Abstract
Working memory deficit is the core neurocognitive disorder in schizophrenia patients. To identify the factors underlying working memory deficit in schizophrenia patients and to explore the implication of possible genes in the working memory using genome-wide association study (GWAS) of schizophrenia, computerized delay-matching-to-sample (DMS) and whole genome genotyping data were obtained from 100 first-episode, treatment-naïve patients with schizophrenia and 140 healthy controls from the Mental Health Centre of the West China Hospital, Sichuan University. A composite score, delay-matching-to-sample total correct numbers (DMS-TC), was found to be significantly different between the patients and control. On associating quantitative DMS-TC with interactive variables of groups × genotype, one SNP (rs1411832), located downstream of YWHAZP5 in chromosome 10, was found to be associated with the working memory deficit in schizophrenia patients with lowest p-value (p = 2.02 × 10−7). ConsensusPathDB identified that genes with SNPs for which p values below the threshold of 5 × 10−5 were significantly enriched in GO:0007155 (cell adhesion, p < 0.001). This study indicates that working memory, as an endophenotype of schizophrenia, could improve the efficacy of GWAS in schizophrenia. However, further study is required to replicate the results from our study.
Keywords: working memory, delayed-matching-to-sample test, schizophrenia, genome-wide association study, pathway
1. Introduction
Schizophrenia is a psychiatric disorder that affects about 1% of the world population [1]. Both win and adoption studies have shown that genetic factors play an important role in the pathogenesis of schizophrenia (heritability close to 0.8) [2]. Schizophrenia, having a high heritability, is a multi-dimensional disorder that cannot be explained by Mendelian genetics. It is now generally agreed that it is caused by combined effects of hundreds or thousands of gene variants with modest effects [3]. Advances in whole-genome genotyping and related analysis methods have enabled identification of these variants. Since 2009, many genome-wide association studies (GWAS), with case-control designs, have been carried out to study the genetic architecture underlying schizophrenia [4,5,6]. The results from a GWA study with the largest sample size so far, identified 108 variants that are associated with schizophrenia. Although some variants, such as ones in ZNF804A and variants in major histocompatibity complex (MHC), have been reported to be replicated, many SNPs were found to be non-overlapping in this study [7]. The inconsistency was attributed to factors such as population stratification, sample size, and clinical heterogeneity, with clinical heterogeneity being the most difficult confounding factor in case-control GWASs. Unlike other complex disorders, diagnosis of schizophrenia lacks reliable biomarkers and animal models. In fact, most of its diagnoses are based on subjective judgments from clinical practitioners. Additionally, lack of objective laboratory measures tends to create a barrier between the disorder diagnosis and its management. Although large sample size is a good strategy to map related variants, many case-control studies still failed to produce satisfactory results. Alternatively, various researchers used quantitative traits (QTs) as endophenotype or intermediate phenotype in order to enhance the efficacy of the GWAS in schizophrenia [8,9]. In comparison to dichotomous phenotype (subjective and random-prone diagnoses), QT is easy to measure and can be standardized. In a continuum from gene to clinical outcome, quantitative pathological changes are supposed to be more proximal for genetic underpinnings of the disease. Various studies have shown that QTs improve the efficacy of GWAS [6,9,10,11,12]. Further, they were also found to be beneficial in mapping the genes associated with schizophrenia. Dickinson et al. [13] used g score, a composite score combining six neuro-cognitive dimensions as QTs in a genome association study to map potential genetic variants associated with schizophrenia. This approach led to unveiling the effects of SCN2A on general cognitive ability, brain physiology, and mRNA expression in schizophrenia. Potkin et al. [14] carried out a genome-wide study on schizophrenia by using QTs with results showing that variants in six genes (POU3F2, TRAF, GPC1, POU3F2, TRAF, and GPC1) were associated significantly with working memory task-related bold signal of functional MRI in schizophrenia, with a p-value threshold of 10−6. Furthermore, they identified these six genes/regions involved in pathways related to neurodevelopment and response to stress. These studies demonstrated the efficacy of using QTs as endophenotype in exploring the pathogenesis of schizophrenia.
Previous studies have demonstrated that working memory deficit exists both in schizophrenic patients and their biological relatives [15,16,17]. In the present study, we used the total correct numbers of delay-matching-to-sample (DMS-TC) as QTs, which is a composite score generated from delay-matching-to-sample (DMS) test in the Cambridge Neuropsychological Test Automated Battery (CANTAB), to explore common genetic variants underlying the working memory deficit in schizophrenia by using a hypothesis-free GWAS analysis. Furthermore, we used ConsensusPathDB (available on line: http://consensuspathdb.org/), one of the most widely applied pathway databases, to analyze pathway over-representation of genes that the associated variants are located in or close to. It is assumed that this downstream strategy will increase validity of the study and might shed light on the mechanism of how these biological pathways bridge genes with neurocognitive deficits in schizophrenia.
2. Results
2.1. Demographic Characteristics and Delayed-Matching-to-Sample (DMS) Test
A hundred and forty first-episode and drug-naive patients with schizophrenia and 100 healthy controls were included in the study. There was no difference between patients and controls in term of sex, age, and education years (Table 1). Nineteen scores of 125 individuals were successfully uploaded into the result dataset (67 controls, 58 cases) after DMS test. Of these 19 measurement scores from DMS, 10 scores remained significantly different between patients and healthy controls with age, sex, and education year being adjusted for. However, after multiple testing, only one of these scores, DMS-TC, survived Bonferroni correction, as shown in Table 1 (p < 0.05). From the study, it was observed that DMS-TC score can be used for QTs in subsequent analysis.
Table 1.
Summary of Demographic characteristics and Delayed-matching-to-sample (DMS) results.
| Demographic Characteristics and DMS Measures | Patients | Controls | Statistic Significance |
|---|---|---|---|
| Race (% Chinese) | 100 | 140 | - |
| Gender (% male) | 44.29 | 55.71 | 0.848 |
| Education (year) | 10.60 ± 5.117 | 11.86 ± 5.416 | 0.071 |
| Age | 21.57 ± 12.850 | 21.08 ± 10.776 | 0.755 |
| DMS-TC | 18.31 (58) | 15.593 (67) | 0.049 |
DMS-TC: Delay-matching-to-sample total correct numbers.
2.2. Analysis of Quantitative Traits (QTs) and Over-Representation Study
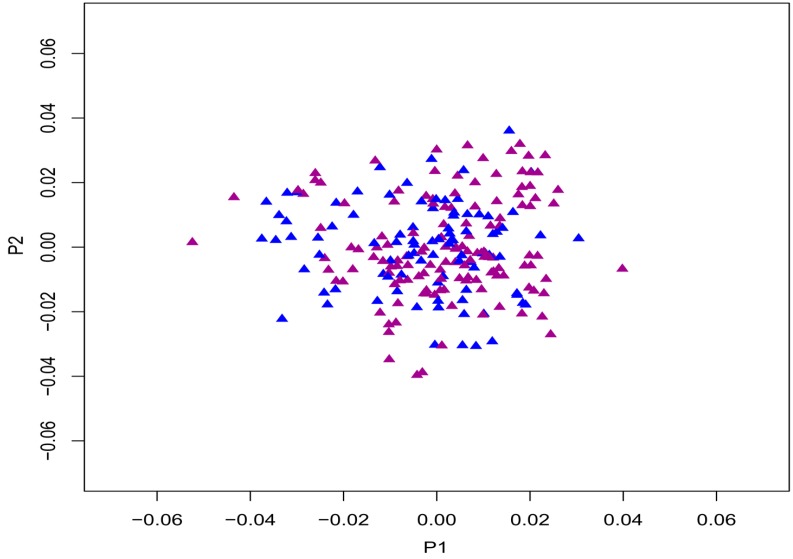
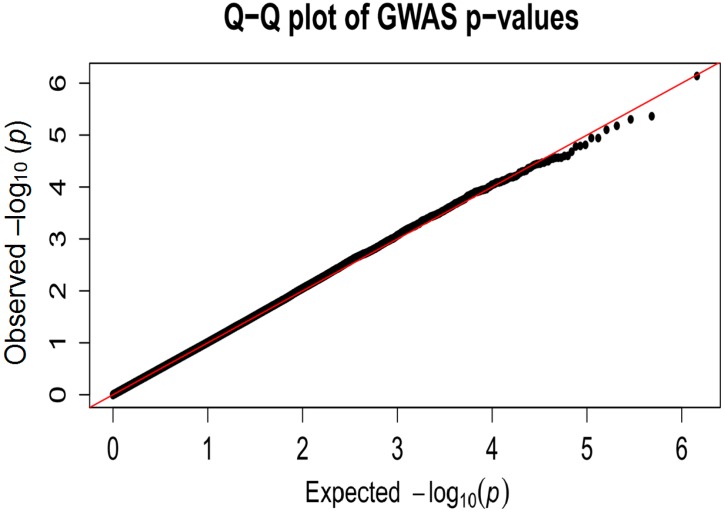
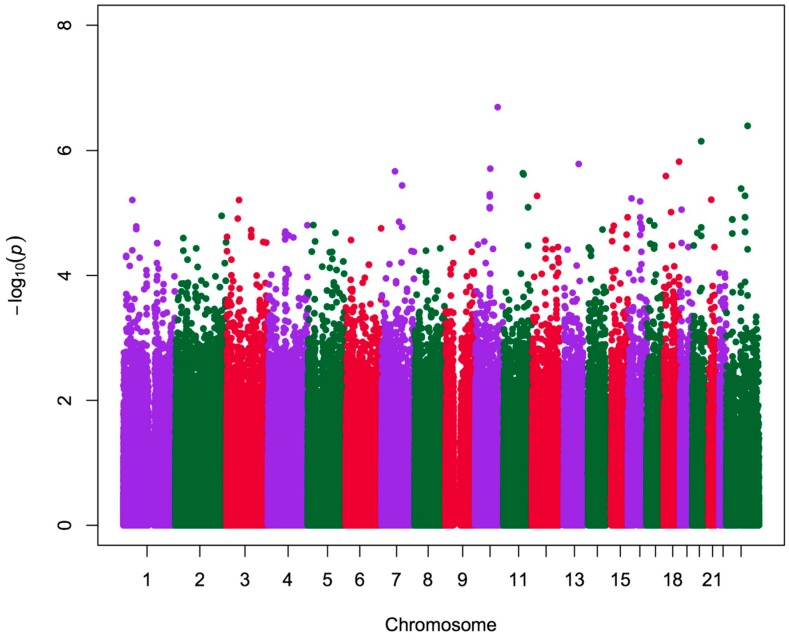
A total of 742,805 SNPs passed the quality control, with a mean call rate of 98.9%. However, seven patients and six controls failed to pass the quality control and cryptic relatedness, and thus were excluded from subsequent analysis. Multidimensional scaling (MDS) in PLINK showed that individuals were tightly clustered, indicating that individuals were of the same ancestral Chinese Han origin (Figure 1). Following quality control, inflation factor (λ), generated from PLINK analysis was found to be 1.0099, which showed that confounding factors were well-adjusted (Figure 2). Since there was no conspicuous population structure among the study samples and no significant deviation of the observed distribution, principal components from MDS was not chosen as a covariate for the linear regression analysis. Finally, genotyping data of 93 patients and 134 controls passed the quality control and were included for the subsequent association study. Results from the association test of genotype × group and DMS-TC QTs demonstrated that rs1411832 was found to be the most significant SNP, which was located at the downstream of YWHAZP5 (p = 2.02 × 10−7) (Table 2). 121 autosomal variants, located in or within flanking areas of 46 genes, passed the significant threshold of 5 × 10−5 using DMS-TC as quantitative trait (Figure 3). These annotated genes were used to map the significant pathways for the study. Pathway over-representation analysis of annotated genes by ConsensusPathDB showed that nine genes (SCARB1, DSCAM, LGALS4, COL14A1, PTPRT, IGSF11, DSCAML1, SMOC2, and FAT3) were significantly over-represented in one GO, cell adhesion (GO:0007155, p < 0.001).
Figure 1.
Multidimensional scaling plot of first two multidimensional scaling (MDS) components. Blue = control; Dark magenta = case.
Figure 2.
Q-Q plot of genome-wide association study (GWAS) on schizophrenia using DMS-TC (DMS-total correct numbers) as quantitative trait.
Table 2.
Significant interaction of SNPs × diagnosis and quantitative trait (DMS-TC) in DMS.
| CHR | SNP | Position | Type | Gene | Traits | p |
|---|---|---|---|---|---|---|
| 10 | rs1411832 | 107886255 | Intergenic | Downstream of YWHAZP5 | DMS-TC | 2.02 × 10−7 |
| 20 | rs61131853 | 41749812 | Intron | PTPRT | DMS-TC | 7.10 × 10−7 |
| 18 | rs79589976 | 73305576 | Intron | TADA2L | DMS-TC | 1.51 × 10−6 |
| 13 | rs74108723 | 90358757 | Intergenic | N/A | DMS-TC | 1.64 × 10−6 |
| 10 | rs10999524 | 72525761 | intergenic | Upstream of C10orf27 | DMS-TC | 1.95 × 10−6 |
| 7 | rs4718138 | 64303065 | Intron | ZNF138 | DMS-TC | 2.15 × 10−6 |
| 11 | rs1552511 | 92605986 | Intron | FAT3 | DMS-TC | 2.31 × 10−6 |
| 11 | rs555329 | 95993708 | Intron | FAT3 | DMS-TC | 2.41 × 10−6 |
| 18 | rs2868934 | 10204383 | Intron | TADA2L | DMS-TC | 2.56 × 10−6 |
| 7 | rs60569161 | 98199903 | intergenic | Upstream of NPTX2 | DMS-TC | 3.62 × 10−6 |
| 10 | rs7899885 | 70143774 | Intron | RUFY2 | DMS-TC | 5.02 × 10−6 |
| 12 | rs12811916 | 25550866 | Intron | DSCAML1 | DMS-TC | 5.33 × 10−6 |
| 10 | rs2281698 | 70104320 | Intron | RUFY2 | DMS-TC | 5.42 × 10−6 |
| 16 | rs4780688 | 17567540 | intergenic | Downstream of XYLT1 | DMS-TC | 5.84 × 10−6 |
| 21 | rs76659985 | 26786931 | Intron | LINC00158 | DMS-TC | 6.13 × 10−6 |
| 1 | rs3738516 | 43440201 | Intron | Downstream of SLC2A1 | DMS-TC | 6.19 × 10−6 |
| 3 | rs17609699 | 60487578 | Intron | FHIT | DMS-TC | 6.20 × 10−6 |
| 16 | rs4785000 | 58963145 | intergenic | Upstream of LOC100132798 | DMS-TC | 6.51 × 10−6 |
| 11 | rs630024 | 117534353 | Intron | MAML2 | DMS-TC | 8.09 × 10−6 |
| 10 | rs3781567 | 70105178 | Intron | RUFY2 | DMS-TC | 8.11 × 10−6 |
| 10 | rs1162753 | 70105560 | synonymous | RUFY2 | DMS-TC | 8.11 × 10−6 |
| 10 | rs17297439 | 70103461 | Intron | RUFY2 | DMS-TC | 8.11 × 10−6 |
| 10 | rs3199937 | 70102749 | Intron | RUFY2 | DMS-TC | 8.11 × 10−6 |
| 10 | rs3781568 | 70105286 | Intron | HNRNPH3 | DMS-TC | 8.11 × 10−6 |
| 10 | rs7897488 | 70179746 | Intron | RUFY2 | DMS-TC | 8.11 × 10−6 |
| 10 | rs7071140 | 70100250 | Intron | HNRNPH3 | DMS-TC | 8.42 × 10−6 |
| 19 | rs7249563 | 7009813 | intergenic | Downstream of VAPA | DMS-TC | 8.85 × 10−6 |
| 18 | rs652630 | 34899378 | Intron | CELF4 | DMS-TC | 9.70 × 10−6 |
| 2 | rs520102 | 220529636 | intergenic | Downstream of SLC4A3 | DMS-TC | 1.11 × 10−5 |
| 15 | rs2453034 | 101241344 | Intron | NPAS3 | DMS-TC | 1.17 × 10−5 |
| 16 | rs6499996 | 58993725 | intergenic | Downstream of XYLT1 | DMS-TC | 1.17 × 10−5 |
| 3 | rs6773944 | 54516884 | Intron | CACNA2D3 | DMS-TC | 1.23 × 10−5 |
| 17 | rs2322973 | 13699188 | intergenic | Downstream of LOC644649 | DMS-TC | 1.33 × 10−5 |
| 7 | rs58908055 | 83788680 | Intron | SEMA3A | DMS-TC | 1.38 × 10−5 |
| 16 | rs9930442 | 58978493 | intergenic | Downstream of LOC644649 | DMS-TC | 1.45 × 10−5 |
| 5 | rs59017736 | 26250801 | intergenic | Downstream of MSNL1 | DMS-TC | 1.56 × 10−5 |
| 4 | rs4470690 | 188721512 | intergenic | Downstream of LOC644325 | DMS-TC | 1.57 × 10−5 |
| 17 | rs8067120 | 35783565 | Intron | TADA2A | DMS-TC | 1.58 × 10−5 |
| 15 | rs1458888 | 35406129 | Intron | NPAS3 | DMS-TC | 1.60 × 10−5 |
| 1 | rs1286830 | 62270494 | Intron | INADL | DMS-TC | 1.64 × 10−5 |
| 16 | rs8060933 | 65891108 | intergenic | Downstream of LOC644649 | DMS-TC | 1.67 × 10−5 |
| 7 | rs705337 | 98226627 | intergenic | Upstream of NPTX2 | DMS-TC | 1.68 × 10−5 |
| 20 | rs6030661 | 41748131 | Intron | PTPRT | DMS-TC | 1.70 × 10−5 |
| 6 | rs4708759 | 169005721 | Intron | SMOC2 | DMS-TC | 1.76 × 10−5 |
| 16 | rs12373039 | 65894066 | intergenic | Downstream of LOC644649 | DMS-TC | 1.80 × 10−5 |
| 1 | rs1286831 | 62271962 | Intron | INADL | DMS-TC | 1.81 × 10−5 |
| 14 | rs78636353 | 88681552 | Intron | KCNK10 | DMS-TC | 1.84 × 10−5 |
| 3 | rs614673 | 118539108 | intergenic | Upstream of IGSF11 | DMS-TC | 1.87 × 10−5 |
| 15 | rs28436697 | 27624233 | Intron | GABRG3 | DMS-TC | 1.92 × 10−5 |
| 4 | rs77470375 | 83823703 | Intron | THAP9 | DMS-TC | 1.98 × 10−5 |
| 5 | rs2066960 | 131994435 | Intron | IL13 | DMS-TC | 2.08 × 10−5 |
| 20 | rs6132627 | 23444688 | Intron | LGALS4 | DMS-TC | 2.08 × 10−5 |
| 4 | rs62303604 | 82397503 | intergenic | Downstream of RASGEF1B | DMS-TC | 2.17 × 10−5 |
| 4 | rs16998600 | 82403384 | intergenic | Downstream of RASGEF1B | DMS-TC | 2.17 × 10−5 |
| 4 | rs62302363 | 82417252 | intergenic | Downstream of RASGEF1B | DMS-TC | 2.17 × 10−5 |
| 4 | rs17005142 | 82402588 | intergenic | Downstream of RASGEF1B | DMS-TC | 2.17 × 10−5 |
| 4 | rs17005144 | 82404274 | intergenic | Downstream of RASGEF1B | DMS-TC | 2.17 × 10−5 |
| 4 | rs6819741 | 82403942 | intergenic | Downstream of RASGEF1B | DMS-TC | 2.17 × 10−5 |
| 15 | rs2575426 | 96405795 | intergenic | Downstream ofLOC441722 | DMS-TC | 2.22 × 10−5 |
| 16 | rs12931857 | 58967112 | intergenic | Upstream of CDH5 | DMS-TC | 2.25× 10−5 |
| 3 | rs80028372 | 118805552 | Intron | IGSF11 | DMS-TC | 2.26 × 10−5 |
| 4 | rs7438406 | 99702148 | intergenic | Downstream of BTF3L3 | DMS-TC | 2.28 × 10−5 |
| 20 | rs2325606 | 41738666 | Intron | PTPRT | DMS-TC | 2.33 × 10−5 |
| 3 | rs17659192 | 3108711 | Intron | IL5RA | DMS-TC | 2.41 × 10−5 |
| 3 | rs12630657 | 118842442 | Intron | IGSF11 | DMS-TC | 2.43 × 10−5 |
| 4 | rs10034975 | 122175499 | intergenic | Upstream of GPR103 | DMS-TC | 2.46 × 10−5 |
| 9 | rs7041922 | 34938198 | intergenic | Upstream of KIAA1045 | DMS-TC | 2.49 × 10−5 |
| 2 | rs6544074 | 37634473 | intergenic | Downstream of QPCT | DMS-TC | 2.52 × 10−5 |
| 4 | rs10015146 | 82401332 | intergenic | Downstream of RASGEF1B | DMS-TC | 2.65 × 10−5 |
| 6 | rs2744229 | 25341580 | Intron | LRRC16A | DMS-TC | 2.71 × 10−5 |
| 12 | rs12318900 | 66044284 | intergenic | Upstream of KRAS | DMS-TC | 2.72 × 10−5 |
| 15 | rs9708085 | 27619217 | Intron | GABRG3 | DMS-TC | 2.83 × 10−5 |
| 5 | rs6894424 | 34732456 | Intron | RAI14 | DMS-TC | 2.85 × 10−5 |
| 10 | rs2503870 | 43796180 | Intron | HNRNPH3 | DMS-TC | 2.86 × 10−5 |
| 3 | rs6808187 | 175861931 | intergenic | Upstream of LOC730168 | DMS-TC | 2.91 × 10−5 |
| 2 | rs755300 | 241652703 | Intron | KIF1A | DMS-TC | 2.94 × 10−5 |
| 3 | rs4687154 | 190304172 | Intron | IL1RAP | DMS-TC | 2.98 × 10−5 |
| 19 | rs1353166 | 6992943 | intergenic | BRUNOL4 | DMS-TC | 3.02 × 10−5 |
| 1 | rs347272 | 162318498 | Intron | NOS1AP | DMS-TC | 3.04 × 10−5 |
| 1 | rs11577628 | 162319524 | Intron | NOS1AP | DMS-TC | 3.04 × 10−5 |
| 1 | rs347273 | 162317513 | Intron | NOS1AP | DMS-TC | 3.04 × 10−5 |
| 17 | rs8065154 | 17614947 | Intron | RAI1 | DMS-TC | 3.11 × 10−5 |
| 17 | rs6502615 | 17612023 | Intron | TADA2L | DMS-TC | 3.11 × 10−5 |
| 10 | rs12268934 | 13581758 | intergenic | Downstream of RASGEF1A | DMS-TC | 3.21 × 10−5 |
| 17 | rs11263747 | 35742069 | Intron | RAI1 | DMS-TC | 3.30 × 10−5 |
| 17 | rs11263750 | 35816826 | Intron | RAI1 | DMS-TC | 3.30 × 10−5 |
| 17 | rs11868171 | 35816330 | Intron | C17orf78 | DMS-TC | 3.30 × 10−5 |
| 17 | rs2898656 | 35806418 | Intron | ACACA | DMS-TC | 3.30 × 10−5 |
| 20 | rs2024886 | 5700696 | Intron | PTPRT | DMS-TC | 3.31 × 10−5 |
| 11 | rs583983 | 117525125 | Intron | DSCAML1 | DMS-TC | 3.32 × 10−5 |
| 18 | rs72899323 | 39845104 | Intron | LINC00907 | DMS-TC | 3.34 × 10−5 |
| 19 | rs1035525 | 39299362 | Intron | LGALS4 | DMS-TC | 3.50 × 10−5 |
| 12 | rs10846743 | 125310305 | Intron | SCARB1 | DMS-TC | 3.51 × 10−5 |
| 21 | rs7283946 | 42143503 | Intron | DSCAM | DMS-TC | 3.51 × 10−5 |
| 14 | rs10131813 | 23745533 | Intron | HOMEZ | DMS-TC | 3.58 × 10−5 |
| 14 | rs10144278 | 23749595 | Intron | HOMEZ | DMS-TC | 3.58 × 10−5 |
| 15 | rs72633609 | 96389640 | intergenic | Upstream of LOC100132798 | DMS-TC | 3.66 × 10−5 |
| 15 | rs11858405 | 96331346 | Intron | GABRG3 | DMS-TC | 3.66 × 10−5 |
| 2 | rs6544072 | 99112745 | Intron | INPP4A | DMS-TC | 3.68 × 10−5 |
| 8 | rs10111291 | 121266654 | Intron | COL14A1 | DMS-TC | 3.68 × 10−5 |
| 17 | rs58509949 | 35016090 | Intron | TADA2L | DMS-TC | 3.70 × 10−5 |
| 12 | rs7137152 | 66063249 | Intron | SCARB1 | DMS-TC | 3.75 × 10−5 |
| 12 | rs17120580 | 66018826 | intergenic | Upstream of LOC204010 | DMS-TC | 3.75 × 10−5 |
| 10 | rs2349764 | 85675348 | intergenic | At upstream of PRPF18 | DMS-TC | 3.76 × 10−5 |
| 14 | rs8005082 | 28802074 | Intron | HOMEZ | DMS-TC | 3.78 × 10−5 |
| 12 | rs7968661 | 99137384 | Intron | ANKS1B | DMS-TC | 3.81 × 10−5 |
| 13 | rs9315422 | 37055511 | intergenic | Downstream of CCNA1 | DMS-TC | 3.86 × 10−5 |
| 1 | rs4660239 | 43431528 | Intron | SLC2A1-AS1 | DMS-TC | 3.95 × 10−5 |
| 14 | rs1958005 | 33646022 | Intron | HOMEZ | DMS-TC | 3.97 × 10−5 |
| 2 | rs6544072 | 37619430 | intergenic | Downstream of QPCT | DMS-TC | 4.01 × 10−5 |
| 8 | rs7829966 | 53081817 | Intron | ST18 | DMS-TC | 4.01 × 10−5 |
| 7 | rs7794560 | 143029983 | Intron | CLCN1 | DMS-TC | 4.06 × 10−5 |
| 3 | rs17659353 | 3111436 | Intron | IL5RA | DMS-TC | 4.09 × 10−5 |
| 5 | rs10059239 | 18879807 | intergenic | Downstream of LOC646241 | DMS-TC | 4.15 × 10−5 |
| 7 | rs3298 | 154685873 | Intron | DPP6 | DMS-TC | 4.17 × 10−5 |
| 16 | rs9924423 | 64526807 | intergenic | Upstream of CDH5 | DMS-TC | 4.18 × 10−5 |
| 9 | rs12237468 | 125491634 | Intergenic | Downstream of OR1L4 | DMS-TC | 4.19 × 10−5 |
| 5 | rs56196053 | 115759247 | intergenic | Upstream of SEMA6A | DMS-TC | 4.20 × 10−5 |
| 5 | rs1480583 | 105022790 | intergenic | Downstream of RAB9P1 | DMS-TC | 4.25 × 10−5 |
| 14 | rs912857 | 33657980 | Intron | NPAS3 | DMS-TC | 4.85 × 10−5 |
| 1 | rs12024045 | 14297220 | Intron | KAZN | DMS-TC | 4.86 × 10−5 |
CHR: Chromosome; DMS-TC: Delay-matching-to-sample total correct numbers; SNP: Single nucleotide polymorphism.
Figure 3.
Manhattan plots of genome-wide association of all SNPs with DMS-TC. SNPs were plotted on the x axis according to their position on each chromosome represented by difference color bars against association of DMS-TC on the y axis (shown as −log10P value).
3. Discussion
3.1. Association Study of Working Memory Deficit as QT
The current study is the first GWAS utilizing one of the composite scores of the working memory task paradigm as QT to map common variants associated with schizophrenia. In this study, significant differences in computerized DMS were found between schizophrenia patients and healthy controls. One Composite scores, DMS-TC, was therefore chosen as QTs to associate with interaction of genotypes × group. Additionally, genes containing variants of significant importance were mapped by one GO term using pathway database. Results of the study highlighted the validity of the method used for genetic underpinning in the pathology of schizophrenia.
We found rs1411832 with the smallest p value in one of the intergenic regions downstream of gene YWHAZP5 (tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta pseudogene 5). Most pseudogenes are very similar protein-coding genes and some are believed to be extra copies of genes; a recent study demonstrated that some pseudogenes are functional [18]. YWHAZ has been well-studied in schizophrenia [19,20,21], its gene product belongs to the 14-3-3 family of proteins which mediate signal transduction by binding to phosphoserine-containing proteins. 14-3-3 is abundant in brain which points to its critical role in neuronal functions [22,23]. Initial finding of association can be followed up by (i) sequence similarity between YWHAZ and YWHAZP5; (ii) better defining the nature of the potential association between YWHAZ and YWHAZP5; and (iii) attempting to replicate it in two additional samples.
Furthermore, some studies have shown that intergenic variants might play an important role in regulating expression of nearby genes [24,25] and they are also often in linkage disequilibrium with causal variants in the gene. Further study should be conducted on causal variants in LD with this intergenic variant. Meanwhile, Since the study was trait-associated GWAS and both environment and gene can contribute to complex traits, it is likely that this significant variant regulates the expression of YWHAZP5 through epigenetic progress. Thus further studies are required to validate this presumption.
3.2. Over-Representation of Genes in Single Pathway
Pathway over-representation analysis of annotated genes by ConsensusPathDB showed that nine genes (SCARB1, DSCAM, LGALS4, COL14A1, PTPRT, IGSF11, DSCAML1, SMOC2, and FAT3) were significantly over-represented in one GO, cell adhesion (GO:0007155, p < 0.001). Of these nine genes, DSCAM and DSCAML1 (Down’s syndrome cell adhesion molecule gene) are associated with Down’s syndrome. Various studies have demonstrated a relationship between DSCAM and neurobehavioral phenotype of Down’s syndrome which includes working memory deficit [26,27,28,29]. Recently, exome sequencing has detected genetic overlapping between schizophrenia and neurodevelopmental disorder [30,31]. From the above studies, we concluded that there should at least be a subgroup in schizophrenia that shares the same molecular pathological pathway with neurodevelopmental disorder. Beside this, PTPRT was often found to be related to neurodevelopment and cell growth [31,32]. A neurodevelopment model of schizophrenia has been frequently verified in studies involving different research strategies. Findings from our study add further evidence that neurodevelopmental deficit is associated with the pathogenesis of schizophrenia.
3.3. Functional Characterization of Genes Post GWAS
In recent years, various arguments were made on the extrapolation of the GWAS results in order to clarify the current vague picture. After GWAS, a posteriori functional pathway is one of the most authentic strategies to study the role of mapped genes in the complex disorder [33,34,35]. Pathway approaches have been adopted in many studies [36,37] and are shown to be robust to detect the joint action of variants of small effect clustering within biological pathways that play a major role in predisposing to complex genetic disorders and they can increase power by summarizing combined effects of all SNPs within a pathway in attempt to make biological meaningful interpretations of the data [38,39,40]. Even very large GWAS may lack power to identify small SNP effects, but these may be detectable at a pathway level.
In our study, subsequent functional enrichment from ConsensusPathDB highlighted that the tagged genes were mostly enriched in cell adhesion (GO:0007155). Cell adhesion molecules in nervous system and neural cell adhesion molecules (NCAMs) play a critical role in neural development such as cell adhesion, growth, and migration as scaffold for novel binding proteins [41,42,43], and various studies have demonstrated changes in NCAMs in schizophrenia patients. Although NCAM abnormalities in schizophrenia patients have been widely described, very few studies focus on the relationship between extracellular matrix and paradigm of the visual working memory test.
4. Experimental Section
4.1. Participants
One hundred first-episode schizophrenia patients (recruited from West China Hospital of Sichuan University, Chengdu, China) and 140 demographically matched healthy controls without family history of psychological disorders (recruited from the local neighborhood), were included in the study. Patients were interviewed with SCID (structured clinical interview) by trained psychiatrists to ensure the diagnosis was based on DSM-IV (Diagnostic and Statistical Manual of Mental Disorders, fourth edition) criterion, whereas healthy controls were screened with structured clinical interview-non-patient (SCID-NP) to ensure the absence of psychiatric illness. Patients were followed up to 6 months in order to confirm the diagnosis. No anti-psychotic medication was administered to patients at the time of clinical and neuro-cognitive evaluation. Both patients and controls were excluded if they had one of following conditions: (i) organic cerebral diseases; (ii) neurological diseases; (iii) severe endocrine diseases; (iv) axis I and II diagnosis other than schizophrenia according to DSM-IV; and (v) single or double limb palsy. Informed written consent was obtained from all the participants after explaining the study. This study was approved by the Institutional Review Broad (IRB) of West China Hospital, Sichuan University.
4.2. DMS Test Paradigm
DMS is one of the computerized tests in the Cambridge Neuropsychological Test Automated Battery (CANTAB) (available on line: http://www.cantab.com), which is used to assess working memory. The paradigm of the DMS test is described elsewhere in detail [44]. DMS test data was obtained for 125 out of 227 genotyped individuals (58 patients and 67 healthy controls). There are 19 scores from the DMS test which belongs to three categories: correctness in terms of number and percentage (total correct percentage, correct percentage on simultaneous level, correct percentage on all delays, correct percentage on 0 ms delay, correct percentage on 4000 ms delay, correct percentage on 12,000 ms delay, total correct numbers, total correct numbers on all delays, total correct numbers on simultaneous level, total correct numbers on 0 ms delay, total correct numbers on 4000 ms delay, total correct numbers on 12,000 ms delay); latency at both simultaneous and delayed level (mean correct latency, mean correct latency on all delays, mean correct latency on simultaneous level); and statistical analysis of discrimination between signal and noise (prob error given correct, prob error given error, DMS A’, DMS B’). DMS-TC, we are choosing here as QTs for association study, belongs to correctness in terms of number and percentage. It indicates the total number of trials in which subjects selected correct stimulus as their first response and is calculated by combination of total correct numbers on simultaneous level and total correct numbers on all delays.
4.3. Statistical Analysis
4.3.1. DMS Test
Analysis of variance (ANOVA) using statistical package for the social sciences (SPSS21.0, SPSS, Inc., Chicago, IL, USA) was used to compare DMS test scores between patients and healthy controls, adjusting age, education years, and gender as covariates. The multiple test was accounted for by using Bonferroni correction. To justify the inter-group differences, statistically significant values (p-value) were set to p < 0.05 (Bonferroni correction).
4.3.2. Genotyping and Quality Control
DNA was extracted from whole blood samples using the standard isolation method and the genotyping was performed on the HumanOmniZhongHua-8 Bead Chip platform. Genotyping data were systematically filtered according to their genotyping rates (per sample and per marker), minor allele frequency (MAF), and Hardy-Weinberg equilibrium tests (only in controls) in PLINK. Participants with low genotyping rate (<97%), markers with missing rate >5% per individual and/or with MAF >0.05, and markers that failed to pass Hardy-Weinberg equilibrium tests (p ≤ 0.00001) were excluded from the study. Furthermore, gender of each participant, with genotyping data on gender-specific loci, was confirmed by genotyping platform and the participants with the lower genotype call rate were excluded if pairs of participants with identical genotypes were found. After systematic filtering, 109,923 SNPs and four individuals (two patients and two healthy controls) were excluded based on the set threshold value.
4.3.3. Correction for Population Stratification
For further analysis of the genetic relationship between SNPs and population structure within the sample, remaining SNPs were further pruned to ensure linkage equilibrium between the SNPs. SNPs with r2 > 0.5, were consequently excluded from the study (PLINK command—indep-pairwise 50 10 0.5). The multi-dimensional scaling algorithm (MDS) for cryptic relatedness in PLINK was applied to a pruned sample. Matrix generated from MDS was visualized by plotting R package (available on line: http://www.r-project.org). Thirty eight thousand five hundred and fifty seven SNPs and nine subjects (five cases and four healthy controls) were excluded after failing the genetic relationship test and cryptic relatedness. Inflation factor, lambda (λ) was estimated by calculating the mean of observed and expected chi-square test statistics to analyze the results of whole-genome association studies for over-dispersion due to population substructure and other confounding factors.
4.3.4. Linear Regression Analysis
DMS-TC scores, which significantly differed between patients and health controls, were used as QTs in regression analysis. Of the total of 227 subjects with high-quality genotyping data, 125 with DMS-TC were included for QT analysis. The QT analysis was based on comparing the differential effects of SNPs on the DMS QTs. Out of the four possible models (additive, co-dominant, dominant, and recessive), additive components (reflecting the additive contribution of risks for complex diseases) were included in the linear model. Multivariate linear regression model in PLINK1.07 [45] was used to assess the correlation of QT and interaction of group and additive genetic risk of minor allele of 742,805 SNPs which passed genetic quality control while adjusting for age, sex, and education year. The group was labeled as discrete variable (1 = patients, 2 = healthy controls) to define the interaction with the genetic risk of allele. Finally, a Manhattan plot was plotted using R to illustrate the whole genome-wide association scans.
4.4. Over-Representation Study
To map the pathway over-represented by genes from association study, genes with SNPs passing a threshold of 5 × 10−5 were further explored by web server of ConsensusPathDB (available on line: http://cpdb.molgen.mpg.de/CPDB) to detect the set most enriched by these genes. Hyper-geometric test detect the gene set in predefined lists of functionally associated genes (pathways, gene ontology categories, and neighborhood-based entity sets) with p-value indicating the extent of corresponding enrichment. Previous studies shown that the p value of 5 × 10−8 is rather conservative which may lead to excessive false negative especially for gene- or pathway- based analysis in smaller sample size [46]. Some genes missed in the single-locus association test can be detected significantly when integrated into pathway association analysis [47,48,49]. In addition, Nicolae et al. [50] found that trait-associated SNPs below 5 × 10−4 are more likely to be eQTLs, and these signals from GWA study are not exhausted after fully investigating the results of expression quantitative trait locus (eQTL) studies in lymphoblastoid cell lines from HapMap samples. Enlightened by these studies, we set 5 × 10−5 as the empirical cut-off p value to identify potential pathways involved in working memory deficit in schizophrenia.
5. Conclusions
In our study, GWAS was conducted to find the genes that were significantly associated with working memory deficit of schizophrenia. The results showed that variants near one gene, which encodes YWHAZP5, were most significantly associated with working memory deficits in schizophrenia patients. Additionally, downstream pathway analysis showed that NCAM is essential in working memory-related pathogenesis of schizophrenia.
Although the study was promising in various aspects, there are some limitations associated with it. First, we chose only one part of results from DMS, i.e., DMS-TC, as our QTs. We are fully aware that there are scores of many kinds generated from DMS of CANTAB, we multiple-corrected all p-values generated from DMS test using Bonferroni correction, and only the total correct numbers of the DMS could survive after the correction, which imply the strongest effect size in all indicators of the DMS. The other indicators could not survive after Bonferroni correction, so they were not included in subsequent analysis.
Second, this study is an explorative study of methodology involving variant mapping and annotation of its related biological pathway. SNP with the most significant p-value in our study is rs1411832 in chromosome 10 (p < 2.02 × 10−7), which is acceptable considering our sample size and missing phenotype data [51]. However, given the limitations of GWAS and its sample size, larger sample size with inclusion of both common and rare variants is required to validate the results of this study. Additionally, patients included in the study were all first-episode and drug-naïve. Some of them even reported difficulty in completing the task, thus resulting in missing phenotype data; given the study design, this is an inevitable trade-off for not being cofounded by medication and chronicity of the disease. In summary, this study showed that working memory might be one of the endophenotypes of schizophrenia and these endophenotypes can increase the efficiency of GWAS in neuropsychiatric disorders such as schizophrenia.
Acknowledgments
This work was partly funded by National Nature Science Foundation of China (81130024, 30530300 and 30125014, Tao Li); the National Basic Research Program of China (973 Program 2007CB512301, Tao Li), the Ph.D. Programs Foundation of Ministry of Education of China (20110181110014, Tao Li), National Key Technology R & D Program of the Ministry of Science and Technology of China during the 12th Five-Year Plan (2012BAI01B06, Tao Li), NARSAD Independent Investigator Award (Tao Li), and the Wellcome Trust (International Collaborative award to Tao Li and DA Collier); National Nature Science Foundation of China (30971056 and 81271479, Qiang Wang).
Author Contributions
The whole study was conceived and designed by Hongyan Ren, Qiang Wang and Tao Li. Recruitment of patients and healthy controls was carried out by Hongyan Ren, Chengcheng Zhang, Chaohua Huang, Na Li, Mingli Li, Yinfei Li, Deng Wei and Xiaohong Ma. Both genotypic data and cognitive data were analyzed by Hongyan Ren, Chengcheng Zhang, Qiang Wang and Bo Xiang Extraction of DNA was implemented by X.M. and Bo Xiang. The manuscript was written by Hongyan Ren, Qiang Wang and Tao Li.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Van Os J., Kapur S. Schizophrenia. Lancet. 2009;374:635–645. doi: 10.1016/S0140-6736(09)60995-8. [DOI] [PubMed] [Google Scholar]
- 2.McGuffin P., Farmer A.E., Gottesman II, Murray R.M., Reveley A.M. Twin concordance for operationally defined schizophrenia. Confirmation of familiality and heritability. Arch. Gen. Psychiatry. 1984;41:541–545. doi: 10.1001/archpsyc.1984.01790170015002. [DOI] [PubMed] [Google Scholar]
- 3.Flint J., Munafo M. Schizophrenia: Genesis of a complex disease. Nature. 2014;511:412–413. doi: 10.1038/nature13645. [DOI] [PubMed] [Google Scholar]
- 4.Schizophrenia Psychiatric Genome-Wide Association Study (GWAS) Genome-wide association study identifies five new schizophrenia loci. Nat. Genet. 2011;43:969–976. doi: 10.1038/ng.940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yue W.-H., Wang H.-F., Sun L.-D., Tang F.-L., Liu Z.-H., Zhang H.-X., Li W.-Q., Zhang Y.-L., Zhang Y., Ma C.-C. Genome-wide association study identifies a susceptibility locus for schizophrenia in han chinese at 11p11. 2. Nat. Genet. 2011;43:1228–1231. doi: 10.1038/ng.979. [DOI] [PubMed] [Google Scholar]
- 6.Zhou X., Stephens M. Genome-wide efficient mixed-model analysis for association studies. Nat. Genet. 2012;44:821–824. doi: 10.1038/ng.2310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schizophrenia Working Group of the Psychiatric Genomics Consortium (GWAS) Biological insights from 108 schizophrenia-associated genetic loci. Nature. 2014;511:421–427. doi: 10.1038/nature13595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Potkin S.G., Turner J., Fallon J., Lakatos A., Keator D., Guffanti G., Macciardi F. Gene discovery through imaging genetics: Identification of two novel genes associated with schizophrenia. Mol. Psychiatry. 2008;14:416–428. doi: 10.1038/mp.2008.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Potkin S.G., Guffanti G., Lakatos A., Turner J.A., Kruggel F., Fallon J.H., Saykin A.J., Orro A., Lupoli S., Salvi E. Hippocampal atrophy as a quantitative trait in a genome-wide association study identifying novel susceptibility genes for alzheimerʼs disease. PLoS One. 2009;4:e6501. doi: 10.1371/journal.pone.0006501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Miller B.H., Schultz L.E., Long B.C., Pletcher M.T. Quantitative trait locus analysis identifies gabra3 as a regulator of behavioral despair in mice. Mamm. Genome. 2010;21:247–257. doi: 10.1007/s00335-010-9266-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Visscher P.M., Andrew T., Nyholt D.R. Genome-wide association studies of quantitative traits with related individuals: Little (power) lost but much to be gained. Eur. J. Hum. Genet. 2008;16:387–390. doi: 10.1038/sj.ejhg.5201990. [DOI] [PubMed] [Google Scholar]
- 12.Manolio T.A. Cohort studies and the genetics of complex disease. Nat. Genet. 2009;41:5–6. doi: 10.1038/ng0109-5. [DOI] [PubMed] [Google Scholar]
- 13.Dickinson D., Straub R.E., Trampush J.W., Gao Y., Feng N., Xie B., Shin J.H., Lim H.K., Ursini G., Bigos K.L. Differential effects of common variants in SCN2A on general cognitive ability, brain physiology, and messenger rna expression in schizophrenia cases and control individuals. JAMA Psychiatry. 2014;71:647–656. doi: 10.1001/jamapsychiatry.2014.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Potkin S.G., Turner J.A., Guffanti G., Lakatos A., Fallon J.H., Nguyen D.D., Mathalon D., Ford J., Lauriello J., Macciardi F. A genome-wide association study of schizophrenia using brain activation as a quantitative phenotype. Schizophr. Bull. 2009;35:96–108. doi: 10.1093/schbul/sbn155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Silver H., Feldman P., Bilker W., Gur R.C. Working memory deficit as a core neuropsychological dysfunction in schizophrenia. Am. J. Psychiatry. 2003;160:1809–1816. doi: 10.1176/appi.ajp.160.10.1809. [DOI] [PubMed] [Google Scholar]
- 16.Park S., Holzman P.S., Goldman-Rakic P.S. Spatial working memory deficits in the relatives of schizophrenic patients. Arch. Gen. Psychiatry. 1995;52:821–828. doi: 10.1001/archpsyc.1995.03950220031007. [DOI] [PubMed] [Google Scholar]
- 17.Conklin H.M., Curtis C.E., Katsanis J., Iacono W.G. Verbal working memory impairment in schizophrenia patients and their first-degree relatives: Evidence from the digit span task. Am. J. Psychiatry. 2000;157:275–277. doi: 10.1176/appi.ajp.157.2.275. [DOI] [PubMed] [Google Scholar]
- 18.Liu H., Heath S.C., Sobin C., Roos J.L., Galke B.L., Blundell M.L., Lenane M., Robertson B., Wijsman E.M., Rapoport J.L. Genetic variation at the 22q11 PRODH2/DGCR6 locus presents an unusual pattern and increases susceptibility to schizophrenia. Proc. Natl. Acad. Sci. USA. 2002;99:3717–3722. doi: 10.1073/pnas.042700699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jia Y., Yu X., Zhang B., Yuan Y., Xu Q., Shen Y., Shen Y. An association study between polymorphisms in three genes of 14-3-3 (tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein) family and paranoid schizophrenia in northern chinese population. Eur. Psychiatry. 2004;19:377–379. doi: 10.1016/j.eurpsy.2004.07.006. [DOI] [PubMed] [Google Scholar]
- 20.Duan S., Gao R., Xing Q., Du J., Liu Z., Chen Q., Wang H., Feng G., He L. A family-based association study of schizophrenia with polymorphisms at three candidate genes. Neurosci. Lett. 2005;379:32–36. doi: 10.1016/j.neulet.2004.12.040. [DOI] [PubMed] [Google Scholar]
- 21.Silberberg G., Baruch K., Navon R. Detection of stable reference genes for real-time PCR analysis in schizophrenia and bipolar disorder. Anal. Biochem. 2009;391:91–97. doi: 10.1016/j.ab.2009.05.026. [DOI] [PubMed] [Google Scholar]
- 22.Fu H., Subramanian R.R., Masters S.C. 14-3-3 proteins: Structure, function, and regulation. Annu. Rev. Pharmacol. Toxicol. 2000;40:617–647. doi: 10.1146/annurev.pharmtox.40.1.617. [DOI] [PubMed] [Google Scholar]
- 23.Berg D., Holzmann C., Riess O. 14-3-3 proteins in the nervous system. Nat. Rev. Neurosci. 2003;4:752–762. doi: 10.1038/nrn1197. [DOI] [PubMed] [Google Scholar]
- 24.Dixon A.L., Liang L., Moffatt M.F., Chen W., Heath S., Wong K.C., Taylor J., Burnett E., Gut I., Farrall M. A genome-wide association study of global gene expression. Nat. Genet. 2007;39:1202–1207. doi: 10.1038/ng2109. [DOI] [PubMed] [Google Scholar]
- 25.Sadee W. Measuring cis-acting regulatory variants genome-wide: New insights into expression genetics and disease susceptibility. Genome Med. 2009;1 doi: 10.1186/gm116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pennington B.F., Moon J., Edgin J., Stedron J., Nadel L. The neuropsychology of down syndrome: Evidence for hippocampal dysfunction. Child. Dev. 2003;74:75–93. doi: 10.1111/1467-8624.00522. [DOI] [PubMed] [Google Scholar]
- 27.Pereira P.L., Magnol L., Sahún I., Brault V., Duchon A., Prandini P., Gruart A., Bizot J.-C., Chadefaux-Vekemans B., Deutsch S. A new mouse model for the trisomy of the ABCG1-U2AF1 region reveals the complexity of the combinatorial genetic code of down syndrome. Hum. Mol. Genet. 2009;18:4756–4769. doi: 10.1093/hmg/ddp438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Thomas J., Garg M.L., Smith D.W. Altered expression of histone and synaptic plasticity associated genes in the hippocampus of streptozotocin-induced diabetic mice. Metab. Brain Dis. 2013;28:613–618. doi: 10.1007/s11011-013-9418-y. [DOI] [PubMed] [Google Scholar]
- 29.Jarrold C., Baddeley A., Hewes A. Genetically dissociated components of working memory: Evidence from downs and williams syndrome. Neuropsychologia. 1999;37:637–651. doi: 10.1016/S0028-3932(98)00128-6. [DOI] [PubMed] [Google Scholar]
- 30.Walsh T., McClellan J.M., McCarthy S.E., Addington A.M., Pierce S.B., Cooper G.M., Nord A.S., Kusenda M., Malhotra D., Bhandari A. Rare structural variants disrupt multiple genes in neurodevelopmental pathways in schizophrenia. Science. 2008;320:539–543. doi: 10.1126/science.1155174. [DOI] [PubMed] [Google Scholar]
- 31.Gulsuner S., Walsh T., Watts A.C., Lee M.K., Thornton A.M., Casadei S., Rippey C., Shahin H., Nimgaonkar V.L., Go R.C. Spatial and temporal mapping of de novo mutations in schizophrenia to a fetal prefrontal cortical network. Cell. 2013;154:518–529. doi: 10.1016/j.cell.2013.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Besco J.A., Hooft van Huijsduijnen R., Frostholm A., Rotter A. Intracellular substrates of brain-enriched receptor protein tyrosine phosphatase RHO (RPTPP/PTPRT) Brain Res. 2006;1116:50–57. doi: 10.1016/j.brainres.2006.07.122. [DOI] [PubMed] [Google Scholar]
- 33.Luo L., Peng G., Zhu Y., Dong H., Amos C.I., Xiong M. Genome-wide gene and pathway analysis. Eur. J. Hum. Genet. 2010;18:1045–1053. doi: 10.1038/ejhg.2010.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Peng G., Luo L., Siu H., Zhu Y., Hu P., Hong S., Zhao J., Zhou X., Reveille J.D., Jin L. Gene and pathway-based second-wave analysis of genome-wide association studies. Eur. J. Hum. Genet. 2009;18:111–117. doi: 10.1038/ejhg.2009.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ramanan V.K., Shen L., Moore J.H., Saykin A.J. Pathway analysis of genomic data: Concepts, methods, and prospects for future development. TRENDS Genet. 2012;28:323–332. doi: 10.1016/j.tig.2012.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Eleftherohorinou H., Wright V., Hoggart C., Hartikainen A.-L., Jarvelin M.-R., Balding D., Coin L., Levin M. Pathway analysis of gwas provides new insights into genetic susceptibility to 3 inflammatory diseases. PLoS One. 2009;4:e8068. doi: 10.1371/journal.pone.0008068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pharoah P.D., Tyrer J., Dunning A.M., Easton D.F., Ponder B.A., Investigators S. Association between common variation in 120 candidate genes and breast cancer risk. PLoS Genet. 2007;3:e42. doi: 10.1371/journal.pgen.0030042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.OʼDushlaine C., Kenny E., Heron E.A., Segurado R., Gill M., Morris D.W., Corvin A. The SNP ratio test: Pathway analysis of genome-wide association datasets. Bioinformatics. 2009;25:2762–2763. doi: 10.1093/bioinformatics/btp448. [DOI] [PubMed] [Google Scholar]
- 39.Lesnick T.G., Papapetropoulos S., Mash D.C., Ffrench-Mullen J., Shehadeh L., de Andrade M., Henley J.R., Rocca W.A., Ahlskog J.E., Maraganore D.M. A genomic pathway approach to a complex disease: Axon guidance and parkinson disease. PLoS Genet. 2007;3:e98. doi: 10.1371/journal.pgen.0030098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang K., Li M., Bucan M. Pathway-based approaches for analysis of genomewide association studies. Am. J. Hum. Genet. 2007;81:1278–1283. doi: 10.1086/522374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Poltorak M., Hemperly J.J., Williams J.R., El-Mallakh R., Freed W.J. Disturbances in cell recognition molecules (N-CAM and L1 antigen) in the CSF of patients with schizophrenia. Exp. Neurol. 1995;131:266–272. doi: 10.1016/0014-4886(95)90048-9. [DOI] [PubMed] [Google Scholar]
- 42.Conant K., Herman M.M., van Kammen D.P., Sedvall G. Characterization of human cleaved N-Cam and association with schizophrenia. Exp. Neurol. 2001;172:29–46. doi: 10.1006/exnr.2001.7790. [DOI] [PubMed] [Google Scholar]
- 43.Brennaman L.H., Maness P.F. Structure and Function of the Neural Cell Adhesion Molecule Ncam. Springer; NY, USA: 2010. NCAM in neuropsychiatric and neurodegenerative disorders; pp. 299–317. [DOI] [PubMed] [Google Scholar]
- 44.Vance A., Hall N., Casey M., Karsz F., Bellgrove M.A. Visuospatial memory deficits in adolescent onset schizophrenia. Schizophr. Res. 2007;93:345–349. doi: 10.1016/j.schres.2007.02.020. [DOI] [PubMed] [Google Scholar]
- 45.Purcell S., Neale B., Todd-Brown K., Thomas L., Ferreira M.A., Bender D., Maller J., Sklar P., de Bakker P.I., Daly M.J. Plink: A tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ioannidis J.P., Tarone R., McLaughlin J.K. The false-positive to false-negative ratio in epidemiologic studies. Epidemiology. 2011;22:450–456. doi: 10.1097/EDE.0b013e31821b506e. [DOI] [PubMed] [Google Scholar]
- 47.Jia P., Wang L., Meltzer H.Y., Zhao Z. Common variants conferring risk of schizophrenia: A pathway analysis of gwas data. Schizophr. Res. 2010;122:38–42. doi: 10.1016/j.schres.2010.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhong H., Yang X., Kaplan L.M., Molony C., Schadt E.E. Integrating pathway analysis and genetics of gene expression for genome-wide association studies. Am. J. Hum. Genet. 2010;86:581–591. doi: 10.1016/j.ajhg.2010.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Menashe I., Maeder D., Garcia-Closas M., Figueroa J.D., Bhattacharjee S., Rotunno M., Kraft P., Hunter D.J., Chanock S.J., Rosenberg P.S. Pathway analysis of breast cancer genome-wide association study highlights three pathways and one canonical signaling cascade. Cancer Res. 2010;70:4453–4459. doi: 10.1158/0008-5472.CAN-09-4502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nicolae D.L., Gamazon E., Zhang W., Duan S., Dolan M.E., Cox N.J. Trait-associated SNPs are more likely to be eqtls: Annotation to enhance discovery from gwas. PLoS Genet. 2010;6:e1000888. doi: 10.1371/journal.pgen.1000888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Spencer C.C., Su Z., Donnelly P., Marchini J. Designing genome-wide association studies: Sample size, power, imputation, and the choice of genotyping chip. PLoS Genet. 2009;5:e1000477. doi: 10.1371/journal.pgen.1000477. [DOI] [PMC free article] [PubMed] [Google Scholar]