google Scholar]
- 55.Sztolsztener M.E., Brewinska A., Guiard B., Chacinska A. Disulfide bond formation: Sulfhydryl oxidase ALR controls mitochondrial biogenesis of human MIA40. Traffic. 2013;14:309–320. doi: 10.1111/tra.12030. [DOI] [PubMed] [Google Scholar]
- 56.Faccio G., Nivala O., Kruus K., Buchert J., Saloheimo M. Sulfhydryl oxidases: Sources, properties, production and applications. Appl. Microbiol. Biotechnol. 2011;91:957–966. doi: 10.1007/s00253-011-3440-y. [DOI] [PubMed] [Google Scholar]
- 57.Gross E., Sevier C.S., Vala A., Kaiser C. A., Fass D. A new FAD-binding fold and intersubunit disulfide shuttle in the thiol oxidase Erv2p. Nat. Struct. Biol. 2002;9:61–67. doi: 10.1038/nsb740. [DOI] [PubMed] [Google Scholar]
- 58.Hatahet F., Ruddock L.W. Protein disulfide isomerase: A critical evaluation of its function in disulfide bond formation. Antioxid. Redox Signal. 2009;11:2807–2850. doi: 10.1089/ars.2009.2466. [DOI] [PubMed] [Google Scholar]
- 59.Kawagoe Y., Suzuki K., Tasaki M., Yasuda H., Akagi K., Katoh E., Nishizawa N.K., Ogawa M., Takaiwa F. The critical role of disulfide bond formation in protein sorting in the endosperm of rice. Plant Cell. 2005;17:1141–1153. doi: 10.1105/tpc.105.030668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Levitan A., Danon A., Lisowsky T. Unique features of plant mitochondrial sulfhydryl oxidase. J. Biol. Chem. 2004;279:20002–20008. doi: 10.1074/jbc.M312877200. [DOI] [PubMed] [Google Scholar]
- 61.Dabir D.V., Leverich E.P., Kim S.K., Tsai F.D., Hirasawa M., Knaff D.B., Koehler C.M. A role for cytochrome c and cytochrome c peroxidase in electron shuttling from Erv1. EMBO J. 2007;26:4801–4811. doi: 10.1038/sj.emboj.7601909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Baena-Gonzalez E., Aro E.M. Biogenesis, assembly and turnover of photosystem II units. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2002;357:1451–1459. doi: 10.1098/rstb.2002.1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Onda Y. Oxidative protein-folding systems in plant cells. Int. J. Cell Biol. 2013;2013 doi: 10.1155/2013/585431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wittenberg G., Danon A. Disulfide bond formation in chloroplasts: Formation of disulfide bonds in signaling chloroplast proteins. Plant Sci. 2008;175:459–466. doi: 10.1016/j.plantsci.2008.05.011. [DOI] [Google Scholar]
- 65.Goodstadt L., Ponting C.P. Vitamin K epoxide reductase: homology, active site and catalytic mechanism. Trends Biochem. Sci. 2004;29:289–292. doi: 10.1016/j.tibs.2004.04.004. [DOI] [PubMed] [Google Scholar]
- 66.Tie J.K., Jin D.Y., Stafford D.W. Human vitamin K epoxide reductase and its bacterial homologue have different membrane topologies and reaction mechanisms. J. Biol. Chem. 2012;287:33945–33955. doi: 10.1074/jbc.M112.402941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Li W., Schulman S., Dutton R.J., Boyd D., Beckwith J., Rapoport T.A. Structure of a bacterial homologue of vitamin K epoxide reductase. Nature. 2010;463:507–512. doi: 10.1038/nature08720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Karamoko M., Cline S., Redding K., Ruiz N., Hamel P.P. Lumen thiol oxidoreductase1, a disulfide bond-forming catalyst, is required for the assembly of photosystem II in Arabidopsis. Plant Cell. 2011;23:4462–4475. doi: 10.1105/tpc.111.089680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Furt F., Oostende C., Widhalm J.R., Dale M.A., Wertz J., Basset G.J. A bimodular oxidoreductase mediates the specific reduction of phylloquinone (vitamin K1) in chloroplasts. Plant J. 2010;64:38–46. doi: 10.1111/j.1365-313X.2010.04305.x. [DOI] [PubMed] [Google Scholar]
- 70.Schulman S., Wang B., Li W., Rapoport T.A. Vitamin K epoxide reductase prefers ER membrane-anchored thioredoxin-like redox partners. Proc. Natl. Acad. Sci. USA. 2010;107:15027–15032. doi: 10.1073/pnas.1009972107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lu Y., Hall D.A., Last R.L. A small zinc finger thylakoid protein plays a role in maintenance of photosystem II in Arabidopsis thaliana. Plant Cell. 2011:1861–1875. doi: 10.1105/tpc.111.085456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Muranaka A., Watanabe S., Sakamoto A., Shimada H. Arabidopsis cotyledon chloroplast biogenesis factor CYO1 uses glutathione as an electron donor and interacts with PSI (A1 and A2) and PSII (CP43 and CP47) subunits. J. Plant Physiol. 2012;169:1212–1215. doi: 10.1016/j.jplph.2012.04.001. [DOI] [PubMed] [Google Scholar]
- 73.Argyrou A., Blanchard J.S. Flavoprotein disulfide reductases: Advances in chemistry and function. Prog. Nucleic Acid Res. Mol. Biol. 2004;78:89–142. doi: 10.1016/S0079-6603(04)78003-4. [DOI] [PubMed] [Google Scholar]
- 74.Chivers P.T., Laboissiere M.C., Raines R.T. The CXXC motif: imperatives for the formation of native disulfide bonds in the cell. EMBO J. 1996;15:2659–2667. [PMC free article] [PubMed] [Google Scholar]
- 75.Heckler E.J., Rancy P.C., Kodali V.K., Thorpe C. Generating disulfides with the Quiescin-sulfhydryl oxidases. Biochim. Biophys. Acta. 2008;1783:567–577. doi: 10.1016/j.bbamcr.2007.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Thorpe C., Hoober K.L., Raje S., Glynn N.M., Burnside J., Turi G.K., Coppock D.L. Sulfhydryl oxidases: emerging catalysts of protein disulfide bond formation in eukaryotes. Arch. Biochem. Biophys. 2002;405:1–12. doi: 10.1016/S0003-9861(02)00337-5. [DOI] [PubMed] [Google Scholar]
- 77.Raje S., Thorpe C. Inter-domain redox communication in flavoenzymes of the quiescin/sulfhydryl oxidase family: Role of a thioredoxin domain in disulfide bond formation. Biochemistry. 2003;42:4560–4568. doi: 10.1021/bi030003z. [DOI] [PubMed] [Google Scholar]
- 78.Meyer S., Lorenz C., Baser B., Wordehoff M., Jager V., van den Heuvel J. Multi-host expression system for recombinant production of challenging proteins. PLoS One. 2013;8:e68674. doi: 10.1371/journal.pone.0068674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Overton T.W. Recombinant protein production in bacterial hosts. Drug Discov. Today. 2014;19:590–601. doi: 10.1016/j.drudis.2013.11.008. [DOI] [PubMed] [Google Scholar]
- 80.Laitinen O.H., Airenne K.J., Hytonen V.P., Peltomaa E., Mahonen A.J., Wirth T., Lind M.M., Makela K.A., Toivanen P.I., Schenkwein D., Heikura T., Nordlund H.R., Kulomaa M.S., Yla-Herttuala S. A multipurpose vector system for the screening of libraries in bacteria, insect and mammalian cells and expression in vivo. Nucleic Acids Res. 2005;33:e42. doi: 10.1093/nar/gni042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.De Marco A. Recombinant polypeptide production in E. coli: Towards a rational approach to improve the yields of functional proteins. Microb. Cell Fact. 2013;12 doi: 10.1186/1475-2859-12-101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Liu L., Yang H., Shin H.D., Chen R.R., Li J., Du G., Chen J. How to achieve high-level expression of microbial enzymes: Strategies and perspectives. Bioengineered. 2013;4:212–223. doi: 10.4161/bioe.24761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Brondyk W.H. Selecting an appropriate method for expressing a recombinanat protein. In: Burgess R.R., Deutscher M.P., editors. Guide to Protein Purification. 2nd ed. Volume 463. Gulf Professional Publishing; San Diego, CA, USA: 2009. p. 131. [DOI] [PubMed] [Google Scholar]
- 84.Klint J.K., Senff S., Saez N.J., Seshadri R., Lau H.Y., Bende N.S., Undheim E.A., Rash L.D., Mobli M., King G.F. Production of recombinant disulfide-rich venom peptides for structural and functional analysis via expression in the periplasm of E. coli. PLoS One. 2013;8:e63865. doi: 10.1371/journal.pone.0063865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Nakamoto H., Bardwell J.C. Catalysis of disulfide bond formation and isomerization in the Escherichia coli periplasm. Biochim. Biophys. Acta. 2004;1694:111–119. doi: 10.1016/j.bbamcr.2004.02.012. [DOI] [PubMed] [Google Scholar]
- 86.Gorman C.M., Groskreutz D.J., Marriott D. Method and host cells for producing two-chain relaxin polypeptide. 1,099,758. Eur. Patent. 2001 May 16;
- 87.Chen X., Bai Y., Zaro J.L., Shen W.C. Design of an in vivo cleavable disulfide linker in recombinant fusion proteins. BioTechniques. 2010;49:513–518. doi: 10.2144/000113450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Jia X.Y., Guo Z.Y., Wang Y., Xu Y., Duan S.S., Feng Y.M. Peptide models of four possible insulin folding intermediates with two disulfides. Protein Sci. 2003;12:2412–2419. doi: 10.1110/ps.0389303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Bessette P.H., Aslund F., Beckwith J., Georgiou G. Efficient folding of proteins with multiple disulfide bonds in the Escherichia coli cytoplasm. Proc. Natl. Acad. Sci. USA. 1999;96:13703–13708. doi: 10.1073/pnas.96.24.13703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Berkmen M. Production of disulfide-bonded proteins in Escherichia coli. Protein Expr. Purif. 2012;82:240–251. doi: 10.1016/j.pep.2011.10.009. [DOI] [PubMed] [Google Scholar]
- 91.Walsh G. Biopharmaceutical benchmarks 2010. Nat. Biotechnol. 2010;28:917–924. doi: 10.1038/nbt0910-917. [DOI] [PubMed] [Google Scholar]
- 92.Walsh G. Biopharmaceutical benchmarks 2014. Nat. Biotechnol. 2014;32:992–1000. doi: 10.1038/nbt.3040. [DOI] [PubMed] [Google Scholar]
- 93.Clark E.D.B. Refolding of recombinant proteins. Curr. Opin. Biotechnol. 1998;9:9157–9163. doi: 10.1016/s0958-1669(98)80109-2. [DOI] [PubMed] [Google Scholar]
- 94.Rudolph R., Lilie H. In vitro folding of inclusion body proteins. FASEB J. 1996;10:49–56. [PubMed] [Google Scholar]
- 95.Helenbeck R., Koths K.E., Cowgill C., Laird W.J. Production of purified, biologically active, bacterially produced recombinant human CSF-1. 4,929,700. US Patent. 1990 May 29;
- 96.Koths K.E., Halenbeck R.F. Method for promoting disulfide bond formation in recombinant proteins. 4,572,798. US Patent. 1986 Feb 25;
- 97.Freedman R.B. The formation of protein disulphide bonds. Curr. Opin. Struct. Biol. 1995;5:85–91. doi: 10.1016/0959-440X(95)80013-Q. [DOI] [PubMed] [Google Scholar]
- 98.Frand A.R., Cuozzo J.W., Kaiser C.A. Pathways for protein disulphide bond formation. Trends Cell Biol. 2000;10:203–210. doi: 10.1016/S0962-8924(00)01745-1. [DOI] [PubMed] [Google Scholar]
- 99.Luan C., Zhang H., Song D., Xie Y., Feng J., Wang Y. Expressing antimicrobial peptide cathelicidin-BF in Bacillus subtilis using SUMO technology. Appl. Microbiol. Biotechnol. 2014;98:3651–3658. doi: 10.1007/s00253-013-5246-6. [DOI] [PubMed] [Google Scholar]
- 100.Alibolandi M., Mirzahoseini H. Chemical assistance in refolding of bacterial inclusion bodies. Biochem. Res. Int. 2011;2011 doi: 10.1155/2011/631607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Sandee D., Tungpradabkul S., Kurokawa Y., Fukui K., Takagi M. Combination of Dsb coexpression and an addition of sorbitol markedly enhanced soluble expression of single-chain Fv in Escherichia coli. Biotechnol. Bioeng. 2005;91:418–424. doi: 10.1002/bit.20524. [DOI] [PubMed] [Google Scholar]
- 102.Kondo A., Kohda J., Endo Y., Shiromizu T., Kurokawa Y., Nishihara K., Yanagi H., Yura T., Fukuda H. Improvement of productivity of active horseradish peroxidase in Escherichia coli by coexpression of Dsb proteins. J. Biosci. Bioeng. 2000;90:600–606. doi: 10.1016/S1389-1723(00)90003-3. [DOI] [PubMed] [Google Scholar]
- 103.Derman A., Prinz W., Belin D., Beckwith J. Mutations that allow disulfide bond formation in the cytoplasm of Escherichia coli. Science. 1993;262:1744–1747. doi: 10.1126/science.8259521. [DOI] [PubMed] [Google Scholar]
- 104.Kaomek M., Mizuno K., Fujimura T., Sriyotha P., Cairns J.R.K. Cloning, expression, and characterization of an antifungal chitinase from Leucaena leucocephala de Wit. Biosci. Biotechnol. Biochem. 2003;67:667–676. doi: 10.1271/bbb.67.667. [DOI] [PubMed] [Google Scholar]
- 105.Shimizu T., Shibata H., Araya T., Nakatsu T., Miyairi K., Okuno T., Kato H. Expression, purification, and crystallization of endopolygalacturonase from a pathogenic fungus, Stereum purpureum, in Escherichia coli. Protein Expr. Purif. 2005;44:130–135. doi: 10.1016/j.pep.2005.06.001. [DOI] [PubMed] [Google Scholar]
- 106.Bar M., Bar-Ziv R., Scherf T., Fass D. Efficient production of a folded and functional, highly disulfide-bonded β-helix antifreeze protein in bacteria. Protein Expr. Purif. 2006;48:243–252. doi: 10.1016/j.pep.2006.01.025. [DOI] [PubMed] [Google Scholar]
- 107.Berndt C., Lillig C.H., Holmgren A. Thioredoxins and glutaredoxins as facilitators of protein folding. Biochim. Biophys. Acta. 2008;1783:641–650. doi: 10.1016/j.bbamcr.2008.02.003. [DOI] [PubMed] [Google Scholar]
- 108.Jurado P., de Lorenzo V., Fernandez L.A. Thioredoxin fusions increase folding of single chain Fv antibodies in the cytoplasm of Escherichia coli: Evidence that chaperone activity is the prime effect of thioredoxin. J. Mol. Biol. 2006;357:49–61. doi: 10.1016/j.jmb.2005.12.058. [DOI] [PubMed] [Google Scholar]
- 109.Heo M.A., Kim S.H., Kim S.Y., Kim Y.J., Chung J., Oh M.K., Lee S.G. Functional expression of single-chain variable fragment antibody against c-Met in the cytoplasm of Escherichia coli. Protein Expr. Purif. 2006;47:203–209. doi: 10.1016/j.pep.2005.12.003. [DOI] [PubMed] [Google Scholar]
- 110.De Marco A. Strategies for successful recombinant expression of disulfide bond-dependent proteins in Escherichia coli. Microb. Cell Fact. 2009;8 doi: 10.1186/1475-2859-8-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Huber D., Boyd D., Xia Y., Olma M. H., Gerstein M., Beckwith J. Use of thioredoxin as a reporter to identify a subset of Escherichia coli signal sequences that promote signal recognition particle-dependent translocation. J. Bacteriol. 2005;187:2983–2991. doi: 10.1128/JB.187.9.2983-2991.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Mergulhao F.J., Monteiro G.A., Larsson G., Bostrom M., Farewell A., Nystrom T., Cabral J.M., Taipa M.A. Evaluation of inducible promoters on the secretion of a ZZ-proinsulin fusion protein in Escherichia coli. Biotechnol. Appl. Biochem. 2003;38:87–93. doi: 10.1042/BA20030043. [DOI] [PubMed] [Google Scholar]
- 113.Obukowicz M.G., Gustafson M.E., Junger K.D., Leimgruber R.M., Wittwer A.J., Wun T.C., Warren T.G., Bishop B.F., Mathis K.J., McPherson D.T., et al. Secretion of active kringle-2-serine protease in Escherichia coli. Biochemistry. 1990;29:9737–9745. doi: 10.1021/bi00493a033. [DOI] [PubMed] [Google Scholar]
- 114.Pasek M., Boeggeman E., Ramakrishnan B., Qasba P.K. Galectin-1 as a fusion partner for the production of soluble and folded human beta-1,4-galactosyltransferase-T7 in E. coli. Biochem. Biophys. Res. Commun. 2010;394:679–684. doi: 10.1016/j.bbrc.2010.03.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Thie H., Schirrmann T., Paschke M., Dübel S., Hust M. SRP and Sec pathway leader peptides for antibody phage display and antibody fragment production in E. coli. New Biotechnol. 2008;25:49–54. doi: 10.1016/j.nbt.2008.01.001. [DOI] [PubMed] [Google Scholar]
- 116.Malik A., Rudolph R., Söhling B. A novel fusion protein system for the production of native human pepsinogen in the bacterial periplasm. Protein Expr. Purif. 2006;47:662–671. doi: 10.1016/j.pep.2006.02.018. [DOI] [PubMed] [Google Scholar]
- 117.Maskos K., Huber-Wunderlich M., Glockshuber R. DsbA and DsbC-catalyzed oxidative folding of proteins with complex disulfide bridge patterns in vitro and in vivo. J. Mol. Biol. 2003;325:495–513. doi: 10.1016/S0022-2836(02)01248-2. [DOI] [PubMed] [Google Scholar]
- 118.Xu Y., Yasin A., Tang R., Scharer J., Moo-Young M., Chou C.P. Heterologous expression of lipase in Escherichia coli is limited by folding and disulfide bond formation. Appl. Microbiol. Biotechnol. 2008;81:79–87. doi: 10.1007/s00253-008-1644-6. [DOI] [PubMed] [Google Scholar]
- 119.Sklar J.G. Defining the roles of the periplasmic chaperones SurA, Skp, and DegP in Escherichia coli. Genes Dev. 2007;21:2473–2484. doi: 10.1101/gad.1581007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Knoblauch N.T., Rudiger S., Schonfeld H.J., Driessen A.J., Schneider-Mergener J., Bukau B. Substrate specificity of the SecB chaperone. J. Biol. Chem. 1999;274:34219–34225. doi: 10.1074/jbc.274.48.34219. [DOI] [PubMed] [Google Scholar]
- 121.Mapa K., Tiwari S., Kumar V., Jayaraj G.G., Maiti S. Information encoded in non-native states drives substrate-chaperone pairing. Structure. 2012;20:1562–1573. doi: 10.1016/j.str.2012.06.014. [DOI] [PubMed] [Google Scholar]
- 122.Martin C., Rojas G., Mitchell J., Vincent K., Wu J., McCafferty J., Schofield D. A simple vector system to improve performance and utilisation of recombinant antibodies. BMC Biotechnol. 2006;6 doi: 10.1186/1472-6750-6-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Schmoldt H.-U., Wentzel A., Becker S., Kolmar H. A fusion protein system for the recombinant production of short disulfide bond rich cystine knot peptides using barnase as a purification handle. Protein Expr. Purif. 2005;39:82–89. doi: 10.1016/j.pep.2004.09.016. [DOI] [PubMed] [Google Scholar]
- 124.Niemann H.H., Schmoldt H.-U., Wentzel A., Kolmar H., Heinz D.W. Barnase fusion as a tool to determine the crystal structure of the small disulfide-rich protein McoEeTI. J. Mol. Biol. 2006;356:1–8. doi: 10.1016/j.jmb.2005.11.005. [DOI] [PubMed] [Google Scholar]
- 125.Schäffner J., Winter J., Rudolph R., Schwarz E. Cosecretion of chaperones and low-molecular-size medium additives increases the yield of recombinant disulfide-bridged proteins. Appl. Environ. Microbiol. 2001;67:3994–4000. doi: 10.1128/AEM.67.9.3994-4000.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Mogk A., Mayer M.P., Deuerling E. Mechanisms of protein folding: molecular chaperones and their application in biotechnology. ChemBioChem. 2002;3:807–814. doi: 10.1002/1439-7633(20020902)3:9<807::AID-CBIC807>3.0.CO;2-A. [DOI] [PubMed] [Google Scholar]
- 127.Bandyopadhyay A., Saxena K., Kasturia N., Dalal V., Bhatt N., Rajkumar A., Maity S., Sengupta S., Chakraborty K. Chemical chaperones assist intracellular folding to buffer mutational variations. Nat. Chem. Biol. 2012;8:238–245. doi: 10.1038/nchembio.768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Barth S., Huhn M., Matthey B., Klimka A., Galinski E.A., Engert A. Compatible-solute-supported periplasmic expression of functional recombinant proteins under stress conditions. Appl. Environ. Microbiol. 2000;66:1572–1579. doi: 10.1128/AEM.66.4.1572-1579.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Andrews H., Li Z., Altuve-Blanco A., Rivera M., Burnap R.L. Expression, mutagenesis, and characterization of recombinant low-potential cytochrome c550 of photosystem II. Biochemistry. 2005;44:6092–6100. doi: 10.1021/bi0500473. [DOI] [PubMed] [Google Scholar]
- 130.Winter J., Neubauer P., Glockshuber R., Rudolph R. Increased production of human proinsulin in the periplasmic space of Escherichia coli by fusion to DsbA. J. Biotechnol. 2001;84:175–185. doi: 10.1016/S0168-1656(00)00356-4. [DOI] [PubMed] [Google Scholar]
- 131.Shimamoto S., Katayama H., Okumura M., Hidaka Y. Chemical methods and approaches to the regioselective formation of multiple disulfide bonds. Curr. Protoc. Protein Sci. 2014;76:28.8.1–28.8.28. doi: 10.1002/0471140864.ps2808s76. [DOI] [PubMed] [Google Scholar]
- 132.Okumura M., Shimamoto S., Hidaka Y. Chemical methods for producing disulfide bonds in peptides and proteins to study folding regulation. Curr. Protoc. Protein Sci. 2014;76:28.7.1–28.7.13. doi: 10.1002/0471140864.ps2807s76. [DOI] [PubMed] [Google Scholar]
- 133.Hidaka Y. Overview of the regulation of disulfide bond formation in Peptide and protein folding. Curr. Protoc. Protein Sci. 2014;76:28.6.1–28.6.6. doi: 10.1002/0471140864.ps2806s76. [DOI] [PubMed] [Google Scholar]
- 134.Harada T., Kurimoto E., Moriyama Y., Ejima D., Sakai T., Nohara D., Kato K. Application of combined reagent solution to the oxidative refolding of recombinant human interleukin 6. Chem. Pharm. Bull. 2001;49:1128–1131. doi: 10.1248/cpb.49.1128. [DOI] [PubMed] [Google Scholar]
- 135.Ito L., Okumura M., Tao K., Kasai Y., Tomita S., Oosuka A., Yamada H., Shibano T., Shiraki K., Kumasaka T., et al. Glutathione ethylester, a novel protein refolding reagent, enhances both the efficiency of refolding and correct disulfide formation. Protein J. 2012;31:499–503. doi: 10.1007/s10930-012-9427-4. [DOI] [PubMed] [Google Scholar]
- 136.Arai K., Noguchi M., Singh B.G., Priyadarsini K.I., Fujio K., Kubo Y., Takayama K., Ando S., Iwaoka M. A water-soluble selenoxide reagent as a useful probe for the reactivity and folding of polythiol peptides. FEBS Open Biol. 2013;3:55–64. doi: 10.1016/j.fob.2012.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]