Abstract
Some cytochrome P450 (CYP) genes are known for their rapid up-regulation in response to insecticide exposures in insects. To date, however, limited information is available with respect to the relationships among the insecticide type, insecticide concentration, exposure duration and the up-regulated CYP genes. In this study, we examined the transcriptional response of eight selected CYP genes, including CYP4G7, CYP4Q4, CYP4BR3, CYP12H1, CYP6BK11, CYP9D4, CYP9Z5 and CYP345A1, to each of four insecticides in the red flour beetle, Tribolium castaneum. Reverse transcription quantitative PCR (RT-qPCR) revealed that CYP4G7 and CYP345A1 can be significantly up-regulated by cypermethrin (1.97- and 2.06-fold, respectively), permethrin (2.00- and 2.03-fold) and lambda-cyhalothrin (1.73- and 1.81-fold), whereas CYP4BR3 and CYP345A1 can be significantly up-regulated by imidacloprid (1.99- and 1.83-fold) when 20-day larvae were exposed to each of these insecticides at the concentration of LC20 for 24 h. Our studies also showed that similar levels of up-regulation can be achieved for CYP4G7, CYP4BR3 and CYP345A1 by cypermethrin, permethrin, lambda-cyhalothrin or imidacloprid with approximately one fourth of LC20 in 6 h. Our study demonstrated that up-regulation of these CYP genes was rapid and only required low concentrations of insecticides, and the up-regulation not only depended on the CYP genes but also the type of insecticides. Our results along with those from previous studies also indicated that there were no specific patterns for predicting the up-regulation of specific CYP gene families based on the insecticide classification.
Keywords: cytochrome P450, insecticides, Tribolium castaneum, up-regulation
1. Introduction
Cytochrome P450 (CYP) genes constitute one of the largest gene superfamilies, with representatives in all living organisms, including bacteria, fungi, plants, and animals [1]. In insects, CYP enzymes are commonly involved in the metabolism of either endogenous or exogenous compounds. Although physiological significance of up-regulation or overexpression in insects is uncertain, the up-regulation is thought to provide versatility in environmental adaptation [2] or as a protective mechanism whereby the organism can detoxify xenobiotics [3]. Indeed, CYP-mediated detoxification is an important resistance mechanism that can cause a significantly high level of resistance to many insecticides in insect populations [4]. Besides detoxification, CYP genes are also involved in insect growth, development and nutrition [5]. It is believed that the diverse functions of cytochrome P450s are primarily due to the diversity of CYP genes [6]. To date, thousands of CYP genes in total have been identified in insects [7], and the number is still growing rapidly as more insect genomes are sequenced [8].
The up-regulation of CYP genes mediated by insecticides and other xenobiotic compounds have been reported in many insect species (references as presented in Table 1). The availability of genome sequences in many insect species has facilitated the identification of new CYP genes and the characterization of up-regulated CYP genes at the genomic scale [9]. For example, several microarray-based studies on Drosophila melanogaster have identified xenobiotic-inducible CYP genes [10,11]. These genes belong to CYP3, CYP4 and mitochondrial clans. The use of microarrays on insecticide-resistant mosquitoes, including Anopheles gambiae [12], Aedes aegypti [13] and Culex quinquefasciatus [14], have collectively identified a relatively small number of up-regulated CYP genes after exposures of the mosquitoes to different concentrations of insecticides. More recently, Zhu et al. [15] identified six CYP genes up-regulated in deltamethrin-resistant strain (QTC279) of Tribolium castaneum. Among them, CYP6BQ9, a brain-specific gene, showed over 200-fold constitutive overexpression, and can be up-regulated when the insects were exposured to deltamethrin [16].
Table 1.
Comparisons of selected cytochrome P450 (CYP) genes from T. castaneum with those known to be up-regulated by chemicals and/or overexpressed in insecticide resistant strains of other insects.
| T. castaneum CYP Genes | Most Similar CYP Genes Found in Other Insect Species by BLASTP Search | ||||
|---|---|---|---|---|---|
| Species | CYP Genes | Identity (%) a | Overexpression Related to Insecticide Resistance b | Up-Regulation Mediated by Insecticides and Other Chemicals | |
| CYP12H1 | M. domestica | CYP12A1 | 37 | Pyrethroids [11] | Pyrethroids [17] |
| D. melanogaster | CYP12D1 | 37 | DDT [18] | pyrethrum [19] | |
| D. melanogaster | CYP12A4 | 39 | Lufenuron [20] | - | |
| A. gambiae | CYP12F1 | 36 | DDT [12] | - | |
| A. aegypti | CYP12F8 | 40 | - | Fluoranthene [21,22] | |
| CYP4G7 | M. domestica | CYP4G2 | 48 | - | Permethrin [23] |
| B. germanica | CYP4G19 | 51 | Pyrethroids [24] | - | |
| C. tentans | CYP4G33 | 54 | - | Atrazine [25] | |
| B. mori | CYP4G25 | 52 | - | Diazinon, permethrin [26] | |
| A. aegypti | CYP4G36 | 51 | - | Imidacloprid [27] | |
| CYP4BR3 | A. gambiae | CYP4H15 | 39 | DDT [12] | - |
| A. aegypti | CYP4H28 | 38 | - | Permethrin [28] | |
| C. pallens | CYP4H21 | 37 | Deltamethrin [29] | - | |
| C. quinquefasciatus | CYP4H34 | 38 | Permethrin [14] | - | |
| D. melanogaster | CYP4E2 | 41 | - | Phenobarbital, caffeine [30] | |
| D. melanogaster | CYP4E3 | 41 | - | Phenobarbital, caffeine [30] | |
| CYP4Q4 | M. sexta | CYP4M1 | 44 | Alkaloids, nicotine [31] | - |
| B. mori | CYP4M5 | 43 | - | Dichlorvos, deltamethrin [32] | |
| H. armigera | CYP4M6 | 43 | Deltamethrin [33] | - | |
| D. virgifera virgifera | CYP4AJ1 | 45 | Parathion, carbaryl [34] | - | |
| CYP6BK11 | M. domestica | CYP6A36 | 49 | Pyrethroids [35] | - |
| D. melanogaster | CYP6A8 | 46 | DDT, malathion [36] | Phenobarbital [30,37] | |
| A. gambiae | CYP6P3 | 43 | Permethrin [38] | - | |
| A. gambiae | CYP6M2 | 43 | Permethrin [39] | - | |
| A. aegypti | CYP6M11 | 41 | Deltamethrin [40] | Permethrin [21] | |
| P. xylostella | CYP6BG1 | 39 | Cypermethrin [41] | Permethrin [5] | |
| CYP345A1 | D. melanogaster | CYP6G1 | 43 | DDT, imidacloprid [42] | DDT, caffeine [37] |
| M. domestica | CYP6D3 | 38 | Pyrethroids [43] | Phenobarbital [44] | |
| C. quinquefasciatus | CYP6F1 | 37 | Permethrin [45] | - | |
| A. aegypti | CYP6AL1 | 37 | Fluoranthene [21] | - | |
| H. zea | CYP6B8 | 36 | Cypermethrin [46] | Chlorogenic acid [47] | |
| CYP9D4 | C. quinquefasciatus | CY9M10 | 37 | Permethrin [48] | - |
| H. armigera | CYP9A12 | 46 | Pyrethroids [49] | - | |
| H. armigera | CYP9A14 | 43 | Pyrethroids [50] | - | |
| A. aegypti | CYP9J27 | 41 | Pyrethroids [13] | - | |
| A. aegypti | CYP9J32 | 44 | Pyrethroids [13] | - | |
| CYP9Z5 | A. mellifera | CYP9Q1 | 38 | Acaricides [51] | - |
| B. mori | CYP9A19 | 45 | - | - | |
| B. mori | CYP9A20 | 45 | - | Dichlorvos, deltamethrin [32] | |
| D. melanogaster | CYP9F2 | 41 | Pyrethrum [19] | - | |
a The identity level is based on the deduced amino acid sequence of each CYP gene in T. castaneum against that of other insect species; b The number(s) in the brackets refer to reference numbers listed at the end of the paper.
The up-regulation of CYP genes could potentially have a significant impact on insect’s ability to metabolize xenobiotics, which may lead to the detoxification of insecticides and even the development of insecticide resistance in the insect populations. To date, however, limited information is available with respect to the relationship between CYP genes and the type of insecticides. In addition, little is known about the level of the up-regulation of CYP genes in relation to the insecticide concentration and the exposure duration in insects. The objectives of this study were to: (1) evaluate transcriptional responses of eight representative CYP genes, including CYP4G7, CYP4Q4, CYP4BR3, CYP12H1, CYP6BK11, CYP9D4, CYP9Z5 and CYP345A1, to four selected insecticides, including cypermethrin, permethrin, lambda-cyhalothrin and imidacloprid, in T. castaneum; (2) examine the up-regulation responses of different CYP genes in relation to their classification to see whether the up-regulation is restricted to certain specific CYP families, or whether the up-regulation can be mediated by different insecticides within the same class; and (3) investigate the effect of insecticide concentrations and exposure duration on the level of the CYP up-regulation. This study is expected to help researchers better evaluate and understand the transcriptional responses of CYP genes to different insecticides in insects, and also provide useful information for future research to evaluate the role of CYP genes in insecticide detoxification and resistance in insects.
2. Results and Discussion
2.1. Phylogenetic Analysis of Deduced Amino Acid Sequences of T. castaneum CYP Genes
Phylogenetic analysis showed that CYP12H1 (Figure 1A), CYP4BR3 and CYP4G7 (Figure 1B) and CYP345 (Figure 1C) from T. castaneum were clustered in distinct clades with the CYPs from other insect species in the phylogenetic trees, and the heme-binding motifs of these clades were conserved. Noticeably, CYPs from CYP6 and CYP9 family were clustered in one clade within T. castaneum rather than with any other species. These clustered CYP genes may have similar roles, and therefore can help us select the representative CYP genes for further analyses in T. castaneum. For example, several genes in CYP6 and CYP9 gene families, which account for nearly half of all CYP genes in T. castaneum, have been implicated in the insecticide-mediated up-regulation and insecticide resistance [16].
Figure 1.
Neighbor-joining phylogenetic trees of three CYP clans. (A) mitochondrial; (B) CYP4; and (C) CYP3. The trees were constructed by using MEGA 5 based on the full-length amino acid sequences deduced from the cDNA or genomic DNA sequences of T. castaneum (Tc), D. melanogaster (Dm), A. gambiae (Ag), Musca domestica (Md), A. aegypti (Aa), C. pipiens pallens (Cp), C. quinquefasciatus (Cq), Blattella germanica (Bg), Helicoverpa armigera (Ha), Diabrotica virgifera virgifera (Dv), Manduca sexta (Ms), Bombyx mori (Bm), H. Zea (Hz), Plutella xylostella (Px), Chironomus tentans (Ct), and Apis mellifera (Am). The accession number of each gene from NCBI is shown in bold at the end of the gene name. All nodes have significant bootstrap support based on 3000 replicates. The trees were constructed with cut-off value of 50%. The CYPs known to be implicated in insecticide resistance and up-regulation were indicated with a black triangle and a red dot, respectively, or both. In addition, sequence logos, which were predicted by WebLogo tool (http://weblogo.berkeley.edu/logo.cgi), depicted the conservation of amino acid residues in CYP heme-binding motif of each clustered clade. The letter size is proportional to the degree of amino acid conservation. Eight CYPs selected for this study were boxed.
2.2. Selection of CYP Genes for Studying Insecticide-Mediated Up-Regulation
We selected eight CYP genes from T. castaneum based on their representations in the phylogenetic trees and their similarities of amino acid sequences to those of other insect CYP genes known to be capable of up-regulation by insecticides (Table 1). The amino acid sequence identities of CYPs among T. castaneum and other insects range from 35%–54%, and seldom beyond 50% only if the comparisons were made within the same subfamily. For example, CYP4G7 in T. castaneum shows the identities of 51% to CYP4G19 from B. germanica, 54% to CYP4G33 from C. tentans, 52% to CYP4G25 from B. mori, and 51% to CYP4G36 from A. aegypti. In order to select a manageable number of the CYP genes for subsequent analyses, the CYPs with the highest identities from eight major subfamilies (Table 1) were selected as representative genes. These genes include CYP4G7, CYP4Q4, CYP4BR3, CYP12H1, CYP6BK11, CYP9D4, CYP9Z5 and CYP345A1. In the process of selecting the representative genes, we also considered the factor of which their homologous genes in other insect species have been reported in insecticide and other chemical-mediated up-regulation and/or insecticide resistance. This strategy has been successfully used by Poupardin et al. on A. aegypti [21].
2.3. Stage and Tissue Dependent Expression Patterns of Eight CYP Genes
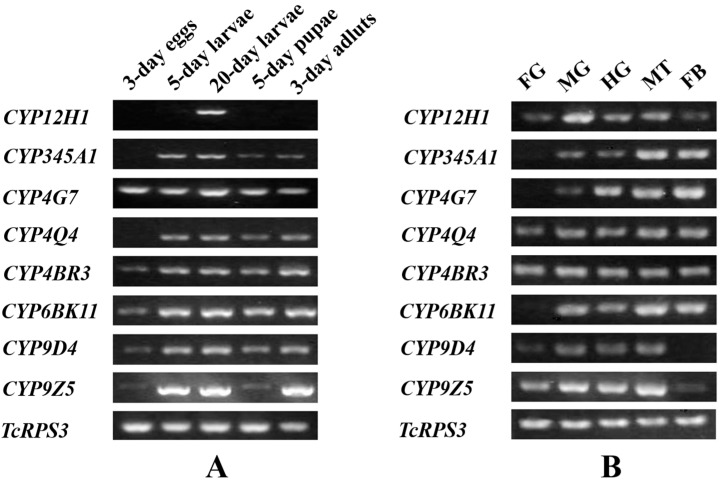
For the stage-dependent expression pattern of the eight CYP genes, we found that almost all these genes were expressed in 20-day larvae, 5-day pupae and 3-day adults except CYP12H1 which appeared to be only expressed in 20-day larvae (Figure 2A). The CYP12H1 expression pattern is consistent with that of the insecticide resistant strain (QTC279) of T. castaneum reported by Zhu et al. [16]. On the other hand, CYP9Z5 showed high expression in larval and adult stages but very low expression in egg and pupal stages. Its high expression appeared to associate with insect feeding. In contrast, CYP12H1, CYP345A1 and CYP4Q4 did not show detectable expression in eggs. However, the remaining five CYPs were constitutively expressed in all life stages. For the tissue-dependent expression pattern, CYP12H1, CYP4Q4, CYP4BR3 and CYP9Z5 were expressed in all the examined tissues (Figure 2B), and all the eight genes were expressed in midgut, hindgut and Malpighian tubules. However, the expression was undetectable for CYP345A1, CYP4G7 and CYP6BK11 in foregut and for CYP9D4 in fat bodies.
Figure 2.
Stage-dependent (A) and tissue-dependent (B) expression patterns of eight selected CYP genes in T. castaneum (Georgia-1 strain). The expression profiles were evaluated by reverse transcription PCR (RT-PCR). The expression patterns of five different tissues, including foregut (FG), midgut (MG), hindgut (HG), Malpighian tubules (MT), fat bodies (FB) were derived from 20-day larvae, and TcRPS3 was used as an internal reference gene.
Different expression patterns of CYP genes in different developmental stages of an insect suggest the diverse roles of these genes during the insect development [52]. For example, CYP12H1 is only expressed in 20-day larvae, which implies its role restricted to this stage [16]. On the other hand, all the eight selected CYP genes were expressed in the midgut, Malpighian tubules and fat bodies (except CYP9D4), which are considered as major tissues involved in metabolism of xenobiotics in insects [53,54]. Therefore, such tissue-specific expression patterns of these CYP genes in T. castaneum may reflect their roles in metabolism of endogeneous and exogenous substances. Since all the eight CYP genes were expressed in 20-day larvae, we used this larval stage in our subsequent studies.
2.4. Selection of Insecticide Concentrations to Mediate Up-Regulation of CYP Genes
The lethal concentrations to kill 20% (LC20) and 50% (LC50) of the insect population, and their 95% confidence intervals (95% CI) for each of the four insecticides were evaluated in 20-day larvae (Table 2, Supplementary Figure S1). The order from the most to least toxic of the four insecticides is lambda-cyhalothrin, cypermethrin, imidacloprid and permethrin. Imidacloprid is a neonicotinoid whereas the remaining three are pyrethroids. Based on these results, we selected two concentrations approximate to the LC20 and one fourth of the LC20 of each insecticide to evaluate possible up-regulation of the eight CYP genes mediated by these insecticides. The approximate LC20 concentrations were 2 μg/mL for cypermethrin and lambda-cyhalothrin, and 16 μg/mL for permethrin and imidacloprid. The concentrations for the one fourth of approximate LC20 were 0.5 μg/mL for cypermethrin and lambda-cyhalothrin, and 4 μg/mL for permethrin and imidacloprid.
Table 2.
Summary of the lethal concentrations to kill 20% (LC20) and 50% (LC50) of the insect population, and their 95% confidence intervals (95% CI) for each of the four insecticides determined in 20-day larvae of T. castaneum (Georgia-1 strain).
| Insecticides | LC20, μg/mL (95% CI) | LC50, μg/mL (95% CI) | Slope a | Intercept a | χ2 | p b |
|---|---|---|---|---|---|---|
| Cypermethrin | 2.27 (1.6–3.0) | 7.77 (6.5–9.4) | 3.66 ± 0.67 | 1.94 ± 0.94 | 5.13 | 1 |
| Lambda-cyhalothrin | 1.76 (1.6–3.3) | 4.24 (3.6–5.0) | 3.49 ± 0.40 | 2.98 ± 0.32 | 7.16 | 0.85 |
| Permethrin | 23.73 (15.3–32) | 77.46 (63.6–94.9) | 3.51 ± 0.80 | 1.63 ± 1.25 | 41.86 | 0.08 |
| Imidacloprid | 14.25 (12.0–16.5) | 48.65 (44.9–53.4) | 1.80 ± 0.49 | 2.02 ± 0.84 | 28.48 | 0.15 |
a Slope and intercept were derived from the logarithm of concentration-probit mortality curve that generated by probit analysis; b p-Value > 0.05 indicates a significant fit between the observed and expected regression lines in a probit analysis.
2.5. Evaluation of CYP Gene Up-Regulation by Insecticides
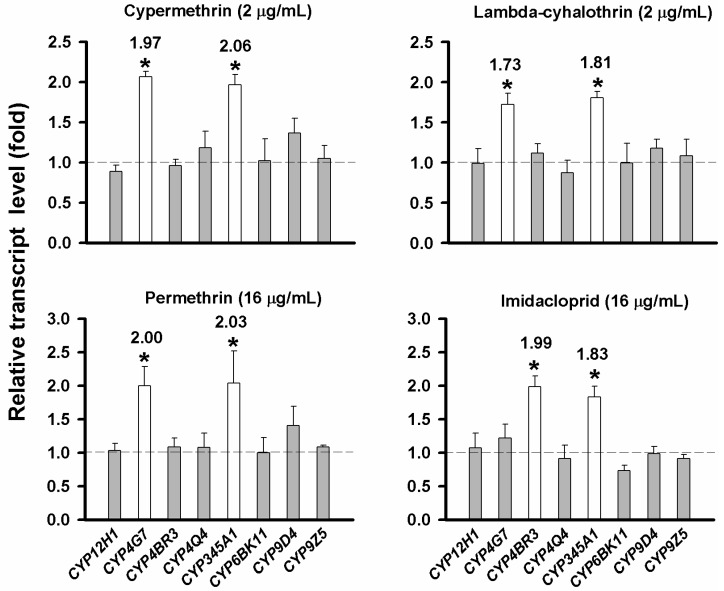
In order to examine which CYP genes can be significantly up-regulated by insecticides, reverse transcription quantitative PCR (RT-qPCR) was performed to determine the change of transcript level for each of the eight CYP genes after the 20-day larvae were exposed to cypermethrin, lambda-cyhalothrin, permethrin or imidacloprid at their approximate LC20 concentrations for 24 h (Figure 3). CYP345A1 was up-regulated by all the four insecticides tested, whereas CYP4G7 was up-regulated by all the three pyrethroids but not by imidacloprid. However, CYP4BR3 was up-regulated only by imidacloprid. Overall, only CYP4G7 and CYP345A1 can be up-regulated by the three pyrethroids, and only CYP4BR3 and CYP345A1 can be up-regulated by imidacloprid. The levels of the up-regulation range from 1.73–2.06-fold.
Figure 3.
Up-regulation of CYP genes in T. castaneum after exposed to different insecticides. Dash lines represent relative transcript level of the control (larvae treated with the insecticide solvent only) as 1.0. The up-regulation fold was acquired by comparing the transcript levels of each CYP between the treated and the control insects. The CYP genes with a statistically significant up-regulation are marked with asterisks (Student’s t test, * p < 0.05).
2.6. Concentration- and Time-Dependent Effect on CYP Up-Regulation
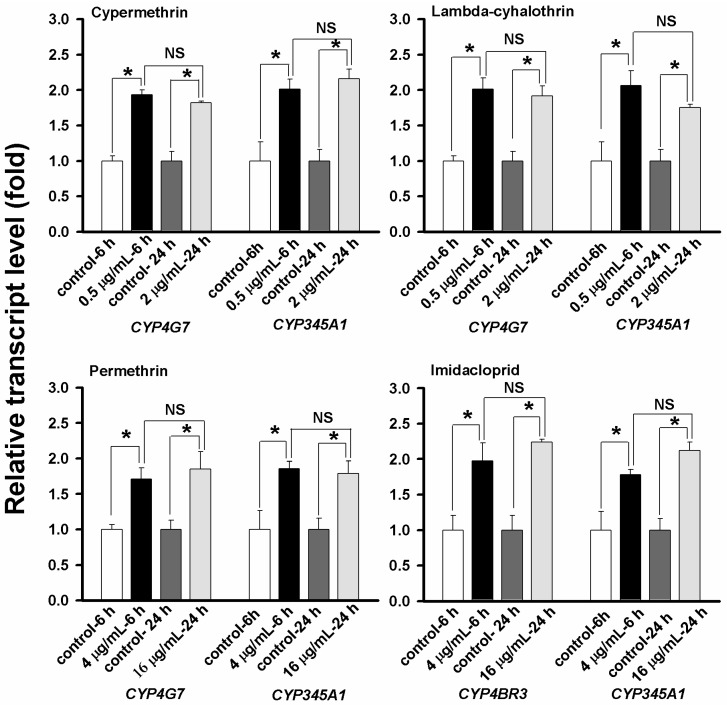
Because only CYP4G7 and CYP345A1 can be up-regulated by the three pyrethroids, and only CYP4BR3 and CYP345A1 can be up-regulated by imidacloprid (Figure 3), our studies on the concentration and time-dependent effect on the up-regulation focused on only three CYP genes (CYP4G7, CYP345A1 and CYP4BR3). Two different concentrations of cypermethrin, 2 μg/mL (approximate LC20) and 0.5 μg/mL (one fourth of the approximate LC20), were used to expose 20-day larvae for 24 h followed by RT-qPCR analysis of CYP4G7 and CYP345A1. The insects exposed to the two concentrations showed approximately 2-fold up-regulation in both genes, and the levels of the up-regulation did not show significant differences between the two insecticide concentrations (Figure 4A). Therefore, we used cypermethrin at 0.5 μg/mL for subsequent analyses.
Figure 4.
Cypermethrin concentration and time dependent up-regulation of CYP4G7 and CYP345A1 in 20-day larvae. Controls were normalized as 1.0, and the relative transcript levels of CYP genes were calculated based on their corresponding controls. (A) Cypermethrin concentration dependent up-regulation as measured at 2 and 0.5 μg/mL with the exposure time of 24 h. Different letters above the standard error bars indicate significant differences based on the one-way ANOVA followed by Tukey’s HSD multiple comparison test (p < 0.05); (B) Time dependent up-regulation of CYP4G7 by cypermethrin (0.5 μg/mL) as measured at 6, 12, 24 and 48 h; and (C) Time dependent up-regulation of CYP345A1 by cypermethrin (0.5 μg/mL) as measured at 6, 12, 24 and 48 h. Dash lines represent relative transcript level of the control (larvae treated with the insecticide solvent only) as 1.0. Statistical analysis was conducted to compare the expression levels between the control and the insecticide-treated insects within the same time duration by using Student’s t test. Asterisk above the standard error bars indicates significant difference whereas NS indicates no significant difference.
The time-dependent effect on the CYP up-regulation was analyzed after 20-day larvae were exposed to cypermethrin at the concentration of 0.5 μg/mL for 6, 12, 24 and 48 h. Both CYP4G7 (Figure 4B) and CYP345A1 (Figure 4C) showed significant up-regulations compared to their corresponding controls (i.e., solvent exposures for the same durations) at 6, 12 and 24 h. However, the levels of such up-regulations began to decrease and did not show significant differences at 48 h compared with those of their controls for both CYP4G7 and CYP345A1. These results indicated that the up-regulation of these two CYP genes by cypermethrin occurred at early stages of insecticide exposures (e.g., from 6–24 h). Based on this finding with cypermethrin, we used 6 h as the exposure time.
Because a 6-h exposure to cypermethrin at 0.5 μg/mL resulted in up-regulations of both CYP4G7 and CYP345A1, and such up-regulations were not significantly different from those with longer exposure times (i.e., 12, 24 and 48 h) and higher concentration of the insecticide (i.e., 2 μg/mL), we compared the up-regulation of the three genes between the exposure times (i.e., 6 and 24 h) and between two insecticide concentrations (i.e., 2 and 0.5 μg/mL for cypermethrin and lambda-cyhalothrin, and 16 and 4 μg/mL for permethrin and imidacloprid). As shown in Figure 5, all the exposures resulted in significant up-regulations of the CYP genes as compared with their corresponding controls. There were no statistical differences between the exposures at 0.5 μg/mL for 6 h and at 2 μg/mL for 24 h for cypermethrin and lambda-cyhalothrin, and between the exposures at 4 μg/mL for 6 h and at 16 μg/mL for 24 h for permethrin and imidacloprid.
Figure 5.
Insecticide concentration and time-dependent effect on the up-regulation of the CYP genes in 20-day larvae. The fold changes were also statistically compared between the two treatment combinations (i.e., 0.5 μg/mL for 6 h against 2 μg/mL for 24 h for cypermethrin and lambda-cyhalothrin, and 4 μg/mL for 6 h against 16 μg/mL for 24 h for permethrin and imidacloprid) by Student’s t test. An asterisk above the standard error bars indicates significant difference whereas NS indicates no significant difference.
2.7. Discussion
By using phylogenetic analysis and protein sequence comparisons of all the 143 CYP genes in T. castaneum retrieved from cytochrome P450 homepage (http://drnelson.uthsc.edu/CytochromeP450.html) along with those found in other insect species, we selected eight CYP genes, including CYP4G7, CYP4Q4, CYP4BR3, CYP12H1, CYP6BK11, CYP9D4, CYP9Z5 and CYP345A1, from T. castaneum for detailed studies on their up-regulation mediated by different insecticides. These genes showed the highest identity levels at amino acid sequence levels with those known to be up-regulated by various chemicals including insecticides and/or involved in insecticide resistance in other insect species (Table 1). The overall amino acid identities of the CYPs between T. castaneum and other insect species ranged from 35%–54%, and seldom beyond 50%; only if the comparisons were made within the same subfamily. Although the strategies that we used to select representative CYP genes from T. castaneum for this study are justifiable, the overall low level of the amino acid sequence identities among the diverse CYPs from different insect species remains to be a challenge for selecting a manageable number of the CYP genes for detailed analyses. Nevertheless, our research provided useful information regarding the developmental stage and tissue-dependent expression patterns, the insecticide concentration and exposure time-dependent up-regulations, and the capability of different insecticides to mediate the up-regulation of various CYP genes representing eight different subfamilies in T. castaneum.
Our results showed that the up-regulation of CYP4G7, CYP4BR3 and CYP345A1 was relatively fast (6 h) and required only low concentrations of insecticides (e.g., one fourth of the LC20). The levels of the up-regulations in 20-day larvae of T. castaneum exposed to insecticides at one fourth of the approximate LC20 for 6 h were not significantly different from those of the larvae exposed to the same insecticides at approximate LC20 for 24 h. Our results suggested that relatively low concentrations of insecticides were effective for the up-regulation of CYP genes, and increasing the insecticide concentration may not necessarily enhance the up-regulation, possibly due to an increased toxic stress to the insects. However, several studies have shown that the up-regulation of a specific CYP gene can be influenced by chemical concentrations and exposure durations. In D. melanogaster, up-regulations of CYP genes increased gradually as the phenobarbital concentration and exposure duration increased [30]. In P. xylostella, Baek et al. [41] found that cypermethrin was able to up-regulate the CYPs transcription under different conditions. However, they found that the up-regulations were more effective when low sublethal concentrations and short exposure durations were used than those of high concentrations (e.g., LD50 or LC50) and long exposure duration. Reduced levels of up-regulations of CYP genes by insecticides at high concentrations could be due to increased stress of the insects caused by the insecticides. These results suggest that appropriate concentrations of an insecticide must be carefully pre-determined to evaluate the insecticide-mediated up-regulation of CYP genes in insects.
The level of the CYP up-regulation mediated by insecticides can also be affected by physiological status of insects. In P. xylostella, Bautista et al. [5] found that three out of six CYP genes were significantly up-regulated by permethrin in permethrin-susceptible strain, but only one of the six genes was moderately up-regulated by permethrin in permethrin-resistant strain although different concentrations of permethrin were used to expose the susceptible and resistant strains. In fact, four out of the six CYP genes in permethrin-resistant strain were significantly down-regulated by permethrin at 100 ppm. Furthermore, significant up-regulation of CYP genes by permethrin in the susceptible strain did not result in a decreased toxicity of permethrin to the insect. These results clearly demonstrated that the up-regulation of CYP genes by insecticides may not necessarily reflect their roles in insecticide detoxification. In fact, CYP genes involved in the detoxification of insecticides are often constitutively up-regulated in resistant insects. These genes may be less likely to be up-regulated by the insecticides.
Our results also indicated that there were no specific patterns related to the CYP gene families or subfamilies in their insecticide-mediated up-regulations in T. castaneum. First, only three out of the eight selected CYP genes (i.e., CYP4G7, CYP4BR3 and CYP345A1) showed their up-regulation when the insects were exposed to each of the four insecticides, although these genes were selected from the CYP gene families (CYP12, CYP4, CYP6 and CYP9) known to be likely involved in insecticide metabolism and/or resistance [16]. None of the selected genes from the CYP6 and CYP9 families exhibited any insecticide-mediated up-regulation in this study. Secondly, previous studies showed that CYP genes which can be up-regulated or exhibited constitutive overexpression mediated by insecticides were mainly from CYP6B subfamily in T. castaneum [15,16]. However, our studies showed that the up-regulation of CYP genes was not restricted to a specific CYP family, as CYP4G7 and CYP4BR3 from CYP4 family, and CYP345A1 from CYP345 family showed significant insecticide-mediated up-regulations.
The insecticide-mediated up-regulation of CYP genes in T. castaneum was insecticide specific. For example, both CYP4G7 and CYP4BR3 are from CYP4 family, but the former was significantly up-regulated by all three pyrethroids whereas the latter was uniquely up-regulated by imidacloprid. In contrast, CYP345A1 was up-regulated by all the four insecticides, suggesting CYP345A1 was responsive to a relatively broad spectrum of insecticides. However, the remaining five CYP genes were not significantly up-regulated by any of the four insecticides. Nevertheless, this does not necessarily mean that these CYP genes cannot be up-regulated by any insecticide. As a matter of fact, the up-regulation of CYP genes by specific insecticides has also been seen in other insect species. For instance, microarray analyses of the detoxification genes in D. melanogaster showed that spinosad, diazinon, nitenpyram, lufenuron and dicyclanil did not significantly increase the expression of any detoxification gene, but DDT induced only a single CYP gene (CYP12D1) among a total of 89 CYP genes [55]. In Lymantria dispar, 12 CYP genes exhibited different expression patterns (some up-regulated whereas others down-regulated) when the insects were exposed to different insecticides including deltamethrin, carbaryl and omethoate [56]. It was also noticed that the same CYP gene responded quite differently to different insecticides [56]. Thus, our results along with those from previous studies suggest that there is no general pattern for predicting the up-regulation of CYP genes based on the insecticide classifications.
3. Experimental Section
3.1. Insect Culture
The Georgia-1 (GA-1) insecticide-susceptible strain of T. castaneum was reared on whole-wheat flour containing 5% (w/w) of brewers’ yeast at 30 °C and 65% RH (relative humidity) in the growth chamber in Insect Toxicology Laboratory at Kansas State University (Manhattan, KS, USA).
3.2. Total RNA Isolation and First Strand cDNA Synthesis
Total RNA was isolated from each T. castaneum sample by using TRIzol reagent (Life Technologies, Carlsbad, CA, USA). Total RNA (2.0 μg) was first treated with DNase I (Fermentas, Glen Burnie, MD, USA) to remove potential genomic DNA contamination. The cDNAs were synthesized using EasyScript cDNA Synthesis SuperMix kit (Applied Biological Materials, Richmond, BC, Canada) with oligo(dT)18 as primer.
3.3. Phylogenetic Tree Construction
The deduced CYP amino acid sequences of T. castaneum and other insect species were retrieved from the National Center for Biotechnology Information (NCBI) (http://www.ncbi.nlm.nih.gov/) and Cytochrome P450 homepage (http://drnelson.uthsc.edu/CytochromeP450.html). The sequences were analyzed using ClustalW alignment with Molecular Evolutionary Genetic Analysis software version 5 (MEGA 5) (http://www.megasoftware net). The pair-wise alignments were performed with the gap opening penalty at 10 and the gap extension penalty at default 0.1. The multiple alignments were conducted with the gap opening penalty at 3 and the gap extension penalty at 1.8. The sites containing obvious missing data or alignment gaps were eliminated in a pair-wise manner. The phylogenetic tree was constructed using neighbor-joining algorithm with a total of 3000 bootstrap replications. Ultimately, the tree was created with cut off value of 50%. Sequence logos, which were predicted by WebLogo tool (http://weblogo.berkeley.edu/logo.cgi), depicted the conservation of amino acids in CYP heme-binding motif of each specific clade. The letter size is proportional to the degree of the conservation for amino acid residues of the motif.
3.4. Selection of Representative CYP Genes
The selections of CYP genes for studying insecticide-mediated up-regulation in T. castaneum were based on the representation of the genes in different CYP families, including CYP12, CYP4G, CYP4B, CYP4Q, CYP6B, CYP345, CYP9D and CYP9Z, and the identity levels of the deduced amino acid sequences of these genes compared with those of homologous genes known to be inducible by insecticides and other chemical substances and/or involved in insecticide resistance in other insect species. Only the genes showing lowest E-values and at least 35% identities from the same CYP family were selected from T. castaneum for further analyses.
3.5. Reverse Transcription Quantitative PCR (RT-qPCR) Analyses
The RT-qPCR was performed with EvaGreen qPCR MasterMix-iCycler (Applied Biological Materials) by using the Bio-Rad iCycler iQTM multi-color real-time PCR detection system (Bio-Rad Laboratories, Hercules, CA, USA). A gene encoding ribosomal protein S3, TcRPS3, was used as an internal reference [57]. Primers for RT-qPCR were designed by Beacon Designer™ (Table 3). RT-qPCR was performed with 3-step amplification protocol with 40 cycles of 95 °C for 15 s, 55 °C for 30 s and 70 °C for 30 s. At the end of the run, amplification specificity was verified by obtaining the dissociation curve, in which the samples were cooled to 55 °C after denaturing and then the melting curves were obtained by increasing 0.5 °C/10 s for each cycle with a total of 80 cycles until reaching 95 °C to denature the double-stranded DNA. The specificity of each reaction was evaluated based on the melting temperatures of the PCR products. The RT-qPCR was performed with three biological replications, and relative transcript levels of each gene were calculated according to the 2−ΔΔCt method [58,59].
Table 3.
Primers used to analyze transcript levels of CYP genes in T. castenuem.
| Primers | Sequence (5'–3') | Tm (°C) | Product Length (bp) |
|---|---|---|---|
| TcCYP12H1-F | AACCGCAAAAACTGATACGG | 60.0 | 299 |
| TcCYP12H1-R | ACCGGTCGTGTCTATTCCTG | 60.0 | |
| TcCYP4G7-F | CGCTGCCAACAGAGACATTA | 60.0 | 207 |
| TcCYP4G7-F | AATGACCCTGAAACCGTCAG | 60.0 | |
| TcCYP4BR3-F | CATCGGTTGTACCCTCCTGT | 59.9 | 168 |
| TcCYP4BR3-R | GAATCGGTCAGGGTCAAAGA | 59.8 | |
| TcCYP4Q4-F | TGGTTCCAATCACCCAATTT | 60.0 | 203 |
| TcCYP4Q4-R | TTTTTGCTCTTTGCGACCTT | 59.5 | |
| TcCYP345A1-F | TTTTTCGATTTTCGGTGGAG | 60.0 | 120 |
| TcCYP345A1-R | TTCGCGAAGGAAGTTGCTAT | 60.0 | |
| TcCYP6BK11-F | GTCAATTTGCGGAAACAGGT | 60.1 | 167 |
| TcCYP6BK11-R | CTACGTCCGTAAACCCGAAA | 60.3 | |
| TcCYP9D4-F | GTGGCACAACTAGCTCCACA | 59.9 | 172 |
| TcCYP9D4-R | GTTTTCCTTTACGGGCTTCC | 60.0 | |
| TcCYP9Z5-F | AGTCATGCAAAACTGCAACG | 59.9 | 250 |
| TcCYP9Z5-R | GTCCGGATTGGGGAAGTATT | 60.0 | |
| TcRPS3-F | CCGTCGTATTCGTGAATTGACTT | 59.3 | 143 |
| TcRPS3-R | TCTAAGAGACTCTGCTTGTGCAATG | 60.8 |
F: Forward; R: Reverse.
3.6. Stage and Tissue-Dependent Expression Patterns of CYP Genes
For analyses of developmental stage-dependent expression patterns, total RNA for each replication was isolated from 250–350 of 3-day old (3-day) eggs, 200–250 of 5-day larvae, 20–25 of 20-day larvae, 20–25 of 5-day pupae and 20–25 of 3-day adults. For analyses of tissue-dependent expression patterns, total RNA for each replication was isolated from each of five tissues (foregut, midgut, hindgut, Malpighian tubules and fat bodies) dissected from 80–100 of 20-day larvae. The selection of these tissues was mainly based on previous research showing abundant expressions of CYP genes in midgut, Malpighian tubules and fat bodies. Both the stage and tissue-dependent expression patterns of the eight CYP genes were examined using reverse transcription PCR (RT-PCR), which consisted of an initial denaturation at 95 °C for 3 min followed by 30 cycles of 95 °C for 30 s, 55 °C for 30 s and 70 °C for 30 s, and finished with a final extension step of 72 °C for 5 min. TcRPS3 was used as reference gene, and samples of 10-μL PCR products were analyzed on 2% agarose gel.
3.7. Insecticide Bioassay
Four insecticides, including cypermethrin (purity: 98%), lambda-cyhalothrin (96.8%), permethrin (97.5%) and imidacloprid (95.5%), were obtained from Chem Service (West Chester, PA, USA). Glass scintillation vials (20-mL) were internally coated with 0.5 mL of acetone containing each insecticide by using a RoTo-Torque rotator (Cole Parmer Instrument, Vernon Hills, IL, USA). At least five different concentrations of each insecticide, each with three replicates, were prepared for each bioassay. A group of 15 larvae (20-day) was transferred into each glass and larval mortality was assessed after the larvae were maintained in the vials at 30 °C and 65% RH (without flour) for 24 h. Larvae were considered dead if they were not able to move when gently touched with a brush. Data were analyzed by probit analysis using the procedure PROC PROBIT from SAS 9.3 (SAS Institute, Cary, NC, USA).
3.8. Evaluation of Up-Regulation of CYP Genes Mediated by Insecticides
The approximate LC20 of each of the four insecticides (2 μg/mL for either cypermethrin or lambda-cyhalothrin, 16 μg/mL for either permethrin or imidacloprid) was first used to expose 20-day larvae as described in Section 3.7. After the larvae were treated for 24 h, 4–5 surviving insects were collected form each replicate for total RNA extraction as described in Section 3.2. After we found each tested insecticide at LC20 could mediate significant up-regulations of the selected CYP genes, we included the one fourth of the approximate LC20 for each insecticide to compare the concentration-dependent effect of each insecticide on the CYP up-regulation. In addition, time-dependent effects were also evaluated using four different time points (6, 12, 24 and 48 h). After exposure, 4–5 surviving larvae were collected from each of 3 biological replicates for total RNA extraction, which was subsequently used to assess the expression levels of CYP genes using RT-qPCR as described in Section 3.5.
4. Conclusions
Many CYP genes are known for their rapid up-regulation in response to exposure to xenobiotics in insects. To date, however, limited information is available with respect to the relationship between the insecticide type, insecticide concentration, exposure duration and the up-regulated CYP genes. Our studies showed that CYP4G7 and CYP345A1 can be significantly up-regulated by cypermethrin, permethrin and lambda-cyhalothrin, whereas CYP4BR3 and CYP345A1 can be significantly up-regulated by imidacloprid in the eight selected CYP genes in 20-day larvae of T. castenuem. The levels of the up-regulation ranged from 1.73–2.06-folds. There were no significant differences in the level of up-regulation either between the two insecticide concentrations (i.e., approximate LC20 and the one fourth of the approximate LC20) or between the two insecticide exposure times (i.e., 24 and 6 h). Our study demonstrated that up-regulation of these CYP genes was rapid and only required low concentrations of insecticides. The up-regulation not only depended on the CYP genes but also the type of insecticides. Our results along with those from previous studies also indicated that there were no specific patterns for predicting the up-regulation of specific CYP gene families based on the insecticide classification.
Acknowledgments
This research was partially supported by the Kansas Agricultural Experiment Station, Kansas State University and the USA Department of Agriculture (USDA/NIFA 2014-51102-21017) to Kun Yan Zhu, and the China Scholarship Council to Xiao Liang. Mention of trade names or commercial products in this publication is solely for the purpose of providing specific information and does not imply recommendation or endorsement by Kansas State University. This manuscript is contribution No. 15-183-J from the Kansas Agricultural Experiment Station. The T. castaneum voucher specimens (voucher No. 159) are located in the Kansas State University Museum of Entomological and Prairie Arthropod Research, Manhattan, KS, USA.
Supplementary Materials
Supplementary materials can be found at http://www.mdpi.com/1422-0067/16/01/2078/s1.
Author Contributions
Xiao Liang, Guonian Zhu, Jianxiu Yao and Kun Yan Zhu conceived and designed the experiments; Xiao Liang, Da Xiao and Yanping He performed the experiments; Xiao Liang and Da Xiao analyzed the data; Kun Yan Zhu contributed reagents/materials/analysis tools; Xiao Liang and Kun Yan Zhu wrote the paper; and Xiao Liang, Jianxiu Yao and Kun Yan Zhu contributed with revisions.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Werck-Reichhart D., Feyereisen R. Cytochromes P450: A success story. Genome Biol. 2000;1:1–9. doi: 10.1186/gb-2000-1-6-reviews3003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Terriere L.C. Induction of detoxication enzymes in insects. Annu. Rev. Entomol. 1984;29:71–88. doi: 10.1146/annurev.en.29.010184.000443. [DOI] [PubMed] [Google Scholar]
- 3.Londono D.K., Siqueira H.A., Wang H., Sarath G., Lydy M.J., Siegfried B.D. Cloning and expression of an atrazine inducible cytochrome P450, CYP4G33, from Chironomus tentans (Diptera: Chironomidae) Pestic. Biochem. Physiol. 2007;89:104–110. doi: 10.1016/j.pestbp.2007.04.001. [DOI] [Google Scholar]
- 4.Scott J.G. Cytochrome P450 and insecticide resistance. Insect Biochem. Mol. Biol. 1999;29:71–78. doi: 10.1016/S0965-1748(99)00038-7. [DOI] [PubMed] [Google Scholar]
- 5.Bautista M.A.M., Tanaka T., Miyata T. Identification of permethrin-inducible cytochrome P450s from the diamondback moth, Plutella xylostella (L.) and the possibility of involvement in permethrin resistance. Pestic. Biochem. Physiol. 2007;87:85–93. doi: 10.1016/j.pestbp.2006.06.004. [DOI] [Google Scholar]
- 6.Feyereisen R. Insect cytochrome P450. Compr. Mol. Insect Sci. 2005;4:1–77. [Google Scholar]
- 7.Urlacher V.B., Girhard M. Cytochrome P450 monooxygenases: An update on perspectives for synthetic application. Trends Biotechnol. 2012;30:26–36. doi: 10.1016/j.tibtech.2011.06.012. [DOI] [PubMed] [Google Scholar]
- 8.Feyereisen R. Insect CYP Genes and P450 Enzymes. In: Lawrence I.G., editor. Insect Molecular Biology and Biochemistry. Elsevier; Oxford, UK: 2011. pp. 236–316. [Google Scholar]
- 9.Guo Y.Q., Zhang J.Z., Yang M.L., Yan L.Z., Zhu K.Y., Guo Y.P., Ma E.B. Comparative analysis of cytochrome P450-like genes from Locusta migratoria manilensis: Expression profiling and response to insecticide exposure. Insect Sci. 2012;19:75–85. doi: 10.1111/j.1744-7917.2011.01450.x. [DOI] [Google Scholar]
- 10.Giraudo M., Unnithan G.C., le Goff G., Feyereisen R. Regulation of cytochrome P450 expression in Drosophila: Genomic insights. Pestic. Biochem. Physiol. 2010;97:115–122. doi: 10.1016/j.pestbp.2009.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Le Goff G., Hilliou F., Siegfried B.D., Boundy S., Wajnberg E., Sofer L., Audant P., Feyereisen R. Xenobiotic response in Drosophila melanogaster: Sex dependence of P450 and GST gene induction. Insect Biochem. Mol. Biol. 2006;36:674–682. doi: 10.1016/j.ibmb.2006.05.009. [DOI] [PubMed] [Google Scholar]
- 12.David J.P., Strode C., Vontas J., Nikou D., Vaughan A., Pignatelli P.M., Louis C., Hemingway J., Ranson H. The Anopheles gambiae detoxification chip: A highly specific microarray to study metabolic-based insecticide resistance in malaria vectors. Proc. Natl. Acad. Sci. USA. 2005;102:4080–4084. doi: 10.1073/pnas.0409348102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Strode C., Wondji C.S., David J.P., Hawkes N.J., Lumjuan N., Nelson D.R., Drane D.R., Karunaratne S., Hemingway J., Black W.C., IV Genomic analysis of detoxification genes in the mosquito Aedes aegypti. Insect Biochem. Mol. Biol. 2008;38:113–123. doi: 10.1016/j.ibmb.2007.09.007. [DOI] [PubMed] [Google Scholar]
- 14.Komagata O., Kasai S., Tomita T. Overexpression of cytochrome P450 genes in pyrethroid-resistant Culex quinquefasciatus. Insect Biochem. Mol. Biol. 2010;40:146–152. doi: 10.1016/j.ibmb.2010.01.006. [DOI] [PubMed] [Google Scholar]
- 15.Zhu F., Parthasarathy R., Bai H., Woithe K., Kaussmann M., Nauen R., Harrison D.A., Palli S.R. A brain-specific cytochrome P450 responsible for the majority of deltamethrin resistance in the QTC279 strain of Tribolium castaneum. Proc. Natl. Acad. Sci. USA. 2010;107:8557–8562. doi: 10.1073/pnas.1000059107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhu F., Moural T.W., Shah K., Palli S.R. Integrated analysis of cytochrome P450 gene superfamily in the red flour beetle, Tribolium castaneum. BMC Genomics. 2013;14:174. doi: 10.1186/1471-2164-14-174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guzov V.M., Unnithan G.C., Chernogolov A.A., Feyereisen R. CYP12A1, a mitochondrial cytochrome P450 from the house fly. Arch. Biochem. Biophys. 1998;359:231–240. doi: 10.1006/abbi.1998.0901. [DOI] [PubMed] [Google Scholar]
- 18.Brandt A., Scharf M., Pedra J., Holmes G., Dean A., Kreitman M., Pittendrigh B.R. Differential expression and induction of two Drosophila cytochrome P450 genes near the Rst(2)DDT locus. Insect Mol. Biol. 2002;11:337–341. doi: 10.1046/j.1365-2583.2002.00344.x. [DOI] [PubMed] [Google Scholar]
- 19.Jensen H., Scott I., Sims S., Trudeau V., Arnason J. The effect of a synergistic concentration of a Piper nigrum extract used in conjunction with pyrethrum upon gene expression in Drosophila melanogaster. Insect Mol. Biol. 2006;15:329–339. doi: 10.1111/j.1365-2583.2006.00648.x. [DOI] [PubMed] [Google Scholar]
- 20.Bogwitz M.R., Chung H., Magoc L., Rigby S., Wong W., O’Keefe M., McKenzie J.A., Batterham P., Daborn P.J. CYP12A4 confers lufenuron resistance in a natural population of Drosophila melanogaster. Proc. Natl. Acad. Sci. USA. 2005;102:12807–12812. doi: 10.1073/pnas.0503709102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Poupardin R., Riaz M., Vontas J., David J., Reynaud S. Transcription profiling of eleven cytochrome P450s potentially involved in xenobiotic metabolism in the mosquito Aedes aegypti. Insect Mol. Biol. 2010;19:185–193. doi: 10.1111/j.1365-2583.2009.00967.x. [DOI] [PubMed] [Google Scholar]
- 22.Poupardin R., Reynaud S., Strode C., Ranson H., Vontas J., David J.P. Cross-induction of detoxification genes by environmental xenobiotics and insecticides in the mosquito Aedes aegypti: Impact on larval tolerance to chemical insecticides. Insect Biochem. Mol. Biol. 2008;38:540–551. doi: 10.1016/j.ibmb.2008.01.004. [DOI] [PubMed] [Google Scholar]
- 23.Zhu F., Li T., Zhang L., Liu N. Co-up-regulation of three P450 genes in response to permethrin exposure in permethrin resistant house flies, Musca domestica. BMC Physiol. 2008;8:18. doi: 10.1186/1472-6793-8-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pridgeon J.W., Zhang L., Liu N. Overexpression of CYP4G19 associated with a pyrethroid-resistant strain of the German cockroach, Blattella germanica (L.) Gene. 2003;314:157–163. doi: 10.1016/S0378-1119(03)00725-X. [DOI] [PubMed] [Google Scholar]
- 25.Londono D.K., Siegfried B.D., Lydy M.J. Atrazine induction of a family 4 cytochrome P450 gene in Chironomus tentans (Diptera: Chironomidae) Chemosphere. 2004;56:701–706. doi: 10.1016/j.chemosphere.2003.12.001. [DOI] [PubMed] [Google Scholar]
- 26.Yamamoto K., Ichinose H., Aso Y., Fujii H. Expression analysis of cytochrome P450s in the silkmoth, Bombyx mori. Pestic. Biochem. Physiol. 2010;97:1–6. doi: 10.1016/j.pestbp.2009.11.006. [DOI] [Google Scholar]
- 27.Riaz M.A., Poupardin R., Reynaud S., Strode C., Ranson H., David J.P. Impact of glyphosate and benzo[a]pyrene on the tolerance of mosquito larvae to chemical insecticides: Role of detoxification genes in response to xenobiotics. Aquat. Toxicol. 2009;93:61–69. doi: 10.1016/j.aquatox.2009.03.005. [DOI] [PubMed] [Google Scholar]
- 28.Saavedra-Rodriguez K., Suarez A.F., Salas I.F., Strode C., Ranson H., Hemingway J., Black I. Transcription of detoxification genes after permethrin selection in the mosquito Aedes aegypti. Insect Mol. Biol. 2012;21:61–77. doi: 10.1111/j.1365-2583.2011.01113.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shen B., Dong H.Q., Tian H.S., Ma L., Li X.L., Wu G.L., Zhu C.L. Cytochrome P450 genes expressed in the deltamethrin-susceptible and-resistant strains of Culex pipiens pallens. Pestic. Biochem. Physiol. 2003;75:19–26. doi: 10.1016/S0048-3575(03)00014-2. [DOI] [Google Scholar]
- 30.Misra J.R., Horner M.A., Lam G., Thummel C.S. Transcriptional regulation of xenobiotic detoxification in Drosophila. Genes Dev. 2011;25:1796–1806. doi: 10.1101/gad.17280911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Snyder M.J., Stevens J.L., Andersen J.F., Feyereisen R. Expression of cytochrome P450 genes of the CYP4 family in midgut and fat body of the tobacco hornworm, Manduca sexta. Arch. Biochem. Biophys. 1995;321:13–20. doi: 10.1006/abbi.1995.1362. [DOI] [PubMed] [Google Scholar]
- 32.Zhao G.D., Zhao S.S., Gao R.N., Wang R.X., Zhang T., Ding H., Li B., Lu C.D., Shen W.D., Wei Z.G. Transcription profiling of eight cytochrome P450s potentially involved in xenobiotic metabolism in the silkworm, Bombyx mori. Pestic. Biochem. Physiol. 2011;100:251–255. doi: 10.1016/j.pestbp.2011.04.009. [DOI] [Google Scholar]
- 33.Brun-Barale A., Héma O., Martin T., Suraporn S., Audant P., Sezutsu H., Feyereisen R. Multiple P450 genes overexpressed in deltamethrin-resistant strains of Helicoverpa armigera. Pest Manag. Sci. 2010;66:900–909. doi: 10.1002/ps.1960. [DOI] [PubMed] [Google Scholar]
- 34.Scharf M., Parimi S., Meinke L.J., Chandler L., Siegfried B.D. Expression and induction of three family 4 cytochrome P450 (CYP4) genes identified from insecticide-resistant and susceptible western corn rootworms, Diabrotica virgifera virgifera. Insect Mol. Biol. 2001;10:139–146. doi: 10.1046/j.1365-2583.2001.00248.x. [DOI] [PubMed] [Google Scholar]
- 35.Zhu F., Feng J.N., Zhang L., Liu N. Characterization of two novel cytochrome P450 genes in insecticide-resistant house-flies. Insect Mol. Biol. 2008;17:27–37. doi: 10.1111/j.1365-2583.2008.00777.x. [DOI] [PubMed] [Google Scholar]
- 36.Waters L.C., Zelhof A.C., Shaw B.J., Chang L.Y. Possible involvement of the long terminal repeat of transposable element 17.6 in regulating expression of an insecticide resistance-associated P450 gene in Drosophila. Proc. Natl. Acad. Sci. USA. 1992;89:4855–4859. doi: 10.1073/pnas.89.11.4855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Morra R., Kuruganti S., Lam V., Lucchesi J., Ganguly R. Functional analysis of the cis-acting elements responsible for the induction of the CYP6A8 and CYP6G1 genes of Drosophila melanogaster by DDT, phenobarbital and caffeine. Insect Mol. Biol. 2010;19:121–130. doi: 10.1111/j.1365-2583.2009.00954.x. [DOI] [PubMed] [Google Scholar]
- 38.Mueller P., Chouaibou M., Pignatelli P., Etang J., Walker E.D., Donnelly M.J., Simard F., Ranson H. Pyrethroid tolerance is associated with elevated expression of antioxidants and agricultural practice in Anopheles arabiensis sampled from an area of cotton fields in Northern Cameroon. Mol. Ecol. 2008;17:1145–1155. doi: 10.1111/j.1365-294X.2007.03617.x. [DOI] [PubMed] [Google Scholar]
- 39.Müller P., Donnelly M.J., Ranson H. Transcription profiling of a recently colonised pyrethroid resistant Anopheles gambiae strain from Ghana. BMC Genomics. 2007;8:36. doi: 10.1186/1471-2164-8-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Marcombe S., Mathieu R.B., Pocquet N., Riaz M.A., Poupardin R., Sélior S., Darriet F., Reynaud S., Yébakima A., Corbel V. Insecticide resistance in the dengue vector Aedes aegypti from Martinique: Distribution, mechanisms and relations with environmental factors. PLoS One. 2012;7:e30989. doi: 10.1371/journal.pone.0030989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Baek J.H., Clark J.M., Lee S.H. Cross-strain comparison of cypermethrin-induced cytochrome P450 transcription under different induction conditions in diamondback moth. Pestic. Biochem. Physiol. 2010;96:43–50. doi: 10.1016/j.pestbp.2009.08.014. [DOI] [Google Scholar]
- 42.Festucci-Buselli R., Carvalho-Dias A., Oliveira-Andrade D., Caixeta-Nunes C., Li H.M., Stuart J., Muir W., Scharf M., Pittendrigh B. Expression of CYP6G1 and CYP12D1 in DDT resistant and susceptible strains of Drosophila melanogaster. Insect Mol. Biol. 2005;14:69–77. doi: 10.1111/j.1365-2583.2005.00532.x. [DOI] [PubMed] [Google Scholar]
- 43.Kamiya E., Yamakawa M., Shono T., Kono Y. Molecular cloning, nucleotide sequences and gene expression of new cytochrome P450s (CYP6A24, CYP6D3v2) from the pyrethroid resistant housefly, Musca domestica L: Diptera: Muscidae. Appl. Entomol. Zool. 2001;36:225–229. doi: 10.1303/aez.2001.225. [DOI] [Google Scholar]
- 44.Kasai S., Scott J.G. Expression and regulation of CYP6D3 in the house fly, Musca domestica (L.) Insect Biochem. Mol. Biol. 2001;32:1–8. doi: 10.1016/S0965-1748(01)00073-X. [DOI] [PubMed] [Google Scholar]
- 45.Kasai S., Weerashinghe I.S., Shono T., Yamakawa M. Molecular cloning, nucleotide sequence and gene expression of a cytochrome P450 (CYP6F1) from the pyrethroid-resistant mosquito, Culex quinquefasciatus Say. Insect Biochem. Mol. Biol. 2000;30:163–171. doi: 10.1016/S0965-1748(99)00114-9. [DOI] [PubMed] [Google Scholar]
- 46.Hopkins B.W., Longnecker M.T., Pietrantonio P.V. Transcriptional overexpression of CYP6B8/CYP6B28 and CYP6B9 is a mechanism associated with cypermethrin survivorship in field-collected Helicoverpa zea (Lepidoptera: Noctuidae) moths. Pest Manag. Sci. 2011;67:21–25. doi: 10.1002/ps.2034. [DOI] [PubMed] [Google Scholar]
- 47.Li X., Schuler M.A., Berenbaum M.R. Jasmonate and salicylate induce expression of herbivore cytochrome P450 genes. Nature. 2002;419:712–715. doi: 10.1038/nature01003. [DOI] [PubMed] [Google Scholar]
- 48.Hardstone M., Komagata O., Kasai S., Tomita T., Scott J. Use of isogenic strains indicates CYP9M10 is linked to permethrin resistance in Culex pipiens quinquefasciatus. Insect Mol. Biol. 2010;19:717–726. doi: 10.1111/j.1365-2583.2010.01030.x. [DOI] [PubMed] [Google Scholar]
- 49.Zhou X., Ma C., Li M., Sheng C., Liu H., Qiu X. CYP9A12 and CYP9A17 in the cotton bollworm, Helicoverpa armigera: Sequence similarity, expression profile and xenobiotic response. Pest Manag. Sci. 2010;66:65–73. doi: 10.1002/ps.1832. [DOI] [PubMed] [Google Scholar]
- 50.Yang Y., Chen S., Wu S., Yue L., Wu Y. Constitutive overexpression of multiple cytochrome P450 genes associated with pyrethroid resistance in Helicoverpa armigera. J. Econ. Entomol. 2006;99:1784–1789. doi: 10.1093/jee/99.5.1784. [DOI] [PubMed] [Google Scholar]
- 51.Mao W., Schuler M.A., Berenbaum M.R. CYP9Q-mediated detoxification of acaricides in the honey bee (Apis mellifera) Proc. Natl. Acad. Sci. USA. 2011;108:12657–12662. doi: 10.1073/pnas.1109535108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chung H., Sztal T., Pasricha S., Sridhar M., Batterham P., Daborn P.J. Characterization of Drosophila melanogaster cytochrome P450 genes. Proc. Natl. Acad. Sci. USA. 2009;106:5731–5736. doi: 10.1073/pnas.0812141106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chahine S., O’Donnell M.J. Interactions between detoxification mechanisms and excretion in Malpighian tubules of Drosophila melanogaster. J. Exp. Biol. 2011;214:462–468. doi: 10.1242/jeb.048884. [DOI] [PubMed] [Google Scholar]
- 54.Mittapalli O., Bai X., Mamidala P., Rajarapu S.P., Bonello P., Herms D.A. Tissue-specific transcriptomics of the exotic invasive insect pest emerald ash borer (Agrilus planipennis) PLoS One. 2010;5:e13708. doi: 10.1371/journal.pone.0013708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Willoughby L., Chung H., Lumb C., Robin C., Batterham P., Daborn P.J. A comparison of Drosophila melanogaster detoxification gene induction responses for six insecticides, caffeine and phenobarbital. Insect Biochem. Mol. Biol. 2006;36:934–942. doi: 10.1016/j.ibmb.2006.09.004. [DOI] [PubMed] [Google Scholar]
- 56.Sun L., Wang Z., Zou C., Cao C. Transcription profiling of 12 asian gypsy moth (Lymantria dispar) cytochrome p450 genes in response to insecticides. Arch. Insect Biochem. Physiol. 2014;85:181–194. doi: 10.1002/arch.21152. [DOI] [PubMed] [Google Scholar]
- 57.Xiao D., Liang X., Gao X., Yao J., Zhu K.Y. The lethal giant larvae gene in Tribolium castaneum: Molecular properties and roles in larval and pupal development as revealed by RNA interference. Int. J. Mol. Sci. 2014;15:6880–6896. doi: 10.3390/ijms15046880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Togawa T., Dunn W.A., Emmons A.C., Nagao J., Willis J.H. Developmental expression patterns of cuticular protein genes with the R&R Consensus from Anopheles gambiae. Insect Biochem. Mol. Biol. 2008;38:508–519. doi: 10.1016/j.ibmb.2007.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pfaffl M.W. A new mathematical model for relative quantification in real-time RT–PCR. Nucleic Acids Res. 2001;29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]