Abstract
Interleukin-10 (IL-10) single nucleotide polymorphisms (SNPs) have been indicated to be correlated with Non-Hodgkin’s lymphoma (NHL) susceptibility. However, the results of these studies on the association remain inconsistent. This meta-analysis was conducted to derive a more accuracy estimation of the association between the common SNPs (rs1800890, rs1800896, rs1800871 and rs1800872) in IL-10 and NHL risk. Meta-analyses were performed on 21 studies with 7,749 cases and 8584 controls. Odds ratios (ORs) with 95% confidence intervals (CIs) were used to evaluate the NHL risk. Meta-analyses showed that rs1800890, rs1800871 and rs1800872 polymorphisms had no association with NHL risk. However, rs1800896 polymorphism has association with NHL risk based on the following comparison models (G vs. A: OR = 1.14, 95% CI = 1.00-1.29; AG vs. AA: OR = 1.20, 95% CI = 1.05-1.37; GG+AG vs. AA: OR = 1.22, 95% CI = 1.08-1.39). In the ethnic subgroup analysis, rs1800896 had an increased NHL risk in Caucasians based on the heterozygote model (OR = 1.21, 95% CI = 1.04-1.41) and dominant model (OR = 1.22, 95% CI = 1.00-1.48). When stratified by subtypes, rs1800890, rs1800896 and rs1800872 polymorphisms were found significant association with an increased risk of diffuse large B-cell Lymphoma (DLBCL) in different comparison models, whereas negative results were obtained for Follicular Lymphoma (FL) and chronic lymphocytic Leukemia/small lymphocytic Lymphoma (CLL/SLL) in all genetic models. Our meta-analysis suggested that the rs1800896 polymorphism had an increased risk with NHL susceptibility, where as the rs1800890, rs1800871 and rs1800872 had no association with NHL risk. Among the common subtypes of NHL, three polymorphisms (rs1800890, rs1800896 and rs1800872) had significant association with DLBCL risk.
Keywords: Non-Hodgkin’s lymphoma, interleukin-10, polymorphism, meta-analysis
Introduction
Non-Hodgkin’s lymphoma (NHL) is a group of heterogeneous disorders that origin of the lymphatic system. In the past 20 years, the incidence of NHL rose rapidly to become one of the major diseases that threaten human health. Immune dysfunction is considered to be related to NHL [1], suggesting that immune-related genes play an important role in the pathogenesis of NHL. Genetic variation of some immunomodulatory cytokine may lead to different subtypes of lymphoma predisposition [2,3], respectively, these cytokine polymorphisms gradually become a hot topic in etiology, progression and prognosis of NHL.
Interleukin-10 (IL-10) is a Th2 cytokine that has strong anti-inflammatory effect and it mainly produced by immune cells, such as Th2 cells, Tr1 cells, monocytes, some subsets of dendritic cells and appropriately stimulated macrophages [4,5]. Studies have shown that IL-10 could regulate Th1 cells [6], promote B cell proliferation and antibody production [7]. Numerous studies show that IL-10 single nucleotide polymorphisms (SNPs) and lymphoma susceptibility [8-25], which often involves four functional SNPs, -3575 T>A (rs1800890), -1082 A>G (rs1800896), -819 T>C (rs1800871), and -592 A>C (rs1800872).
Although previous studies have shown that IL-10 gene promoter polymorphisms play an important immunoregulatory role on non-Hodgkin’s lymphoma [26], the results were inconsistent and conflicted. Some studies have found associations between IL-10rs1800890 as well as rs1800896 and the NHL predisposition [8,13,14], while other investigations have failed to identify these correlations [10,11,19]. Therefore, we conduct the present meta-analysis on all eligible case-control studies to evaluate the association between IL10 SNPs (rs1800890, rs1800896, rs1800871 and rs1800872) and susceptibility of NHL.
Materials and methods
Publication search
Check all the literature published by online electronic databases (PubMed, Embase, Web of Science and CNKI), keyword searched for: interleukin-10 or IL10 or IL-10, polymorphism or mutation or variant, Non-Hodgkin’s lymphoma OR lymphoid neoplasm OR lymphoid malignancy. All qualified studies were searched up to June 30, 2014. In order to identify the relevant publications, reference lists of research papers were also reviewed by manual search.
Selection criteria
The inclusion criteria for the meta-analysis study: (1) association studies between IL-10 SNPs (rs1800890, rs1800896, rs1800871, rs1800872) and NHL risk; (2) case-control studies; (3) full-text published articles and published in English; (4) included detailed genotype distribution data; (5) fulfilling Hardy-Weinberg equilibrium (P>0.05).
Data extraction
Two investigators independently extracted the information from all eligible publications according to the inclusion criteria and reached a consensus on all items. The following information were collected: first author, year of publication, country of origin, ethnicity, study design, genotype methods, source of control, numbers of cases and controls, total number of cases and controls, as well as number of cases and controls with the different genotypes. Different ethnicities were categorized as Caucasian, Asian, African and mixed. The controls had no present evidence of any malignant disease. The Newcastle-Ottawa Scale (NOS) is developed to assess the quality of nonrandom studies with its design, including case-control and cohort studies, content and ease of use directed to the task of incorporating the quality assessments in the interpretation of meta-analytic results [27]. Five independent researches from Rothman’s report, including Rothman-Canada, Rothman-Italy, Rothman-UCSF and Rothman-UK, Rothman-Spain case-control res- earches, were selected respectively [13]. Pathology histologic subtypes of NHL, including diffuse large B-cell Lymphoma (DLBCL), Follicular Lymphoma (FL), chronic lymphocytic Leukemia/small lymphocytic Lymphoma (CLL/SLL), were classified for analyses based on the World Health Organization classification for lymphoma diagnoses [28].
Statistical analysis
Based on the genotype frequencies in cases and controls of each study, odds ratios (ORs) together with their 95% confidence intervals (95% CIs) were calculated to assess the association strength. The significance of the pooled ORs was determined by the Z test. Statistical heterogeneity between studies was evaluated by chi-square based Q-test and I2 statistics [29]. If the p value of the heterogeneity test was more-than 0.1, the pooled OR estimate of the study was calculated by the fixed effects model. Otherwise, the random-effects model was used. Sensitivity analysis was performed to assess the stability of the final results. In order to assess the influence of each study to the pooled OR, risk assessment was tested by sequentially omitting one individual study at a time. Sensitivity analysis determines whether the individual data in fact have a major effect on the results of the review. Publication bias was evaluated by the visual inspection of asymmetry in Begg’s funnel plots and further assessed by the method of Egger’s test [30]. The meta-analysis assessed the following genetic models: dominant model (BB+AB vs. AA), recessive model (BB vs. AA+AB), homozygote comparison (BB vs. AA), heterozygote comparison (AB vs. AA) and allele comparison (B vs. A), the A represents the major allele, and the B represents the minor allele. All statistical analyses were calculated with the Review Manage 5.0 (The Cochrane Collaboration, Oxford, United Kingdom) and STATA 12.0 (Stata Corp, College Station, TX). A P value less-than 0.05 was considered statistically significant.
Results
Characteristics of eligible studies
There were 126 articles relevant to the search words and manual search. The flow chart for studies search strategy was showed as Figure 1. Finally, 17 research literatures selected for this meta-analysis [8-25], including a total of 21 researches (which included five independent researches from Rothman’s report). Among of these studies, 17 studies based on Caucasian, 1 Asian and 3 mixed ethnicities. There were a total with 7749 cases and 8584 controls enrolled for this analysis (Table 1), in which fourteen studies with 6428 cases and 7300 controls for rs1800890, eleven studies with 2637 cases and 2410 controls for rs1800896, seven studies with 2565 cases and 2233 controls for rs1800872 and six studies with 1520 cases and 1430 controls for rs1800871, respectively.
Figure 1.
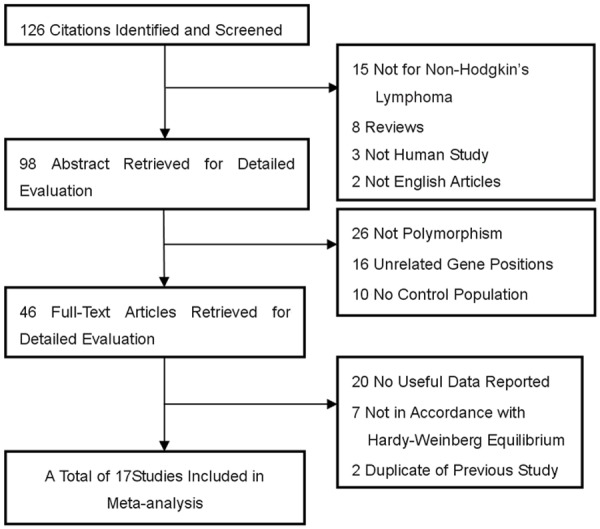
Studies identified with criteria for inclusion and exclusion.
Table 1.
Characteristics of the studies included in the meta-analysis
| First author | Year | Country | Ethnicity | Study design | Genotyping method | Source of control | Total sample size(case/control) | SNP No. | NOS score |
|---|---|---|---|---|---|---|---|---|---|
| Cunningham | 2003 | Australia | Caucasian | CC | SSOP | PB | 109/164 | 2 | 8 |
| Lech-Maranda | 2004 | France | Caucasian | CC | PCR-RFLP | PB | 199/112 | 2;3;4 | 8 |
| Guzowski | 2005 | USA | Mix | CC | Taqman | PB | 17/25 | 2;3;4 | 8 |
| Berglund | 2005 | Sweden | Caucasian | CC | NR | PB | 244/195 | 2 | 6 |
| Wang | 2006 | USA | Mix | CC | Taqman | PB | 1133/936 | 1 | 9 |
| Lan | 2006 | USA | Mix | CC | Taqman | Female | 510/597 | 1;2;3 | 7 |
| Nieters | 2006 | Germany | Caucasian | CC | Taqman | PB | 507/661 | 1 | 9 |
| Rothman | 2006a | Canada | Caucasian | CC | Taqman | PB | 85/349 | 1 | 8 |
| 2006b | Italy | Caucasian | CC | Taqman | PB | 59/112 | 1 | 8 | |
| 2006c | Spain | Caucasian | CC | Taqman | HB | 77/554 | 1 | 7 | |
| 2006d | USA | Caucasian | CC | Taqman | PB | 93/677 | 1 | 8 | |
| 2006e | UK | Caucasian | CC | Taqman | PB | 262/459 | 1 | 8 | |
| Purdue | 2007 | USA | Caucasian | CC | Taqman | PB | 524/475 | 1;2;3;4 | 9 |
| Lech-Maranda | 2007 | France | Caucasian | CC | PCR-RFLP | PB | 175/112 | 2;3;4 | 8 |
| Kube | 2008 | Germany | Caucasian | CC | Taqman | HB | 500/236 | 1;2;4 | 7 |
| Maria | 2008 | Italy | Caucasian | CC | Taqman | PB | 39/112 | 1;2 | 7 |
| Liang | 2009 | USA | Caucasian | CC | OPA | PB | 39/102 | 1 | 7 |
| Fernberg | 2010 | Sweden | Caucasian | CC | MassArray | PB | 2312/1838 | 1 | 9 |
| Zhang | 2012 | China | Asian | CC | Taqman | PB | 514/557 | 4 | 8 |
| Lech-Maranda | 2013 | Poland | Caucasian | CC | Taqman | PB | 290/192 | 1;2 | 8 |
| Talaat | 2014 | Egypt | Caucasian | CC | SSP-PCR | PB | 100/119 | 2;3 | 7 |
CC: case-control; SNP: single-nucleotide polymorphisms; SNP No.1: -3575 T>A (rs1800890); 2: -1082 A>G (rs1800896); 3: -819 T>C (rs1800871); 4: -592 A>C (rs1800872); PB: population based; HB: hospital based; NR: not reported; PCR-RFLP: polymerase chain reaction and restriction fragment length polymorphism; OPA: Oligo Pool Assay; SSOP: sequence specific oligo probing; SSC-PCR: sequence-specific primers polymerase chain reaction; NOS: the Newcastle-Ottawa Scale.
The Newcastle-Ottawa Scale (NOS) was used for assessing the quality of each included literature, the results was showed as the last column of Table 1. The NOS score of all articles are not less than 6scores that mean each included literature was a high-quality study. The genotype distributions for four SNPs of IL-10 are shown in Table 2, and the frequency of the minor allele was diverse widely across the twenty-one eligible studies, ranging from 0.24 to 0.44 (rs1800890), 0.36 to 0.54 (rs1800896), 0.67 to 0.80 (rs1800871), 0.35 to 0.80 (rs1800872). The average frequency of the minor allele in the four polymorphisms was 0.37, 0.46, 0.73, and 0.34, respectively.
Table 2.
IL-10 polymorphisms Genotype Distribution and Allele Frequency in Cases and Controls
| First author | Genotype (N) | MAF | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
| |||||||||||||
| Case | Control | Case | Control | ||||||||||
|
| |||||||||||||
| total | AA | AB | BB | total | AA | AB | BB | A | B | A | B | ||
| rs1800890 | |||||||||||||
| Wang 2006 | 1133 | 447 | 521 | 165 | 936 | 387 | 417 | 132 | 1415 | 851 | 1191 | 681 | 0.38 |
| Lan 2006 | 510 | 188 | 244 | 78 | 597 | 261 | 280 | 56 | 620 | 400 | 802 | 392 | 0.39 |
| Nieters 2006 | 507 | 223 | 207 | 77 | 661 | 262 | 306 | 93 | 653 | 361 | 830 | 492 | 0.36 |
| Rothman 2006a | 85 | 28 | 48 | 9 | 349 | 119 | 179 | 51 | 104 | 66 | 417 | 281 | 0.39 |
| Rothman 2006b | 59 | 34 | 22 | 3 | 112 | 73 | 37 | 2 | 90 | 28 | 183 | 41 | 0.24 |
| Rothman 2006c | 77 | 34 | 39 | 4 | 554 | 271 | 233 | 50 | 107 | 47 | 775 | 333 | 0.31 |
| Rothman 2006d | 93 | 35 | 41 | 17 | 677 | 238 | 343 | 96 | 111 | 75 | 819 | 535 | 0.40 |
| Rothman 2006e | 262 | 82 | 128 | 52 | 459 | 174 | 212 | 73 | 292 | 232 | 560 | 358 | 0.44 |
| Purdue 2007 | 524 | 165 | 255 | 104 | 475 | 159 | 233 | 83 | 585 | 463 | 551 | 399 | 0.44 |
| Kube 2008 | 500 | 177 | 249 | 74 | 236 | 81 | 122 | 33 | 603 | 397 | 284 | 188 | 0.40 |
| Maria 2008 | 37 | 18 | 18 | 1 | 112 | 73 | 37 | 2 | 54 | 20 | 183 | 41 | 0.27 |
| Liang 2009 | 39 | 19 | 16 | 4 | 102 | 33 | 48 | 21 | 54 | 24 | 114 | 90 | 0.31 |
| Fernberg 2010 | 2312 | 847 | 1092 | 373 | 1838 | 695 | 865 | 278 | 2786 | 1838 | 2255 | 1421 | 0.40 |
| Lech-Maranda 2013 | 290 | 112 | 132 | 46 | 192 | 69 | 85 | 38 | 356 | 224 | 223 | 161 | 0.39 |
| rs1800896 | |||||||||||||
| Cunningham 2003 | 109 | 37 | 46 | 26 | 164 | 41 | 82 | 41 | 120 | 98 | 164 | 164 | 0.45 |
| Lech-Maranda 2004 | 199 | 55 | 100 | 44 | 112 | 45 | 47 | 20 | 210 | 188 | 137 | 87 | 0.47 |
| Guzowski 2005 | 17 | 4 | 9 | 4 | 25 | 9 | 12 | 4 | 17 | 17 | 30 | 20 | 0.50 |
| Berglund 2005 | 244 | 70 | 136 | 38 | 195 | 60 | 89 | 46 | 276 | 212 | 209 | 181 | 0.43 |
| Lan 2006 | 510 | 137 | 260 | 113 | 587 | 184 | 305 | 98 | 534 | 486 | 673 | 501 | 0.48 |
| Purdue 2007 | 532 | 108 | 269 | 155 | 477 | 111 | 239 | 127 | 485 | 579 | 461 | 493 | 0.54 |
| Lech-Maranda 2007 | 175 | 56 | 87 | 32 | 112 | 45 | 47 | 20 | 210 | 188 | 137 | 87 | 0.43 |
| Kube 2008 | 500 | 134 | 253 | 113 | 236 | 81 | 122 | 33 | 521 | 479 | 284 | 188 | 0.48 |
| Maria 2008 | 39 | 15 | 20 | 4 | 111 | 61 | 43 | 7 | 50 | 28 | 165 | 57 | 0.36 |
| Lech-Maranda 2013 | 292 | 82 | 152 | 58 | 192 | 48 | 94 | 50 | 316 | 268 | 190 | 194 | 0.46 |
| Talaat 2014 | 100 | 28 | 54 | 18 | 119 | 43 | 61 | 15 | 110 | 90 | 147 | 91 | 0.45 |
| rs1800871 | |||||||||||||
| Lech-Maranda 2004 | 199 | 11 | 81 | 107 | 112 | 13 | 46 | 53 | 103 | 295 | 72 | 152 | 0.74 |
| Guzowski 2005 | 17 | 2 | 6 | 9 | 25 | 1 | 10 | 14 | 10 | 24 | 10 | 40 | 0.71 |
| Lan 2006 | 491 | 26 | 191 | 274 | 574 | 34 | 211 | 329 | 243 | 739 | 279 | 869 | 0.75 |
| Purdue 2007 | 538 | 21 | 175 | 342 | 488 | 23 | 170 | 295 | 217 | 859 | 216 | 760 | 0.80 |
| Lech-Maranda 2007 | 175 | 15 | 68 | 92 | 112 | 13 | 46 | 53 | 98 | 252 | 72 | 152 | 0.72 |
| Talaat 2014 | 100 | 6 | 54 | 40 | 119 | 5 | 52 | 62 | 66 | 134 | 62 | 176 | 0.67 |
| rs1800872 | |||||||||||||
| Lech-Maranda 2004 | 199 | 11 | 81 | 107 | 112 | 13 | 46 | 53 | 103 | 295 | 72 | 152 | 0.74 |
| Guzowski 2005 | 17 | 2 | 5 | 10 | 25 | 2 | 11 | 12 | 9 | 25 | 15 | 35 | 0.74 |
| Wang 2006 | 1120 | 93 | 426 | 601 | 938 | 81 | 342 | 515 | 612 | 1628 | 504 | 1372 | 0.73 |
| Purdue 2007 | 540 | 21 | 176 | 343 | 489 | 23 | 169 | 297 | 218 | 862 | 215 | 763 | 0.80 |
| Lech-Maranda 2007 | 175 | 15 | 68 | 92 | 112 | 13 | 46 | 53 | 98 | 252 | 72 | 152 | 0.72 |
| Kube 2008 | 500 | 26 | 196 | 278 | 236 | 14 | 98 | 124 | 248 | 752 | 126 | 346 | 0.35 |
| Zhang 2012 | 514 | 60 | 228 | 226 | 557 | 53 | 235 | 269 | 348 | 680 | 341 | 773 | 0.66 |
A represents the major allele, B represents the minor allele. MAF: minor allele frequencies.
Meta-analysis results
IL-10-3575T>A polymorphism (rs1800890)
There was no association between rs1800890 and NHL susceptibility in any comparison model. In the subgroup analysis by ethnicity, no significant correlation was observed between rs1800890 and an increased risk of NHL in Caucasians. According the origin of lymphoma cell, there were significantly associated in different comparison models (AA vs. TT+TA: OR = 1.15, 95% CI = 1.03-1.28; AA+TA vs. TT: OR = 1.11, 95% CI = 1.03-1.20; A vs. T: OR = 1.09, 95% CI = 1.01-1.19) between rs1800890 and B-NHL risk, where as there were no significant association between rs1800890 and T-NHL predisposition. Among the common subtypes, the rs1800890 was found significant association with an increased risk of DLBCL in all different comparison models (TA vs. TT: OR = 1.15, 95% CI = 1.03-1.28, Figure 2; AA vs. TT: OR = 1.35, 95% CI = 1.16-1.57; AA vs. TT+TA: OR = 1.25, 95% CI = 1.09-1.44; AA+TA vs. TT: OR = 1.20, 95% CI = 1.08-1.33; A vs. T: OR = 1.16, 95% CI = 1.08-1.25), whereas there were no statistical significance between rs1800890 and CLL/SLL or FL risk (Table 3).
Figure 2.
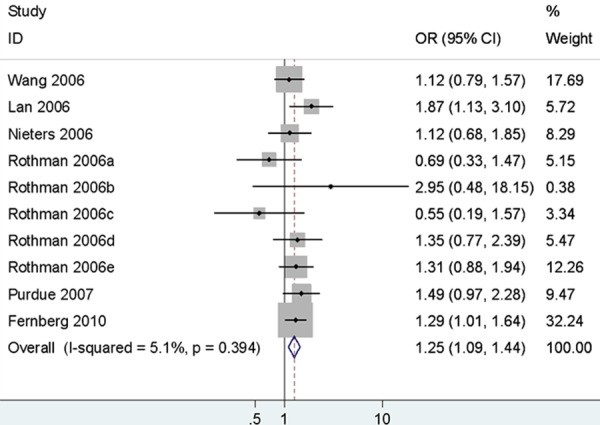
Forest plots of IL-10 rs1800890 polymorphism and diffuse large B-cell lymphoma risk (AA vs. TT+TA).
Table 3.
Meta-analysis results Between IL-10 Polymorphisms and NHL Risk
| Comparisons | B vs. A | BB vs. AA | AB vs. AA | BB vs. AA+AB | AB+BB vs. AA | |||||
|---|---|---|---|---|---|---|---|---|---|---|
|
| ||||||||||
| OR (95% CI) | P | OR (95% CI) | P | OR (95% CI) | P | OR (95% CI) | P | OR (95% CI) | P | |
| rs1800890 | 1.06 (0.98-1.14) | 0.15 | 1.12 (0.96-1.31) | 0.16 | 1.04 (0.97-1.13) | 0.29 | 1.10 (1.00-1.22) | 0.06 | 1.06 (0.99-1.14) | 0.10 |
| Caucasian | 1.03 (0.97-1.10) | 0.29 | 1.07 (0.94-1.21) | 0.29 | 1.02 (0.93-1.11) | 0.74 | 1.07 (0.95-1.20) | 0.26 | 1.03 (0.95-1.12) | 0.50 |
| B-NHL | 1.09 (1.01-1.19) | 0.03 | 1.18 (0.99-1.41) | 0.06 | 1.08 (1.00-1.18) | 0.06 | 1.15 (1.03-1.28) | 0.01 | 1.11 (1.03-1.20) | 0.008 |
| T-NHL | 0.91 (0.76-1.08) | 0.27 | 0.78 (0.53-1.16) | 0.22 | 0.97 (0.75-1.24) | 0.80 | 0.79 (0.55-1.14) | 0.21 | 0.93 (0.73-1.17) | 0.52 |
| DLBCL | 1.16 (1.08-1.25) | 0.0001 | 1.35 (1.16-1.57) | 0.0001 | 1.15 (1.03-1.28) | 0.02 | 1.25 (1.09-1.44) | 0.001 | 1.20 (1.08-1.33) | 0.001 |
| CLL/SLL | 0.99 (0.90-1.10) | 0.91 | 0.97 (0.79-1.20) | 0.81 | 1.00 (0.86-1.16) | 0.96 | 0.99 (0.82-1.20) | 0.92 | 0.99 (0.86-1.14) | 0.94 |
| FL | 1.07 (0.97-1.18) | 0.19 | 1.14 (0.93-1.40) | 0.19 | 1.04 (0.90-1.21) | 0.57 | 1.12 (0.93-1.35) | 0.22 | 1.07 (0.93-1.23) | 0.34 |
| rs1800896 | 1.14 (1.00-1.29) | 0.02 | 1.26 (0.96-1.64) | 0.09 | 1.20 (1.05-1.37) | 0.007 | 1.12 (0.90-1.40) | 0.313 | 1.22 (1.08-1.39) | 0.002 |
| Caucasian | 1.12 (0.96-1.30) | 0.16 | 1.20 (0.88-1.64) | 0.25 | 1.21 (1.04-1.41) | 0.01 | 1.07 (0.83-1.38) | 0.62 | 1.22 (1.00-1.48) | 0.048 |
| B-NHL | 1.12 (0.98-1.29) | 0.09 | 1.23 (0.93-1.62) | 0.15 | 1.22 (1.06-1.40) | 0.007 | 1.09 (0.86-1.38) | 0.49 | 1.23 (1.08-1.41) | 0.002 |
| DLBCL | 1.15 (0.97-1.36) | 0.11 | 1.32 (0.91-1.91) | 0.14 | 1.26 (1.05-1.52) | 0.01 | 1.14 (0.83-1.56) | 0.41 | 1.29 (1.08-1.53) | 0.004 |
| CLL/SLL | 1.17 (0.70-1.94) | 0.55 | 0.84 (0.53-1.35) | 0.48 | 1.15 (0.80-1.66) | 0.45 | 0.81 (0.54-1.20) | 0.29 | 1.08 (0.76-1.52) | 0.67 |
| FL | 1.14 (0.97-1.34) | 0.12 | 1.27 (0.92-1.76) | 0.15 | 1.07 (0.81-1.41) | 0.64 | 1.25 (0.95-1.63) | 0.11 | 1.14 (0.88-1.47) | 0.33 |
| rs1800871 | 1.05 (0.93-1.18) | 0.43 | 1.24 (0.90-1.70) | 0.19 | 1.22 (0.88-1.69) | 0.23 | 1.03 (0.89-1.19) | 0.69 | 1.24 (0.91-1.69) | 0.18 |
| Caucasian | 1.11 (0.95-1.29) | 0.18 | 1.38 (0.92-2.06) | 0.12 | 1.29 (0.86-1.94) | 0.23 | 1.09 (0.91-1.32) | 0.36 | 1.35 (0.91-2.00) | 0.13 |
| B-NHL | 1.04 (0.91-1.18) | 0.56 | 1.33 (0.94-1.87) | 0.10 | 1.36 (0.96-1.93) | 0.08 | 1.00 (0.85-1.16) | 0.96 | 1.35 (0.96-1.88) | 0.08 |
| DLBCL | 1.03 (0.79-1.35) | 0.80 | 1.43 (0.89-2.30) | 0.14 | 1.52 (0.94-2.47) | 0.08 | 0.97 (0.70-1.34) | 0.84 | 1.48 (0.93-2.36) | 0.10 |
| FL | 0.99 (0.82-1.20) | 0.95 | 1.16 (0.70-1.95) | 0.56 | 1.18 (0.70-1.98) | 0.53 | 0.96 0.76-1.21) | 0.71 | 1.17 (0.71–1.92) | 0.54 |
| rs1800872 | 1.02 (0.93-1.11) | 0.71 | 1.05 (0.85-1.29) | 0.66 | 1.07 (0.86-1.32) | 0.55 | 1.01 0.91-1.13) | 0.85 | 1.06 (0.87-1.29) | 0.59 |
| Caucasian | 1.06 (0.97-1.17) | 0.21 | 1.18 (0.93-1.50) | 0.17 | 1.17 (0.91-1.49) | 0.22 | 1.05 (0.93–1.19) | 0.39 | 1.18 (0.93-1.48) | 0.17 |
| B-NHL | 1.18 (0.99-1.40) | 0.06 | 1.55 (1.00-2.41) | 0.05 | 1.36 (0.87-2.14) | 0.17 | 1.17 (0.95-1.44) | 0.15 | 1.47 (0.96-2.26) | 0.08 |
| DLBCL | 1.28 (1.01-1.62) | 0.04 | 2.06 (1.06-3.99) | 0.03 | 1.77 (0.91-3.48) | 0.10 | 1.26 (0.95-1.68) | 0.11 | 1.95 (1.02-3.72) | 0.04 |
| FL | 1.08 (0.85-1.36) | 0.54 | 1.19 (0.65-2.16) | 0.58 | 1.03 (0.56-1.91) | 0.92 | 1.09 (0.82-1.44) | 0.57 | 1.12 (0.62-2.01) | 0.71 |
A: the major allele; B: the minor allele; CI: confidence interval; OR: odds ratio; B-NHL: B-cell non-Hodgkin’s Lymphoma; T-NHL: T-cell non-Hodgkin’s Lymphoma; DLBCL: diffuse large B-cell Lymphoma; FL: Follicular Lymphoma; CLL/SLL: chronic lymphocytic Leukemia/small lymphocytic Lymphoma.
IL-10-1082A>G polymorphism (rs1800896)
Significant increased risk of NHL was observed in different comparison models (AG vs. AA: OR = 1.20, 95% CI = 1.05-1.37, Figure 3; GG+AG vs. AA: OR = 1.22, 95% CI = 1.08-1.39; G vs. A: OR = 1.14, 95% CI = 1.00-1.29). When stratified by ethnicity, statistically significant was examined in two comparison models in Caucasians (AG vs. AA: OR = 1.21, 95% CI = 1.04-1.41; GG+AG vs. AA: OR = 1.22, 95% CI = 1.00-1.48). In addition, the association was detected between rs1800896 and risk of B-NHL and DLBCL (B-NHL: AG vs. AA: OR = 1.22, 95% CI = 1.06-1.40; GG+AG vs. AA: OR = 1.23, 95% CI = 1.08-1.41; DLBCL: AG vs. AA: OR= 1.26, 95% CI = 1.05-1.52; GG+AG vs. AA: OR = 1.29, 95% CI = 1.08-1.53). However, there were no significant increased CLL/SLL and FL risk in any gene distribution model of rs1800896. The results are presented in Table 3.
Figure 3.
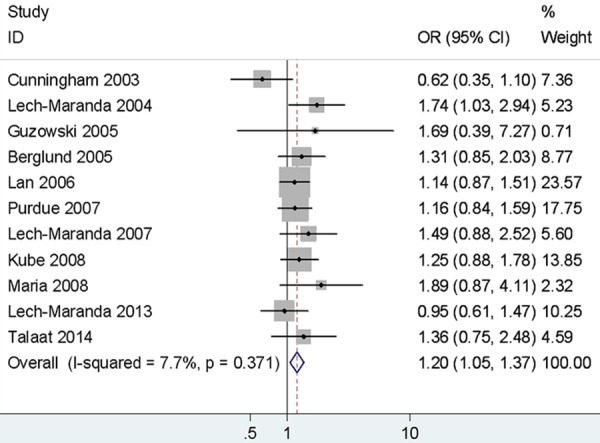
Forest plots of IL-10 rs1800896 polymorphism and non-Hodgkin’s lymphoma risk (AG vs. AA).
IL-10-819T>C polymorphism (rs1800871)
As shown in Table 3 and Figure 4, the rs1800871 variant was found no significant association with NHL susceptibility, whether it is stratified by ethnicity, or in the classification by NHL subtypes.
Figure 4.
Forest plots of IL-10 rs1800871 polymorphism and non-Hodgkin’s Lymphoma risk (CC vs. TT).
IL-10-592A>C polymorphism (rs1800872)
The rs1800872 polymorphism had no association with NHL predisposition based on all genetic models. Similarly, negative results were obtained for Caucasians in all genetic models when carried out stratified analysis by ethnicity. However, there was significant association between the rs1800872 polymorphism and DLBCL risk in homozygote, dominant and haploid models (CC vs. AA: OR = 2.06, 95% CI = 1.06-3.99, Figure 5; CC+AC vs. AA: OR = 1.95, 95% CI = 1.02-3.72; C vs. A: OR = 1.28, 95% CI = 1.01-1.62), whereas there was no association between the rs1800872 variation and FL susceptibility. The results are showed as Table 3.
Figure 5.
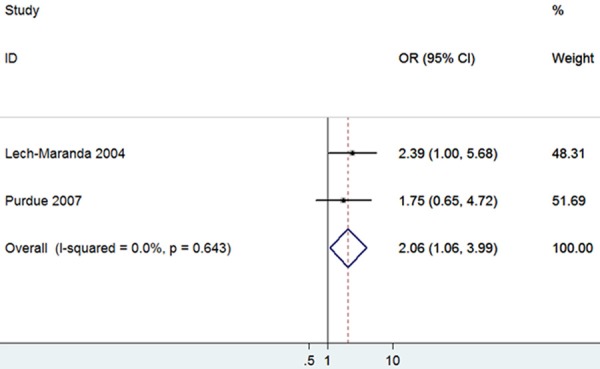
Forest plots of IL-10 rs1800872 polymorphism and non-Hodgkin’s Lymphoma risk in Caucasians (CC vs. AA).
Tests of heterogeneity and sensitivity analysis
Statistically significant heterogeneity was observed between trials of the following analyses using Q statistic. As show in Table 4, when the p value of the heterogeneity test was more than 0.1, a fixed-effects model was performed. Otherwise, the random-effects model was used.
Table 4.
Heterogeneity-analysis results
| Comparisons | B vs. A | BB vs. AA | AB vs. AA | BB vs. AA+AB | AB+BB vs. AA | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
| |||||||||||||||
| I2 | P | EM | I2 | P | EM | I2 | P | EM | I2 | P | EM | I2 | P | EM | |
| Rs1800890 | 39% | 0.07 | R | 35% | 0.09 | R | 14% | 0.30 | F | 23% | 0.21 | F | 30% | 0.14 | F |
| Caucasian | 25% | 0.20 | F | 8% | 0.36 | F | 18% | 0.27 | F | 0% | 0.50 | F | 25% | 0.20 | F |
| B-NHL | 37% | 0.09 | R | 38% | 0.08 | R | 0% | 0.57 | F | 28% | 0.16 | F | 16% | 0.28 | F |
| T-NHL | 0% | 0.75 | F | 11% | 0.34 | F | 0% | 0.42 | F | 49% | 0.12 | F | 0% | 0.76 | F |
| DLBCL | 0% | 0.71 | F | 2% | 0.42 | F | 0% | 0.95 | F | 5% | 0.39 | F | 0% | 0.91 | F |
| CLL/SLL | 41% | 0.13 | F | 17% | 0.31 | F | 4% | 0.39 | F | 3% | 0.40 | F | 28% | 0.23 | F |
| FL | 29% | 0.23 | F | 26% | 0.25 | F | 22% | 0.27 | F | 3% | 0.39 | F | 29% | 0.23 | F |
| rs1800896 | 52% | 0.02 | R | 53% | 0.02 | R | 8% | 0.37 | F | 50% | 0.03 | R | 29% | 0.17 | F |
| Caucasian | 59% | 0.01 | R | 58% | 0.01 | R | 24% | 0.23 | F | 59% | 0.03 | R | 42% | 0.09 | R |
| B-NHL | 52% | 0.02 | R | 52% | 0.02 | R | 0% | 0.58 | F | 51% | 0.03 | R | 23% | 0.23 | F |
| DLBCL | 56% | 0.04 | R | 59% | 0.02 | R | 0% | 0.44 | F | 59% | 0.02 | R | 30% | 0.20 | F |
| CLL/SLL | 65% | 0.06 | R | 49% | 0.14 | F | 23% | 0.27 | F | 18% | 0.30 | F | 53% | 0.12 | F |
| FL | 0% | 0.86 | F | 0% | 0.82 | F | 0% | 0.57 | F | 0% | 0.99 | F | 0% | 0.64 | F |
| rs1800871 | 38% | 0.16 | F | 8% | 0.37 | F | 0% | 0.72 | F | 21% | 0.28 | F | 0% | 0.52 | F |
| Caucasian | 48% | 0.12 | F | 22% | 0.28 | F | 0% | 0.64 | F | 44% | 0.15 | F | 0% | 0.44 | F |
| B-NHL | 37% | 0.16 | F | 1% | 0.41 | F | 0% | 0.70 | F | 24% | 0.25 | F | 0% | 0.55 | F |
| DLBCL | 59% | 0.06 | R | 26% | 0.26 | F | 0% | 0.73 | F | 58% | 0.07 | R | 0% | 0.46 | F |
| FL | 25% | 0.27 | F | 0% | 0.91 | F | 0% | 0.71 | F | 53% | 0.12 | F | 0% | 0.95 | F |
| rs1800872 | 26% | 0.23 | F | 20% | 0.27 | F | 0% | 0.66 | F | 3% | 0.41 | F | 2% | 0.41 | F |
| Caucasian | 0% | 0.40 | F | 0% | 0.44 | F | 0% | 0.75 | F | 0% | 0.56 | F | 0% | 0.55 | F |
| B-NHL | 0% | 0.80 | F | 0% | 0.68 | F | 0% | 0.59 | F | 0% | 0.88 | F | 0% | 0.63 | F |
| DLBCL | 0% | 0.70 | F | 0% | 0.64 | F | 0% | 0.61 | F | 0% | 0.88 | F | 0% | 0.64 | F |
| FL | 0% | 0.33 | F | 0% | 0.67 | F | 0% | 0.84 | F | 22% | 0.26 | F | 0% | 0.87 | F |
A: the major allele; B: the minor allele; EM: Effects model; F: fixed effects model; R: random effects model; B-NHL: B-cell non-Hodgkin’s Lymphoma; T-NHL: T-cell non-Hodgkin’s Lymphoma; DLBCL: diffuse large B-cell Lymphoma; FL: Follicular Lymphoma; CLL/SLL: chronic lymphocytic Leukemia/small lymphocytic Lymphoma.
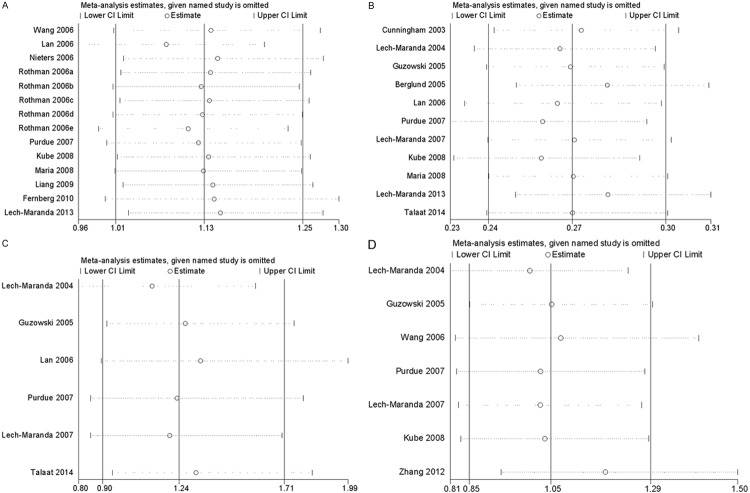
Sensitivity analysis was performed by sequentially omitting one individual study at a time, in order to reflect the influence of each study on the overall meta-analysis. As show in Figure 6, sensitivity tests suggested that no single study greatly influenced the estimates of overall risk for the four IL-10 polymorphisms respectively and the results of our meta-analysis were stable.
Figure 6.
Sensitivity analysis of association between the polymorphisms and non-Hodgkin’s Lymphoma risk. A. rs1800896; B. rs1800896; C. rs1800871; D. rs1800872.
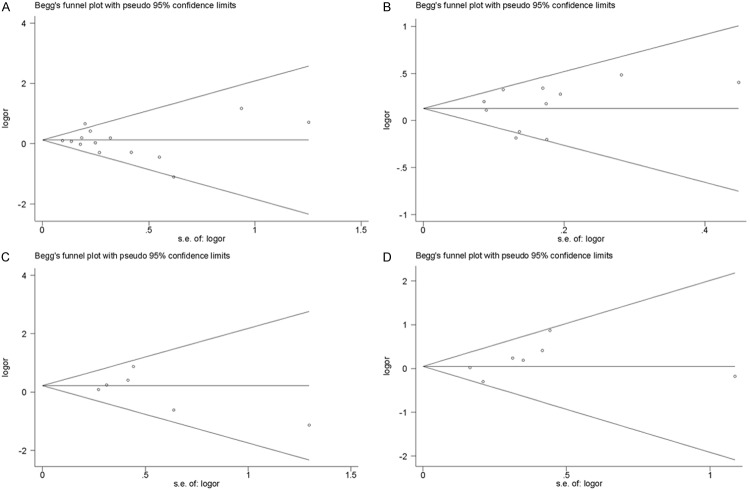
Publication bias
In this meta-analysis, we performed Begg’s funnel plot and Egger’s test to access the publication bias. As show in Figure 7, the funnel plots could not show any obvious asymmetry in all genotypes in the overall population. Therefore, the results of Egger’s test revealed that publication bias was not significant in this meta-analysis (P>0.05).
Figure 7.
Funnel plot assessing evidence of publication bias from the eligible studies. A. rs1800896; B. rs1800896; C. rs1800871; D. rs1800872.
Discussion
Malignant diseases of the lymphoid system, which originated in the process of lymphocyte proliferation and differentiation, was a multifactorial disease cau- sed by complex inherited genetic variations and environmental factors. Immune dysfunction was expected to be the potential basis of lymphomagenesis, expression of key cytokines might play an important part in the pathogenesis of NHL. IL-10, an immunoregulatory cytokine, was considered promoting the occurrence of cancer by immunosuppression effects and suppressing the occurrence of cancer by anti-angiogenic functions, therefore, which were double-edged properties of immunosuppression and immune stimulation [31]. Genotypic variations in the promoter sequences of the IL-10 gene may influence in IL-10 production of different individual [32,33] and play a certain role in the susceptibility and clinical progression of lymphoma [34,35].
In present meta-analysis, we analyzed the relationship between the four common IL-10 SNPs (rs1800890, rs1800896, rs1800871 and rs1800872) and NHL susceptibility including twenty-one studies with 7749 cases and 8360 controls for this meta-analysis. To our known, this was the first meta-analysis which researched the association between the four common IL-10 SNPs and risk of NHL.
Overall, our research indicated, populations with IL-10 rs1800896 G allele were associated with a 14% increased risk of NHL (P = 0.02). Similarly, rs1800896 was found 20% increased (P = 0.007) and 22% increased (p = 0.002) susceptibility of NHL in heterozygote and dominant genetic model in the overall population, respectively. However, statistically significant was not examined between the three polymorphisms (rs1800890, rs1800871 and rs1800872) and NHL susceptibility. Our results were inconsistent with a previous single study, which found aggressive lymphoma had a higher frequency of IL-10-3575A and IL-10-1082A haploid genotype [8]. The reason may be that it included fewer cases and controls. Ethnic origin analysis, significant association was detected between rs1800896 variation and NHL susceptibility in heterozygote and dominant models in Caucasians (AG vs. AA: OR = 1.21, P = 0.01; GG+AG vs. AA: OR = 1.21, p = 0.008), whereas other three SNPs (rs1800890, rs1800872 and rs1800871) did not find statistical significance with NHL risk in Caucasians. Previous studies found IL-10 gene polymorphisms associated with the risk of NHL [9,13,36], others shown that IL-10 was relative to the NHL’s course [8,37], IL-10 genetic loci haplotypes AGCC/TATA (-3575, -1082, -819, -592) and ATA/ACC (-1082, -819, -592) was relative to NHL predisposition and invasiveness [10,19]. That supported IL-10 polymorphisms was associated with NHL susceptibility.
According the origin of lymphoma cell, the rs1800890 variation had increased B-NHL risk in recessive and dominant comparison models (AA vs. TT+TA: OR = 1.15, P = 0.01; AA+TA vs. TT: OR = 1.11, P = 0.008), whereas the rs1800896 polymorphism had increased B-NHL risk in heterozygote and dominant comparison models (AG vs. AA: OR = 1.22, P = 0.007; GG+AG vs. AA: OR = 1.23, P = 0.002), respectively. Our study also studied the rs1800890 variation and T-NHL predisposition, no correlation was observed in end, which may be related to involve less number of subjects. In addition, the current meta-analysis results indicated that the rs1800871 and rs1800872 polymorphisms had no association with B-NHL predisposition based on all genetic models.
Among the common subtypes of NHL, there were significant association between the three polymorphisms (rs1800890, rs1800896 and rs1800872) and DLBCL risk in different genetic models. Specifically, the rs1800890A allele may be associated with substantial increased DLBCL risk in overall populations, our results was consistent with a previous meta-analysis by Rothman [13]. The rs1800896 variant were found to be significantly associated with an increased DLBCL risk in heterozygote and dominant comparison models (AG vs. AA: OR = 1.26, P = 0.01; GG+AG vs. AA: OR = 1.29, P = 0.004), respectively. In addition, the rs1800872 polymorphism was showed statistical significance with DLBCL risk in the following comparison models (homozygote model: CC vs. AA: OR = 2.06, P = 0.03; dominant model: CC+AC vs. AA: OR = 1.95, P = 0.04; allele comparison: C vs. A: OR = 1.28, P = 0.04). However, no significant association was detected between rs1800871 polymorphism and DLBCL risk in any comparison model. The results of the meta-analysis [36] suggested that there were significant association between the three polymorphisms (rs1800890, rs1800896 and rs1800872) and DLBCL risk, which was consistent with our investigation. In addition, the four polymorphisms (rs1800890, rs1800896, rs1800871 and rs1800872) were found no correlation with FL predisposition. Studies suggested that FL has a familial aggregation [2,39], that mean FL was associated with genetic factor. However, case-control studies were not found IL-10 SNPs increase the risk of FL [10,20]. Similarly, this study also found that the rs1800890 and rs1800896 polymorphisms unrelated with CLL/SLL susceptibility, which was consistent with previous data [14,21]. Whereas a previous cohort study, CLL/SLL patients with the IL-10-1082AA genotype have better over survival (OS) compared with those individuals with AA and AC carriers [22], which indicated rs1800896 variant may predict clinical prognosis of CLL/SLL.
In summary, our study include the meta-analysis approach we took to evaluating the four important genetic polymorphisms (rs1800890, rs1800896, rs1800871 and rs1800872) and NHL predisposition in the present study that IL-10 polymorphisms had an association with the risk of NHL subtypes, particularly B-cell malignancy, and may contribute to the pathogenesis of DLBCL.
Several limitations of our meta-analysis should be attention. First, our meta-analysis was based on unadjusted estimates; thus, we could not assess the risk of lymphoma according to stratification of age, environmental factors, and other risk factors of NHL. The lack of such data for the analysis may cause serious confounding bias. Second, there was only one eligible study researched the association of IL-10 polymorphisms and risk of NHL in Asian. Therefore, more case-control studies are needed to further identify the association among Asians. Third, since some of the studies had a relatively small sample size, the results did not have adequate power to definitely confirm the association. Further large-scale studies with more detailed individual data, with different environmental factors are warranted to further precise gene-gene and gene-environment interactions on IL-10 polymorphisms and NHL risk.
Conclusion
In conclusion, our present meta-analysis indicated that IL-10 rs1800896 polymorphism was associated with NHL risk, further studies showed that rs1800896 polymorphism had an increased risk of NHL in Caucasians. Among the common NHL subtypes, significant association was found between rs1800890, rs1800896 as well as rs1800872 polymorphisms and the susceptibility of DLBCL. Further large-scale multicenter epidemiological studies were needed to confirm our findings of IL-10 polymorphisms in Non-Hodgkin lymphoma carcinogenesis.
Disclosure of conflict of interest
None.
References
- 1.Anderson LA, Gadalla S, Morton LM, Landgren O, Pfeiffer R, Warren JL, Berndt SI, Ricker W, Parsons R, Engels EA. Population-based study of autoimmune conditions and the risk of specific lymphoid malignancies. Int J Cancer. 2009;125:398–405. doi: 10.1002/ijc.24287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Altieri A, Bermejo JL, Hemminki K. Familial risk for non-Hodgkin lymphoma and other lymphoproliferative malignancies by histopathologic subtype: the Swedish Family-Cancer Database. Blood. 2005;106:668–72. doi: 10.1182/blood-2005-01-0140. [DOI] [PubMed] [Google Scholar]
- 3.Chatterjee N, Hartge P, Cerhan JR, Cozen W, Davis S, Ishibe N, Colt J, Goldin L, Severson RK. Risk of non-Hodgkin’s lymphoma and family history of lymphatic, hematologic, and other cancers. Cancer Epidemiol Biomarkers Prev. 2004;13:1415–21. [PubMed] [Google Scholar]
- 4.Villalta SA, Rinaldi C, Deng B, Liu G, Fedor B, Tidball JG. Interleukin-10 reduces the pathology of mdx muscular dystrophy by deactivating M1 macrophages and modulating macrophage phenotype. Hum Mol Genet. 2011;20:790–805. doi: 10.1093/hmg/ddq523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.O’Garra A, Vieira P. T(H)1 cells control themselves by producing interleukin-10. Nat Rev Immunol. 2007;7:425–428. doi: 10.1038/nri2097. [DOI] [PubMed] [Google Scholar]
- 6.Sojka DK, Fowell DJ. Regulatory T cells inhibit acute IFN-γ synthesis without blocking T-helper cell type 1 (Th1) differentiation via a compartmentalized requirement for IL-10. Proc Natl Acad Sci U S A. 2011;108:18336–18341. doi: 10.1073/pnas.1110566108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cottrez F, Hurst SD, Coffman RL, Groux H. T regulatory cells 1 inhibit a Th2-specific response in vivo. J Immunol. 2000;165:4848–4853. doi: 10.4049/jimmunol.165.9.4848. [DOI] [PubMed] [Google Scholar]
- 8.Cunningham LM, Chapman C, Dunstan R, Bell MC, Joske DJ. Polymorphisms in the interleukin 10 gene promoter are associated with susceptibility to aggressive non-Hodgkin’s lymphoma. Leuk Lymphoma. 2003;44:251–255. doi: 10.1080/1042819021000035590. [DOI] [PubMed] [Google Scholar]
- 9.Lech-Maranda E, Baseggio L, Bienvenu J, Charlot C, Berger F, Rigal D, Warzocha K, Coiffier B, Salles G. Interleukin-10 gene promoter polymorphisms influence the clinical outcome of diffuse B-cell lymphoma. Blood. 2004;103:3529–3534. doi: 10.1182/blood-2003-06-1850. [DOI] [PubMed] [Google Scholar]
- 10.Lan Q, Zheng T, Rothman N, Zhang Y, Wang SS, Shen M, Berndt SI, Zahm SH, Holford TR, Leaderer B, Yeager M, Welch R, Boyle P, Zhang B, Zou K, Zhu Y, Chanock S. Cytokine polymorphisms in the Th1/Th2 pathway and susceptibility to non-Hodgkin lymphoma. Blood. 2006;107:4101–4108. doi: 10.1182/blood-2005-10-4160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nieters A, Beckmann L, Deeg E, Becker N. Gene polymorphisms in Toll-like receptors, interleukin-10, and interleukin-10 receptor alpha and lymphoma risk. Genes Immun. 2006;7:615–624. doi: 10.1038/sj.gene.6364337. [DOI] [PubMed] [Google Scholar]
- 12.Wang SS, Cerhan JR, Hartge P, Davis S, Cozen W, Severson RK, Chatterjee N, Yeager M, Chanock SJ, Rothman N. Common genetic variants in proinflammatory and other immunoregulatory genes and risk for non-Hodgkin lymphoma. Cancer Res. 2006;66:9771–9780. doi: 10.1158/0008-5472.CAN-06-0324. [DOI] [PubMed] [Google Scholar]
- 13.Rothman N, Skibola CF, Wang SS, Morgan G, Lan Q, Smith MT, Spinelli JJ, Willett E, De Sanjose S, Cocco P, Berndt SI, Brennan P, Brooks-Wilson A, Wacholder S, Becker N, Hartge P, Zheng T, Roman E, Holly EA, Boffetta P, Armstrong B, Cozen W, Linet M, Bosch FX, Ennas MG, Holford TR, Gallagher RP, Rollinson S, Bracci PM, Cerhan JR, Whitby D, Moore PS, Leaderer B, Lai A, Spink C, Davis S, Bosch R, Scarpa A, Zhang Y, Severson RK, Yeager M, Chanock S, Nieters A. Genetic variation in TNF and IL10 and risk of non-Hodgkin lymphoma: a report from the InterLymph Consortium. Lancet Oncol. 2006;7:27–38. doi: 10.1016/S1470-2045(05)70434-4. [DOI] [PubMed] [Google Scholar]
- 14.Fernberg P, Chang ET, Duvefelt K, Hjalgrim H, Eloranta S, Sørensen KM, Porwit A, Humphreys K, Melbye M, Ekström Smedby K. Genetic variation in chromosomal translocation breakpoint and immune function genes and risk of non-Hodgkin lymphoma. Cancer Causes Control. 2010;21:759–769. doi: 10.1007/s10552-010-9504-y. [DOI] [PubMed] [Google Scholar]
- 15.Wang SS, Cozen W, Cerhan JR, Colt JS, Morton LM, Engels EA, Davis S, Severson RK, Rothman N, Chanock SJ, Hartge P. Immune mechanisms in non-Hodgkin lymphoma: joint effects of the TNF G308A and IL10 T3575A polymorphisms with non-Hodgkin lymphoma risk factors. Cancer Res. 2007;67:5042–5054. doi: 10.1158/0008-5472.CAN-06-4752. [DOI] [PubMed] [Google Scholar]
- 16.Purdue MP, Lan Q, Kricker A, Grulich AE, Vajdic CM, Turner J, Whitby D, Chanock S, Rothman N, Armstrong BK. Polymorphisms in immune function genes and risk of non-Hodgkin lymphoma: findings from the New South Wales non-Hodgkin Lymphoma Study. Carcinogenesis. 2007;28:704–712. doi: 10.1093/carcin/bgl200. [DOI] [PubMed] [Google Scholar]
- 17.Liang XS, Caporaso N, McMaster ML, Ng D, Landgren O, Yeager M, Chanock S, Goldin LR. Common genetic variants in candidate genes and risk of familial lymphoid malignancies. Br J Haematol. 2009;146:418–423. doi: 10.1111/j.1365-2141.2009.07790.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guzowski D, Chandrasekaran A, Gawel C, Palma J, Koenig J, Wang XP, Dosik M, Kaplan M, Chu CC, Chavan S, Furie R, Albesiano E, Chiorazzi N, Goodwin L. Analysis of single nucleotide polymorphisms in the promoter region of interleukin-10 by denaturing high-performance liquid chromatography. J Biomol Tech. 2005;16:154–166. [PMC free article] [PubMed] [Google Scholar]
- 19.Kube D, Hua TD, von Bonin F, Schoof N, Zeynalova S, Klöss M, Gocht D, Potthoff B, Tzvetkov M, Brockmöller J, Löffler M, Pfreundschuh M, Trümper L. Effect of interleukin-10 gene polymorphisms on clinical outcome of patients with aggressive non-Hodgkin’s lymphoma: an exploratory study. Clin Cancer Res. 2008;14:3777–3784. doi: 10.1158/1078-0432.CCR-07-5182. [DOI] [PubMed] [Google Scholar]
- 20.Lech-Maranda E, Baseggio L, Charlot C, Rigal D, Berger F, Jamroziak K, Warzocha K, Coiffier B, Salles G. Genetic polymorphisms in the proximal IL-10 promoter and susceptibility to non-Hodgkin lymphoma. Leuk Lymphoma. 2007;48:2235–8. doi: 10.1080/10428190701615926. [DOI] [PubMed] [Google Scholar]
- 21.Maria GE, Moore PS, Zucca M, Angelucci E, Cabras MG, Melis M, Gabbas A, Serpe R, Madeddu C, Scarpa A, Cocco P. Interleukin-1B (IL1B) and interleukin-6 (IL6) gene polymorphisms are associated with risk of chronic lymphocytic leukaemia. Hematol Oncol. 2008;26:98–103. doi: 10.1002/hon.843. [DOI] [PubMed] [Google Scholar]
- 22.Lech-Maranda E, Mlynarski W, Grzybowska-Izydorczyk O, Borowiec M, Pastorczak A, Cebula-Obrzut B, Klimkiewicz-Wojciechowska G, Wcislo M, Majewski M, Kotkowska A, Robak T, Warzocha K. Polymorphisms of TNF and IL-10 genes and clinical outcome of patients with chronic lymphocytic leukemia. Genes Chromosomes Cancer. 2013;52:287–296. doi: 10.1002/gcc.22028. [DOI] [PubMed] [Google Scholar]
- 23.Berglund M, Thunberg U, Roos G, Rosenquist R, Enblad G. The interleukin-10 gene promoter polymorphism (-1082) does not correlate with clinical outcome in diffuse large B-cell lymphoma. Blood. 2005;105:4894–4895. doi: 10.1182/blood-2004-12-4814. [DOI] [PubMed] [Google Scholar]
- 24.Zhang Y, Wang MY, He J, Wang JC, Yang YJ, Jin L, Chen ZY, Ma XJ, Sun MH, Xia KQ, Hong XN, Wei QY, Zhou XY. Tumor necrosis factor-α induced protein 8 polymorphism and risk of non-Hodgkin’s lymphoma in a Chinese population: a case-control study. PLoS One. 2012;7:e37846. doi: 10.1371/journal.pone.0037846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Talaat RM, Abdel-Aziz AM, El-Maadawy EA, et al. Interleukin 10 gene promoter polymorphism and risk of diffuse large B cell lymphoma (DLBCL) Egyptian Journal of Medical Human Genetics. 2014;15:7–13. [Google Scholar]
- 26.Domingo-Domènech E, Benavente Y, González-Barca E, Montalban C, Gumà J, Bosch R, Wang SS, Lan Q, Whitby D, Fernández de Sevilla A, Rothman N, de Sanjosé S. Impact of interleukin-10 polymorphisms (-1082 and -3575) on the survival of patients with lymphoid neoplasms. Haematologica. 2007;92:1475–1481. doi: 10.3324/haematol.11350. [DOI] [PubMed] [Google Scholar]
- 27.Stang A. Critical evaluation of the Newcastle-Ottawa scale forthe assessment of the quality of nonrandomized studies in meta-analyses. Eur J Epidemiol. 2010;25:603–5. doi: 10.1007/s10654-010-9491-z. [DOI] [PubMed] [Google Scholar]
- 28.Morton LM, Turner JJ, Cerhan JR, Linet MS, Treseler PA, Clarke CA, Jack A, Cozen W, Maynadié M, Spinelli JJ, Costantini AS, Rüdiger T, Scarpa A, Zheng T, Weisenburger DD. Proposed classification of lymphoid neoplasms for epidemiologic research from the Pathology Working Group of the International Lymphoma Epidemiology Consortium (InterLymph) Blood. 2007;110:695–708. doi: 10.1182/blood-2006-11-051672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Higgins JP, Thompson SG. Quantifying heterogeneity in a meta-analysis. Stat Med. 2002;21:1539–1558. doi: 10.1002/sim.1186. [DOI] [PubMed] [Google Scholar]
- 30.Egger M, Davey Smith G, Schneider M, Minder C. Bias in meta-analysis detected by a simple, graphical test. BMJ. 1997;315:629–634. doi: 10.1136/bmj.315.7109.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mocellin S, Marincola F, Rossi CR, Nitti D, Lise M. The multifaceted relationship between IL-10 and adaptive immunity: putting together the pieces of a puzzle. Cytokine Growth Factor Rev. 2004;15:61–76. doi: 10.1016/j.cytogfr.2003.11.001. [DOI] [PubMed] [Google Scholar]
- 32.Turner DM, Williams DM, Sankaran D, Lazzarus M, Sinnott PJ, Hutchinson IV. An investigation of polymorphism in the interleukin-10 gene promoter. Eur J Immunogenet. 1997;24:1–8. doi: 10.1111/j.1365-2370.1997.tb00001.x. [DOI] [PubMed] [Google Scholar]
- 33.Mormann M, Rieth H, Hua TD, Assohou C, Roupelieva M, Hu SL, Kremsner PG, Luty AJ, Kube D. Mosaics of gene variations in the Interleukin-10 gene promoter affect interleukin-10 production depending on the stimulation used. Genes Immun. 2004;5:246–55. doi: 10.1038/sj.gene.6364073. [DOI] [PubMed] [Google Scholar]
- 34.Blay JY, Burdin N, Rousset F, Lenoir G, Biron P, Philip T, Banchereau J, Favrot MC. Serum interleukin-10 in non-Hodgkin’s lymphoma: a prognostic factor. Blood. 1993;82:2169–2174. [PubMed] [Google Scholar]
- 35.Lech-Maranda E, Bienvenu J, Michallet AS, Houot R, Robak T, Coiffier B, Salles G. Elevated IL-10 plasma levels correlate with poor prognosis in diffuse large B-cell lymphoma. Eur Cytokine Netw. 2006;17:60–66. [PubMed] [Google Scholar]
- 36.Purdue MP, Lan Q, Kricker A, Grulich AE, Vajdic CM, Turner J, Whitby D, Chanock S, Rothman N, Armstrong BK. Polymorphisms in immune function genes and risk of non-Hodgkin lymphoma: findings from the New South Wales non-Hodgkin Lymphoma Study. Carcinogenesis. 2006;28:704–712. doi: 10.1093/carcin/bgl200. [DOI] [PubMed] [Google Scholar]
- 37.Lee JJ, Kim DH, Lee NY, Sohn SK, Kim JG, Kim HJ, Do YR, Park YH. Interleukin-10 gene polymorphism influences the prognosis of T cell non-Hodgkin lymphomas. Br J Haematol. 2007;137:329–336. doi: 10.1111/j.1365-2141.2007.06570.x. [DOI] [PubMed] [Google Scholar]
- 38.Cao HY, Zou P, Zhou H. Genetic association of interleukin-10 promoter polymorphisms and susceptibility to diffuse large B-cell lymphoma: a meta-analysis. Gene. 2013;519:288–94. doi: 10.1016/j.gene.2013.01.066. [DOI] [PubMed] [Google Scholar]
- 39.Goldin LR, Bjorkholm M, Kristinsson SY, Turesson I, Landgren O. Highly increased familial risks for specific lymphoma subtypes. Br J Haematol. 2009;146:91–4. doi: 10.1111/j.1365-2141.2009.07721.x. [DOI] [PMC free article] [PubMed] [Google Scholar]