Abstract
Gene expression profiles and chromosome 3 copy number divide uveal melanomas into two distinct classes correlating with prognosis1–3. Using exome sequencing, we identified recurrent somatic mutations in EIF1AX and SF3B1, specifically occurring in uveal melanomas with disomy 3, which rarely metastasize. Targeted resequencing showed that 24 of 31 tumors with disomy 3 (77%) had mutations in either EIF1AX (15; 48%) or SF3B1 (9; 29%). Mutations were infrequent (2/35; 5.7%) in uveal melanomas with monosomy 3, which are associated with poor prognosis2. Resequencing of 13 uveal melanomas with partial monosomy 3 identified 8 tumors with a mutation in either SF3B1 (7; 54%) or EIF1AX (1; 8%). All EIF1AX mutations caused in-frame changes affecting the N terminus of the protein, whereas 17 of 19 SF3B1 mutations encoded an alteration of Arg625. Resequencing of ten uveal melanomas with disomy 3 that developed metastases identified SF3B1 mutations in three tumors, none of which targeted Arg625.
Uveal melanoma is the most frequent malignant tumor of the eye. Notably, risk of metastatic disease and, thus, survival are strongly associated with the molecular characteristics of the primary tumor. Specifically, two major classes of uveal melanoma have been proposed on the basis of gene expression profile analyses1,3. Gene expression profile–based classification is almost congruent with chromosome 3 status and patient survival. Metastatic disease, to which about half of all individuals with uveal melanoma succumb, is strongly associated with loss of one chromosome 3 (monosomy 3) in the tumor. In contrast, tumors with disomy 3 rarely metastasize2,4,5. The proportion of individuals with tumors showing partial monosomy 3 has been controversial, with figures varying from 0 to 48% (ref. 6). Most recent studies consistently show that these tumors, in contrast to those with monosomy 3, are associated with good prognosis5,6.
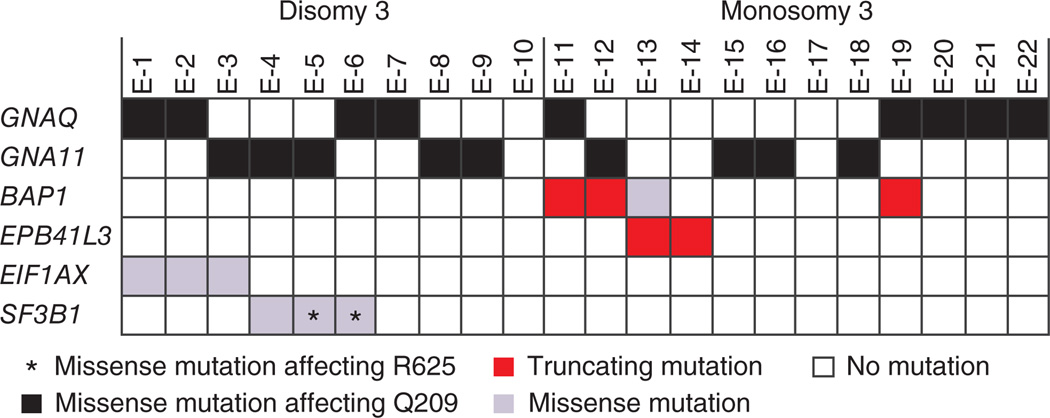
To capture the genetic differences underlying these diverse classes of uveal melanoma, we performed exome sequencing on a series of primary uveal melanomas and matched blood samples from 22 affected individuals randomly chosen from our cohort of 374 individuals treated by enucleation5. On average, we identified 33 amino acid–changing sequence variations per tumor exome not present in either dbSNP, data from the 1000 Genomes Project or exomes from the matched blood samples (Supplementary Tables 1 and 2). Six genes were recurrently mutated (Fig. 1). All mutations in these genes were verified by Sanger sequencing. Four of these genes (GNAQ, GNA11, BAP1 and SF3B1) are known targets of recurrent somatic mutations in uveal melanoma (Fig. 1). Activating mutations in GNAQ or GNA11 are found in >80% of all uveal melanomas, irrespective of tumor class, and are also frequent in blue nevi7,8. Therefore, it was suggested that these mutations occur early in tumor progression but are not sufficient for malignant transformation9. We also identified an inactivating hemizygous BAP1 mutation in four uveal melanomas with monosomy 3, a finding in line with that of a previous study focusing on chromosome 3 genes10. Exome sequencing also detected SF3B1 mutations in three tumors, two of which harbored a heterozygous missense mutation altering Arg625. Recently, mutations at this codon of SF3B1 have been identified in low-grade uveal melanoma with good prognosis11. Our exome analysis also identified two other genes, EPB41L3 and EIF1AX, as recurrent mutation targets. EPB41L3, located at 18p11, harbored a heterozygous mutation resulting in premature termination in two tumors with monosomy 3. This gene was found mutated at low frequency in other tumor entities (Catalogue of Somatic Mutations in Cancer (COSMIC))12. The EIF1AX gene, located at Xp22, has not previously been reported as a target of recurrent mutations in tumors. We detected a hemizygous EIF1AX missense mutation in three uveal melanomas. These tumors belonged to the group with disomy 3 and did not have SF3B1 mutations (Fig. 1).
Figure 1.
Exome sequencing of uveal melanomas. Sequencing of 22 uveal melanomas (E-1 to E-22) that were randomly chosen from a cohort of 374 uveal melanomas5 identified 6 recurrently mutated genes. Tumors are grouped according to their chromosome 3 status. All mutations identified in either GNAQ or GNA11 were missense mutations affecting Gln209. All mutations were verified by Sanger sequencing and are listed in Supplementary Table 3.
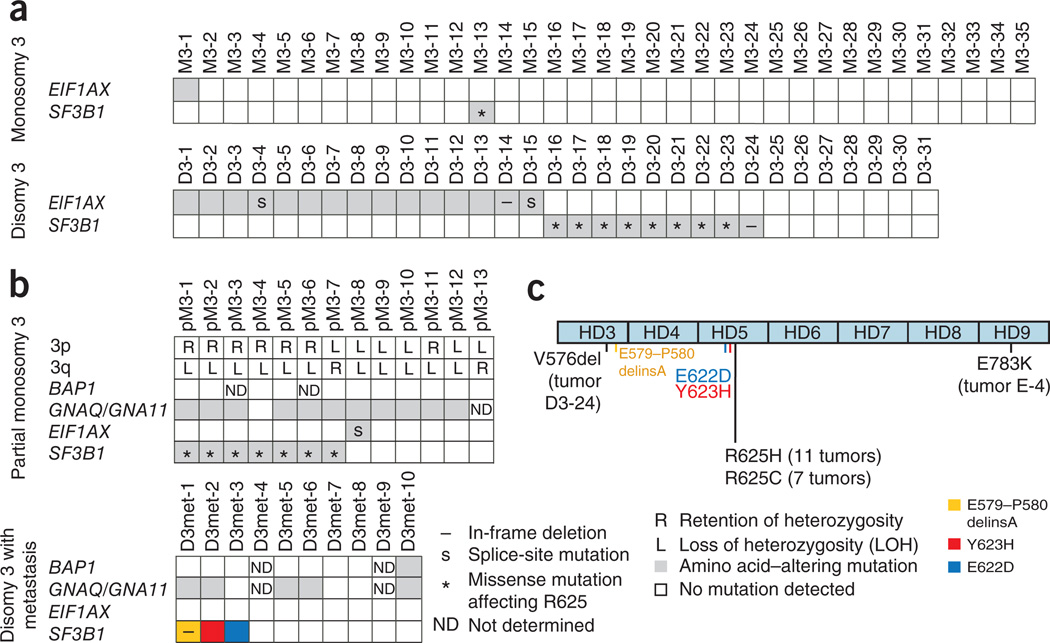
To further explore the pattern and frequency of SF3B1, EIF1AX and EPB41L3 mutations in uveal melanoma, we resequenced these genes in tumor samples from another 66 affected individuals randomly chosen from our cohort of 374 individuals, excluding tumors with partial monosomy 3 and those with disomy 3 that had led to metastases5. Resequencing of all coding exons of EPB41L3 did not identify any additional tumors with a somatic mutation in this gene. SF3B1 mutations have previously been detected in various hematological and lymphoid malignancies and have clear mutational hotspots at specific codons in the region encoding the HEAT repeats (HD) 4–9 (refs. 13,14). We therefore sequenced exons 12 to 16 of SF3B1 (corresponding to Ala514 to Lys790), which code for HD3 to HD8 and the N-terminal portion of HD9 (ref. 15). We detected a heterozygous mutation of the SF3B1 gene in 10 tumors (15%), including in 9 of 31 uveal melanomas with disomy 3 (29%) and 1 of 35 uveal melanomas with monosomy 3 (3%) (Fig. 2 and Supplementary Table 3). Nine of these mutations resulted in a missense change at Arg625 to either cystine or histidine. The SF3B1 gene product is an essential component of both major (U2-like) and minor (U12-like) spliceosomes and is involved in binding of these complexes to the intron near the branch point16,17. It was suggested that missense mutations targeting the HEAT repeats do not impair the general function of the SF3B1 product but alter the splicing of numerous target genes14,18. In uveal melanomas with disomy 3, the SF3B1 mutation pattern is uniform and distinct from that of neoplasms of hematopoietic and lymphoid origin, which involves a key mutational hotspot at codon 700 (reviewed in ref. 15).
Figure 2.
Results of targeted resequencing. Resequencing of EIF1AX (exons 1 and 2) and SF3B1 (exons 12–16) in 89 uveal melanomas with monosomy 3 (n = 35), disomy 3 (n = 31), partial monosomy 3 (n = 13) and disomy 3 with metastasis (n = 10). All mutations identified in tumor DNA were also analyzed in matched blood DNA samples, but no germline mutations were detected. (a) Sixty-six uveal melanomas randomly chosen from our cohort of 374 subjects, excluding tumors with partial monosomy 3 and disomy 3 with metastasis. (b) Results of EIF1AX and SF3B1 resequencing of all tumors with partial monosomy 3 and disomy 3 with metastasis in this cohort from which DNA was available. BAP1 mutation screening was performed in all tumors with partial monosomy 3 and disomy 3 with metastasis from which DNA was available. Exons 4 and 5 of GNAQ and GNA11 were sequenced in all tumors to detect recurrent mutations affecting Arg183 and Gln209. All mutations identified in either GNAQ or GNA11 were missense mutations affecting Gln209. (c) Distribution of SF3B1 mutations in uveal melanoma. Arg625 in the HD5 repeat was altered in 18 uveal melanomas. All non–codon 625 mutations in SF3B1 were found in only one tumor. Distinct SF3B1 mutations outside of codon 625 in tumors from individuals who died because of uveal melanoma metastases are colored in blue, yellow and red.
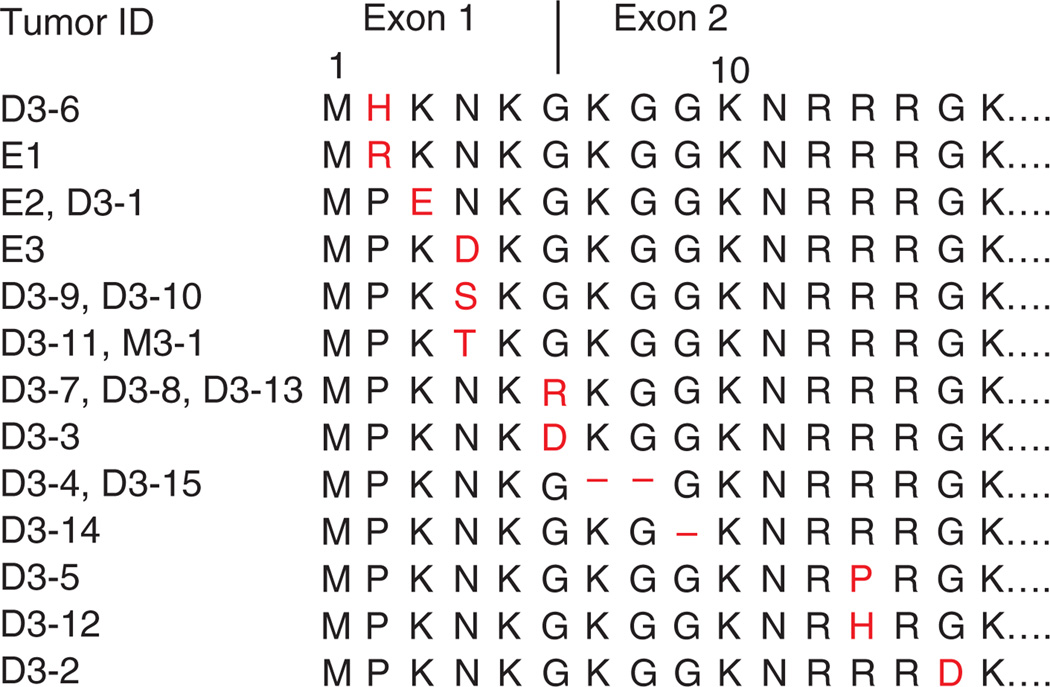
Resequencing of the coding regions and the flanking splice sites of all 7 EIF1AX exons identified mutations in another 16 of 66 tumors (24%), including in 15 of 31 uveal melanomas with disomy 3 (48%) and 1 of 35 uveal melanomas with monosomy 3 (3%) (Fig. 2 and Supplementary Table 3). Most EIF1AX mutations resulted in amino acid substitutions. However, two mutations (in D3-4 and D3-15) affected the invariant AG of the intron 1 splice acceptor site. Altered splicing is also a potential consequence of substitutions of the last (D3-7, D3-8 and D3-13) or first (D3-3) base of an exon, which were found in four tumors. To evaluate the consequences of these mutations on EIF1AX mRNA and also to determine whether the active allele was mutated in tumors from females with uveal melanoma, we sequenced the entire EIF1AX coding region in cDNA from 20 tumors with disomy 3 from which mRNA was available (Supplementary Table 3). In all but the two uveal melanomas with mutations affecting the splice acceptor site (D3-4 and D3-15), only normally spliced transcripts were found. Both tumors with a splice-site mutation showed the same mutant EIF1AX cDNA with a deletion of the first six nucleotides of exon 2, which can be explained by activation of a cryptic splice acceptor site in exon 2. Therefore, all 16 EIF1AX mutations led to amino acid substitutions or short deletions of one or two amino acids in the unstructured N-terminal tail (NTT) of the EIF1AX product (Fig. 3), leaving the core protein with its RNA-binding surface unchanged19. In all tumors with EIF1AX mutation, including those with potential exonic splice-site mutations, cDNA sequencing only showed mutant EIF1AX transcript. This finding implies that all mutations in females with uveal melanoma target the active allele, whereas the other allele is silenced as the result of X-chromosome inactivation. The anticorrelation of SF3B1 and EIF1AX mutations (Fig. 2) suggests that the effect of a mutation in one of these genes can substitute for the other. Given the role of the SF3B1 protein in pre-mRNA splicing, mutant forms might induce alternative splicing of EIF1AX pre-mRNA. We sequenced EIF1AX cDNA in SF3B1-mutant tumors (Supplementary Table 3) but did not detect any mis-spliced transcripts that would support this conjecture. None of the tumors showed an overt loss-of-function mutation in EIF1AX. This is in line with results obtained from immunohistochemistry analysis carried out on sections of primary tumor tissue. Staining with antibody to EIF1AX was observed in the cytoplasm of all tumors analyzed, regardless of their mutation status (data not shown). EIF1AX encodes eukaryotic translation initiation factor 1A (eIF1A), which stimulates the transfer of methionyl initiator tRNA (Met-tRNAi) to the small (40S) ribosomal subunit20. The segment of EIF1AX harboring the mutations is highly conserved in eukaryotes (Supplementary Fig. 1), and substitutions in the corresponding portion of yeast eIF1A reduce bulk translation in a manner rescued by overexpressing eIF1 (ref. 21), which binds cooperatively with eIF1A to the 40S subunit during the assembly of the preinitiation complex (PIC)22. These and other findings implicated the eIF1A NTT in promoting PIC assembly23, possibly through interactions with eIF3 and the eIF2-GTP-Met-tRNAi ternary complex24. Thus, EIF1AX mutations associated with uveal melanoma could diminish the rate of bulk translation and might also induce transcription factors whose mRNA translation is inversely coupled to ternary complex concentration by a specialized reinitiation mechanism25,26. Other findings implicate yeast eIF1A NTT in regulating the accuracy of start codon recognition, as NTT substitutions suppress initiation at triplets with one-base mismatches from AUG and cause the PIC to bypass the first AUG encountered while scanning from the 5′ end of the mRNA23. Hence, EIF1AX mutations might suppress the recognition of near-cognate initiation sites, which seem to be more prevalent than previously suspected27,28, or of 5′ proximal AUG codons to alter the relative use of different start codons in mRNAs encoded by tumor-promoting or tumor-suppressing genes.
Figure 3.
Alignment of EIF1AX amino acid sequences from 19 uveal melanomas with an EIF1AX mutation. The first 15 N-terminal residues are shown. Amino acids affected by mutations are restricted to positions 2–15 in the unstructured NTT. Codon 6 (Gly6) is split by intron 1. All EIF1AX mutations lead to amino acid substitutions (red) or short deletions of one or two amino acids.
Few tumors from affected individuals in our cohort had lost only portions of chromosome 3 (partial monosomy 3), and some individuals with disomy 3 tumors died from metastases5. We next sequenced SF3B1 (exons 12–16), EPB41L3 (entire coding region) and EIF1AX (exons 1 and 2) in 13 tumors with partial monosomy 3 and 10 tumors with disomy 3 that developed metastases. EPB41L3 mutations were not detected. We found SF3B1 mutations in 7 of 13 samples with partial monosomy 3 (54%), and all mutations changed codon 625. We detected an EIF1AX mutation affecting the intron 1 splice acceptor site in one tumor with partial monosomy 3 (8%) (Fig. 2). Notably, mutations in SF3B1 or EIF1AX preferentially occurred in tumors with loss of 3q and retention of 3p. Resequencing of all coding exons of BAP1 in uveal melanomas with partial monosomy 3 and disomy 3 leading to metastasis identified a mutation in one of these tumors (D3met-10). These results suggest that the biology of SF3B1- and EIF1AX-mutant uveal melanomas with partial monosomy 3, which comprise about 60% of all uveal melanomas with partial monosomy 3 in our cohort, is similar to that of uveal melanomas with disomy 3. This observation agrees with our previous finding that both uveal melanomas with disomy 3 and those with partial monosomy 3 are associated with good prognosis5. Resequencing of SF3B1 and EIF1AX in ten uveal melanomas with disomy 3 leading to metastasis identified no mutation in EIF1AX; however, three tumors (30%) harbored a mutation in SF3B1 (Fig. 2 and Supplementary Table 3). Notably, these mutations did not affect codon 625 of SF3B1. This observation suggests that the spectrum of SF3B1 mutations in these rare tumors with disomy 3 leading to metastasis is distinct from that in the majority of uveal melanomas with disomy 3. In view of the distinct mutational spectra of SF3B1 in various malignancies, it is plausible that some functional diversity of the mutant protein depends on the location of the mutation. It is conceivable that spliceosomes with the various mutant forms of the SF3B1 protein act differently on pre-mRNAs from different genes, possibly depending on sequence features present in the pre-mRNAs that are to be processed. As SF3B1 is part of a trans-acting complex, it is possible that the transcripts of many genes are affected by SF3B1 alterations. We also found SF3B1 mutations outside of codon 625 in two uveal melanomas with disomy 3 that did not develop metastases. It remains to be shown whether these non-Arg625 alterations in SF3B1 are also associated with a higher risk of metastatic disease.
We tested whether the mutation states of SF3B1 and EIF1AX were associated with clinical features of the affected individuals. Kaplan-Meier analysis showed that individuals with SF3B1- and EIF1AX-mutant tumors (excluding those with partial monosomy 3 and disomy 3 leading to metastasis) had better prognoses than those with tumors that lacked mutation in these genes (Supplementary Fig. 2). We also found that tumors with mutations in either of the two genes were more frequent among males with uveal melanoma (P = 0.0012, Fisher’s exact test) (Supplementary Fig. 3 and Supplementary Table 4). To evaluate whether alterations of EIF1AY, a Y-chromosomal paralog of EIF1AX, might contribute to this sex imbalance, we compared EIF1AX and EIF1AY expression in 24 tumors with disomy 3 or partial monosomy 3 from males and found that EIF1AY was expressed at levels comparable to those of EIF1AX in 16 of these tumors (Supplementary Table 3). We also sequenced the entire EIF1AY coding region in cDNA from EIF1AY-expressing samples, without detecting sequence alterations or mis-spliced transcripts. We also found SF3B1 or EIF1AX mutations in 2 of 47 uveal melanomas with monosomy 3 (4%); both tumors developed metastases and had a BAP1 mutation. This finding shows that, in rare cases, mutations in both genes might also occur in the class of uveal melanoma characterized by monosomy 3.
In summary, the newly identified driver mutations in EIF1AX and SF3B1, specifically found in uveal melanomas with disomy 3, confirm and extend the established classification model of uveal melanoma. Uveal melanomas with partial monosomy 3 also have mutations in these genes, placing this subgroup of tumors into the class of uveal melanomas with disomy 3. Moreover, as SF3B1 mutations in metastasized and non-metastasized uveal melanomas with disomy 3 are distinct, a further subgrouping according to mutation type may help to identify high-risk patients.
ONLINE METHODS
Subjects
All tumor and blood samples analyzed in this study were obtained from individuals with uveal melanoma who had been treated by primary enucleation without previous radiation at the Department of Ophthalmology of the University Hospital Essen between January 1998 and December 2007. All tumor samples were chosen from the cohort of 374 affected individuals published previously5. For exome sequencing, we randomly chose 22 tumors and matched blood DNA samples. For validation by targeted Sanger sequencing, we randomly chose another 66 tumor samples from this cohort, initially excluding tumors with partial monosomy 3 or allelic imbalance and those with disomy 3 that developed metastases. In addition, we selected an additional 23 tumors from this cohort for resequencing, including 13 tumors with partial monosomy 3 and 10 tumors with disomy 3 from individuals who died from uveal melanoma metastases. Follow-up data were available from all subjects. Clinical diagnosis with uveal melanoma was histopathologically confirmed using sections stained with hematoxylin and eosin and antibodies to HMB45, S100, Ki-67 and Melan-A. Each tumor was classified according to the modified Callender system recognizing spindle, epithelioid and mixed cell type29. Blood samples were obtained at the time of surgery. This study was conducted in accordance with the Declaration of Helsinki and Good Clinical Practice Guidelines and was approved by the institutional ethical review board (11-14690). All subjects provided informed consent.
Microsatellite analysis
DNA extraction and microsatellite analysis (MSA) were slightly modified from the analyses described in a previous study30. Briefly, DNA was extracted from tumor tissue using a conventional phenolchloroform procedure and from blood using the FlexiGene kit (Qiagen). To remove melanin, which impairs PCR, 6 µg of tumor DNA was purified using the QIAamp tissue kit (Qiagen) according to the manufacturer’s instructions and was eluted twice with 150 µl of water. The following chromosome 3 polymorphic microsatellite loci were analyzed: D3S3050-HEX, D3S1263-FAM, D3S1481-FAM, D3S2406-TET, D3S3045-FAM, D3S1744-TET, D3S2421-FAM and D3S1311-HEX. PCR and automated fragment length analysis were performed on a 3130xl Genetic Analyzer (Applied Biosystems) as described30.
Sanger sequencing
Coding exons of EIF1AX, SF3B1, BAP1 and EPB41L3 were sequenced using PCR-based capillary Sanger sequencing. Oligonucleotides were ordered from Eurogentec. PCR was performed in a reaction volume of 25 µl containing 20–30 ng of genomic DNA, 1.25 U of GoTaq polymerase (Promega), 5 µl of 5 × GoTaq PCR Buffer (7.5 mM MgCl2), 4 µl of dNTP (1.25 mM each; PEQLAB) and 5 pmol of each of the primers. The thermal cycling profile was as follows: initial denaturation at 95 °C for 2 min and 35 rounds of amplification at 95 °C for 30 s, 58 °C for 30 s and 72 °C for 1 min. A final extension step at 72 °C for 5 min was added. PCR products were purified using ExoSAP-IT (Affymetrix) according to the manufacturer’s protocol. Sequencing of the PCR products was carried out using the BigDye Terminator v1.1 Cycle Sequencing kit (Applied Biosystems) following the manufacturer’s instructions. Automated electrophoreses was performed on an ABI PRISM 3100 or 3010xl Genetic Analyzer (Applied Biosystems). Sequencing data were analyzed using Geneious Pro software (5.6.4., Biomatters). All primer sequences for amplification and sequencing are given in Supplementary Table 5. Sequence alterations were classified as somatic mutations only if sequencing showed that they were absent in constitutive DNA from the corresponding individual.
Exome capture
Exome capture was performed on 13 tumor and matched blood DNA samples with the NimbleGen SeqCap EZ Human Exome Library SR kit (designated V1) (UM-E2, UM-E4, UM-E5, UM-E7, UM-E10, UM-E11, UM-E14, UM-E15, UM-E16, UM-E17, UM-E18, UM-E20 and UM-E22). Nine tumor and matched blood samples were captured using NimbleGen SeqCap EZ Human Exome Library v2.0 (designated V2) (UM-E1, UM-E3, UM-E6, UM-E8, UM-E9, UM-E12, UM-E13, UM-E19 and UM-E21) (Roche NimbleGen).
Genomic library preparation
For exome enrichment using the V1 NimbleGen Capture kit, 5 µg of genomic DNA was fragmented by nebulization using a nebulizer device (Invitrogen) according to the manufacturer’s instructions. Fragmented DNA was purified using a QIAquick PCR Purification column (Qiagen) and quantified on a Nanodrop Photometer. Genomic libraries were prepared using the Illumina Paired-End Sample Prep kit following the manufacturer’s instructions, which involves size selection of DNA fragments of ~200–400 bp in length on a 2% agarose gel. Exome enrichment was performed with 1 µg of the adaptor-ligated DNA sample library using the NimbleGen SeqCap EZ SR kit according to the manufacturer’s instructions. Enrichment using the V2 NimbleGen Capture kit was performed on 1 µg of DNA fragmented by adaptive focused acoustics on a Covaris S220 for 60 s with duty cycle of 10%, intensity of 5 and cycles per burst of 200. For library preparation of fragmented DNA, we used TruSeq Sample Preparation kit v2 (Illumina), following the low-throughput and gel-free protocols. Exome enrichment of library fragments was performed using the NimbleGen SeqCap EZ v2.0 kit according to the manufacturer’s protocol, taking into consideration the technical note Targeted Sequencing with NimbleGen SeqCap EZ Libraries and Illumina TruSeq DNA Sample Preparation kits released by NimbleGen. All samples were analyzed on a Bioanalyzer using the Agilent DNA 1000 kit before sequencing. Sequence analysis of library fragments enriched by the NimbleGen V1 and V2 protocols was performed on the Illumina Genome Analyzer IIx or HiSeq 2000 platform, using a paired-end sequencing protocol (2 × 100 bp).
Data analysis
Sequence reads were mapped to the human reference genome assembly GRCh37 (ref. 31) using Burrows-Wheeler Aligner (BWA) version 0.5.9-r18-dev32 (see URLs); PCR duplicates were removed using Picard 1.63 (see URLs). The Genome Analysis Toolkit (GATK) version 1.4-33-g051b450 was used for realignment of insertions and deletions, quality recalibration and variant calling (UnifiedGenotyper)33. NimbleGen V1 raw data sets had on average 10.5 Gb of sequence; NimbleGen V2 raw data sets had on average 17.4 Gb of sequence. After filtering out PCR duplicates and non-mappable and non-exonic reads (within 4 bp of exon borders), variants were annotated using Ensembl gene annotation track v63 and were kept if they were nonsynonymous, insertions and/or deletions, missense, nonsense or in the vicinity of a splice site (within 4 bp of an exon-intron boundary). Sequence variations detected in blood samples were added to our in-house database, which totaled 70 non-tumor exomes.
Filtering
To identify somatic mutations, we set the thresholds for the variant quality score (as reported by UnifiedGenotyper) to 70 and 200 for samples captured by NimbleGen V1 and V2, respectively. We filtered out all variants that occurred in the in-house database or in dbSNP135 (but did not filter out those marked as ‘clinical’, ‘precious’ or ‘contained in a locus-specific database’) and those occurring in mucin and mitochondrial genes or pseudogenes. After filtering, a total of 832 sequence variations were identified in all 22 tumor DNA samples. In sample UM-E7, the number of variations (n = 131) was much higher than the average, owing to poor coverage of the matched blood DNA sample. Excluding this sample, we found, on average, 33 candidate genes per sample with a somatic mutation (Supplementary Tables 1 and 2). We identified 43 genes (shown in bold in Supplementary Tables 1 and 2) with sequence alterations in at least 2 different tumor samples. Careful inspection of the reads from tumor and blood samples using the Integrative Genomics Viewer (IGV)34 excluded another 34 genes, mostly owing to the presence of the sequence variation in the matched blood DNA sample. We reassessed the sequence alterations of the remaining nine genes by Sanger sequencing and confirmed the mutations in the six genes presented in Figure 1.
Statistical analysis
Mosaic plots were used to visualize the association between two categorical variables. Significance of association was tested using Fisher’s exact test for count data. Statistical analyses were performed with R (version 2.15.2; see URLs) or with JMP7 (SAS Institute).
Supplementary Material
ACKNOWLEDGMENTS
We thank M. Klutz and D. Falkenstein for technical assistance and H.-J. Lüdecke for elaborate discussion. This work was supported by the Deutsche Krebshilfe (DKF, 108612), the Mercator Research Center Ruhr (Pr-2010-0016) and the Wilhelm Sander Stiftung (2012.006.1) and by the Intramural Research Program of the US National Institutes of Health.
Footnotes
URLs. Human reference genome, ftp://ftp.1000genomes.ebi.ac.uk/vol1/ftp/technical/reference/human_g1k_v37.fasta.gz; Picard 1.63, http://picard.sourceforge.net/; R (version 2.15.2), http://cran.r-project.org/; Ensembl database, http://www.ensembl.org/index.html; accession for this study at the European Nucleotide Archive (ENA), http://www.ebi.ac.uk/ena/data/view/PRJEB3979.
Accession codes. Exome sequences generated in this study are available at ENA under accession ERP003230. Transcript sequences are available at Ensembl under the following accessions: EIF1AX, ENST00000379607; EIF1AY, ENST00000361365; and SF3B1, ENST00000335508.
Note: Supplementary information is available in the online version of the paper.
AUTHOR CONTRIBUTIONS
M.Z. designed and supervised the study, and wrote the manuscript. C.M. and N.B. provided the samples and clinical data of the affected individuals. J.v.d.N. performed immunohistochemistry. L.K.-H. performed next-generation sequencing. M.M. and S.R. performed bioinformatic analysis of exome sequencing data. L.M. performed exome capture of tumor and blood samples. C.M. and P.T. performed Sanger resequencing and analysis of the sequencing data. B.H. and D.R.L. participated in the design of the study and revised the manuscript. D.R.L. performed statistical analysis. A.G.H. provided background on eIF1A function.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
References
- 1.Onken MD, Worley LA, Ehlers JP, Harbour JW. Gene expression profiling in uveal melanoma reveals two molecular classes and predicts metastatic death. Cancer Res. 2004;64:7205–7209. doi: 10.1158/0008-5472.CAN-04-1750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Prescher G, et al. Prognostic implications of monosomy 3 in uveal melanoma. Lancet. 1996;347:1222–1225. doi: 10.1016/s0140-6736(96)90736-9. [DOI] [PubMed] [Google Scholar]
- 3.Tschentscher F, et al. Tumor classification based on gene expression profiling shows that uveal melanomas with and without monosomy 3 represent two distinct entities. Cancer Res. 2003;63:2578–2584. [PubMed] [Google Scholar]
- 4.Damato B, Dopierala JA, Coupland SE. Genotypic profiling of 452 choroidal melanomas with multiplex ligation-dependent probe amplification. Clin. Cancer Res. 2010;16:6083–6092. doi: 10.1158/1078-0432.CCR-10-2076. [DOI] [PubMed] [Google Scholar]
- 5.Thomas S, et al. Prognostic significance of chromosome 3 alterations determined by microsatellite analysis in uveal melanoma: a long-term follow-up study. Br. J. Cancer. 2012;106:1171–1176. doi: 10.1038/bjc.2012.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Abdel-Rahman MH, et al. Frequency, molecular pathology and potential clinical significance of partial chromosome 3 aberrations in uveal melanoma. Mod. Pathol. 2011;24:954–962. doi: 10.1038/modpathol.2011.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Van Raamsdonk CD, et al. Frequent somatic mutations of GNAQ in uveal melanoma and blue naevi. Nature. 2009;457:599–602. doi: 10.1038/nature07586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Van Raamsdonk CD, et al. Mutations in GNA11 in uveal melanoma. N. Engl. J. Med. 2010;363:2191–2199. doi: 10.1056/NEJMoa1000584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bauer J, et al. Oncogenic GNAQ mutations are not correlated with disease-free survival in uveal melanoma. Br. J. Cancer. 2009;101:813–815. doi: 10.1038/sj.bjc.6605226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Harbour JW, et al. Frequent mutation of BAP1 in metastasizing uveal melanomas. Science. 2010;330:1410–1413. doi: 10.1126/science.1194472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Harbour JW, et al. Recurrent mutations at codon 625 of the splicing factor SF3B1 in uveal melanoma. Nat. Genet. 2013;45:133–135. doi: 10.1038/ng.2523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Forbes SA, et al. The Catalogue of Somatic Mutations in Cancer (COSMIC) Curr. Protoc. Hum. Genet. 2008;Chapter 10(Unit 10.11) doi: 10.1002/0471142905.hg1011s57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rossi D, et al. Mutations of the SF3B1 splicing factor in chronic lymphocytic leukemia: association with progression and fludarabine-refractoriness. Blood. 2011;118:6904–6908. doi: 10.1182/blood-2011-08-373159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yoshida K, et al. Frequent pathway mutations of splicing machinery in myelodysplasia. Nature. 2011;478:64–69. doi: 10.1038/nature10496. [DOI] [PubMed] [Google Scholar]
- 15.Hahn CN, Scott HS. Spliceosome mutations in hematopoietic malignancies. Nat. Genet. 2012;44:9–10. doi: 10.1038/ng.1045. [DOI] [PubMed] [Google Scholar]
- 16.Corrionero A, Minana B, Valcarcel J. Reduced fidelity of branch point recognition and alternative splicing induced by the anti-tumor drug spliceostatin A. Genes Dev. 2011;25:445–459. doi: 10.1101/gad.2014311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Folco EG, Coil KE, Reed R. The anti-tumor drug E7107 reveals an essential role for SF3b in remodeling U2 snRNP to expose the branch point–binding region. Genes Dev. 2011;25:440–444. doi: 10.1101/gad.2009411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Quesada V, et al. Exome sequencing identifies recurrent mutations of the splicing factor SF3B1 gene in chronic lymphocytic leukemia. Nat. Genet. 2012;44:47–52. doi: 10.1038/ng.1032. [DOI] [PubMed] [Google Scholar]
- 19.Battiste JL, Pestova TV, Hellen CU, Wagner G. The eIF1A solution structure reveals a large RNA-binding surface important for scanning function. Mol. Cell. 2000;5:109–119. doi: 10.1016/s1097-2765(00)80407-4. [DOI] [PubMed] [Google Scholar]
- 20.Chaudhuri J, Si K, Maitra U. Function of eukaryotic translation initiation factor 1A (eIF1A) (formerly called eIF-4C) in initiation of protein synthesis. J. Biol. Chem. 1997;272:7883–7891. doi: 10.1074/jbc.272.12.7883. [DOI] [PubMed] [Google Scholar]
- 21.Fekete CA, et al. The eIF1A C-terminal domain promotes initiation complex assembly, scanning and AUG selection in vivo. EMBO J. 2005;24:3588–3601. doi: 10.1038/sj.emboj.7600821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Maag D, Lorsch JR. Communication between eukaryotic translation initiation factors 1 and 1A on the yeast small ribosomal subunit. J. Mol. Biol. 2003;330:917–924. doi: 10.1016/s0022-2836(03)00665-x. [DOI] [PubMed] [Google Scholar]
- 23.Fekete CA, et al. N- and C-terminal residues of eIF1A have opposing effects on the fidelity of start codon selection. EMBO J. 2007;26:1602–1614. doi: 10.1038/sj.emboj.7601613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Olsen DS, et al. Domains of eIF1A that mediate binding to eIF2, eIF3 and eIF5B and promote ternary complex recruitment in vivo. EMBO J. 2003;22:193–204. doi: 10.1093/emboj/cdg030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hinnebusch AG. Translational regulation of GCN4 and the general amino acid control of yeast. Annu. Rev. Microbiol. 2005;59:407–450. doi: 10.1146/annurev.micro.59.031805.133833. [DOI] [PubMed] [Google Scholar]
- 26.Baird TD, Wek RC. Eukaryotic initiation factor 2 phosphorylation and translational control in metabolism. Adv. Nutr. 2012;3:307–321. doi: 10.3945/an.112.002113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ingolia NT, Lareau LF, Weissman JS. Ribosome profiling of mouse embryonic stem cells reveals the complexity and dynamics of mammalian proteomes. Cell. 2011;147:789–802. doi: 10.1016/j.cell.2011.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee S, Liu B, Huang SX, Shen B, Qian SB. Global mapping of translation initiation sites in mammalian cells at single-nucleotide resolution. Proc. Natl. Acad. Sci. USA. 2012;109:E2424–E2432. doi: 10.1073/pnas.1207846109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McLean IW, Foster WD, Zimmerman LE. Uveal melanoma: location, size, cell type, and enucleation as risk factors in metastasis. Hum. Pathol. 1982;13:123–132. doi: 10.1016/s0046-8177(82)80116-0. [DOI] [PubMed] [Google Scholar]
- 30.Tschentscher F, Prescher G, Zeschnigk M, Horsthemke B, Lohmann DR. Identification of chromosomes 3, 6, and 8 aberrations in uveal melanoma by microsatellite analysis in comparison to comparative genomic hybridization. Cancer Genet. Cytogenet. 2000;122:13–17. doi: 10.1016/s0165-4608(00)00266-1. [DOI] [PubMed] [Google Scholar]
- 31.Church DM, et al. Modernizing reference genome assemblies. PLoS Biol. 2011;9:e1001091. doi: 10.1371/journal.pbio.1001091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.DePristo MA, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat. Genet. 2011;43:491–498. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Thorvaldsdóttir H, Robinson JT, Mesirov JP. Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief. Bioinform. 2013;14:178–192. doi: 10.1093/bib/bbs017. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.