Abstract
Background
Bladder urothelial carcinoma is the most common genitourinary system cancer in China. The objective of this study was to investigate whether the miR-9 can regulate the invasion ability of human bladder transitional cell carcinoma cells by down-regulation of CBX7.
Material/Methods
The expression of miR-9 was detected by quantitative real-time PCR in bladder transitional cell carcinomas (TCC) and normal bladder transitional cell (NBTC) samples. Bioinformatics software was used to predict some potential target genes of miR-9. T24 cells were transfected with pre-miR-9, and the CBX7 protein expression was detected by Western blot. Luciferase activities assay was selected to verify that CBX7 was a direct and specific gene of miR-9. T24 cells were transfected with pcDNA-CBX7, and the expression of CBX7 gene was detected. Then, the transwell assay was used to detect the invasion ability of T24 cells with CBX7 over-expression.
Results
The expression of miR-9 increased significantly in human TCC specimens compared to that in NBTC specimens. TargetScan and PicTar software programs predicted CBX7 gene was a target gene of miR-9. The pre-miR-9 could up-regulate the miR-9 expression and down-regulate CBX7 protein expression. The luciferase activities assay verified that CBX7 gene was a direct and specific target gene of miR-9. The pcDNA-CBX7 transfection could up-regulate the CBX7 protein expression, and the invasion ability of T24 cells with CBX7 over-expression decreased significantly.
Conclusions
Aberrantly expressed miR-9 contributes to T24 cells invasion, partly through directly down-regulating CBX7 protein expression in TCC. This miRNA signature offers a new potential therapeutic target for TCC.
MeSH Keywords: MicroRNAs, Neoplasm Invasiveness, Urinary Bladder Neoplasms
Background
Bladder urothelial carcinoma is the 9th most common malignancy worldwide [1,2]. It is also the most common genitourinary system cancer in China; 98% are epithelial malignancies, with the vast majority being transitional cell carcinomas (TCC) [3]. TCC occurs about 3 times more commonly in men than in women. The biological behavior of this disease shows numerous patterns, such as easy relapse, and multiple invasion and metastasis. Chinese and international data show the high incidence and mortality rates of bladder urothelial carcinoma, with an increasing trend in incidence in recent years. Surgical resection is still the most effective therapy for non-invasive TCC patients. Although chemotherapy based on BCG may reduce recurrence in patients, approximately 70% will recur and 25±30% will eventually progress to muscle invasive disease or distant metastasis [4,5]. Thus, there is an urgent need to find new therapeutic targets and strategies, which may be helpful in understanding the molecular mechanisms and improving TCC therapy.
Recently, microRNAs (miRNAs) have emerged as key regulators of gene expression stability, providing a new dimension of tumor research [6,7]. These small, non–protein-coding RNAs are molecules of 22 nucleotides transcribed from primary transcripts (pri-miRNAs) and precursor miRNA (pre-miRNA) cleaved by Dicer. The mature miRNAs incorporate into the RNA-induced silencing complex (RISC) that regulates the expression of target gene at post-transcriptional level. Through this mechanism, miRNAs influence various cellular activities, including cell differentiation, proliferation, and apoptosis under normal and disease conditions [8,9]. A growing body of evidence indicates that approximately 20–30% of human genes are regulated by miRNAs. Around 580 miRNAs have been identified and more than 1000 are predicted to exist in humans [10–12]. Recent studies have reported that some miRNAs act as oncogenes or tumor-suppressing genes, which could be involved in the genesis, progression, and metastasis of tumors [13–15]. Genome-wide miRNA expression profiling found aberrant expression of miR-9 in some kinds of malignant tumors, including gastric cancer and breast cancer [16,17]. However, there have been no reports on the expression and function changes of miR-9 in TCC, and the molecular regulative mechanism of miR-9 to TCC progression is unknown.
Our previous microarray assays found that miR-9 had higher expression in TCC samples than in controls, suggesting that miR-9 might be an oncogene in tumorigenesis of bladder carcinoma. In the present study, we investigated the molecular mechanism of miR-9 regulates invasion ability of bladder carcinoma cells. Further related research might be helpful in developing new therapies against TCC.
Material and Methods
Clinical specimens
A total of 57 TCC and 32 NBTC specimens were surgically collected after informed consent from patients ages 47–62 years. The non-muscle-invasive bladder cancer samples were collected by transurethral resection of bladder tumour (TUR-BT), and the muscle-invasive bladder cancer tissue was collected by radical cystectomy. Patients in the same age group undergoing suprapubic transvesical prostatectomy, lithocystotomy, and cystostomy for nonmalignant conditions served as NBTC controls. About 500 mg of tissue was collected from each subject. Histological identification of TCC was confirmed according to the World Health Organization criteria. Participants provided written informed consent to participate in this study. Written informed consent (as outlined in the PLOS consent form) was obtained to publish these case details. All protocols were reviewed and approved by the Ethics Committee of China Medical University.
Quantitative real-time PCR
After total RNA was extracted from tissue and cell samples, cDNA was synthesized and used to detect the mRNA expression using a miRNA detection kit (Invitrogen, USA) [18]. The samples were normalized to 18s and the 18<CT<30 were calculated with 2−ΔΔCT using the Applied Biosystems 7500 device.
Cell culture
The human TCC cell line T24 was stored by our laboratory. The cells were cultured in DMEM medium supplemented with 10% fetal calf serum and incubated at 37°C with 5% CO2.
Transfection
An appropriate concentration (about 80%) of resuspended T24 cells were seeded into a 24-well plate for transfection assay using Lipofectamine™ 2000 Reagent (Invitrogen, USA) in accordance with the manufacturer’s instructions. A total of 100 nM microRNA mimics or controls were used for the functional assays. Suitable amounts of pcDNA-CBX7 and the same procedures were used for CBX7 up-regulation with Transmessenger Regent, following the manufacturer’s instructions.
Western blotting
Total protein was extracted using RAPI buffer. Constant 50 μg protein was loaded into SDS-PAGE 8–10% for electrophoresis, transferred onto PVDF membranes, and hybridized with a primary antibody, followed by a horseradish peroxidase-conjugated secondary antibody. After ECL reaction, gray assay were performed by ImageQuant 5.2 software. All procedures were done as describe previously [19]. β-actin was used for the reference.
Vector construction and Luciferase reporter assay
CBX7 3′UTR luciferase reporters were constructed by Life Technology Company (USA). In brief, EST clone containing the 3′UTR sequences of CBX7 mRNA was constructed into the wild-type (Wt) vector, and the corresponding nonsense point mutations were drawn into the specific binding site (seed zone) to construct the mutation type of vector (Mut). After cells were seeded in a 24-well plate for 24 h, 100 ng CBX7 3′UTR luciferase construct and 400 ng microRNA mimics were cotransfected using Lipofectamine™ 2000 Reagent. After 48 h, the luciferase activity was detected with the Dual-Glo™ Luciferase Assay System (Promega, USA). To normalize the transfection efficiency, the β-galactosidase expression vector was transfected in each experiment. The relative luciferase activity was calculated by luciferase activity of firefly to Renilla.
Invasion assays
After a pretreatment with miRNA mimics or siRNA for 3 days, transwell assay was carried out by using a Transwell chamber (Qiagen, German) with pore size of 8.0 μm. The Transwell chamber was coated with Matrigel. Total 2×105 cells were suspended in 200 μl serum-free medium and seeded in the upper compartment of the chamber. The lower compartment was loaded with 750 μl full culture medium containing 10% FBS. After being incubated at 37° for 12 h, the membrane was fixed with formaldehyde, and stained with hematoxylin. Then, the trans-membrane cells were counted.
Statistical analysis
All experiments were done 3 times. All numerical data are presented as mean ± standard deviation (SD) and processed with SPSS 13.0 software (SPSS, USA). Differences were evaluated with ANOVA in nonparametric statistics and t test. When P value was lower than 0.01, there was statistical significance, indicated by (*).
Results
MiR-9 up-regulated in TCC specimens
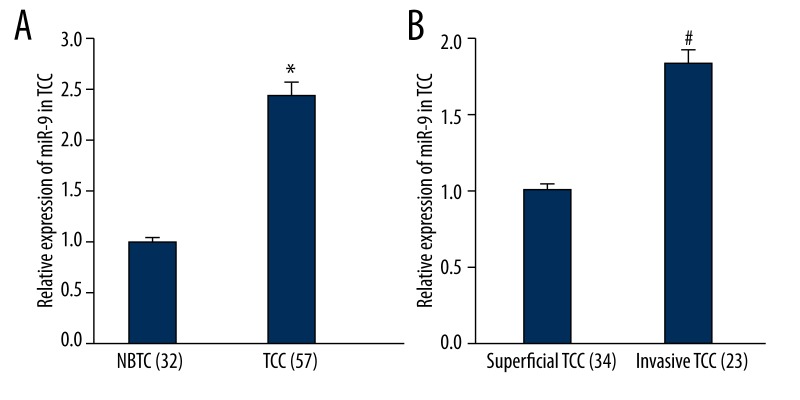
To search for the possible effect of miR-9 in TCC, the expression level of miR-9 was first detected in TCC and NBTC by SYBR-Green quantitative real-time PCR. The experiment results showed that, in contrast with NBTC tissues, the miR-9 expression in TCC tissues were significantly (p<0.01) up-regulated (Figure 1A). Those results indicate that miR-9 might be involved in TCC development. Further, the expression of miR-9 in invasive TCC tissue was significantly higher than that of superficial TCC (p<0.01) (Figure 1B).
Figure 1.
qRT-PCR analysis for the expression of miR-9 in NTBC and TCC samples. (A) Compared to NTBC, the expression of miR-9 was higher in TCC, * P<0.01 vs. NTBC. (B) Compared to superficial TCC, the expression of miR-9 was higher in invasive TCC, * P<0.01 vs. superficial TCC.
MiR-9 represses CBX7 expression in T24 cells
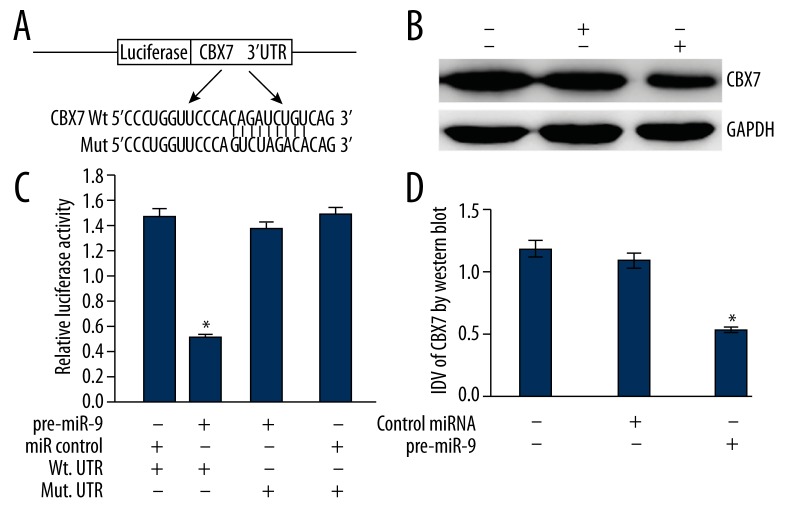
TargetScan and PicTar predicted that the CBX7 gene was a target gene of miR-9. To determine the interaction between miR-9 and CBX7 gene, the luciferase reporter assay was used. The pre-miR-9 and reporter vectors were cotransfected into T24 cells. The relative luciferase activity in T24 cells decreased with WT vector by enhanced miR-9 level, and the above-mentioned inhibitive effect could be recovered with Mut vector. These results suggest that the CBX7 gene was a specific and direct target gene of miR-9 (Figure 2A, 2B).
Figure 2.
(A) 3′UTR of CBX7 is a target of miR-9 predicted by TargetScan and PicTar. (B) The luciferase reporter assay results, with each bar representing values from 3 independent experiments. The transfection efficiency was normalized by co-transfected Renilla luciferase and the light units were calculated by relative luciferase activity of firefly to Renilla. * P<0.01. (C) Representative image of the protein level of CBX7. GAPDH was used as a reference control. (D) Quantitative analysis of the relative protein levels of CBX7 normalized to those of GAPDH is shown. Data are mean ± SD of 3 independent experiments. * P<0.01.
To ascertain the regulative effects of miR-9 to endogenous CBX7 expression, the pre-miR-9 was transfected to T24 cells, and the CBX7 expression was detected by quantitative real-time PCR and Western blot. In these experiments, the blank T24 cells and T24 cells transfected with nonsense microRNA control were selected to be controls. The results showed that, in contrast to controls, the expression level of CBX7 mRNA was not significantly altered in T24 cells (data not shown), whereas the expression of CBX7 protein was significantly down-regulated in T24 cells with over-expression of miR-9 (p<0.01) (Figure 2C, 2D). These results verified that miR-9 could inhibit the expression of CBX7 at the post-translational level.
Over-expression of CBX7 inhibited T24 cells invasion
The pCDNA-CBX7 was transfected into T24 cells to up-regulate the CBX7 expression. The TCC T24 cell line was selected because it imitates TCC biology better than other TCC cell lines, especially cell invasion ability. After transfection with pCDNA-CBX7, Western blot was used to confirm the over-expression of CBX7 protein (Figure 3). Then, transwell assay was used to detect the invasion ability of T24 cells. The results show that the invasion ability of T24 cells with CBX7 over-expression decreased remarkably. These results suggest that the over-expression of CBX7 suppresses T24 cells invasion.
Figure 3.
Effects of CBX7 over-expression on T24 cell invasion. (A–C) Are photographs of invasive cells on the membrane (400×). The membrane was formaldehyde-fixed and hematoxylin-stained. Randomly, 5 fields were selected to calculate the trans-membrane cells, which represented the invasion ability of T24 cells. The trans-membrane cells of 3 groups (A–C) were 26.1±2.7, 24.3±2.4, and 12.5±2.1, respectively.
Discussion
The expression of target genes related to various tumor cellular activities, such as apoptosis, invasion, and malignant proliferation, are inhibited by miRNAs. Recent studies have reported the abnormal expression and functions of miRNAs in human cancers. Several studies have shown that up-regulation of miR-9 gene expression was followed by enhanced tumor cell invasion and poor prognosis [20,21]. In the present study, miR-9 was up-regulated in TCC, suggesting that miR-9 might play a role in TCC tumorigenesis, but its role in this is not yet entirely clear. Aberrant miRNA expression can regulate critical biological processes, including cell proliferation and invasion, which may promote TCC development and lead to poor prognosis [22]. Further, the expression of miR-9 in invasive TCC tissue is significantly higher than that of superficial TCC. Therefore, our results indicate that miR-9 is critical for the development of human TCC, especially the invasion ability of TCC.
Because miRNAs target different genes, such as miR-34a, Tgif2, miR-320a, and ITGB3 [23,24], we speculate that miR-9 can regulate some single genes to modify the regulation network and trigger cell invasion in TCC. Computational algorithms are effective tools to predict and validate the miRNA gene targets. Through analysis using TargetScan and PicTar, a number of important candidate targets for miR-9 were predicted. Among these potential targets, CBX7 is the most intriguing target.
CBX7 is a Polycomb protein member of the Polycomb-repressive complex 1 (PRC1), a multiprotein complex that, together with the polycomb repressive complex 2(PRC2), maintains important developmental genes in a transcriptionally repressed state [25]. Moreover, further studies have shown that the correlation of the loss of CBX7 with a highly malignant phenotype and a consequent poor prognosis is a general event in oncology [26,27]. In fact, the loss of CBX7 expression has been recently shown to be associated with increasing malignancy grade in several human carcinomas, whereas the retention of CBX7 expression correlates with a longer survival of patients with colon and pancreatic cancer [28,29]. Therefore, it is reasonable to hypothesize that the loss of expression of CBX7 may play a key role in cancer progression. Consistently, CBX7 is able to counteract the decreased expression of the E-cadherin gene, whose loss of expression is a feature of the epithelial-mesenchymal transition, playing a critical role in maintaining normal epithelial cell morphology [30]. Therefore, the CBX7 gene acts as an anti-oncogene to repress the epithelial-mesenchymal transition, which was proven very important for dissemination of tumor cells.
Our results obtained from gain-of-function approaches confirmed that CBX7 is a specific and direct target gene of miR-9. Firstly, over-expression of miR-9 remarkably reduced the luciferase activity in T24 cells transfected with WT vector and pre-miR-9. Secondly, the nonsense point mutations at seed zone of 3′UTR of CBX7 could be restored by the inhibitive effect of miR-9 enhancement. Thirdly, miR-9 over-expression repressed the expression of CBX7 protein post-transcriptionally. In summary, these results affirmed that CBX7 is a specific and direct target gene of miR-9.
In addition, our studies have found that pcDNA-CBX7 could up-regulate the CBX7 expression, and could sharply inhibit the invasion ability of T24 cells. All these results indicate the following: miR-9 functions as an endogenous siRNA for CBX7 in TCC, up-regulation of miR-9 in TCC could inhibit the expression of CBX7, and low-expression of CBX7 lost its inhibitive effects on invasion. Thus, the identification of CBX7 as a target gene of miR-9 could explain why the over-expression of miR-9 functions as an oncogene in TCC. However, further research is necessary to gain a full understanding of the underlying molecular mechanisms involved.
Conclusions
In conclusion, miR-9, up-regulated in TCC, functions as an oncogene, partly through targeting the CBX7 gene. This adds to our understanding of the molecular mechanism of TCC genesis, and shows us a strategy by which miR-9 might act as a new therapy target for TCC. Nevertheless, further research is essential to identify the detailed molecular mechanism of miR-9 in the genesis and development of bladder carcinoma.
Footnotes
Competing interests
All authors announce that they have no competing interests.
Source of support: This work was supported by the National Nature Science Foundation of China (81301834, 30901480, 81172408)
References
- 1.Zhao J, Xing N. Identification of γ-Synuclein as a Stage-Specific Marker in Bladder Cancer by Immunohistochemistry. Med Sci Monit. 2014;20:2550–55. doi: 10.12659/MSM.892927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Murta-Nascimento C, Schmitz-Dräger BJ, Zeegers MP, et al. Epidemiology of urinary bladder cancer: from tumor development to patient’s death. World J Urol. 2007;25:285–95. doi: 10.1007/s00345-007-0168-5. [DOI] [PubMed] [Google Scholar]
- 3.Li W, Gu M. SULT1A1 Arg213His polymorphism is associated with bladder cancer risk: a meta-analysis. Med Sci Monit. 2014;20:1590–95. doi: 10.12659/MSM.890822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hassen W, Droller MJ. Current concepts in assessment and treatment of bladder cancer. Curr Opin Urol. 2000;10:291–99. doi: 10.1097/00042307-200007000-00002. [DOI] [PubMed] [Google Scholar]
- 5.Lin YL, Xie PG, Ma JG. Aberrant methylation of CDH13 is a potential biomarker for predicting the recurrence and progression of non muscle invasive bladder cancer. Med Sci Monit. 2014;20:1572–77. doi: 10.12659/MSM.892130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Alečković M, Kang Y. Regulation of cancer metastasis by cell-free miRNAs. Biochim Biophys Acta. 2014;1855(1):24–42. doi: 10.1016/j.bbcan.2014.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ullah S, John P, Bhatti A. MicroRNAs with a role in gene regulation and in human diseases. Mol Biol Rep. 2014;41(1):225–32. doi: 10.1007/s11033-013-2855-1. [DOI] [PubMed] [Google Scholar]
- 8.Tessitore A, Cicciarelli G, Del Vecchio F, et al. MicroRNAs in the DNA Damage/Repair Network and Cancer. Int J Genomics. 2014;2014:820248. doi: 10.1155/2014/820248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cheng CJ, Bahal R, Babar IA, et al. MicroRNA silencing for cancer therapy targeted to the tumour microenvironment. Nature. 2014 doi: 10.1038/nature13905. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Krek A, Grün D, Poy MN, et al. Combinatorial microRNA target predictions. Nat Genet. 2005;37(5):495–500. doi: 10.1038/ng1536. [DOI] [PubMed] [Google Scholar]
- 11.Zhang H, Qi F, Cao Y, et al. Down-regulated microRNA-101 in bladder transitional cell carcinoma is associated with poor prognosis. Med Sci Monit. 2014;20:812–17. doi: 10.12659/MSM.890300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chung TK, Lau TS, Cheung TH, et al. Dysregulation of microRNA-204 mediates migration and invasion of endometrial cancer by regulating FOXC1. Int J Cancer. 2011;130(5):1036–45. doi: 10.1002/ijc.26060. [DOI] [PubMed] [Google Scholar]
- 13.Shang C, Lu YM, Meng LR. MicroRNA-125b down-regulation mediates endometrial cancer invasion by targeting ERBB2. Med Sci Monit. 2012;18(4):BR149–55. doi: 10.12659/MSM.882617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang YF, Zhang AR, Zhang BC, et al. MiR-26a regulates cell cycle and anoikis of human esophageal adenocarcinoma cells through Rb1-E2F1 signaling pathway. Mol Biol Rep. 2013;40(2):1711–20. doi: 10.1007/s11033-012-2222-7. [DOI] [PubMed] [Google Scholar]
- 15.Lei Y, Hu X, Li B, et al. miR-150 modulates cisplatin chemosensitivity and invasiveness of muscle-invasive bladder cancer cells via targeting PDCD4 in vitro. Med Sci Monit. 2014;20:1850–57. doi: 10.12659/MSM.891340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Su YH, Zhou Z, Yang KP, et al. MIR-142-5p and miR-9 may be involved in squamous lung cancer by regulating cell cycle related genes. Eur Rev Med Pharmacol Sci. 2013;17(23):3213–20. [PubMed] [Google Scholar]
- 17.Gwak JM, Kim HJ, Kim EJ, et al. MicroRNA-9 is associated with epithelial-mesenchymal transition, breast cancer stem cell phenotype, and tumor progression in breast cancer. Breast Cancer Res Treat. 2014;147(1):39–49. doi: 10.1007/s10549-014-3069-5. [DOI] [PubMed] [Google Scholar]
- 18.Shang C, Guo Y, Hong Y, et al. MiR-21 up-regulation mediates glioblastoma cancer stem cells apoptosis and proliferation by targeting. FASLG Mol Biol Rep. 2014 doi: 10.1007/s11033-014-3820-3. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 19.Kumar M, Singh M, Singh SB. Optimization of conditions for expression of recombinant interferon-g in E. coli. Mol Biol Rep. 2014;41(10):6537–43. doi: 10.1007/s11033-014-3537-3. [DOI] [PubMed] [Google Scholar]
- 20.Sun Z, Han Q, Zhou N, et al. MicroRNA-9 enhances migration and invasion through KLF17 in hepatocellular carcinoma. Mol Oncol. 2013;7(5):884–94. doi: 10.1016/j.molonc.2013.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ye M, Du YL, Nie YQ, et al. Overexpression of activated leukocute cell adhesion molecule in gastric cancer is associated with advanced stages and poor prognosis and miR-9 deregulation. Mol Med Rep. 2015;11(3):2004–12. doi: 10.3892/mmr.2014.2933. [DOI] [PubMed] [Google Scholar]
- 22.Li S, Chen T, Zhong Z, et al. microRNA-155 silencing inhibits proliferation and migration and induces apoptosis by upregulating BACH1 in bladder cancer cells. Mol Med Report. 2012;5(4):949–54. doi: 10.3892/mmr.2012.779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Krzeszinski JY, Wei W, Huynh H, et al. miR-34a blocks osteoporosis and bone metastasis by inhibiting osteoclastogenesis and Tgif2. Nature. 2014;512(7515):431–35. doi: 10.1038/nature13375. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 24.Shang C, Zhang H, Guo Y, et al. MiR-320a down-regulation mediates bladder carcinoma invasion by targeting ITGB3. Mol Biol Rep. 2014;41(4):2521–27. doi: 10.1007/s11033-014-3110-0. [DOI] [PubMed] [Google Scholar]
- 25.Morey L, Aloia L, Cozzuto L, et al. RYBP and Cbx7 define specific biological functions of polycomb complexes in mouse embryonic stem cells. Cell Rep. 2013;3(1):60–69. doi: 10.1016/j.celrep.2012.11.026. [DOI] [PubMed] [Google Scholar]
- 26.Forzati F, Federico A, Pallante P, et al. CBX7 gene expression plays a negative role in adipocyte cell growth and differentiation. Biol Open. 2014;3(9):871–79. doi: 10.1242/bio.20147872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shinjo K, Yamashita Y, Yamamoto E, et al. Expression of chromobox homolog 7 (CBX7) is associated with poor prognosis in ovarian clear cell adenocarcinoma via TRAIL-induced apoptotic pathway regulation. Int J Cancer. 2014;135(2):308–18. doi: 10.1002/ijc.28692. [DOI] [PubMed] [Google Scholar]
- 28.Pallante P, Terracciano L, Carafa V, et al. The loss of the CBX7 gene expression represents an adverse prognostic marker for survival ofcolon carcinoma patients. Eur J Cancer. 2010;46(12):2304–13. doi: 10.1016/j.ejca.2010.05.011. [DOI] [PubMed] [Google Scholar]
- 29.Karamitopoulou E, Pallante P, Zlobec I, et al. Loss of the CBX7 protein expression correlates with a more aggressive phenotype in pancreaticcancer. Eur J Cancer. 2010;46(8):1438–44. doi: 10.1016/j.ejca.2010.01.033. [DOI] [PubMed] [Google Scholar]
- 30.Federico A, Pallante P, Bianco M, et al. Chromobox protein homologue 7 protein, with decreased expression in human carcinomas, positively regulates E-cadherin expression by interacting with the histone deacetylase 2 protein. Cancer Res. 2009;69(17):7079–87. doi: 10.1158/0008-5472.CAN-09-1542. [DOI] [PubMed] [Google Scholar]