Abstract
Nucleocytoplasmic large dsDNA viruses (NCLDVs) encompass an ever-increasing group of large eukaryotic viruses, infecting a wide variety of organisms. The set of core genes shared by all these viruses includes a major capsid protein with a double jelly-roll fold forming an icosahedral capsid, which surrounds a double layer membrane that contains the viral genome. Furthermore, some of these viruses, such as the members of the Mimiviridae and Phycodnaviridae have a unique vertex that is used during infection to transport DNA into the host.
Keywords: assembly, giant virus, NCLDV, unique vertex
Introduction
The characterization of Mimivirus, which was initially identified as a small bacterium, has led to a substantial increase in the number of characterized large viruses (La Scola et al., 2003). The size of Mimivirus particles (0.75 µm) broke the notion established by Pasteur and Chamberland in 1884 that viruses are filterable agents, meaning that they can pass through a 0.45 to 0.20 µm filter and remain infectious. Applying this criterion excluded many large viruses from metagenomic studies. During the last two decades the application of less stringent isolation techniques and improvements of isolation protocols for viruses infecting amoeba (Pagnier et al., 2013) have led to the discovery of many large viruses, especially in aquatic environments (Ghedin and Claverie, 2005; Boyer et al., 2009; Fischer et al., 2010; Thomas et al., 2011; Aherfi et al., 2013; Clouthier et al., 2013). In combination with improvements in sequencing technologies such as pyrosequencing, this allows for fast and effective analysis of the large genomes associated with these viruses and their subsequent classification based on similarities of genetic markers. Almost all of the viruses described by these techniques belong to the group of nucleocytoplasmic large dsDNA viruses (NCLDVs) (Iyer et al., 2001). Members of this clade infect animals and eukaryotes, and encompass members of the Asfarviridae, Ascoviridae, Iridoviridae, Poxviridae, Mimiviridae, Phycodnaviridae and the proposed family of Marseilleviridae (Colson et al., 2013b) (Table 1). It has recently been proposed to group these viruses into the order of Megavirales (Colson et al., 2013a).
Table 1.
Overview of Nucleocytoplasmic dsDNA viruses
| Virus | Host species | Family | Initial isolation | Genome size (in kbp) |
Size (in nm) |
T number |
|---|---|---|---|---|---|---|
| Pandoravirus salinus | Acanthamoeba castellanii | Pandoraviridae | Tunquen River in central Chile | 2474 | 1000 × 500 | n.a. |
| Mimivirus | Acanthamoeba castellanii | Mimiviridae | Cooling tower in Bradford, UK | 1181 | 750 | ~1000 |
| PBCV-1 | Chlorella variabilis strain NC64A | Phycodnaviridae | Paramecium bursaria in the southeastern USA | 330 | 190 | 169 |
| CIV | Galleria mellonella larvae | Iridioviridae | Chilosuppressalis in Japan | 212 | 185 | 147 |
| CroV | Cafeteria roenbergensis | Mimiviridae | Cafeteria roenbergensis in the southern USA | 730 | 300 | unknown |
Some of the NCLDVs have larger genomes than small unicellular organisms. For example the genomes of Pandoravirus salinus (the largest virus known to date), Mimivirus (a virus infecting amoeba) and CroV (Cafeteria roenbergensis virus, a virus infecting marine zooplankton) are 2.5Mbp, 1.2Mbp and 0.75Mbp, respectively (Raoult, 2004; Fischer et al., 2010; Philippe et al., 2013). The genome of all of these viruses is significantly larger than that of the smallest known organism, Mycoplasma genitalium, which has a size of 0.58Mbp (Su and Baseman, 1990). Many of the genes encoded by these viruses show little or no similarity to known proteins from other organisms. For example, only about 24% of the genes present in Mimivirus and 16% of the ones present in Pandoravirus salinus have known homologs in bacteria, archea and eukaryotes (Raoult, 2004; Philippe et al., 2013). Furthermore, these viruses encode many genes that are commonly associated with a cellular lifestyle, such as tRNA synthetases or nucleotide kinases. The reason for this repertoire of genes is not clear and there is an ongoing debate about whether these viruses are degenerated cells that lost some of their cellular functions or superviruses that acquired many genes from microorganisms that cohabitate in amoeba (Raoult, 2004; Filée et al., 2008; Moreira and Brochier-Armanet, 2008; Claverie and Abergel, 2009; 2010).
Another remarkable feature that has not been associated with viruses historically is the presence of so-called virophages for some of these viruses. Virophages need to coinfect cells with their host virus in order to successfully propagate, thereby interfering with the infection process and reducing the yield of the host virus. This is similar to satellite viruses and there is an ongoing debate as to whether these two categories should exist or should be combined (Krupovic and Cvirkaite-Krupovic, 2011; Desnues and Raoult, 2012).
Virus shape and major capsid protein
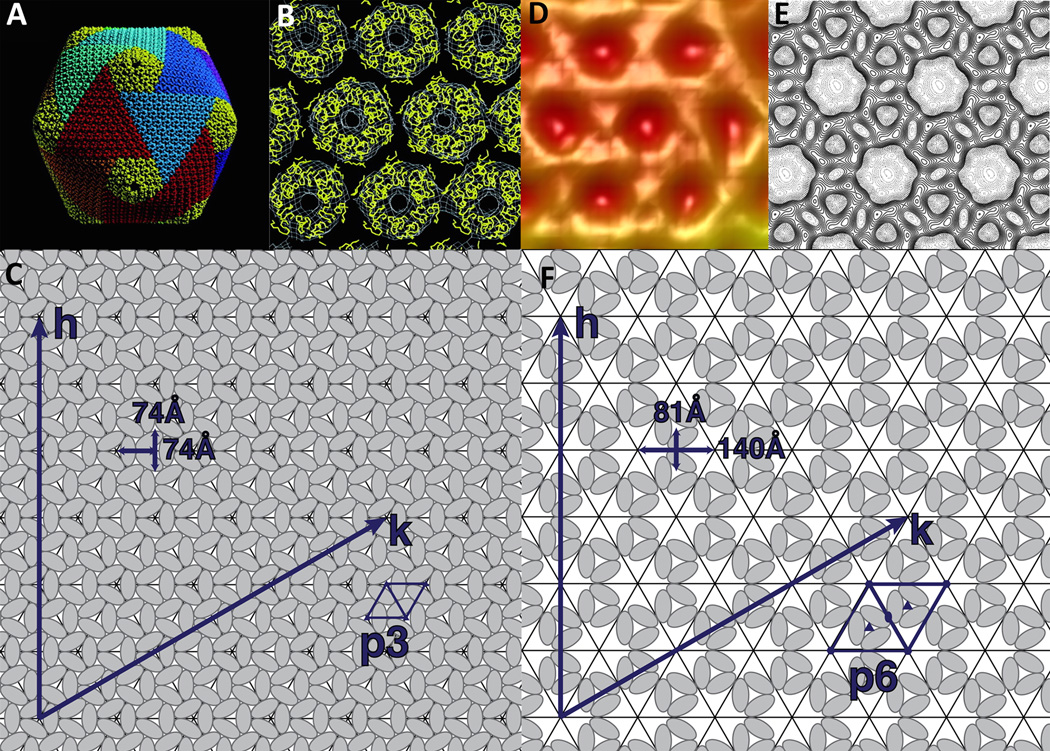
Viruses need to protect their genome between infections from extracellular and intracellular factors, without sacrificing too much of their limited coding capacity for structural proteins. Many roughly spherical shaped viruses, including most members of the NCLDVs, achieve this goal by an icosahedral arrangement of their subunits. As first hypothesized by Crick and Watson (Crick and Watson, 1956) such an icosahedral shell can be formed by 60 subunits, with the capsomers making equivalent contacts. In order to form larger structures, the number of subunits has to be increased, but then not all subunits can have equivalent environments. Instead they have quasi-equivalent environments, a concept established by Caspar and Klug (Caspar and Klug, 1961). Larger icosahedral viruses can essentially be constructed by pentamers and hexamers, with the hexamers occupying the relatively flat side of a triangular facet and the pentamers occupying the 12 fivefold vertices, introducing the necessary curvature in order to form a closed shell. The number of units in an icosahedron can be described by the triangulation number T that is equivalent to T=h2+hk+k2 with h and k being the number of subunits necessary to go from one pentameric vertex to the next in a hexagonal planar lattice arrangement (Figure 1). Therefore, the number of subunits in each icosahedral virus is equivalent to 60T.
Figure 1.
Capsomer arrangement in NCLDVs. (A) Quasi-atomic structure of PBCV-1 obtained by fitting the capsid protein structure into the icosahedral cryo-EM reconstruction of the virus. Trisymmetrons are shown in various colors and pentasymmetrons are colored yellow. (B) Detailed view of the middle of a trisymmetrons with the capsid protein shown as a Cα trace in yellow and the cryo-EM map shown in white. (A) and (B) are adapted from Nandhagopal, N., Simpson, A.A., Gurnon, J.R., Yan, X., Baker, T.S., Graves, M.V., van Etten, J.L., and Rossmann, M.G. (2002). The structure and evolution of the major capsid protein of a large, lipid-containing DNA virus. Proc. Natl. Acad. Sci. USA 99, 14758–14763. Copyright (2002) National Academy of Sciences, U.S.A. (C) Schematic representation of the p3 capsid arrangement in PBCV-1 shown in A and B. (D) Cryo-EM reconstruction of the Mimivirus surface at high magnification (adapted from (Xiao et al., 2009)). (E) Surface representation of the vaccinia virus scaffolding D13 on top of a membrane (modified from (Hyun et al., 2011)). (F) Schematic representation of the p6 protein arrangement in the Mimivirus virion and the vaccinia virus scaffold shown in D and E.
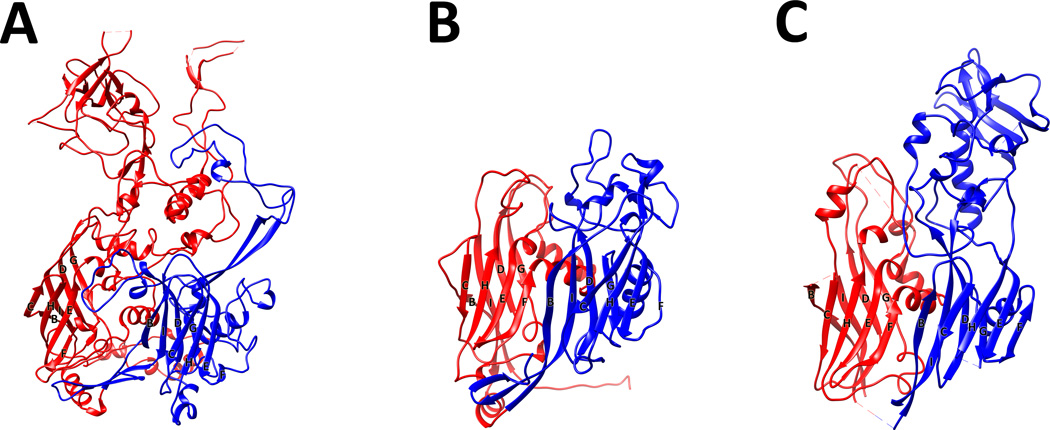
The two common motifs associated with capsid proteins are the HK97 fold (Wikoff et al., 2000; Helgstrand et al., 2003) and the jelly-roll fold (Rossmann and Johnson, 1989). The HK97 fold is exclusively utilized by tailed bacteriophages, with the exception of herpesvirus (Baker et al., 2005), whereas all NCLDVs have capsid proteins composed of a jelly-roll fold. The jelly-roll fold was first described in small RNA plant (Harrison et al., 1978; Abad-Zapatero et al., 1980) and animal viruses (Hogle et al., 1985; Rossmann et al., 1985) and is composed of a wedge-shaped, eight-stranded, anti-parallel, β-barrel. If the β-strands along the capsid protein polypeptide chain are identified as A, B, C, D, E, F, G, H and I, then the two β-sheets in this fold are composed of the strands BIDG and CHEF (Figure 2).
Figure 2.
Structures of double jelly-roll proteins in the NCLDV clade in comparison with the Adenovirus hexon protein. Cartoon representation of the (A) Adenovirus hexon protein (PDB:1P30), (B) PBCV-1 major capsid protein (PDB:1M3Y) and the (C) vaccinia virus scaffolding protein D13 (PDB:3SAQ). The N-terminal jelly-roll is colored red and the C-terminal is colored in blue. The β-strands at the base of the proteins are labeled from B to I. Amino acids that are missing in the structure are represented by dashed lines. Figure was produced with CHIMERA (Pettersen et al., 2004).
The hexameric positions in capsids of the NCLDVs are occupied by trimers in which each monomer is formed by the consecutive arrangement of two jelly-roll folds, producing an overall hexagonal shape. The pentameric positions are occupied by five copies of a separate protein that is composed of a single jelly-roll motif and assembled into a pentamer (Figure 1).
The core structure of the double jelly-roll protein in all NCLDVs is conserved and forms the base of the capsid protein with a height of 75Å and a diameter of 75Å to 85Å. A variable part of this protein is found on the surface and is formed by insertions between β-strands DE and FG. Probably the largest insertions that occur in a double jelly-roll protein are the towers in the adenovirus major capsid protein (MCP) (Figure 2).
Almost all large dsDNA viruses package their capsomers in a hexagonal close arrangement with planar p6 symmetry of the capsomers. The number of single jelly-rolls can be calculated by multiplying the T number of the capsid by 60. However, there are two known exceptions to this rule. One is the structure of the Poxvirus scaffolding protein D13 (Bahar et al., 2011; Hyun et al., 2011) (Figure 1). Although this protein is not part of the mature virion, it forms a hexagonal array around the viral genome during assembly. This hexagonal array has a hole at every third position in the array and is therefore equivalent to a p3 planar arrangement. Second, the 65Å resolution structure of Mimivirus implies that the same is also true for this virus (Xiao et al., 2009). Because of this packaging arrangement, the number of jelly-rolls in Mimivirus is lower than expected and is equal to 2/3*T*60 (Figure 1). Because of the limited resolution it is currently impossible to exactly determine the T number of Mimivirus, but it is likely to be between 972 and 1200 (h=19±1, k=19±1).
Although the T number established by Caspar and Klug describes the arrangement of capsomers in small capsids, it can be difficult to describe the structure of large viruses using this concept. As first observed by Wrigley in negative stain images of Sericesthis iridescent virus, most if not all large viruses possess ordered substructures in their capsid (Wrigley, 1969). These sub-structures were observed by Wrigley in virus preparations as degradation products after prolonged storage (Wrigley, 1969). They can also be observed in cryo-EM reconstructions of these viruses (Yan et al., 2000; 2005; 2009). These sub-structures, which are also potential building blocks, are centered on the threefold and fivefold axes and are termed trisymmetrons and pentasymmetrons, respectively. All NCLDVs described to date can be split into 20 trisymmetrons and 12 pentasymmetrons (Yan et al., 2005). Remarkably, the size of the pentasymmetrons in Paramecium bursaria Chlorella virus-1 (PBCV-1), Chilo iridescent virus (CIV) and Phaeocystis pouchetti virus (PpV01) is exactly the same and is comprised of 30 trimers and 1 pentamer, implying that this is the ideal size to relieve the strain induced by forming a large capsid. The number of trimers in the trisymmetron varies from 91 in PpV01 (Yan et al., 2005), to 66 in PBCV-1 (Nandhagopal et al., 2002; Zhang et al., 2011) and 55 in CIV (Yan et al., 2009).
Minor capsid proteins
High resolution structures of CIV (Yan et al., 2009) and PBCV-1 (Zhang et al., 2011) showed that these viruses have a variety of minor capsid proteins. These proteins can be found underneath the major capsid protein layer and can be broadly classified into two categories: (1) support of the viral capsid shell and connectivity at the “seam” of neighboring trisymmetrons and (2) connection between the outer shell and the inner membrane surrounding the viral genome.
Some of the minor proteins described in CIV or PBCV-1 seem to act as a scaffold to connect capsomers within a trisymmetron and between neighboring trisymmetrons (Cherrier et al., 2009; Yan et al., 2009; Zhang et al., 2011). Some of these minor proteins are missing from the unique vertex of PBCV-1 (Zhang et al., 2011), which may help the virus to partially disassemble upon binding to the host cell wall. This supports the hypothesis that these proteins are necessary for stability. In CIV no such differential distribution has been observed, probably because no unique vertex has been identified in this virus. Another possible function of the proteins that interconnect capsomers could be the equivalent of a “tape measure” protein that regulates the size and shape of the virus. This has been shown in a variant of African swine fever virus (ASFV) with a mutation in a minor capsid proteins (pB438L), which forms filamentous instead of icosahedral particles (Epifano et al., 2006).
Apart from acting as a scaffold underneath the main capsid layer, the minor proteins also seem to hold the membrane surrounding the viral genome in place. Indeed, the membrane in PBCV-1 follows the icosahedral curvature of the virus when it is in a preinfectious state, but becomes round and detaches from the viral shell upon infection (Zhang et al., 2011).
Nucleocapsid/internal membrane
A feature shared by all NCLDVs described to date is an internal membrane sac surrounding and protecting the viral DNA. For some viruses it has been shown that the internal membrane plays an essential role during infection and two examples are described below.
During the infection of Mimivirus, the virus opens at a unique vertex (described later in more detail). The internal membrane then fuses with the phagosomal membrane of the host. Subsequently the viral genome is released into the cell (Suzan-Monti et al., 2007; Zauberman et al., 2008; Mutsafi et al., 2010). Factors required for the fusion between the internal membrane and the phagosomal membrane remain to be elucidated.
Although there is no direct evidence, several experiments indicate that a similar mechanism is employed by PBCV-1. After digestion of the algae cell wall, the internal pressure of the host cell leads to a close contact between the viral and algae membrane (Kuznetsov et al., 2005; Thiel et al., 2010; Zhang et al., 2011). The exact mechanism for how the membranes fuse is also unknown in this case.
Mimivirus as well as PBCV-1 shows a depression of the internal membrane sac opposite of the unique vertex, thereby creating an empty pocket in cryo-EM reconstructions. However, it is rather unlikely that this pocket is purely water. Its components are probably not ordered and can therefore not be visualized when enforcing symmetry during the cryo-EM reconstruction process. Atomic force microscopy (AFM) studies of broken Mimivirus particles showed several long fibers with a diameter of about 60Å and 70Å unit repeats along the fibers (Xiao et al., 2009; Kuznetsov et al., 2010; Kuznetsov and McPherson, 2011). Possibly these fibers support the viral membrane. No internal fibers have been described for PBCV-1.
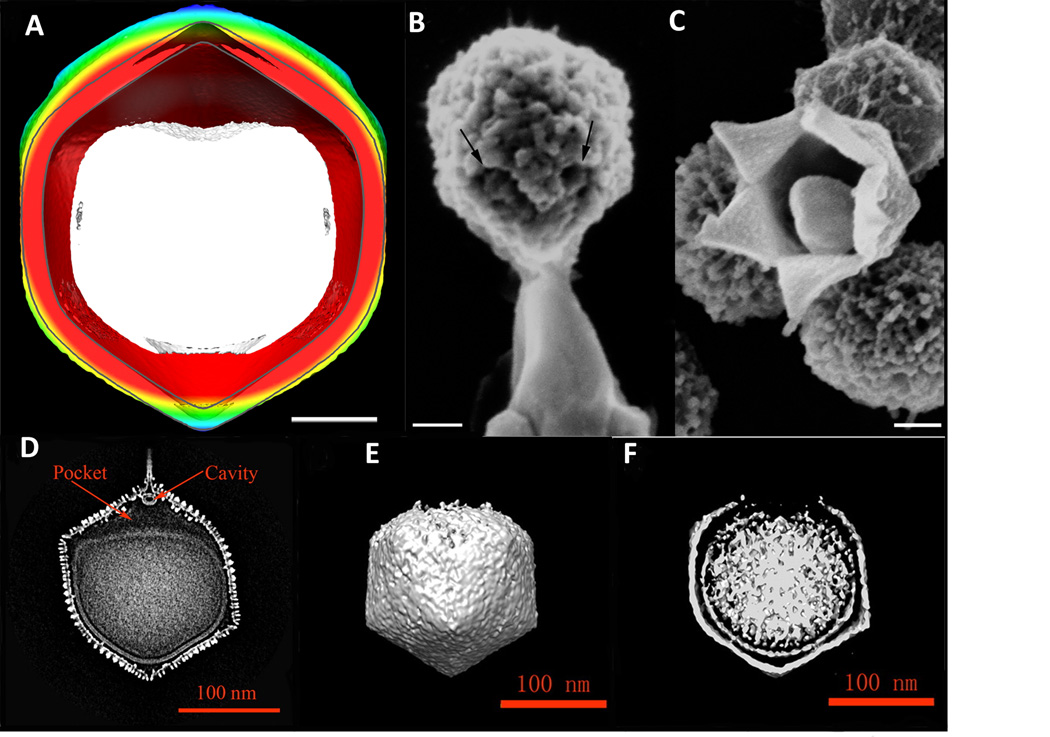
Both Mimivirus and PBCV-1 have a unique vertex where the membrane becomes separated from the capsid (Figure 3). On the other hand viruses that lack a unique vertex, such as CIV, ASFV and PpV01, lack a space between the viral membrane and the outer capsid.
Figure 3.
The unique vertex of Mimivirus and PBCV-1. (A) Central section of Mimivirus. The internal membrane sac is shown in white and the outer shell is colored radially, with red for a small diameter and going through yellow, green and blue for increasing diameter (Adapted from (Xiao et al., 2009)). (B) SEM images of a Mimivirus particle releasing a membranous structure after heating. (C) Mimivirus particle with an opening at one of the fivefold vertices (B and C adapted from (Zauberman et al., 2008)). Scale bars 100 nm. (D) Central section of PBCV-1 showing the unique vertex and the empty pocket between the internal membrane sac and spike assembly. (E) and (F) External view and central section of PBCV-1 after its attachment to cell walls of C. variabilis NC64A. The virus has partially disassembled around the unique vertex and the internal membrane is more rounded compared to the preinfection state shown in (D) (D–F adapted from (Zhang et al., 2011)).
Outer decorations/fibers
Many giant viruses have decoration proteins associated with the viral capsid as, for example, surface fibers.
Mimivirus has a dense layer of fibers that covers the complete virion, with the exception of the special “stargate” vertex (see below) (Xiao et al., 2005; Zauberman et al., 2008; Xiao et al., 2009; Kuznetsov et al., 2010). It is currently not known what role the fibers play in the viral life cycle. However, the viral fibers can be stained Gram-positive and, therefore, mimic the chemical composition of the outer shell of bacteria (Raoult et al., 2007). It has been hypothesized that these properties increase the chance of Mimivirus-like bacteria to be ingested by amoebal hosts. Apart from the actual fiber proteins, additional proteins have been identified on the outside of Mimivirus, namely R135, L725 and L829 (Boyer et al., 2011). R135 has been broadly classified as a glucose-methanol-cholesterol oxidoreductase (Renesto et al., 2006), whereas the remaining two proteins have no known homologs. R135 might also function as the “receptor” of Sputnik, the virophage that coinfects with Mimivirus and there is some evidence that it attaches to the fibers of Mimivirus with the help of R135 (La Scola et al., 2008; Boyer et al., 2011).
Analysis of the Pandoravirus genome does not suggest a homolog to the NCLDV major capsid protein (Philippe et al., 2013). Instead, its oval-shaped outer shell seems to be composed of a homolog to L829 in Mimivirus. As mentioned above, L829 has no known homologs and its structure is unknown.
PBCV-1 does not have a dense layer of fibers around the capsid, but cryo-EM reconstructions and AFM studies have shown that the capsid is decorated with some small fibers (Kuznetsov et al., 2005; Cherrier et al., 2009; Zhang et al., 2011). A fivefold averaged map of the PBCV-1 virion shows that one specific capsomer in each trisymmetron forms the base of a fiber that extends 60Å, then bends at a 90° angle and extends for at least another 130Å. Comparison of this specific capsomer with neighboring capsomers suggests that the protein sitting at the base of the fiber is not identical to the major capsid protein VP54, but likely a homologue with high sequence similarity (Zhang et al., 2011). Although the role of the fibers is not clear, it is likely that the fibers attach to the cell wall of algae during infection. This event is possibly comparable to the initial attachment utilized by several bacteriophages such as T4 (Leiman et al., 2010).
Other members of the NCLDVs also possess fibers. CIV has a 40nm long fiber of unknown identity tightly associated with every capsomer (Yan et al., 2009) and PbV01 has 60 fibers, three per icosahedral facet, that are associated with a unique capsomer (Yan et al., 2005). The exact function of these fibers is unknown, but it is possible that these help the virus during attachment or carry some kind of receptor recognition site(s).
Unique vertices
Studies by cryo-EM or AFM of some giant viruses revealed the presence of a unique fivefold vertex, which plays an important role during infection. The unique vertex in Mimivirus has been termed stargate or starfish, referring to its appearance (Xiao et al., 2005; Zauberman et al., 2008) (Figure 3). As described above, this unique structure is the only part of the virus that is not covered by fibers. Instead it is elevated in comparison to the underlying capsid shell, but seems to be tightly connected to the adjacent capsomers. The seam between the faces at the unique vertex is covered with a protein or protein complex that holds the stargate closed and probably needs to be removed before infection can occur (Kuznetsov et al., 2010). The stargate therefore has to strike the fine balance between a stable structure that is able to protect the genome from an early and unproductive release and the exact timing when the release needs to be triggered. The stargate is also the site of assembly, as has recently been demonstrated by AFM and EM studies (Kuznetsov et al., 2013; Mutsafi et al., 2013). Virus assembly nucleates at the tip of the stargate and the arms are assembled from this initial point. Subsequently the space between the arms and points defining the remaining fivefold vertices of the virus are filled with capsid protein. This order of events makes sense, as it would probably be almost impossible to close an open stargate after packaging the viral genome in a systematic fashion.
The second virus with a well-characterized unique vertex is PBCV-1. The virus has a 34nm-long spike sitting at a fivefold axis that is responsible for binding and attachment. It has been shown that A140/145R is the protein placed on the top of the spike. Parts of this protein are responsible for recognizing sugars on the cell wall of algae. Variations in the sequence of A140/145R and its equivalent proteins in different Phycodnaviridae can be correlated with host specificity (Onimatsu et al., 2004; 2006). A small compartment has been identified below the spike that sits within the capsid shell and likely contains enzymes to digest the cell wall during infection (Zhang et al., 2011) (Figure 3). Additionally, as described above, minor capsid proteins that are distributed throughout the virion and probably aid in stabilizing the capsid shell are missing underneath the unique fivefold axis. The membrane sac containing the viral genome does not extend into this vertex. Upon infection the virus shell disassembles directly underneath the spike and the membrane containing the viral genome is delivered into the host cell. PBCV-1 has therefore evolved a localized weakness in its capsid shell, which enhances infection. Whether this unique vertex is also the point of assembly, as is the case in Mimivirus, remains to be determined.
Outlook
Although our knowledge of the structure and life cycle of NCLDVs has improved significantly during recent years, there are still many remaining questions. What controls the size of the viral capsids and what role do minor capsid proteins play in assembly and stability? Are the internal viral membranes held in place by the capsid protein or does the virus assemble around the membrane with the minor capsid proteins acting as assembly points? How did the viral shell evolve? This is a question that is of special interest in combination with the recent discovery that Pandoravirus does not encode a double jelly-roll fold protein. With the help of future structural investigations and the improvement in cryo-EM techniques it should be possible to answer these question and shed more light on the evolution of these large viruses.
Acknowledgments
The authors are grateful for financial support from the National Institutes of Health (AI011219 to MGR).
References
- Abad-Zapatero C, Abdel-Meguid SS, Johnson JE, Leslie AG, Rayment I, Rossmann MG, Suck D, Tsukihara T. Structure of southern bean mosaic virus at 2.8 A resolution. Nature. 1980;286:33–39. doi: 10.1038/286033a0. [DOI] [PubMed] [Google Scholar]
- Aherfi S, Pagnier I, Fournous G, Raoult D, La Scola B, Colson P. Complete genome sequence of Cannes 8 virus, a new member of the proposed family “Marseilleviridae”. Virus Genes. 2013 doi: 10.1007/s11262-013-0965-4. [DOI] [PubMed] [Google Scholar]
- Bahar MW, Graham SC, Stuart DI, Grimes JM. Insights into the evolution of a complex virus from the crystal structure of vaccinia virus d13. Structure. 2011;19:1011–1020. doi: 10.1016/j.str.2011.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker ML, Jiang W, Rixon FJ, Chiu W. Common ancestry of herpesviruses and tailed DNA bacteriophages. J Virol. 2005;79:14967–14970. doi: 10.1128/JVI.79.23.14967-14970.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyer M, Azza S, Barrassi L, Klose T, Campocasso A, Pagnier I, Fournous G, Borg A, Robert C, Zhang X, et al. Mimivirus shows dramatic genome reduction after intraamoebal culture. Proc Natl Acad Sci USA. 2011;108:10296–10301. doi: 10.1073/pnas.1101118108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyer M, Yutin N, Pagnier I, Barrassi L, Fournous G, Espinosa L, Robert C, Azza S, Sun S, Rossmann MG, et al. Giant Marseillevirus highlights the role of amoebae as a melting pot in emergence of chimeric microorganisms. Proc Natl Acad Sci USA. 2009;106:21848–21853. doi: 10.1073/pnas.0911354106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caspar D, Klug A. Physical principles in the construction of regular viruses. Cold Spring Harb Symp Quant Biol. 1961;27:1–24. doi: 10.1101/sqb.1962.027.001.005. [DOI] [PubMed] [Google Scholar]
- Cherrier MV, Kostyuchenko VA, Xiao C, Bowman VD, Battisti AJ, Yan X, Chipman PR, Baker TS, van Etten JL, Rossmann MG. An icosahedral algal virus has a complex unique vertex decorated by a spike. Proc Natl Acad Sci USA. 2009;106:11085–11089. doi: 10.1073/pnas.0904716106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claverie J-M, Abergel C. Mimivirus and its virophage. Annu. Rev. Genet. 2009;43:49–66. doi: 10.1146/annurev-genet-102108-134255. [DOI] [PubMed] [Google Scholar]
- Claverie J-M, Abergel C. Mimivirus: the emerging paradox of quasi-autonomous viruses. Trends Genet. 2010;26:431–437. doi: 10.1016/j.tig.2010.07.003. [DOI] [PubMed] [Google Scholar]
- Clouthier SC, Vanwalleghem E, Copeland S, Klassen C, Hobbs G, Nielsen O, Anderson ED. A new species of nucleo-cytoplasmic large DNA virus (NCLDV) associated with mortalities in Manitoba lake sturgeon Acipenser fulvescens. Dis. Aquat. Org. 2013;102:195–209. doi: 10.3354/dao02548. [DOI] [PubMed] [Google Scholar]
- Colson P, De Lamballerie X, Yutin N, Asgari S, Bigot Y, Bideshi DK, Cheng X-W, Federici BA, Van Etten JL, Koonin EV, et al. “Megavirales,” a proposed new order for eukaryotic nucleocytoplasmic large DNA viruses. Arch. Virol. 2013a;158:2517–2521. doi: 10.1007/s00705-013-1768-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colson P, Pagnier I, Yoosuf N, Fournous G, La Scola B, Raoult D. “Marseilleviridae,” a new family of giant viruses infecting amoebae. Arch. Virol. 2013b;158:915–920. doi: 10.1007/s00705-012-1537-y. [DOI] [PubMed] [Google Scholar]
- Crick FH, Watson JD. Structure of small viruses. Nature. 1956;177:473–475. doi: 10.1038/177473a0. [DOI] [PubMed] [Google Scholar]
- Desnues C, Raoult D. Virophages question the existence of satellites. Nat Rev Microbiol. 2012;10:234. doi: 10.1038/nrmicro2676-c3. [DOI] [PubMed] [Google Scholar]
- Epifano C, Krijnse-Locker J, Salas ML, Salas J, Rodríguez JM. Generation of filamentous instead of icosahedral particles by repression of African swine fever virus structural protein pB438L. J Virol. 2006;80:11456–11466. doi: 10.1128/JVI.01468-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filée J, Pouget N, Chandler M. Phylogenetic evidence for extensive lateral acquisition of cellular genes by Nucleocytoplasmic large DNA viruses. BMC Evol Biol. 2008;8:320. doi: 10.1186/1471-2148-8-320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer MG, Allen MJ, Wilson WH, Suttle CA. Giant virus with a remarkable complement of genes infects marine zooplankton. Proc Natl Acad Sci USA. 2010;107:19508–19513. doi: 10.1073/pnas.1007615107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghedin E, Claverie J-M. Mimivirus relatives in the Sargasso sea. Virol J. 2005;2:62. doi: 10.1186/1743-422X-2-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison SC, Olson AJ, Schutt CE, Winkler FK, Bricogne G. Tomato bushy stunt virus at 2.9 Å resolution. Nature. 1978;276:368–373. doi: 10.1038/276368a0. [DOI] [PubMed] [Google Scholar]
- Helgstrand C, Wikoff WR, Duda RL, Hendrix RW, Johnson JE, Liljas L. The refined structure of a protein catenane: the HK97 bacteriophage capsid at 3.44 A resolution. J Mol Biol. 2003;334:885–899. doi: 10.1016/j.jmb.2003.09.035. [DOI] [PubMed] [Google Scholar]
- Hogle JM, Chow M, Filman DJ. Three-dimensional structure of poliovirus at 2.9 A resolution. Science. 1985;229:1358–1365. doi: 10.1126/science.2994218. [DOI] [PubMed] [Google Scholar]
- Hyun J-K, Accurso C, Hijnen M, Schult P, Pettikiriarachchi A, Mitra AK, Coulibaly F. Membrane remodeling by the double-barrel scaffolding protein of poxvirus. PLoS Pathog. 2011;7:e1002239. doi: 10.1371/journal.ppat.1002239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer LM, Aravind L, Koonin EV. Common origin of four diverse families of large eukaryotic DNA viruses. J Virol. 2001;75:11720–11734. doi: 10.1128/JVI.75.23.11720-11734.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krupovic M, Cvirkaite-Krupovic V. Virophages or satellite viruses? Nat Rev Microbiol. 2011;9:762–763. doi: 10.1038/nrmicro2676. [DOI] [PubMed] [Google Scholar]
- Kuznetsov YG, McPherson A. Nano-fibers produced by viral infection of amoeba visualized by atomic force microscopy. Biopolymers. 2011;95:234–239. doi: 10.1002/bip.21563. [DOI] [PubMed] [Google Scholar]
- Kuznetsov YG, Gurnon JR, van Etten JL, McPherson A. Atomic force microscopy investigation of a chlorella virus, PBCV-1. J Struct Biol. 2005;149:256–263. doi: 10.1016/j.jsb.2004.10.007. [DOI] [PubMed] [Google Scholar]
- Kuznetsov YG, Klose T, Rossmann M, McPherson A. Morphogenesis of mimivirus and its viral factories: an atomic force microscopy study of infected cells. J Virol. 2013;87:11200–11213. doi: 10.1128/JVI.01372-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuznetsov YG, Xiao C, Sun S, Raoult D, Rossmann M, McPherson A. Atomic force microscopy investigation of the giant mimivirus. Virology. 2010;404:127–137. doi: 10.1016/j.virol.2010.05.007. [DOI] [PubMed] [Google Scholar]
- La Scola B, Audic S, Robert C, Jungang L, de Lamballerie X, Drancourt M, Birtles R, Claverie J-M, Raoult D. A giant virus in amoebae. Science. 2003;299:2033. doi: 10.1126/science.1081867. [DOI] [PubMed] [Google Scholar]
- La Scola B, Desnues C, Pagnier I, Robert C, Barrassi L, Fournous G, Merchat M, Suzan-Monti M, Forterre P, Koonin E, et al. The virophage as a unique parasite of the giant mimivirus. Nature. 2008;455:100–104. doi: 10.1038/nature07218. [DOI] [PubMed] [Google Scholar]
- Leiman PG, Arisaka F, van Raaij MJ, Kostyuchenko VA, Aksyuk AA, Kanamaru S, Rossmann MG. Morphogenesis of the T4 tail and tail fibers. Virol J. 2010;7:355. doi: 10.1186/1743-422X-7-355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreira D, Brochier-Armanet C. Giant viruses, giant chimeras: the multiple evolutionary histories of Mimivirus genes. BMC Evol Biol. 2008;8:12. doi: 10.1186/1471-2148-8-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mutsafi Y, Shimoni E, Shimon A, Minsky A. Membrane Assembly during the Infection Cycle of the Giant Mimivirus. PLoS Pathog. 2013;9:e1003367. doi: 10.1371/journal.ppat.1003367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mutsafi Y, Zauberman N, Sabanay I, Minsky A. Vaccinia-like cytoplasmic replication of the giant Mimivirus. Proc Natl Acad Sci USA. 2010;107:5978–5982. doi: 10.1073/pnas.0912737107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nandhagopal N, Simpson AA, Gurnon JR, Yan X, Baker TS, Graves MV, van Etten JL, Rossmann MG. The structure and evolution of the major capsid protein of a large, lipid-containing DNA virus. Proc Natl Acad Sci USA. 2002;99:14758–14763. doi: 10.1073/pnas.232580699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onimatsu H, Suganuma K, Uenoyama S, Yamada T. C-terminal repetitive motifs in Vp130 present at the unique vertex of the Chlorovirus capsid are essential for binding to the host Chlorella cell wall. Virology. 2006;353:433–442. doi: 10.1016/j.virol.2006.06.010. [DOI] [PubMed] [Google Scholar]
- Onimatsu H, Sugimoto I, Fujie M, Usami S, Yamada T. Vp130, a chloroviral surface protein that interacts with the host Chlorella cell wall. Virology. 2004;319:71–80. doi: 10.1016/j.virol.2003.10.030. [DOI] [PubMed] [Google Scholar]
- Pagnier I, Reteno D-GI, Saadi H, Boughalmi M, Gaia M, Slimani M, Ngounga T, Bekliz M, Colson P, Raoult D, et al. A decade of improvements in mimiviridae and marseilleviridae isolation from amoeba. Intervirology. 2013;56:354–363. doi: 10.1159/000354556. [DOI] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera--a visualization system for exploratory research and analysis. Journal of Computational Chemistry. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- Philippe N, Legendre M, Doutre G, Couté Y, Poirot O, Lescot M, Arslan D, Seltzer V, Bertaux L, Bruley C, et al. Pandoraviruses: amoeba viruses with genomes up to 2.5 Mb reaching that of parasitic eukaryotes. Science. 2013;341:281–286. doi: 10.1126/science.1239181. [DOI] [PubMed] [Google Scholar]
- Raoult D. The 1.2-Megabase Genome Sequence of Mimivirus. Science. 2004;306:1344–1350. doi: 10.1126/science.1101485. [DOI] [PubMed] [Google Scholar]
- Raoult D, La Scola B, Birtles R. The discovery and characterization of Mimivirus, the largest known virus and putative pneumonia agent. Clin. Infect. Dis. 2007;45:95–102. doi: 10.1086/518608. [DOI] [PubMed] [Google Scholar]
- Renesto P, Abergel C, Decloquement P, Moinier D, Azza S, Ogata H, Fourquet P, Gorvel J-P, Claverie J-M. Mimivirus giant particles incorporate a large fraction of anonymous and unique gene products. J Virol. 2006;80:11678–11685. doi: 10.1128/JVI.00940-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossmann MG, Johnson JE. Icosahedral RNA virus structure. Annu Rev Biochem. 1989;58:533–573. doi: 10.1146/annurev.bi.58.070189.002533. [DOI] [PubMed] [Google Scholar]
- Rossmann MG, Arnold E, Erickson JW, Frankenberger EA, Griffith JP, Hecht HJ, Johnson JE, Kamer G, Luo M, Mosser AG. Structure of a human common cold virus and functional relationship to other picornaviruses. Nature. 1985;317:145–153. doi: 10.1038/317145a0. [DOI] [PubMed] [Google Scholar]
- Su CJ, Baseman JB. Genome size of Mycoplasma genitalium. J Bacteriol. 1990;172:4705–4707. doi: 10.1128/jb.172.8.4705-4707.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzan-Monti M, La Scola B, Barrassi L, Espinosa L, Raoult D. Ultrastructural characterization of the giant volcano-like virus factory of Acanthamoeba polyphaga Mimivirus. PLoS ONE. 2007;2:e328. doi: 10.1371/journal.pone.0000328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiel G, Moroni A, Dunigan DD, van Etten JL. Initial Events Associated with Virus PBCV-1 Infection of Chlorella NC64A. Prog Bot. 2010;71:169–183. doi: 10.1007/978-3-642-02167-1_7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas V, Bertelli C, Collyn F, Casson N, Telenti A, Goesmann A, Croxatto A, Greub G. Lausannevirus, a giant amoebal virus encoding histone doublets. Environ Microbiol. 2011 doi: 10.1111/j.1462-2920.2011.02446.x. [DOI] [PubMed] [Google Scholar]
- Wikoff WR, Liljas L, Duda RL, Tsuruta H, Hendrix RW, Johnson JE. Topologically linked protein rings in the bacteriophage HK97 capsid. Science. 2000;289:2129–2133. doi: 10.1126/science.289.5487.2129. [DOI] [PubMed] [Google Scholar]
- Wrigley NG. An electron microscope study of the structure of Sericesthis iridescent virus. J Gen Virol. 1969;5:123–134. doi: 10.1099/0022-1317-5-1-123. [DOI] [PubMed] [Google Scholar]
- Xiao C, Chipman PR, Battisti AJ, Bowman VD, Renesto P, Raoult D, Rossmann MG. Cryo-electron microscopy of the giant Mimivirus. J Mol Biol. 2005;353:493–496. doi: 10.1016/j.jmb.2005.08.060. [DOI] [PubMed] [Google Scholar]
- Xiao C, Kuznetsov YG, Sun S, Hafenstein SL, Kostyuchenko VA, Chipman PR, Suzan-Monti M, Raoult D, McPherson A, Rossmann MG. Structural studies of the giant mimivirus. PLoS Biol. 2009;7:e92. doi: 10.1371/journal.pbio.1000092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan X, Olson NH, van Etten JL, Bergoin M, Rossmann MG, Baker TS. Structure and assembly of large lipid-containing dsDNA viruses. Nat Struct Biol. 2000;7:101–103. doi: 10.1038/72360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan X, Chipman PR, Castberg T, Bratbak G, Baker TS. The marine algal virus PpV01 has an icosahedral capsid with T=219 quasisymmetry. J Virol. 2005;79:9236–9243. doi: 10.1128/JVI.79.14.9236-9243.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan X, Yu Z, Zhang P, Battisti AJ, Holdaway HA, Chipman PR, Bajaj C, Bergoin M, Rossmann MG, Baker TS. The capsid proteins of a large, icosahedral dsDNA virus. J Mol Biol. 2009;385:1287–1299. doi: 10.1016/j.jmb.2008.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zauberman N, Mutsafi Y, Halevy DB, Shimoni E, Klein E, Xiao C, Sun S, Minsky A. Distinct DNA exit and packaging portals in the virus Acanthamoeba polyphaga mimivirus. PLoS Biol. 2008;6:e114. doi: 10.1371/journal.pbio.0060114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Xiang Y, Dunigan DD, Klose T, Chipman PR, van Etten JL, Rossmann MG. Three-dimensional structure and function of the Paramecium bursaria chlorella virus capsid. Proc Natl Acad Sci USA. 2011;108:14837–14842. doi: 10.1073/pnas.1107847108. [DOI] [PMC free article] [PubMed] [Google Scholar]