Abstract
Purpose: In the present study, we constructed an efficient expression system for visfatin and identified the peptide sequences which binding to visfatin. Methods and results: The fusion protein was identified by SDS-PAGE and Western blot. The yield of visfatin was 300 mg/L culture medium with optimal conditions. The recombinant visfatin binds with insulin receptor with a dose-dependent effect. All of the 24 sequence identified by ELISA were able to bind to visfatin specifically. Among the 24 DNA sequences, there were 8 clones of AAKTPTE, 4 clones of ATTVPAS and 4 clones of MSLQQEH. Conclusion: The sequence of AA(X)TPT(X) was the most frequently existed sequence in all of sequences analyzed, which suggests that AA(X)TPT(X) is likely to be the essential motif in peptides which visfatin could bind with.
Keywords: Visfatin, adipokine, gene engineering, fusion expression, phage display
Introduction
Visfatin is a peptide hormone identified most recently; it can promote the uptake of glucose in fat cell and muscle cell, and inhibit the export of hepatic glucose [1-3]. More importantly, visfatin binds with insulin receptor, then stimulates phosphorylation of tyrosine in insulin receptor, insulin receptor substrate-1 (IRS-1) and insulin receptor substrate-2 (IRS-2), subsequently, they activate protein kinase B and mitogen-activated protein kinase signal pathway, which also mediated by insulin [1,4,5].
Visfatin doesn’t change dramatically as the altering of energetic metabolism in the human body, and short-term variance in blood sugar has no impact on it [6]. Up to now, it’s still not clear about the specific mechanism behind the regulation of visfatin, such as if there any intrinsic specific receptor for this peptide hormone? However, it’s certain that visfatin plays an important role in the pathogenesis of type 2 diabetes. On this basis, the study of visfatin has intrigued a new interest in the research area of diabetes and obesity since its discovery [7,8].
In the present study, we constructed a prokaryotic expression system with genetic engineering techniques, and obtained a large amount of recombinant visfatin with high purity, which laid the foundation for the further study of visfatin. On this basis, we screened polypeptide sequence [9] which visfatin specifically binds to by protein - protein interactions with the phage display technology [10]. Our study provide a favorable support for studying the potential binding sites on insulin receptor for visfatin, and even the specific receptor of visfatin according to the analysis of obtained polypeptide sequences.
Material and methods
Reagents
Prokaryotic expression vector pQE-30Xa and E.coli strains M15 [pREP4] were purchased from Qiagen. The enzymes Sal I, Stu I, Pvu II, Ex Taq polymerase and dNTP were purchased from Takara Bio Inc. M-MLV reverse transcriptase and T4 DNA ligase were purchased from Promega. Nickel-nitrilotriacetic acid (Ni-NTA) column was purchased from Gene Script. Factor Xa Cleavage Capture kit was purchased from Novagen. Ph.D.-7 Phage Display Peptide Library Kit was purchased from New England Biolabs. M13 DNA extraction kit was purchased from Omega. Mouse anti-His antibodie, human insulin receptor and goat anti-human insulin R (D-17) antibody were from Santa Cruz.
Plasmid construction
Total RNA was extracted from omental adipose tissue using TRIzol according to the standard protocol. 4 μL total RNA was reverse transcribed using 24 Units of MMLV enzyme. Oligo (dT) 18 was used for priming. The cDNA was then amplified by Polymerase Chain Reaction (PCR) using the primers (Forward: 5’-CCACCCAACACAAGCAAAG-3’, reverse: 5’-CCGTCGACTTA ATGATGTGCTGCTTCCAGTTC-3’) for 40 cycles. The PCR product was then digested with the restriction enzyme Sal I. The vector pQE-30Xa was digested with restriction enzymes Stu I and Sal I. Then PCR product was cloned into the pQE-30Xa vector. The positive clone was confirmed by sequencing. The confirmed constructed sequence was transected into E.coli M15 [pREP4] competent cells.
Recombinant visfatin expression
Transformed E.coli M15 [pREP4] cells were cultured at 37°C in Luria-Bertani (LB) medium with ampicillin (100 mg/L) and kanamycin (25 mg/L). When the absorbance reached to 0.6 at 600 nm, 0.5 mmol/L isopropyl-1-thio-β-D- galactopyranoside (IPTG) was added to induce the expression of recombinant visfatin at 37°C for 6 h. These cultures were collected by centrifugation at 4°C. Pellets were dissolved in buffer and sonication was carried out on ice using ultra-sonicator. After the sonication, pellets and supernatants were separated by centrifugation. The contents of recombinant visfatin in pellets and supernatants were compared by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) analysis.
Recombinant visfatin purification
Inclusion bodies were obtained from pellets. The denatured recombinant visfatin protein was purified with Ni-NTA column. Then ultrafiltration was carried out to concentrate the purified proteins. The His-tag was removed with Factor Xa Cleavage Capture kit according to the instruction.
Activity assay
The activity of recombinant visfatin protein was measured by enzyme-linked immunosorbent assays (ELISA). The 96-well plate was added recombinant visfatin protein with different concentrations of at 4°C. After washed with TBS and blocked with blocking solution, insulin receptor (100 μg/ml) was added into the 96-well plate at room temperature and cultured for 1 h. Then the anti-human insulin receptor antibody and (HRP)-conjugated donkey-anti-goat IgG were added to the culture. Following extensive washes, ABTS (HRP substrate) was added in to detect the absorbance values at 410 nm.
Peptide library screening
Visfatin-binding peptides were identified using a commercially available Ph.D.-7 Phage Display Peptide Library Kit. The target binding, elution, and phage amplification were conducted according to the manufacturer’s instructions. Briefly, the culture-dishes were coated with visfatin solution. After blocked and washed extensively, culture dishes were incubated with 2× 1011 phage particles for 45 min. After non-specifically or weakly bound phage particles were removed, the remaining bound phage particles were eluted. Then the eluted phage was amplified in E.coli ER2738. The entire procedure was repeated three rounds to allow enrichment of tight-binding protein. Randomly selected blued phage plaques was analyzed by DNA sequencing after blue-white screening was performed in LB agar plates containing 5-bromo-4-chloro-3-indolyl-β-D-galactopyranoside (Xgal) and isopropyl-β-D-thiogalactopyranosid (IPTG).
Binding affinity assays
The binding affinity of identified phage clone with visfatin was determined by ELISA. After coated with visfatin solution, the 96-well plate was incubated with the phage. The unbound phage was removed through washing, and the bound phage was measured with (HRP)-conjugated anti-M13 and ABTS reaction as described above.
Results
DNA sequence analysis
pQE-30Xa visfatin plasmid was digested with Pvu II, which could be seen in two fragments around 4544 bp and 317 bp (Figure 1). The results of DNA sequence analysis (data not shown) demonstrated the whole coding sequence of visfatin has been inserted into Stu I and Sal I restriction sites of pQE-30Xa with the frame could be read correctly.
Figure 1.
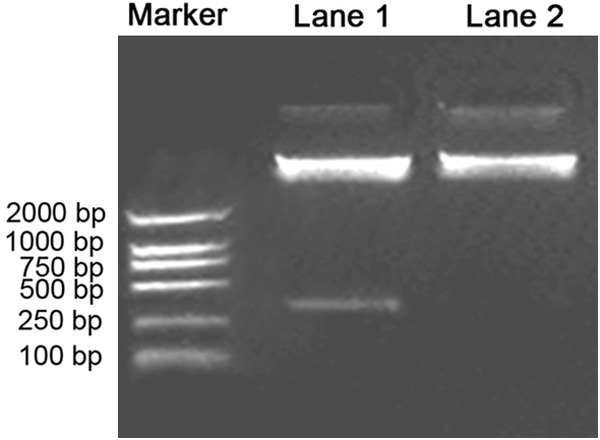
Examining of pQE-30Xa-visfatin plasmid with restriction endonuclease. Lane 1, pQE-30Xa-visfatin plasmid was digested with Pvu II, which could be seen in two fragments around 4544 bp and 317 bp; Lane 2, pQE-30Xa Pvu II enzyme digested products.
Induced expression of visfatin
Taking host strains E.coli M15 [pREP4] with empty plasmid pQE-30 Xa and Xa-visfatin with non-induced pQE-30 as controls, transformed bacteria containing recombinant plasmid pQE-30 Xa-visfatin was induced at 37°C for 6 hours, the bands at 54 kD were observed in both supernatant and precipitation. As a contrast, there’s no obvious band in both empty and non-induced recombinant bacteria (Figure 2). Identified with western blot, a specific band was shown at 54 kD for the expression product of transformed bacteria which contains recombinant plasmid pQE-30 Xa-visfatin M15 [pREP4] (Figure 3). A rough analysis of stained protein electrophoresis with GeneSnap gel analysis systems, and evaluation of expression efficiency of pQE-30Xa/visfatin recombinant plasmid in M15 [pREP4] strains showed the percentage of target protein expressed as high as 42.634%.
Figure 2.
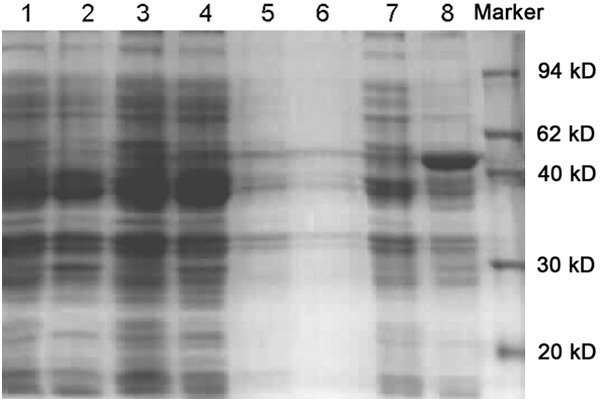
Examining of visfatin expression products with SDS-PAGE. Lane 1, the supernatant expression of M15 induced bacteria; Lane 2, the precipitation expression of M15 induced bacteria; Lane 3, the supernatant expression of induced bacteria after M15 was transformed by empty pQE-30Xa plasmids; Lane 4, the precipitation expression of induced bacteria after M15 was transformed by empty pQE-30Xa plasmids; Lane 5, the supernatant expression of un-induced bacteria after M15 was transformed by recombinant plasmid pQE-30Xa-visfatin; Lane 6, the precipitation expression of un-induced bacteria after M15 was transformed by recombinant plasmid pQE-30Xa-visfatin; Lane 7, the supernatant expression of 6 hours-induced bacteria after M15 was transformed by recombinant plasmid pQE-30Xa-visfatin; Lane 8, the precipitation expression of 6 hours-induced bacteria after M15 was transformed by recombinant plasmid pQE-30Xa-visfatin; Lane 9, marker.
Figure 3.
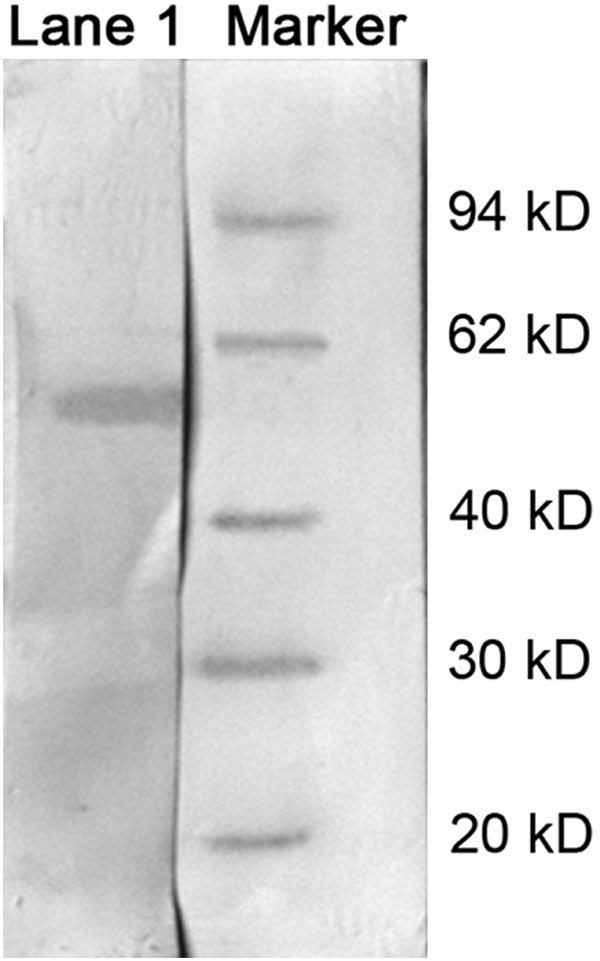
Examining of visfatin with Western Blot, a specific band was shown at about 54 kD in transformed bacteria which contains recombinant plasmid pQE-30 Xa-visfatin M15 [pREP4].
Purification and activity assay of recombinant protein
Under denaturing conditions, the bacterial were dissolved and added to the column of metal chelating chromatography, followed by washing with a gradient of urea-containing solution. At last, the recombinant protein was obtained with the elution buffer, and identified by SDS-PAGE with the target protein at approximately 54 kD (Figure 4). The fusion tags were removed by factor Xa (Figure 5), and the purity of target protein was more than 95% (Figure 6). The concentration of the purified fusion protein was 220 μg/ml with BCA method. The concentration of target protein was 142 μg/ml. After calculation, the expression of visfatin was 300 mg/L in medium. Table 1 presented the biological activity of recombinant visfatin by its binding to the receptor of insulin in a dose-dependent manner.
Figure 4.
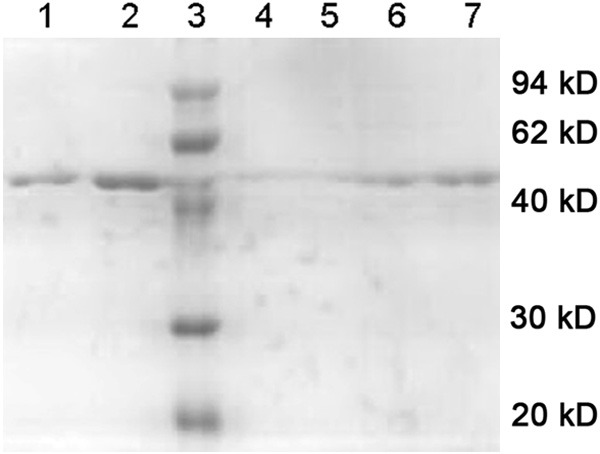
Examining of separation and purification products of visfatin with SDS-PAGE. Lanes 1 and 2, eluted recombinant visfatin protein (initial concentration of imidazole: 20 mM; terminated concentration: 250 mM); Lanes 3: protein marker; Lanes 4 and 5: refolded protein after rinse (initial concentration of urea: 6 M, terminated concentration: 0 M); Lanes 6 and 7, corresponding protein solutions of the peak sample.
Figure 5.
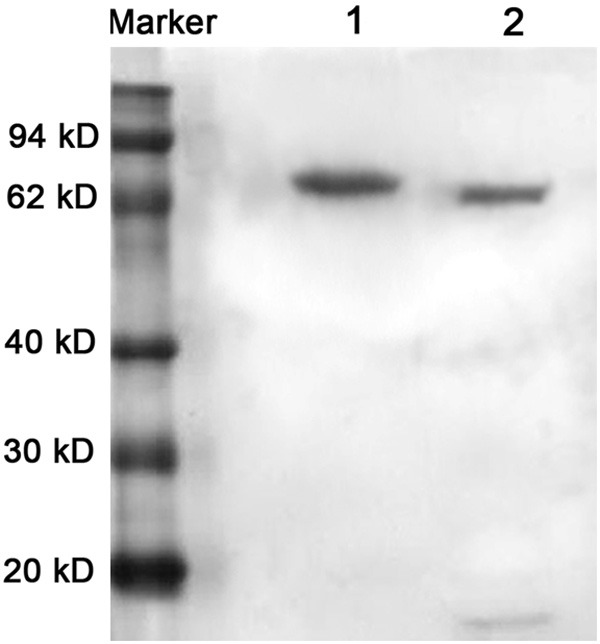
Examining of hydrolyzed products of Xa Factor with SDS-PAGE. Lane 1, purified protein; Lane 2, hydrolyzed protein.
Figure 6.
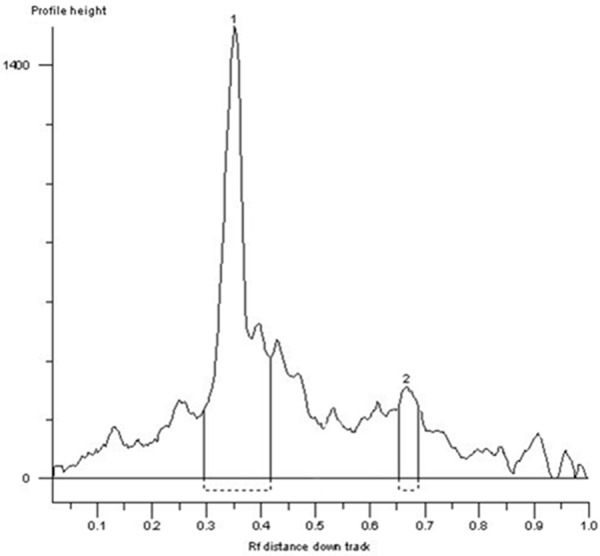
Examining of purity for hydrolyzed products of factor Xa by HPLC. Protein 1: Purified target protein (with fusion tag), the purity reached to 95.629%; Protein 2: Hydrolyzed protein by Xa factor (without fusion tag).
Table 1.
Semi-quantitative measurement of binding affinity of visfatin and insulin receptor examined with ELISA
| Sample | OD410 | ||
|---|---|---|---|
| Visfatin (10 μg/ml) | 0.042 | 0.037 | 0.045 |
| Visfatin (50 μg/ml) | 0.188 | 0.178 | 0.205 |
| Visfatin (100 μg/ml) | 0.311 | 0.301 | 0.408 |
| TBS | 0.013 | 0.019 | 0.011 |
| 6× His/PTHrP | 0.017 | 0.021 | 0.015 |
Determining the sequence that visfatin binds
A total of 24 phages were randomly sequenced against DNA, according to the report of sequencing (data not shown), we derived the sequence of interested peptide (Table 2).
Table 2.
Sequence analysis of the positive phage clones
| Peptide sequence | Clone Identifiers |
|---|---|
| AAKTPTE | 1, 3, 4, 5, 9, 15, 17, 19 |
| ATTVPAS | 11, 14, 16, 24 |
| MSLQQEH | 2, 10, 18, 23 |
| SAPSSKN | 12, 13 |
| LPRTPTD | 6, 22 |
| MHAPPFY | 20 |
| DEQPYDL | 21 |
| TSLSMPS | 7 |
Semi-quantitative measurement of the binding of peptide and visfatin
Visfatin could bind with its nature ligand- HRP, the semi-quantitative measuring was performed to test the binding of visfatin and HRP (existed in second antibody), the absorbance of chromogenic substances of HRP at the 410 nm was proportional to the concentration of protein. 24 clones showed positive with ABTS staining, and the intensity of staining declined with concentration. With benchmarking with BSA-negative as control, the OD410 values for all the wells were more than 0.1, and the data presented a significant concentration gradient, which suggests the efficient binding of the positive clones with visfatin.
Discussion
Since the study of visfatin has just begun, its price is so expensive that seems hard for most of researchers, which has become a bottleneck in the current study. Obtaining large amounts of visfatin with high purity is the prerequisite for further work. Due to the limitation of as low concentration of 10% (fasting) and 3% (postprandial) of insulin level for visfatin in serum, it’s difficult to obtain visfatin through directly purifying. For recombinant proteins, the genetic engineering methods has obvious advantages, such as high yield, lower costs and free of fragment length limitations [11], thus, genetic engineering could be widely used [12,13]. Compared with non-fusion expression, fusion protein expression has its advantages, such as reduced degrading of heterologous proteins in prokaryotes by proteases, enhanced yielding 7and simplified expression of target protein due to the fusion tag [14]. pQE expression system is widely used with the advantages of step purifying the target protein with high yielding, and less interference [15]. After obtaining the target protein visfatin, the 6× His tag can be completely removed using factor Xa, so that the protein contains only visfatin mature peptide sequence.
The peptide library of phage contains more than a hundred millions of short sequence encoding exogenous peptide. Ph.D.-7 phage peptide library kit fused random heptapeptide with the M13 (pIII) phage capsid protein to a combinatorial library. The library has a wide diversity in sequence with no obvious bias to any position. The library contained ~ 2.8×109 electrical conversion sequence (the possible number of random heptapeptide sequence is 207, which means 1.28×109), and were amplified with 10 μl of phage. Each sequence can be generated ~70 copies. The phage display random peptide library can be used in many ways [16], such as the study of protein-protein interactions [17], drawing epitope mapping , immunological diagnostics, vaccine development and drug screening. Phage display technology establishes a direct link between large amounts of random peptide sequences and their DNA coding, which made the quick identification of a variety of targeted polypeptide. Incubation of the phage display peptide library with the targeted molecule in coated plates made phage adsorbed on the target molecule. After washed unbound phage firstly, we eluted specific binding phage. Eluted phage was amplified by infection of E.coli, and then went into the next round of binding/amplification cycles to enrich those bound sequences. After three rounds of selections, the phages with high affinity and specific peptide presenting on the surface were screened from phage expression libraries, and identified with analysis of DNA sequence. Because each target binds to phage peptide at specific structural motif (motif), we could identify which site does the natural receptor binds just as the peptide does by analyzing the characteristics of the structural motif of the polypeptide and comparing with the natural receptor of targeted molecule.
In this study, we covered the dishes with purified visfatin free of fusion-label, after three rounds of screening and continuous cycling of adsorption-elution-amplification-readsorption; we enriched the phages with polypeptides which capable of binding with the targeted protein. At last, we randomly selected 24 blue plaques from less than 100 cultured plates, and extracted phage DNA. The results of phage DNA sequence analysis suggested peptide sequence existed in the phage coat protein. The most frequently appeared sequences are AAKTPTE, ATTVPAS and MSLQQEH, which have 8, 4 and 4 clones respectively. Analyzed of sequencing results demonstrated the appearing of high frequency-sequence of AA(X) TPT(X), which suggests this sequence is probably the basic polypeptide motif that visfatin binds to. The proline residue all appears in multiple sequences, which exists in the secondary structure of β-angle protein. The appearance of proline likely suggests the existence of β-turn in structure, which further indicates the possible biological function. The high frequency of the same sequence occurring suggests the existence of highly conserved sequences, and high binding affinity of visfatin on insulin receptors. The clarification of binding domain is very important for understanding the physiology function and mechanism of visfatin [18]. Thus, the fundamental researches in these areas could provide guidance for understanding the potential treatment of visfatin for patients with resistance and deficiency of insulin. Obtaining peptide sequence could further explore the potential binding sites on insulin receptor even specific receptor for visfatin. For the future work, we shall focus on bioinformatics and identify such binding sites with simulating the protein conformations on computer, as well as finding out any specific receptor for visfatin.
Conclusion
In this study, we successfully constructed the efficient prokaryotic expression system for human visfatin. The expressed recombinant protein has considerable biological activity. The sequence of AA(X)TPT(X) was the most frequently existed sequence in all, which suggests that AA(X)TPT(X) is likely to be the essential motif in peptides which visfatin could bind with.
Disclosure of conflict of interest
None.
References
- 1.Kim da S, Kang S, Moon NR, Park S. Central visfatin potentiates glucose-stimulated insulin secretion and beta-cell mass without increasing serum visfatin levels in diabetic rats. Cytokine. 2014;65:159–166. doi: 10.1016/j.cyto.2013.11.008. [DOI] [PubMed] [Google Scholar]
- 2.Yu-Duan T, Chao-Ping W, Chih-Yu C, Li-Wen L, Tsun-Mei L, Chia-Chang H, Fu-Mei C, Hsien-Chang L, Hsia-Fen H, Yau-Jiunn L, Jer-Yiing H. Elevated plasma level of visfatin/pre-b cell colony-enhancing factor in male oral squamous cell carcinoma patients. Med Oral Patol Oral Cir Bucal. 2013;18:e180–186. doi: 10.4317/medoral.18574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Skoczen S, Tomasik PJ, Gozdzik J, Fijorek K, Krasowska-Kwiecien A, Wiecha O, Czogala W, Dluzniewska A, Sztefko K, Starzyk J, Siedlar M. Visfatin concentrations in children with leukemia before and after stem cell transplantation. Exp Hematol. 2014;42:252–260. doi: 10.1016/j.exphem.2013.12.006. [DOI] [PubMed] [Google Scholar]
- 4.Mannino GC, Sesti G. Individualized therapy for type 2 diabetes: clinical implications of pharmacogenetic data. Mol Diagn Ther. 2012;16:285–302. doi: 10.1007/s40291-012-0002-7. [DOI] [PubMed] [Google Scholar]
- 5.Yamawaki H, Hara N, Okada M, Hara Y. Visfatin causes endothelium-dependent relaxation in isolated blood vessels. Biochem Biophys Res Commun. 2009;383:503–508. doi: 10.1016/j.bbrc.2009.04.074. [DOI] [PubMed] [Google Scholar]
- 6.Pilz S, Mangge H, Obermayer-Pietsch B, Marz W. Visfatin/pre-B-cell colony-enhancing factor: a protein with various suggested functions. J Endocrinol Invest. 2007;30:138–144. doi: 10.1007/BF03347412. [DOI] [PubMed] [Google Scholar]
- 7.Moschen AR, Kaser A, Enrich B, Mosheimer B, Theurl M, Niederegger H, Tilg H. Visfatin, an adipocytokine with proinflammatory and immunomodulating properties. J Immunol. 2007;178:1748–1758. doi: 10.4049/jimmunol.178.3.1748. [DOI] [PubMed] [Google Scholar]
- 8.Adya R, Tan BK, Punn A, Chen J, Randeva HS. Visfatin induces human endothelial VEGF and MMP-2/9 production via MAPK and PI3K/Akt signalling pathways: novel insights into visfatin-induced angiogenesis. Cardiovasc Res. 2008;78:356–365. doi: 10.1093/cvr/cvm111. [DOI] [PubMed] [Google Scholar]
- 9.Sidhu SS, Koide S. Phage display for engineering and analyzing protein interaction interfaces. Curr Opin Struct Biol. 2007;17:481–487. doi: 10.1016/j.sbi.2007.08.007. [DOI] [PubMed] [Google Scholar]
- 10.Cortese R, Monaci P, Nicosia A, Luzzago A, Felici F, Galfre G, Pessi A, Tramontano A, Sollazzo M. Identification of biologically active peptides using random libraries displayed on phage. Curr Opin Biotechnol. 1995;6:73–80. doi: 10.1016/0958-1669(95)80012-3. [DOI] [PubMed] [Google Scholar]
- 11.McGlothlin JR, Gao L, Lavoie T, Simon BA, Easley RB, Ma SF, Rumala BB, Garcia JG, Ye SQ. Molecular cloning and characterization of canine pre-B-cell colony-enhancing factor. Biochem Genet. 2005;43:127–141. doi: 10.1007/s10528-005-1505-2. [DOI] [PubMed] [Google Scholar]
- 12.Nuc P, Nuc K. [Recombinant protein production in Escherichia coli] . Postepy Biochem. 2006;52:448–456. [PubMed] [Google Scholar]
- 13.Sharma SS, Blattner FR, Harcum SW. Recombinant protein production in an Escherichia coli reduced genome strain. Metab Eng. 2007;9:133–141. doi: 10.1016/j.ymben.2006.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Waugh DS. Making the most of affinity tags. Trends Biotechnol. 2005;23:316–320. doi: 10.1016/j.tibtech.2005.03.012. [DOI] [PubMed] [Google Scholar]
- 15.Rao XC, Li S, Hu JC, Jin XL, Hu XM, Huang JJ, Chen ZJ, Zhu JM, Hu FQ. A novel carrier molecule for high-level expression of peptide antibiotics in Escherichia coli. Protein Expr Purif. 2004;36:11–18. doi: 10.1016/j.pep.2004.01.020. [DOI] [PubMed] [Google Scholar]
- 16.Brissette R, Goldstein NI. The use of phage display peptide libraries for basic and translational research. Methods Mol Biol. 2007;383:203–213. doi: 10.1007/978-1-59745-335-6_13. [DOI] [PubMed] [Google Scholar]
- 17.Rowley MJ, O’Connor K, Wijeyewickrema L. Phage display for epitope determination: a paradigm for identifying receptor-ligand interactions. Biotechnol Annu Rev. 2004;10:151–188. doi: 10.1016/S1387-2656(04)10006-9. [DOI] [PubMed] [Google Scholar]
- 18.Arner P. Visfatin--a true or false trail to type 2 diabetes mellitus. J Clin Endocrinol Metab. 2006;91:28–30. doi: 10.1210/jc.2005-2391. [DOI] [PubMed] [Google Scholar]


