Abstract
The AP2/ERF family of plant transcription factors (TFs) regulate a variety of developmental and physiological processes. Here, we report the isolation of six AP2/ERF TF family genes from Chrysanthemum nankingense. On the basis of sequence similarity, one of these belonged to the Ethylene Responsive Factor (ERF) subfamily and the other five to the Dehydration Responsive Element Binding protein (DREB) subfamily. A transient expression experiment showed that all six AP2/ERF proteins localized to the nucleus. A yeast-one hybrid assay demonstrated that CnDREB1-1, 1-2 and 1-3 all function as transactivators, while CnERF1, CnDREB3-1 and 3-2 have no transcriptional activation ability. The transcription response of the six TFs in response to wounding, salinity and low temperature stress and treatment with abscisic acid (ABA), salicylic acid (SA) and jasmonic acid (JA) showed that CnERF1 was up-regulated by wounding and low temperature stress but suppressed by salinity stress. The transcription of CnDREB1-1, 1-2 and 1-3 was down-regulated by ABA and JA to varying degrees. CnDREB3-1 and 3-2 was moderately increased or decreased by wounding and SA treatment, suppressed by salinity stress and JA treatment, and enhanced by low temperature stress and ABA treatment.
Keywords: hormone, PCR, stress response, transcription pattern
1. Introduction
Plants have evolved a diversity of responses to external stress. Many of these rely on signalling provided by the phytohormones abscisic acid (ABA), jasmonate (JA), salicylic acid (SA) or ethylene [1]. A large class of rapid defense responses are mediated by transcription factors (TFs) [2]. The AP2/ERF TFs belong to a particularly diverse family of TFs which are recognized by the presence of a characteristic 57–66 residue long AP2/ERF DNA binding domain [3]. The AP2/ERF family is divided into four major sub-families based on sequence similarity and the number of AP2/ERF domains present; these sub-families are AP2, DREB/ C-repeat binding factor (CBF), ERF and RELATED TO ABI3/VP1 (RAV) [4,5]. DREB and ERF TFs have frequently been implicated in the response to drought, salinity, rapid changes in temperature and disease [6]. AP2 TFs harbor two AP2/ERF domains, and many of them participate in the regulation of development [7]. The RAV subfamily TFs contain one AP2/ERF domain and one B3 domain, interact with either ethylene or brassinosteroids [8], and form part of the response to biotic and abiotic stress [9]. The DREB/CBF and ERF TFs each harbor only one AP2/ERF domain. DREB/CBF TFs are an important component of the plant abiotic stress tolerance [10], while the ERF TFs participate in the response to both biotic and abiotic stress [11]. The Arabidopsis thaliana genome harbors 147 AP2/ERF TFs [12], rice 167 [13], poplar 200 [5], grapevine 132 [4], soybean 148 [14] and bread wheat 117 [15].
The diploid species Chrysanthemum nankingense is native to China [16]. It is used both as a vegetable and as a source of anti-cancer flavonoids and aromatic oils. The species also features a number of genes which have proven to be beneficial in chrysanthemum breeding [17]. As yet, the TF content of this species has not been explored. Here, the aim was to isolate and characterize a number of AP2/ERF TFs from C. nankingense.
2. Results and Discussion
2.1. Identification of Six AP2/ERF TFs in C. nankingense
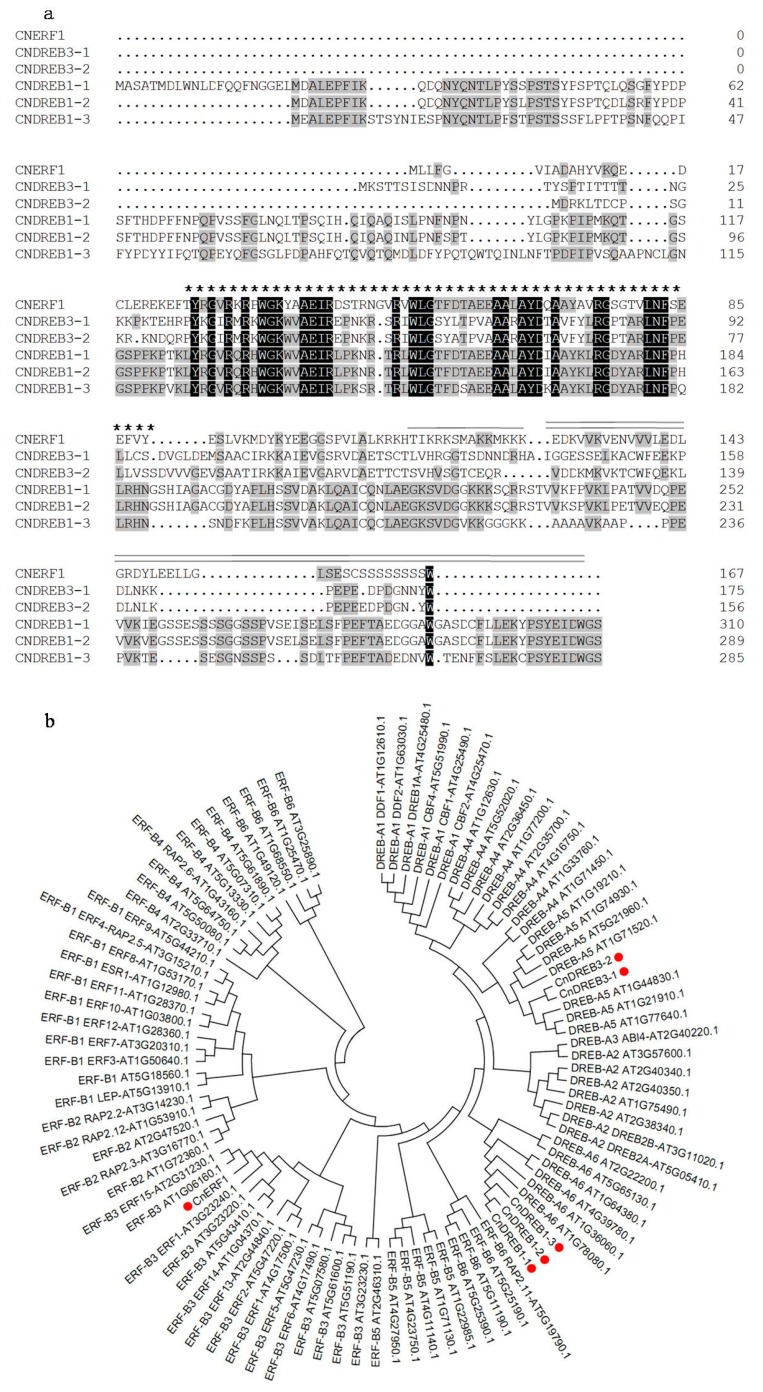
Six AP2/ERF full-length cDNAs were isolated and designated CnERF1, CnDREB1-1, 1-2, 1-3, 3-1 and 3-2. Their length varied from 683 to 1246 bp, and their predicted translation products comprised from 156 to 311 residues. CnERF1 belongs to ERF subfamily group B-3, while CnDREB1-1, 1-2 and 1-3 all belong to DREB subfamily A-6 and CnDREB3-1 and 3-2 to DREB group A-5 (Table 1, Figure 1b). All six enc00oded proteins having a single 64 residue AP2 domain, except for CnERF1 (65 residues) (Figure 1a). Analysis using Smart online software (available online: http://smart.embl-heidelberg.de/) showed that the conserved residues in the CnERF1 AP2/ERF were A14 and D19. The corresponding positions in CnDREB1-1, 1-2 and 1-3 were occupied by, respectively valine and leucine, whereas in CnDREB3-1 and 3-2, the 19th residue was glutamic acid. The 14th and 19th residues are both located on the β-sheet of the AP2/ERF domain.
Table 1.
CnAP2/ERF TF sequences and the identity of likely A. thaliana homologs.
| Gene | GenBank Accession No. | cDNA Length (bp) | Amino Acids Length (aa) | AtAP2/ERF Orthologs | Locus Name | E-Value |
|---|---|---|---|---|---|---|
| CnERF1 | KF986840 | 683 | 167 | ERF1B | AT3G23240 | 1 × 10−38 |
| CnDREB1-1 | KF986841 | 1241 | 311 | RAP2.4 | AT1G78080.1 | 2 × 10−69 |
| CnDREB1-2 | KF986842 | 1164 | 290 | RAP2.4 | AT1G78080.1 | 3 × 10−69 |
| CnDREB1-3 | KF986843 | 1246 | 286 | ERF055 | AT1G78080.1 | 8 × 10−42 |
| CnDREB3-1 | KF986844 | 881 | 175 | ERF008 | AT2G23340.1 | 4 × 10−45 |
| CnDREB3-2 | KF986845 | 658 | 156 | ERF008 | AT2G23340.1 | 1 × 10−48 |
Figure 1.
Deduced peptide sequences of the CnAP2/ERF transcription factor (TF) products and their phylogenetic relationship with A. thaliana homologs. (a) The deduced polypeptide sequences; residues shared by at least three of the six sequences are shown shaded, whereas those conserved across all six polypeptides are marked in dark grey. Asterisks indicate the conserved DNA-binding AP2/ERF domain, a double overline indicates the putative acidic domain and a black underline indicates the putative nuclear localization signal; (b) Phylogeny of the CnAP2/ERF TF products. Dots indicate likely homologs.
Many examples have been provided where the heterologous expression of a TF can enhance abiotic stress tolerance [18,19]. Five of the six C. nankingense TFs isolated here belong to the DREB class, and one to the ERF class; both these types of TF are heavily implicated in the regulation of the stress response [20]. TFs are an attractive target for engineering the stress tolerance of crop plants [21], since a single TF commonly regulates a whole suite of genes, and thus can control a whole adaptive pathway [22].
2.2. Subcellular Localization of Cnap2/Erf Products and the Transcription Activation of the TFs
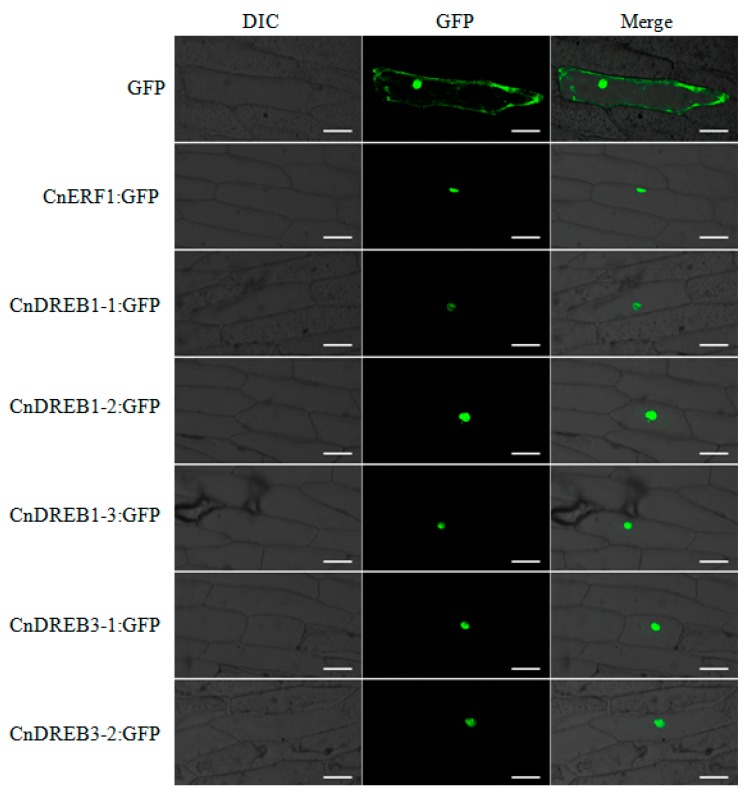
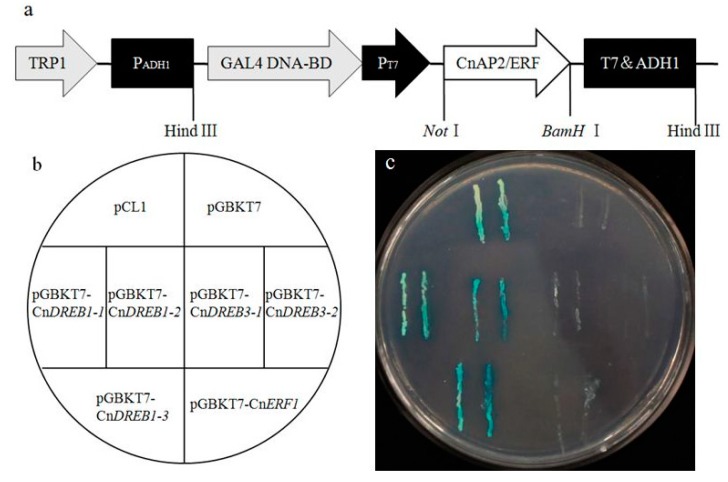
The outcome of the transient transformation of the six CnAP2/ERF TFs is shown in Figure 2. As expected for a TF, the CnAP2/ERF-GFP signal was localized predominantly in the nucleus, while the control GFP only transgene was expressed throughout the cell. Based on the yeast one hybrid assay, CnDREB1-1, 1-2 and 1-3 all showed transcription activation activity in yeast (Figure 3), while CnERF1, CnDREB3-1 and 3-2 did not.
Figure 2.
Localization of transiently expressed CnAP2/ERF TF products in onion epidermal cells. The upper row shows the control 35S::GFP signal, and each of the lower rows the signal from one of the 35S::CnAP2/ERF-GFP transgenes. The left panel shows bright field images, the middle one green fluorescence signals detected at 488 nm and the right one the merged Green Fluorescent Protein (GFP) and bright field images. Bar: 50 μm.
Figure 3.
Transcriptional activation activity of the CnAP2/ERF TFs. (a) The structure of the pGBKT7-CnAP2/ERF plasmid; (b) the arrangement of yeast strains on the plate; (c) the growth of transformed yeast cells on SD/-His-Ade + 20 mg/mL X-α-gal medium. pCL1 and pGBKT7 are positive and negative controls, respectively.
The product of all six of the TFs localized, as would be expected for a TF, in the nucleus (Figure 2), while the test of their transactivation activity was positive for three of them (CnDREB3-1, 3-2 and CnERF1, see Figure 3). The EAR (ERF-associated amphiphilic repressor) motif has been proposed to convert transcriptional activators into dominant repressors in plant cells [23], and this motif is present in both CnDREB3-1 and 3-2, inferring that these two TFs might be repressors. A-5 subgroup of AP2/ERF family contains a clade of proteins that have a functional EAR motif at their C-terminus [24,25]. Moreover, genes encoding these EAR motif-containing proteins are upregulated in transgenic plants overexpressing DREB1A/CBF3 or DREB2A CA [26,27]. Overexpression of these genes results in reduced expression of DREB1/CBF and DREB2 target genes under cold and dehydration, respectively [28,29]. Taken together, it is possible that CnDREB3-1 and 3-2 function in negative feedback regulation of the DREB1/CBF and DREB2 pathways [30]. The product of the tomato TF LeERF1 binds rather weakly to its target GCC box [31], while that of AtERF1 is quite sensitive to changes in the GCC box sequence (AGCCGCC) [32]. The binding activity of CnERF1 to GCC box remained unknown. In addition, CnERF1 does not possess EAR motif; however, whether the CnERF1 acts as transcriptional repressor remains to be studied.
2.3. Transcription of CnAP2/ERF TFs in Response to Abiotic Stress
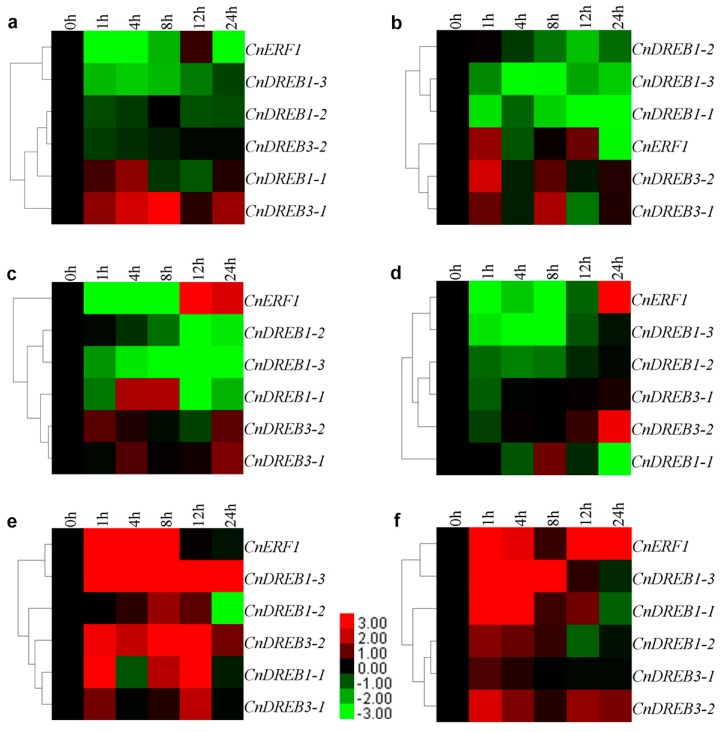
Transcript abundance of CnDREB1-1 was raised by salinity stress, peaking 4 h after the stress was imposed, whereas CnDREB3-1 was up-regulated throughout the salinity stress episode, peaking at 8 h. The other four TFs were down-regulated to differing degrees; thus CnDREB1-3 was down-regulated throughout, while the abundance of CnERF1, CnDREB1-2 and 3-2 transcript recovered to the background level after some time (Figure 4a). CnDREB3-1 and 3-2 were substantially up-regulated after a 1 and 8 h exposure to ABA, but CnDREB1-1, 1-2 and 1-3 were down-regulated throughout the treatment, and CnERF1 was only marginally induced at 1 and 12 h (Figure 4b). The effect of SA treatment was to initially induce CnDREB3-1 and 3-2 transcription, but the transcript abundance then declined, only to increase again by 24 h. CnDREB1-2 and 1-3 were both strongly down-regulated, while CnDREB1-1 was induced initially and CnERF1 at a somewhat later time (Figure 4c). When subjected to JA treatment, the transcription of both CnDREB3-1 and 3-2 was largely unaffected, that of CnERF1 was increased at the end of the period and the other three TFs were all repressed to differing extents (Figure 4d). The low temperature stress induced CnDREB1-3 and 3-2 throughout the treatment, while it caused an oscillation in the transcription of CnDREB1-1 and 3-1. CnERF1 was up-regulated within the first 8 h, but later was suppressed, and CnDREB1-2 behaved similarly (Figure 4e). Wounding increased the transcript abundance of all six TFs initially, with CnDREB1-1 and 1-3 peaking at 4 h, CnERF1 and CnDREB3-2 still substantially showing up-regulation at 24 h, and CnDREB1-3, 1-1, 1-2 and 3-1 being variously suppressed towards the end of the stress episode (Figure 4f). All obtained data were displayed in Table S1.
Figure 4.
Differential transcript abundance of the CnAP2/ERF TFs in response to (a) salinity stress; (b) abscisic acid (ABA) treatment; (c) salicylic acid (SA) treatment; (d) jasmonic acid (JA) treatment; (e) low temperature stress; and (f) wounding. Green cells indicate suppressed and red ones enhanced levels of transcript abundance compared to the relevant control. Black cells represent no significant change of transcript abundance.
The differential responses of the AP2/ERF TFs to the various abiotic stresses and hormone treatments suggested that each TF has a specific physiological role. Emerging evidence suggests that pathways regulated by ABA, SA, JA and ethylene are involved in a substantial amount of crosstalk with biotic and abiotic stress pathways [33]. The SA signal transduction pathway can be antagonistic to the ethylene/JA pathway and JA is considered to be a key regulator of stress-induced gene expression across the plant kingdom [34]. The tomato TF Pti4 and the A. thaliana TF AtERF1 are both induced by SA as well as by JA [32]. AtERF1 plays a positive role in drought, salt and heat stress tolerance by stress-specific gene regulation, which integrates JA and abscisic acid signals [35]. Here, CnERF1 was up-regulated by wounding and low temperature, but suppressed by salinity and ABA (Figure 4), suggesting its involvement in the response to the former two stresses. In case of DREB genes, members of the DREB1/CBF subfamily are rapidly induced in response to low temperature, and when constitutively expressed, enhance freezing tolerance in A. thaliana [36]. Some DREB TFs are regulated by ABA, high-salt and cold [37], as was the case for CnDREB3-1, 3-2 and 3-3; the transcription of these same TFs was also affected by JA. CnDREB1-1 and 1-2 was moderately increased/decreased by wounding and SA treatment, suppressed by salinity stress and JA treatment, and enhanced by low temperature stress and ABA treatment; the implication is that both CnDREB1-1 and 1-2 are involved in the low temperature and the ABA response. Indeed, CnDREB1-1 and 1-3 were more likely involved in the abiotic stress response than CnDREB1-2.
3. Experimental Section
3.1. Plant Materials and Stress Treatments
The accession of C. nankingense used was obtained from the Chrysanthemum Germplasm Resource Preserving Centre, Nanjing Agriculture University, Nanjing, China. The plants were propagated by cutting and grown in a 1:1 (v/v) mixture of garden soil and vermiculite with no additional fertilizer provided. Rooted seedlings were maintained in a greenhouse. Young plants at the 6–8 leaf stage were used to assess the transcription response to abiotic stress and phytohormne treatment. Salinity stress was imposed by transferring young plants for one day to a liquid medium containing 200 mM NaCl [18]; the low temperature stress consisted of an exposure for one day to 4 °C under a 16 h photoperiod (50 µmol·m−2·s−1·light). The second fully expanded leaf counted from the apex was sampled in each case. A wounding treatment was conducted by puncturing three leaves in five places with a size 10 (approximately 0.30 mm diameter) needle, and repeating this with ten further punctures after 24 h. The second true leaf was sampled 2 h after the second wounding event [38]. The phytohormone treatments involved spraying the leaves with either 20 µM ABA, 100 µM methyl JA or 10 µM SA. Control plants were sprayed with distilled water. Leaves were sampled at prior to the application of phytohormones, and then again after 1, 4, 8, 12 and 24 h. each time point stress/phytohormone treatment was imposed on three plants and each treatment was replicated three times. Control plants were kept at 22 °C. The sampled leaf material was snap-frozen in liquid nitrogen and stored at −70 °C.
3.2. Isolation and Sequencing of Full-Length CnAP2/ERF cDNAs
Total RNA was isolated using the RNAiso reagent (TaKaRa, Tokyo, Japan) following the manufacturer’s instructions. The first cDNA strand was synthesized from 1 µg total RNA using an M-MLV RTase cDNA Synthesis kit (TaKaRa) according to the manufacturer’s instructions. Primer pairs were designed (their sequences, along with all other PCR primer sequences used here are listed in Table 2) to amplify CnAP2/ERF fragments based on an EST sequence identified in a C. nankingense transcriptome database [39]. RACE PCR was then used to obtain the full-length cDNA. For the 3' reaction, the first cDNA strand was synthesized using the dT adaptor primer dT-AP, and this was followed by a nested PCR based on a specific primer pair and the adaptor primer AP. For the 5' reaction, the nested PCR used the primers AAP and AUAP provided with the 5' RACE System kit v2.0 (Invitrogen, Camarillo, CA, USA), along with gene-specific primers. For sequences, the amplicons were purified using an AxyPrep DNA Gel Extraction kit (Axygen, Shanghai, China) and cloned into the pMD19-T easy vector (TaKaRa). Finally, pairs of gene-specific primers were designed to amplify each open reading frame (ORF), which was also cloned into the pMD19-T easy vector for sequencing.
Table 2.
PCR primer sequences utilized in this study.
| Primer Name | Sequence (5' to 3') | Annotation |
|---|---|---|
| AP | AAGCAGTGGTATCAACGCAGAGTAC | Universal primers for 3' RACE |
| dT-AP | AAGCAGTGGTATCAACGCAGAGTACTTTTTTTTTTTTTTTT | |
| AUAP | GGCCACGCGTCGACTAGTAC | Universal primers for 5' RACE |
| AAP | GGCCACGCGTCGACTAGTACGGGIIGGGIIGGGIIG | |
| CnERF1-F | ATGCTTCTCTTCGGGGTCATTGCT | ORF of CnAP2/ERFs |
| CnERF1-R | TCACCAACTAGAACTACTGCTGCTGCT | |
| CnDREB1-1-F | ATGGCTTCAGCTACAATGGACTTAT | |
| CnDREB1-1-R | ATATACCCTCATAAACACTGCCACG | |
| CnDREB1-2-F | ATGGATGCACTAGAACCATTCATCAAGC | |
| CnDREB1-2-R | CTAGATAGAACCCCAATCAATCTCGTAC | |
| CnDREB1-3-F | ATGGAAGCACTTGAACCTTTTATCA | |
| CnDREB1-3-R | CTATAATGAACCCCAGTCAATCTCG | |
| CnDREB3-1-F | ATGAAATCCACAACATCCATCAGCG | |
| CnDREB3-1-R | TCACCAATAATTATTTCCATCCGGATC | |
| CnDREB3-2-F | ATGGACAGAAAATTAACAGACTGTCCATC | |
| CnDREB3-2-R | TCACCAATAATTTCCATCCGGATCTTCT | |
| EFIα-F | TTTTGGTATCTGGTCCTGGAG | qRT-PCR for CnEFIα |
| EFIα-R | CCATTCAAGCGACAGACTCA | |
| CnERF1-BamHI-F | CGGGATCCGGATGCTTCTCTTCGGGGTCATTG | Vector construction of CnAP2/ERFs |
| CnERF1-NOTI-R | TTGCGGCCGCGATCACCAACTAGAACTACTGCTGCTG | |
| CnDREB1-1-BamHI-F | CGGGATCCGGATGGCTTCAGCTACAATGGAC | |
| CnDREB1-1-NOTI-R | TTGCGGCCGCGAGATAGAACCCCAATCAATCTCGTA | |
| CnDREB1-2-BamHI-F | CGGGATCCGGATGGATGCACTAGAACCATTCAT | |
| CnDREB1-2-NOTI-R | TTGCGGCCGCGAGATAGAACCCCAATCAATCTCGTA | |
| CnDREB1-3-BamHI-F | CGGGATCCGGATGGAAGCACTTGAACCTTTTATC | |
| CnDREB1-3-NOTI-R | TTGCGGCCGCGATATTGAACCCCAGTCAATCTCGTAC | |
| CnDREB3-1-BamHI-F | CGGGATCCGGATGAAATCCACAACATCCATCAGC | |
| CnDREB3-1-NOTI-R | TTGCGGCCGCGATCACCAATAATTATTTCCATCCGG | |
| CnDREB3-2-BamHI-F | CGGGATCCGGATGGACAGAAAATTAACAGACTGTC | |
| CnDREB3-2-NOTI-R | TTGCGGCCGCGATCACCAATAATTTCCATCCGG |
3.3. Sequence Alignment and Phylogenetic Analysis
The presence of the AP2 domain in the isolated TFs was detected by a Blast search (available online: http://www.ncbi.nlm.nih.gov/index.html). A. thaliana AP2/ERF sequences were downloaded Arabidopsis thaliana transcription factor database [20], and combined with the newly acquired CnAP2/ERF sequences to perform a multiple alignment analysis based on ClustalW software [40]. The subsequent phylogenetic analysis relied on a Neighbor-Joining method, and a graphical representation was produced with the support of MEGA v5 software [41]. Internal branching support was estimated from 1000 bootstrap replicates.
3.4. Subcellular Localization of CnAP2/ERF
The plasmid for transient transformation was generated using the Invitrogen Gateway system, according to the manufacturer’s instructions. The CnAP2/ERF ORFs, lacking their stop codon, were amplified using a Phusion® High-Fidelity PCR kit (New England Biolabs, MA, USA) (primers listed in Table 1), then cloned into the pMD19-T easy vector (Takara) and validated by DNA sequencing. Each of the isolated CnAP2/ERF TFs was inserted into the pENTR™ 1A vector (Invitrogen) using T4 DNA ligase (Fermentas, Burlington, ON, Canada), and the resulting construct recombined with pMDC43 to form the fusion vectors CnERF1-GFP, CnDREB1-1-GFP, CnDREB1-2-GFP, CnDREB1-3-GFP, CnDREB3-1-GFP and CnDREB3-2-GFP using the LR Clonase™ II enzyme mix (Invitrogen). Each plasmid DNA was then transiently introduced into onion epidermal cells, using a He-driven particle accelerator (PDS-1000; Bio-Rad, Hercules, CA, USA) according to the manufacturer’s instructions. As a control, an empty vector containing only the GFP sequence was transformed into similar epidermal cells. After bombardment, the onion peels were incubated for 16 h on Murashige and Skoog plates in the dark [42]. Green fluorescence was monitored by confocal laser microscopy [43].
3.5. Trans-Activation Activity Assay of CnAP2/ERF
The ORF (Open reading frame) of each CnAP2/ERF TF was amplified (primer sequences given in Table 1) and inserted into the NotI/BamHI cloning site of the yeast expression vector pGBKT7 to produce pBD-CnERF1, pBD-CnDREB1-1, pBD-CnDREB1-2, pBD-CnDREB1-3, pBD-CnDREB3-1 and pBD-CnDREB3-2. Either one of these constructs or an empty pGBKT7 (negative control) or pCL1 (positive control) plasmid was introduced into the yeast strain Y2Hgold (Clontech, Mountain View, CA, USA) [18]. Selection for transformants carrying either one of the pBD-CnAP2/ERFs or pGBKT7 was carried out by culturing on SD/-Trp medium, while the pCL1 transformants were selected on SD/-Leu medium. All three classes of transformant cells were transferred to an SD/-His-Ade medium supplemented with 20 mg/mL X-α-gal to observe cell growth. Since the expression of His3 is regulated by the GAL4-BD region, a CnAP2/ERF TF possessing activation ability should bind to the GAL4 BD upstream promoter sequence of His3, thereby activating its expression and enabling the transformed cells to grow on SD/-His-Ade + 20 mg/mL X-α-gal medium [44].
3.6. Transcription Profiling of the CnAP2/ERF TFs
Transcription profiling was achieved using qRT-PCR, based on the CnAP2/ERF-RT-F/R primers listed in Table 1. The chosen reference gene was EF1a (Genbank accession number: KF305681). Each 20 µL reaction contained 10 µL SYBR® Premix Ex Taq™ II (Takara), 0.2 µM of each primer and 10 ng cDNA template. The amplification regime consisted of an initial denaturation (95 °C/2 min), followed by 40 cycles of 95 °C/10 s, 55 °C/15 s, 72 °C/20 s [45]. A melting curve analysis was conducted following each assay to ensure the specificity of the amplicons. The qRT-PCRs were performed on three independent sets of RNA samples. Relative transcript abundances were calculated by the 2−∆∆Ct method [46]. The relative transcription levels of each CnAP2/ERF were log2 transformed, and the profiles compared using Cluster v3.0 software [47] and visualized using Treeview [48]. SPSS v17.0 software (SPSS Inc., Chicago, IL, USA) was used for all statistical analyses.
4. Conclusions
In summary, the six C. nankingense AP2/ERF TFs appear to have distinct roles as they responded differentially to the various stresses and hormonal treatments. Our results lay the basis for further investigation into the regulatory mechanism of CnAP2/ERF genes in various stresses.
Acknowledgments
This study was funded by the National Natural Science Foundation of China (31272202), the Program for New Century Excellent Talents in University of the Chinese Ministry of Education (NCET-10-0492), the Program for Hi-Tech Research, Jiangsu, China (BE2012350, BE2011325), Fund for Independent Innovation of Agricultural Sciences in Jiangsu Province [CX(12)2020], China Postdoctoral Science Foundation (2014M561673), and the Fundamental Research Funds for the Central Universities (KYZ201112).
Supplementary Materials
Supplementary materials can be found at http://www.mdpi.com/1422-0067/16/01/2052/s1.
Author Contributions
Chunyan Gao, Peiling Li and Aiping Song contributed to experimental operation, bioinformatics analysis, and writing of the manuscript. Fadi Chen, Jiafu Jiang and Sumei Chen designed the experiments and contributed to revisions of the manuscript. Haibin Wang and Xiangyu Qi contributed to the phytohormone treatments. Yinjie Wang and Liping Ren contributed to the abiotic stresses treatments. All authors read and approved the final manuscript.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Anderson J.P., Badruzsaufari E., Schenk P.M., Manners J.M., Desmond O.J., Ehlert C., Maclean D.J., Ebert P.R., Kazan K. Antagonistic interaction between abscisic acid and jasmonate-ethylene signaling pathways modulates defense gene expression and disease resistance in Arabidopsis. Plant Cell. 2004;16:3460–3479. doi: 10.1105/tpc.104.025833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ashraf M. Inducing drought tolerance in plants: Recent advances. Biotechnol. Adv. 2010;28:169–183. doi: 10.1016/j.biotechadv.2009.11.005. [DOI] [PubMed] [Google Scholar]
- 3.Okamuro J.K., Caster B., Villarroel R., van Montagu M., Jofuku K.D. The AP2 domain of APETALA2 defines a large new family of DNA binding proteins in Arabidopsis. Proc. Natl. Acad. Sci. USA. 1997;94:7076–7081. doi: 10.1073/pnas.94.13.7076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhuang J., Cai B., Peng R., Zhu B., Jin X., Xue Y., Gao F., Fu X., Tian Y., Zhao W. Genome-wide analysis of the AP2/ERF gene family in Populus trichocarpa. Biochem. Biophys. Res. Commun. 2008;371:468–474. doi: 10.1016/j.bbrc.2008.04.087. [DOI] [PubMed] [Google Scholar]
- 5.Zhuang J., Peng R., Cheng Z.M., Zhang J., Cai B., Zhang Z., Gao F., Zhu B., Fu X., Jin X. Genome-wide analysis of the putative AP2/ERF family genes in Vitis vinifera. Sci. Hortic. 2009;123:73–81. doi: 10.1016/j.scienta.2009.08.002. [DOI] [Google Scholar]
- 6.Qin Q., Liu J., Zhang Z., Peng R., Xiong A., Yao Q., Chen J. Isolation, optimization, and functional analysis of the cDNA encoding transcription factor OsDREB1B in Oryza Sativa L. Mol. Breed. 2007;19:329–340. doi: 10.1007/s11032-006-9065-7. [DOI] [Google Scholar]
- 7.Boutilier K., Offringa R., Sharma V.K., Kieft H., Ouellet T., Zhang L., Hattori J., Liu C.-M., van Lammeren A.A., Miki B.L. Ectopic expression of BABY BOOM triggers a conversion from vegetative to embryonic growth. Plant Cell. 2002;14:1737–1749. doi: 10.1105/tpc.001941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Alonso J.M., Stepanova A.N., Leisse T.J., Kim C.J., Chen H., Shinn P., Stevenson D.K., Zimmerman J., Barajas P., Cheuk R. Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science. 2003;301:653–657. doi: 10.1126/science.1086391. [DOI] [PubMed] [Google Scholar]
- 9.Sohn K.H., Lee S.C., Jung H.W., Hong J.K., Hwang B.K. Expression and functional roles of the pepper pathogen-induced transcription factor RAV1 in bacterial disease resistance, and drought and salt stress tolerance. Plant Mol. Biol. 2006;61:897–915. doi: 10.1007/s11103-006-0057-0. [DOI] [PubMed] [Google Scholar]
- 10.Thomashow M.F. Plant cold acclimation: Freezing tolerance genes and regulatory mechanisms. Annu. Rev. Plant Biol. 1999;50:571–599. doi: 10.1146/annurev.arplant.50.1.571. [DOI] [PubMed] [Google Scholar]
- 11.Xu Z., Xia L., Chen M., Cheng X., Zhang R., Li L., Zhao Y., Lu Y., Ni Z., Liu L. Isolation and molecular characterization of the Triticum aestivum L. ethylene-responsive factor 1 (TaERF1) that increases multiple stress tolerance. Plant Mol. Biol. 2007;65:719–732. doi: 10.1007/s11103-007-9237-9. [DOI] [PubMed] [Google Scholar]
- 12.Nakano T., Suzuki K., Fujimura T., Shinshi H. Genome-wide analysis of the ERF gene family in Arabidopsis and rice. Plant Physiol. 2006;140:411–432. doi: 10.1104/pp.105.073783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Riaño-Pachón D.M., Ruzicic S., Dreyer I., Mueller-Roeber B. PlnTFDB: An integrative plant transcription factor database. BMC Bioinform. 2007;8 doi: 10.1186/1471-2105-8-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang G., Chen M., Chen X., Xu Z., Guan S., Li L., Li A., Guo J., Mao L., Ma Y. Phylogeny, gene structures, and expression patterns of the ERF gene family in soybean (Glycine max L.) J. Exp. Bot. 2008;59:4095–4107. doi: 10.1093/jxb/ern248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhuang J., Chen J., Yao Q., Xiong F., Sun C., Zhou X., Zhang J., Xiong A. Discovery and expression profile analysis of AP2/ERF family genes from Triticum aestivum. Mol. Biol. Rep. 2011;38:745–753. doi: 10.1007/s11033-010-0162-7. [DOI] [PubMed] [Google Scholar]
- 16.Zhao H., Liu Z., Hu X., Yin J., Li W., Rao G., Zhang X., Huang C., Anderson N., Zhang Q. Chrysanthemum genetic resources and related genera of Chrysanthemum collected in China. Genet. Res. Crop. Evol. 2009;56:937–946. doi: 10.1007/s10722-009-9412-8. [DOI] [Google Scholar]
- 17.Cheng X., Chen S., Chen F., Fang W., Deng Y., She L. Interspecific hybrids between Dendranthema morifolium (Ramat.) Kitamura and D. nankingense (Nakai) Tzvel. achieved using ovary rescue and their cold tolerance characteristics. Euphytica. 2010;172:101–108. doi: 10.1007/s10681-009-0056-8. [DOI] [Google Scholar]
- 18.Gao H., Song A., Zhu X., Chen F., Jiang J., Chen Y., Sun Y., Shan H., Gu C., Li P. The heterologous expression in Arabidopsis of a chrysanthemum Cys2/His2 zinc finger protein gene confers salinity and drought tolerance. Planta. 2012;235:979–993. doi: 10.1007/s00425-011-1558-x. [DOI] [PubMed] [Google Scholar]
- 19.Song A., An J., Guan Z., Jiang J., Chen F., Lou W., Fang W., Liu Z., Chen S. The constitutive expression of a two transgene construct enhances the abiotic stress tolerance of chrysanthemum. Plant Physiol. Biochem. 2014;80:114–120. doi: 10.1016/j.plaphy.2014.03.030. [DOI] [PubMed] [Google Scholar]
- 20.Guo A., He K., Liu D., Bai S., Gu X., Wei L., Luo J. DATF: A database of Arabidopsis transcription factors. Bioinformatics. 2005;21:2568–2569. doi: 10.1093/bioinformatics/bti334. [DOI] [PubMed] [Google Scholar]
- 21.Sakuma Y., Liu Q., Dubouzet J.G., Abe H., Shinozaki K., Yamaguchi-Shinozaki K. DNA-Binding Specificity of the ERF/AP2 Domain of Arabidopsis DREBs, Transcription Factors Involved in Dehydration-and Cold-Inducible Gene Expression. Biochem. Biophys. Res. Commun. 2002;290:998–1009. doi: 10.1006/bbrc.2001.6299. [DOI] [PubMed] [Google Scholar]
- 22.Khong G.N., Richaud F., Coudert Y., Pati P.K., Santi C., Périn C., Breitler J.C., Meynard D., Vinh D.N., Guiderdoni E. Modulating rice stress tolerance by transcription factors. Biotechnol. Genet. Eng. 2008;25:381–404. doi: 10.5661/bger-25-381. [DOI] [PubMed] [Google Scholar]
- 23.Hiratsu K., Matsui K., Koyama T., Ohme-Takagi M. Dominant repression of target genes by chimeric repressors that include the EAR motif, a repression domain, in Arabidopsis. Plant J. 2003;34:733–739. doi: 10.1046/j.1365-313X.2003.01759.x. [DOI] [PubMed] [Google Scholar]
- 24.Ohta M., Matsui K., Hiratsu K., Shinshi H., Ohme-Takagi M. Repression domains of class II ERF transcriptional repressors share an essential motif for active repression. Plant Cell. 2001;13:1959–1968. doi: 10.1105/tpc.13.8.1959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen M., Wang Q.-Y., Cheng X.-G., Xu Z.-S., Li L.-C., Ye X.-G., Xia L.-Q., Ma Y.-Z. GmDREB2, a soybean DRE-binding transcription factor, conferred drought and high-salt tolerance in transgenic plants. Biochem. Biophys. Res. Commun. 2007;353:299–305. doi: 10.1016/j.bbrc.2006.12.027. [DOI] [PubMed] [Google Scholar]
- 26.Sakuma Y., Maruyama K., Qin F., Osakabe Y., Shinozaki K., Yamaguchi-Shinozaki K. Dual function of an Arabidopsis transcription factor DREB2A in water-stress-responsive and heat-stress-responsive gene expression. Proc. Nat. Acad. Sci. USA. 2006;103:18822–18827. doi: 10.1073/pnas.0605639103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Maruyama K., Sakuma Y., Kasuga M., Ito Y., Seki M., Goda H., Shimada Y., Yoshida S., Shinozaki K., Yamaguchi‐Shinozaki K. Identification of cold‐inducible downstream genes of the Arabidopsis DREB1A/CBF3 transcriptional factor using two microarray systems. Plant J. 2004;38:982–993. doi: 10.1111/j.1365-313X.2004.02100.x. [DOI] [PubMed] [Google Scholar]
- 28.Tsutsui T., Kato W., Asada Y., Sako K., Sato T., Sonoda Y., Kidokoro S., Yamaguchi-Shinozaki K., Tamaoki M., Arakawa K. DEAR1, a transcriptional repressor of DREB protein that mediates plant defense and freezing stress responses in Arabidopsis. J. Plant Res. 2009;122:633–643. doi: 10.1007/s10265-009-0252-6. [DOI] [PubMed] [Google Scholar]
- 29.Dong C.-J., Liu J.-Y. The Arabidopsis EAR-motif-containing protein RAP2.1 functions as an active transcriptional repressor to keep stress responses under tight control. BMC Plant Biol. 2010;10 doi: 10.1186/1471-2229-10-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mizoi J., Shinozaki K., Yamaguchi-Shinozaki K. AP2/ERF family transcription factors in plant abiotic stress responses. BBA Gene Regul. Mech. 2012;1819:86–96. doi: 10.1016/j.bbagrm.2011.08.004. [DOI] [PubMed] [Google Scholar]
- 31.Tournier B., Sanchez-Ballesta M.T., Jones B., Pesquet E., Regad F., Latché A., Pech J., Bouzayen M. New members of the tomato ERF family show specific expression pattern and diverse DNA-binding capacity to the GCC box element. FEBS Lett. 2003;550:149–154. doi: 10.1016/S0014-5793(03)00757-9. [DOI] [PubMed] [Google Scholar]
- 32.Fujimoto S.Y., Ohta M., Usui A., Shinshi H., Ohme-Takagi M. Arabidopsis ethylene-responsive element binding factors act as transcriptional activators or repressors of GCC box-mediated gene expression. Plant Cell. 2000;12:393–404. doi: 10.1105/tpc.12.3.393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fujita M., Fujita Y., Noutoshi Y., Takahashi F., Narusaka Y., Yamaguchi-Shinozaki K., Shinozaki K. Crosstalk between abiotic and biotic stress responses: A current view from the points of convergence in the stress signaling networks. Curr. Opin. Plant Biol. 2006;9:436–442. doi: 10.1016/j.pbi.2006.05.014. [DOI] [PubMed] [Google Scholar]
- 34.Yan L., Zhai Q., Wei J., Li S., Wang B., Huang T., Du M., Sun J., Kang L., Li C. Role of Tomato Lipoxygenase D in Wound-Induced Jasmonate Biosynthesis and Plant Immunity to Insect Herbivores. PLoS Genet. 2013;9:e1003964. doi: 10.1371/journal.pgen.1003964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cheng M., Liao P., Kuo W., Lin T. The Arabidopsis ETHYLENE RESPONSE FACTOR1 regulates abiotic stress-responsive gene expression by binding to different cis-acting elements in response to different stress signals. Plant Physiol. 2013;162:1566–1582. doi: 10.1104/pp.113.221911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kasuga M., Liu Q., Miura S., Yamaguchi-Shinozaki K., Shinozaki K. Improving plant drought, salt, and freezing tolerance by gene transfer of a single stress-inducible transcription factor. Nat. Biotechnol. 1999;17:287–291. doi: 10.1038/7036. [DOI] [PubMed] [Google Scholar]
- 37.Xu Z., Ni Z., Liu L., Nie L., Li L., Chen M., Ma Y. Characterization of the TaAIDFa gene encoding a CRT/DRE-binding factor responsive to drought, high-salt, and cold stress in wheat. Mol. Genet. Genomics. 2008;280:497–508. doi: 10.1007/s00438-008-0382-x. [DOI] [PubMed] [Google Scholar]
- 38.Moran P.J., Thompson G.A. Molecular responses to aphid feeding in Arabidopsis in relation to plant defense pathways. Plant Physiol. 2001;125:1074–1085. doi: 10.1104/pp.125.2.1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang H., Jiang J., Chen S., Qi X., Peng H., Li P., Song A., Guan Z., Fang W., Liao Y. Next-generation sequencing of the Chrysanthemum nankingense (Asteraceae) transcriptome permits large-scale unigene assembly and SSR marker discovery. PLoS One. 2013;8:e62293. doi: 10.1371/journal.pone.0062293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Larkin M., Blackshields G., Brown N., Chenna R., McGettigan P.A., McWilliam H., Valentin F., Wallace I.M., Wilm A., Lopez R. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 41.Tamura K., Peterson D., Peterson N., Stecher G., Nei M., Kumar S. MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Song A., Zhu X., Chen F., Gao H., Jiang J., Chen S. A chrysanthemum heat shock protein confers tolerance to abiotic stress. Int. J. Mol. Sci. 2014;15:5063–5078. doi: 10.3390/ijms15035063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Song A., Lu J., Jiang J., Chen S., Guan Z., Fang W., Chen F. Isolation and characterisation of Chrysanthemum crassum SOS1, encoding a putative plasma membrane Na+/H+ antiporter. Plant Biol. 2012;14:706–713. doi: 10.1111/j.1438-8677.2011.00560.x. [DOI] [PubMed] [Google Scholar]
- 44.Chen Y., Jiang J., Song A., Chen S., Shan H., Luo H., Gu C., Sun J., Zhu L., Fang W., Chen F. Ambient temperature enhanced freezing tolerance of Chrysanthemum dichrum CdICE1 Arabidopsis via miR398. BMC Biol. 2013;11 doi: 10.1186/1741-7007-11-121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Song A., Li P., Jiang J., Chen S., Li H., Zeng J., Shao Y., Zhu L., Zhang Z., Chen F. Phylogenetic and transcription analysis of chrysanthemum WRKY transcription factors. Int. J. Mol. Sci. 2014;15:14442–14455. doi: 10.3390/ijms150814442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCt Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 47.de Hoon M.J., Imoto S., Nolan J., Miyano S. Open source clustering software. Bioinformatics. 2004;20:1453–1454. doi: 10.1093/bioinformatics/bth078. [DOI] [PubMed] [Google Scholar]
- 48.Eisen M.B., Spellman P.T., Brown P.O., Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc. Natl. Acad. Sci. USA. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]