Abstract
Animals with habitats in the intertidal zone often display biological rhythms that coordinate with both the tidal and the daily environmental cycles. Two recent studies show that the molecular components of the biological clocks mediating tidal rhythms are likely different from the phylogenetically conserved components that mediate circadian (daily) rhythms.
Organisms that live in intertidal habitats are exposed to a complex oscillatory environment that results from the ebb and flow of tidal waters. This pervasive cyclic change between land and water habitats must have exerted a tremendous selective pressure for biological rhythms that adapt to the tides. Indeed, several decades ago it became clear that the physiology and behavior of intertidal animals is temporally organized in harmony with the tidal cycle, and that this organization emerges from circatidal clocks, biological clocks with a period close to the period of the tidal cycle and entrained to environmental cycles associated with tides. For decades, researchers have wondered whether natural selection has recruited similar clock components to build both circadian and circatidal clocks, and until the mid 1970s behavioral studies were the main tool to assess how these two clocks were put together [1]. The cloning of the first circadian clock gene in Drosophila in the 1980s [2,3] set the stage for three decades of remarkable progress in our understanding of the circadian molecular clockwork. In the absence of comparable genetic model systems, the molecular analysis of circatidal clocks lagged behind that of circadian clocks so that the components of circatidal timekeepers are still unknown and whether they share components with circadian clocks remains unanswered. Two recent studies [4,5] published in Cell Reports and Current Biology, respectively, offer the first insight into this question.
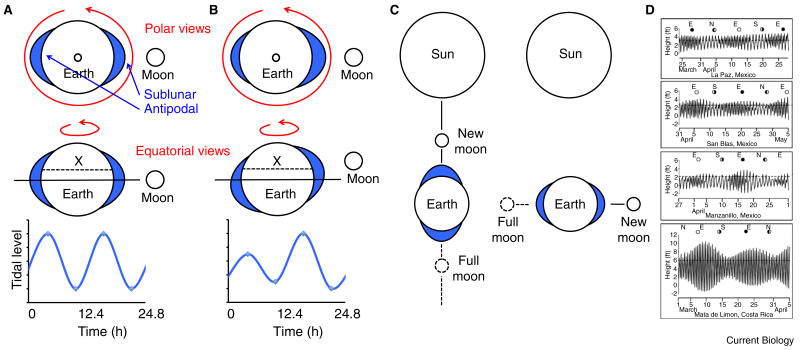
Tides are predominantly the result of the gravitational pull that the moon exerts on the ocean waters. The lunar day (24.8 h) is the result of the combined moon's orbit around earth, which takes about 29 days — a lunar month — and the 24-h earth rotation. Tidal waters rise twice per lunar day, every 12.4 h — the period of the tidal cycle (Figure 1A). Because the moon typically describes orbits around earth that are off the equatorial plane, as the earth rotates on its axis, the tidal bulge will be formed either north or south of the equatorial lane (Figure 1B). Tides on any coast will still occur every 12.4 h; however, the high and low tides on successive cycles can show very different levels, leading to a tidal imbalance known as diurnal inequality (Figure 1B). The moon's gravitational pull is added to that of the sun. Throughout the lunar month the position of the moon changes so that every 14.5 days, during new and full moon days, the earth, moon and sun are aligned on the same axis, leading to maximally high and maximally low tides (Figure 1C). The constant change in the relative position of the earth, moon and sun make the intertidal habitat a highly complex oscillatory environment: the 24-h light–dark (LD) cycle is combined with a lunar LD cycle — prominent only around full moon nights; in turn, these luminance cycles are combined with the ebb and flow of water that show overlaying periodicities of 12.4 h, 24.8 h and 14.5 days. Furthermore, these tidal periodicities can be modulated by local geographical and meteorological conditions, resulting in unique yet harmonically predictable tidal patterns to which organisms are exposed along shores (Figure 1D) [6].
Figure 1.
Organisms in the intertidal habitat are exposed to complex yet predictable tidal regimes.
(A) The gravitational pull of the moon causes high tides (represented by the blue water bulge) on the region closest to the moon (sublunar) as well as on the region diametrically opposed to it (antipodal). As the earth rotates under this deformed shell of water, any particular seacoast will experience two tides per lunar day (i.e., every 12.4 h). (B) The moon typically orbits off the equatorial plane; at some phases of the lunar month it is deviated northward (as in the example shown) and at other phases deviated southward. These deviations lead to tidal bulges that are north or south of the equator. As a consequence, successive low and high tides are asymmetric on seashores that are off the equator — any point on the ‘X’ dotted line as an example, diamonds on curve. (C) The added gravitational pull of the moon and sun lead to maximally high and maximally low tides on the phases of the moon month on which the earth, moon and sun are aligned on the same axis, during full and new moons (left). Conversely, tides are minimally high and minimally low when the earth–moon axis is perpendicular to the earth–sun axis (right). (D) The constant change in the relative position of the earth, moon and sun lead to complex regimes that are predictable and differ among different coasts. The dotted line indicates the level at which the same species of fiddler crab lives on each shore. ‘D’ from [6] with permission.
Could endogenous biological timing systems predict environmental cycles of such complexity? Stillman and Barnwell [6] demonstrated this by transporting fiddler crabs of the same species, living in three of the habitats shown in Figure 1D, to Minneapolis, Minnesota and housing them under constant tidal conditions in a controlled laboratory setting. Crabs could time motor activity in synchrony with the tides at their specific home beach, despite having left them several thousand miles behind. Studies in animals from diverse phyla have further shown remarkably precise circatidal rhythms [7,8]. Other researchers have focused on endogenous rhythms synchronized to the high tides associated with the phases of the lunar month. Intertidal marine invertebrates usually synchronize their mating, spawning or larval hatching with spring tides to maximize reproductive success [7]. These rhythms have been described in insects, crustaceans and annelids and can either be synchronized with both high tides in the lunar month (semilunar rhythms) or with one of them (lunar rhythms). They typically emerge from circasemilunar and circalunar clocks, respectively, and are often synchronized by moonlight [9].
Multiple models have emerged that attempt to explain the presence of circadian, circatidal, circalunar and circasemilunar rhythms within the same species. One of these models proposes that the presence of rhythms with circa-12.4-h periods represents the output of two anti-phase ‘circalunidian’ clocks, namely clocks with a period close to that of the lunar day period (24.8 h) [10]. Because the circadian and circalunidian periods are similar, the circalunidian clock could rely on similar molecular components as phylogenetically conserved circadian clocks. An alternative hypothesis proposes that an independent bona fide circatidal clock, presumably with different molecular mechanisms, is responsible for the generation of circatidal rhythms [11]. Circasemilunar and circalunar clocks have been proposed to rely either on counting circadian cycles or on the presence of a circatidal clock and a circadian clock that resonate every ∼15 days to achieve a specific phase relationship that triggers an event such as reproduction [12]. None of these models have been rigorously tested because the molecular bases of any biological clock in intertidal species are unknown. Drosophila flies that are arrhythmic after deleting their pigment-dispersing factor (pdf) gene, essential for the expression of circadian rhythms, can be rescued by expression of a pdf homolog from an intertidal crab, suggesting at least common clockwork outputs [13]. A recent study using RNA interference (RNAi) against the gene period in the mangrove cricket that exhibits both circatidal and circadian rhythmicity reported that the apparent knock down of period expression interferes with the circadian modulation of motor activity but not with its circatidal rhythmicity [14] (although there are alternative explanations for those results [5]).
In the recent Current Biology paper, Zhang et al. [5] cloned several clock gene homologs in the speckled sea louse, Eurydice pulchra, and show that at least some of them appear to be canonical circadian clock genes. Eurydice lives in the beaches of the Northeast Atlantic, where it is unburied by the surf of the rising tide and remains active foraging and mating during the high tide. When housed under constant laboratory conditions, Eurydice expresses both circadian and circatidal rhythms. Spontaneous motor activity exhibits a clear ∼12.4-h rhythm that can be synchronized by cyclic turbulence that replicates the action of the surf; this rhythm is in turn modulated by a circadian clock that leads to higher activity levels on the nocturnal tidal peak than the diurnal one. Furthermore, Eurydice exhibits a circadian rhythm of dermal chromatophore-pigment dispersion that is synchronized by the LD cycle. In an attempt to perturb the circadian clock — or a putative circatidal clock with similar clock components — the authors used constant light, a stimulus that typically leads to circadian arrhythmicity, and RNAi against the period gene. Surprisingly, both approaches severely disrupted circadian rhythms, including circadian expression of clock genes and pigment dispersion, but spared the circatidal activity rhythm. In contrast to this circadian-specific disruption of the timing systems of Eurydice, pharmacologically decreasing the activity of casein kinase 1 (CK1), an enzyme that phosphorylates the clock protein PERIOD, disrupted circadian rhythms but also lengthened the circatidal period. These results suggest that even if the circadian and circatidal mechanisms are different, they may share common regulators.
In the second study, reported in Cell Reports [4], Zantke and collaborators explored spawning rhythms in the worm Platynereis dumerilii. Adult Platynereis show a circalunar rhythm in sexual maturation that can be synchronized by moonlight. These authors cloned several circadian clock gene homologs and demonstrated that their expression patterns are similar to that of canonical clock components in model organisms. The expression of both behavioral and molecular circadian rhythms is modulated by the circalunar clock in Platynereis. To disrupt the circadian molecular clockwork, the authors also targeted the activity of CK1 pharmacologically; this treatment severely disrupted circadian expression of clock genes but had no measurable effect on the circalunar rhythm of sexual maturation. Thus, interfering with CK1 activity robustly disrupted the circadian system of both Eurydice and Platynereis. In contrast, this treatment had the ability to change the period of Eurydice circatidal rhythms but had no effect on Platynereis circalunar rhythms. Together, these studies suggest that the molecular mechanisms by which intertidal species endogenously time their biology to lunar-day or lunar-month tides can differ from the mechanisms underlying circadian time measurement. Further study of circatidal mechanisms may ultimately find that different intertidal species have enlisted different processes to respond to the selective pressure of the tidal environment. The conservation of circadian molecular clock components among metazoans ranges from annelids to humans and has allowed circadian biologists to use the first identified clock genes in model systems to characterize circadian molecular clocks from diverse creatures. Parsimonious and economical evolutionary processes might have been predicted to co-op the same components to measure tidal time, but evolution appears to have selected distinct clockwork mechanisms to manage cyclic tidal challenges alongside the daily program. Future research will hopefully identify the molecular components of these tidal timekeepers and how they are modulated to provide different patterns along coasts with different tidal regimes. Such studies should also identify how these components interact with the circadian clock machinery. This interaction could take place at the single cell level or it may involve separate circatidal/circalunar neuronal networks that interact with circadian ones. Could common molecular components or regulators such as CK1 couple the circatidal and circadian timekeepers? A deeper understanding of circatidal and circalunar timepieces will be critical to assess how intertidal organisms and their uniquely diverse ecosystems respond to the changes in environmental cycles that they use as cues to entrain their clocks.
Contributor Information
Horacio O. de la Iglesia, Email: horaciod@uw.edu.
Carl Hirschie Johnson, Email: carl.h.johnson@vanderbilt.edu.
References
- 1.Decoursey PJ. Biological Rhythms in the Marine Environment. University of South Carolina Press; 1976. [Google Scholar]
- 2.Reddy P, Zehring WA, Wheeler DA, Pirrotta V, Hadfield C, Hall JC, Rosbash M. Molecular analysis of the period locus in Drosophila melanogaster and identification of a transcript involved in biological rhythms. Cell. 1984;38:701–710. doi: 10.1016/0092-8674(84)90265-4. [DOI] [PubMed] [Google Scholar]
- 3.Bargiello TA, Jackson FR, Young MW. Restoration of circadian behavioural rhythms by gene transfer in Drosophila. Nature. 1984;312:752–754. doi: 10.1038/312752a0. [DOI] [PubMed] [Google Scholar]
- 4.Zantke J, Ishikawa-Fujiwara T, Arboleda E, Lohs C, Schipany K, Hallay N, Straw AD, Todo T, Tessmar-Raible K. Circadian and circalunar clock interactions in a marine annelid. Cell Rep. 2013;5:99–113. doi: 10.1016/j.celrep.2013.08.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang L, Hastings MH, Green EW, Tauber E, Sladek M, Webster SG, Kyriacou CP, Wilcockson DC. Dissociation of circadian and circatidal timekeeping in the marine crustacean Eurydice pulchra. Curr Biol. 2013;23:1979–1989. doi: 10.1016/j.cub.2013.08.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stillman JH, Barnwell FH. Relationship of daily and circatidal activity rhythms of the fiddler crab, Uca princeps, to the harmonic structure of semidiurnal and mixed tides. Marine Biol. 2004;144:473–482. [Google Scholar]
- 7.Naylor E. Chronobiology of Marine Organisms. Cambridge: Cambridge University Press; 2010. [Google Scholar]
- 8.de la Iglesia HO, Hsu YW. Biological clocks and rhythms in intertidal crustaceans. Front Biosci (Elite Ed) 2010;2:1394–1404. doi: 10.2741/e200. [DOI] [PubMed] [Google Scholar]
- 9.Kronfeld-Schor N, Dominoni D, de la Iglesia HO, Levy O, Herzog ED, Dayan T, Helfrich-Forster C. Chronobiology by moonlight. Proc R Soc B. 2013;280:1471–2954. doi: 10.1098/rspb.2012.3088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Palmer JD. Dueling hypotheses: Circatidal versus circalunidian battle basics. Chronobiol Int. 1997;14:337–346. doi: 10.3109/07420529709001455. [DOI] [PubMed] [Google Scholar]
- 11.Naylor E. Crab clocks rewound. Chronobiol Int. 1997;14:427–430. doi: 10.3109/07420529709001462. [DOI] [PubMed] [Google Scholar]
- 12.Soong K, Chang YH. Counting circadian cycles to determine the period of a circasemilunar rhythm in a marine insect. Chronobiol Int. 2012;29:1329–1335. doi: 10.3109/07420528.2012.728548. [DOI] [PubMed] [Google Scholar]
- 13.Beckwith EJ, Lelito KR, Hsu YW, Medina BM, Shafer O, Ceriani MF, de la Iglesia HO. Functional conservation of clock output signaling between flies and intertidal crabs. J Biol Rhythms. 2011;26:518–529. doi: 10.1177/0748730411420242. [DOI] [PubMed] [Google Scholar]
- 14.Takekata H, Matsuura Y, Goto SG, Satoh A, Numata H. RNAi of the circadian clock gene period disrupts the circadian rhythm but not the circatidal rhythm in the mangrove cricket. Biol Lett. 2012b;8:488–491. doi: 10.1098/rsbl.2012.0079. [DOI] [PMC free article] [PubMed] [Google Scholar]