Abstract
BRAF mutations are frequent in cutaneous melanomas and BRAF inhibitors(BRAFi) have shown remarkable clinical efficacy in BRAF mutant melanoma patients. However, acquired drug resistance can occur rapidly and tumor(s) often progress thereafter. Various mechanisms of BRAFi resistance have recently been described; however, the mechanism of resistance remains controversial. In this study we developed BRAFi resistant melanoma cell lines and found that metastasis related EMT properties of BRAFi resistant cells were enhanced significantly. Upregulation of EGFR was observed in BRAFi resistant cell lines and patient tumors due to demethylation of EGFR regulatory DNA elements. EGFR induced PI3K/AKT pathway activation in BRAFi resistant cells through epigenetic regulation. Treatment of EGFR inhibitor was effective in BRAFi resistant melanoma cell lines. The study demonstrates that EGFR epigenetic activation has important implications in BRAFi resistance in melanoma.
Keywords: melanoma, BRAF mutation, EGFR, metastasis, BRAF inhibitor, epigenetics
INTRODUCTION
Despite recent major advances in therapies, the prognosis for melanoma patients with advanced stage disease remains very poor (Gopal et al., 2010; Siegel et al., 2012). Approximately 50% of melanomas harbor an activating BRAF V600 mutation(BRAFmt) (Davies et al., 2002; Jakob et al., 2012; Long et al., 2011), an oncogene known to have an important role in the proliferation and progression of melanoma cells through activation of the RAF/MEK/ERK cell signaling pathway (Gopal et al., 2010). FDA approved BRAF inhibitors (BRAFi) such as vemurafenib (PLX4032, Roche) and recently dabrafenib (GSK2118436, GlaxoSmithKline) were used to inhibit the RAF/MEK/ERK signaling pathway by targeting BRAFmt (Flaherty et al., 2012; Lito et al., 2012). Clinical studies have shown that stage IV BRAFmt melanoma patients respond rapidly to BRAFi therapies with high response rates (Chapman et al., 2011; Falchook et al., 2012; Flaherty et al., 2010; Hauschild et al., 2012; Long et al., 2012). However, a majority of patients eventually develop resistance.
Recently, studies have attempted to identify mechanisms and changes associated with BRAFi resistance. Studies have shown that other signal transduction pathways, such as the IGF1R/PI3K/AKT pathway (Villanueva et al., 2010) and MAPK pathway (Johannessen et al., 2010), were hyperactivated when the RAS/RAF/MAPK/ERK pathway was blocked by BRAFi. Studies have also demonstrated that overexpression of proteins such as CRAF, N-RAS, cyclin D1, FGF Receptor 3, EGFR, and platelet-derived growth factor receptor β (PDGFR β) contribute to melanoma BRAFi resistance (Girotti et al., 2013; Montagut et al., 2008; Nazarian et al., 2010; Shi et al., 2011; Smalley et al., 2008; Yadav et al., 2012). Mutation of PTEN (Paraiso et al., 2011) and gatekeeper residue in BRAF (Whittaker et al., 2010) have also been associated with acquired BRAFi resistance in melanoma. An increase in RTK-ligand levels, through autocrine tumor-cell production, paracrine contribution from tumor stroma or systemic sources, could confer resistance to BRAFi (Wilson et al., 2012). BRAFi resistances have been reported to be developed through NRAS mutations (Nazarian et al., 2010), BRAFV600E mutation amplification (Shi et al., 2012) and BRAFV600E alternative splice variants (Poulikakos et al., 2011). However, the acquired mechanism(s) of BRAFi resistance remains elusive in melanoma patients.
In this study, we found that EGFR expression was significantly upregulated in BRAFi resistant melanoma cells and tumor tissues, and that EGFR/PI3K/AKT pathway is hyperactivated in BRAFi resistant melanoma cells. Our results suggested that in melanoma, BRAFi resistance is caused by enhanced EGFR expression through epigenetic regulation, and that activation of the EGFR/PI3K/AKT pathway contributes to melanoma progression.
RESULTS
Epithelial to mesenchymal transition characterizes melanoma resistance to BRAFi
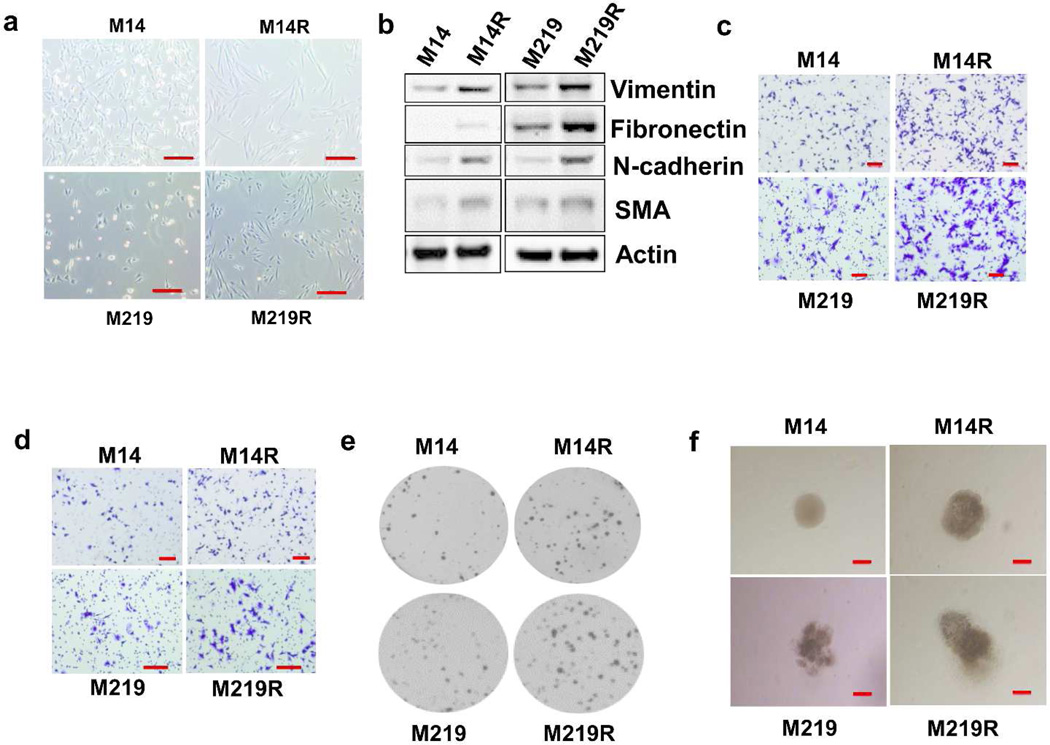
Melanoma, a tumor of neuroectodermal origin, can display epithelial to mesenchymal transition (EMT), which is a complex process associated with invasion and metastasis of various types of solid tumors (Craene and Berx, 2013; Kudo-Saito et al., 2009; Weiss et al., 2012). We developed several BRAFi melanoma cells (see Methods and Materials) and compared their phenotype to respective parental cells. BRAFi resistant melanoma cells were more elongated and fibroblast-like in shape than respective parental cells, and resembled mesenchymal cells (Figure 1A, Supplementary Figure 1). To determine whether expression of known mesenchymal protein markers was changed, we performed immunoblotting on paired cells. The expression of vimentin, fibronectin, α-smooth muscle actin (SMA) and N-cadherin was upregulated in BRAFi resistant cells (Figure 1B). This confirmed that development of BRAFi resistance is associated with an EMT phenotype.
Figure 1. EMT of BRAFi resistant cells. Proliferation, migration and invasion of BRAFi resistant cells.
A. BRAFi resistant melanoma cells (M14R and M219R) were more elongated and fibroblast-like in shape compared to respective parental cells (M14 and M219). Scale bar =100 µm. B. Immunoblots of EMT protein markers (vimentin, fibronectin, N-cadherin and alpha-smooth actin [SMA]). C. Migration of melanoma parental cells (M14 and M219) and BRAFi resistant cells (M14R and M219R). Scale bar = 100 µm. D. Invasion of parental cells and BRAFi resistant cells. Scale bar = 100 µm. E. Cell growth in soft agar. F. Cell growth in 3D matrigel. Migration and invasion were higher in BRAFi resistant cells than in respective parental cells, and BRAF resistant cells grew faster than parental cells in soft agar and 3D matrigel. Scale bar = 100 µm.
Resistant cells have aggressive growth
We characterized BRAFi resistant melanoma cells and showed that migration and invasion were higher in BRAFi resistant cells than in respective parental cells (Figure 1C and D; Supplementary Figures 2, 3). Soft agar colony formation assay was proformed to investigate the cell's ability to grow unattached to a surface suspended in agar. The 3D matrigel assay was used to monitor invasive potential of cells. To compare growth and invasive potential of parental and resistant cells, we performed soft agar colony formation assay and 3D matrigel gel culture. Our results showed that BRAFi resistant cells grew faster than respective parental cells in soft agar and 3D matrigel culture (Figure 1E and F). Furthermore, quantification of colony formation was shown in Supplementary Figure 4A and morphology of M14 to M14R and M219 to M219R in 3D matrigel growth medium (days 1 and 12) was shown for comparison in Supplementary Figure 4B and Figure 1F, respectively. Analysis of cell growth also confirmed faster growth of BRAFi resistant cells than respective parental cells in 2D adherent cell culture (Supplementary Figure 5A, Supplementary Figure 5B). Combining together, this suggested that resistant cells have more aggressive growth than parental cells.
Cyclin D1 activity is related to cell cycle regulation and cell proliferation (Choi et al., 2012; Ray et al., 2010). Immunoblot analysis demonstrated that expression of cyclin D1 was approximately two fold higher in BRAFi resistant cells than in respective parental cells (Supplementary 5C and D). These results supported the aggressive growth properties of BRAFi resistant cells. Upregulated expression of cyclin D1 accelerates G(1)-phase progression in the cell cycle through the RAS/RAF/MEK/ERK pathway (Cai et al., 2013).
Tumorigenicity of parental versus BRAFi resistant melanoma xenografts
To assess the tumorigenic properties of parental versus BRAFi resistant cells in vivo, 1×106 cells from M219, and M219R cell lines were inoculated subdermally into the thigh of male nude-BALB/c mice (6 mice/cell line). BRAFi resistant cells had significantly higher tumorigenicity and faster growth than respective parental cells (Supplementary Figure 6). The xenograft study demonstrated the aggressive growth properties of BRAFi resistant cells (M219R) in vivo, supporting the respective analysis of the in vitro cell line studies (Supplementary Figure 5A). This in vivo xenograft analysis is one paired parental and resistant cell line.
EGFR expression is enhanced in BRAFi resistant melanoma cells
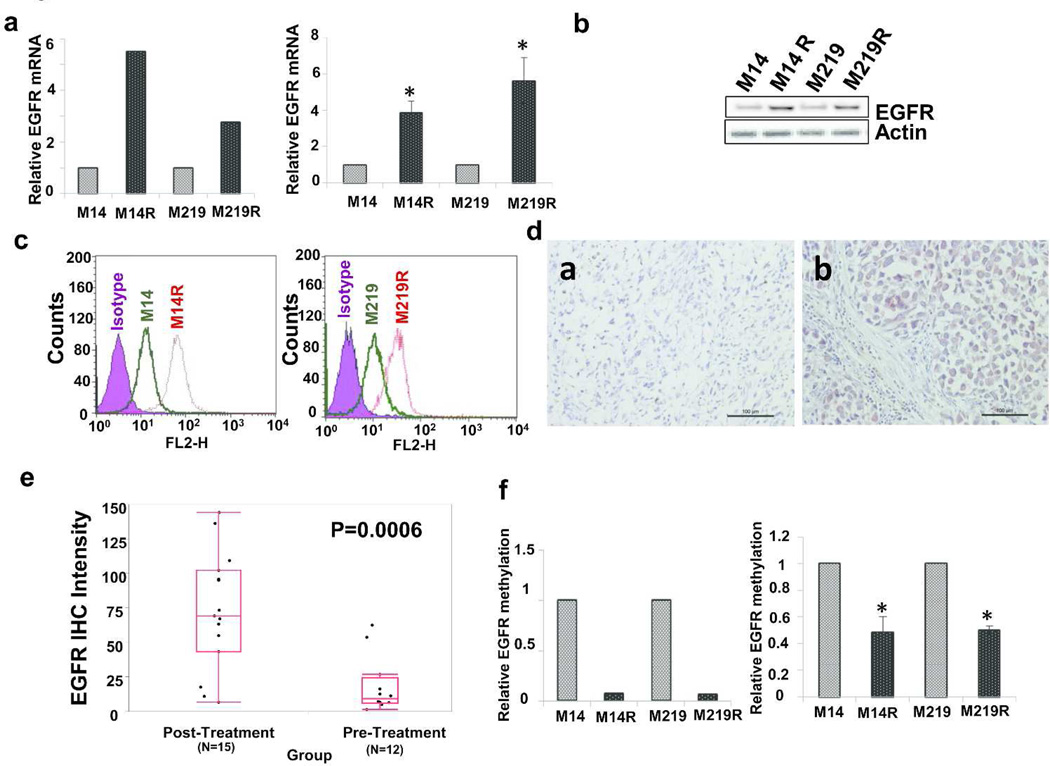
Gene expression profiling by gene expression microarray (Affymetrix array) analysis was performed on M14, M14R, M219, and M219R cell lines to identify major significant changes in BRAFi cells compared to respective parental cells. EGFR mRNA expression level was identified 5.51-fold higher in M14R than in M14 cells, and 2.76-fold higher in M219R than in M219 cells (Figure 2A, left). RNA-sequence analysis of total RNA from M14 and M14R cells verified higher(6.23-fold) RNA expression for EGFR in M14R cells than in M14 cells (Supplementary Figure 7). Quantitative RT-PCR analysis further confirmed higher expression of mRNA EGFR in M14R(3.85-fold higher) and M219R(5.62-fold higher) cells than in respective parental cells (Figure 2A, right).
Figure 2. EGFR expression in metastatic melanoma cells(panels A–C) and metastatic melanoma tissues(panels D–F).
A. EGFR mRNA expression in parental(M14 and M219) and BRAFi resistant(M14R and M219R) cells assessed by gene expression microarray(left) and qRT-PCR(right) analyses. B. Immunoblot of EGFR expression in cells. C. Flow cytometric analysis of EGFR expression in cells. D. Representative IHC stain of EGFR in a metastatic tumor specimen obtained before(a) and after(b) treatment with BRAFi. In picture (b), melanoma has higher EGFR expression than normal cells in tissue. Scale bar = 100 µm. E. Intensity of EGFR IHC staining in melanoma tumors obtained from patients before and after treatment with BRAFi (Three pre-treatment patients weren’t available). F. EGFR methylation array(left) and EGFR methylation-specific PCR(right). Error bars, s.d.(*p <0.05).
To determine whether protein expression of EGFR was enhanced in BRAFi resistant cell lines, we assessed EGFR expression by immunoblotting and flow cytometry. As shown in Figure 2B and 2C, EGFR expression was significantly higher in BRAFi resistant cells than in respective parental cells.
EGFR expression is enhanced in BRAFi resistant melanoma metastases
We assessed EGFR expression level by IHC in AJCC stage III and IV metastatic melanomas using a melanoma tissue microarray (TMA) that was annotated with long-term clinical follow-up data(non BRAFi treatment) (Nguyen et al., 2010). AJCC stage III and IV melanomas generally had low levels of EGFR expression independent of BRAFmt presence (Supplementary Figure 8).
To determine whether EGFR expression was enhanced in BRAFi resistant melanomas, we immunostained melanoma tumor specimens obtained from 15 patients before and after BRAFi treatment with dabrafenib (13 patients) or vemurafenib (2 patients) (Figure 2D, Supplementary Figure 9). Analysis of staining intensity showed that EGFR expression was higher in BRAFi resistant tumors (Figure 2E). To confirm these results, we compared EGFR expression in autologous pairs of BRAFmt melanoma specimens obtained from 12 different patients before and after treatment with BRAFi (Supplementary Figure 10). EGFR expression post-treatment was significantly higher in BRAFi resistant recurrent tumor.
EGFR gene hypomethylation in BRAF resistant cells
Since EGFR was demonstrated to be significantly activated in BRAFi resistant cells, we investigated its expression regulation. Methylation profiling with a HumanMethylation450 BeadChip analysis was carried out to determine if EGFR was epigenetically regulated by CpG site methylation status. The methylation level of the EGFR promoter region was 14.9-fold lower in M14R cells than in M14 cells, and 16.4-fold lower in M219R cells than in M219 cells (Figure 2F, left). Methylation-specific PCR confirmed lower EGFR methylation in M14R and M219R cells than in respective parental cells (Figure 2F, right). To verify whether hypermethylation of EGFR affects its expression, M14 and M219 cells were treated with a 5-Aza demethylation agent and assessed for EGFR protein expression by immunoblotting. EGFR expression was enhanced in cells treated by 5-Aza (Figure 3D, Supplementary Figure 11). These results supported that EGFR expression is regulated by the CpG methylation status in the promoter region.
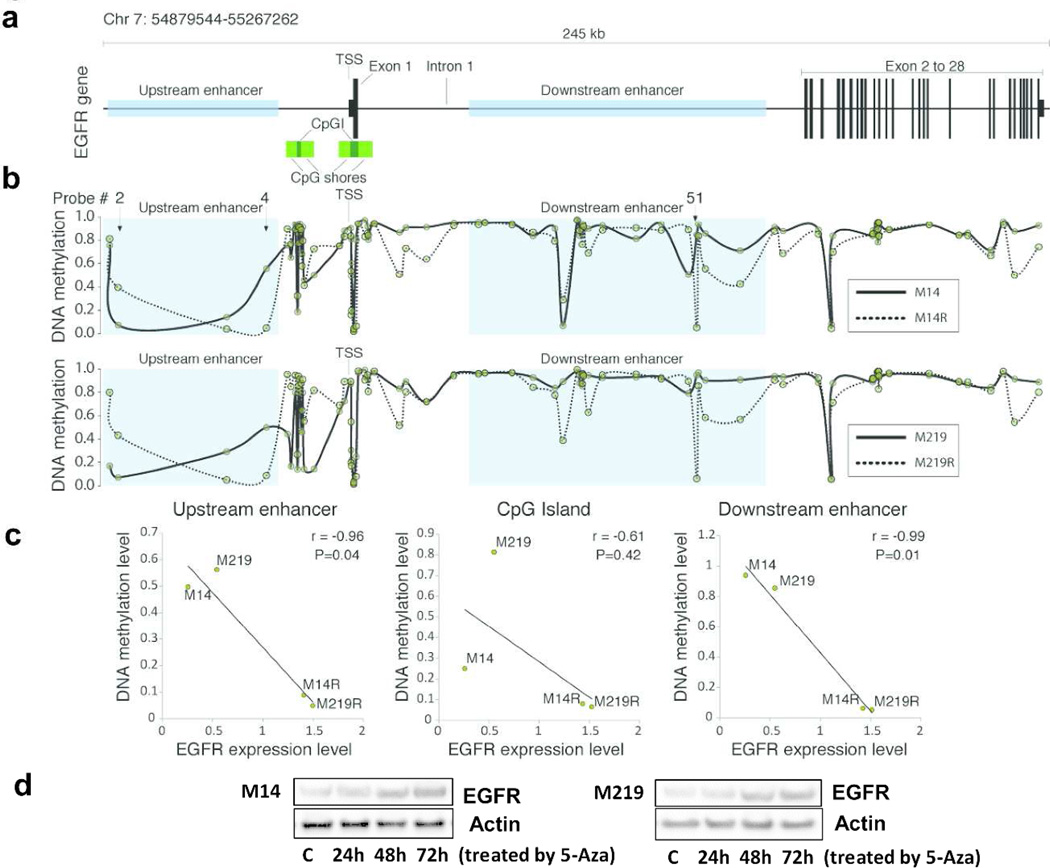
Figure 3. Hypomethylations of EGFR Enhancers in BRAFi-resistant melanoma cells.
A. The 245 kb region surrounding EGFR gene(65 kb upstream and 180 kb downstream to the transcription start site; TSS). B. Profiles of DNA methylation at region surrounding EGFR gene. One enhancer region located upstream of the TSS, is represented by the probe #4, and the other located downstream of the TSS, specifically in the first intron of the gene, represented by probe #51 defined in HumanMethylation450K BeadChip array. C. There was significant correlation between DNA methylation and expression levels were located in enhancer elements. D. Immunoblot of EGFR expression in M14 and M219 treated with 5-Aza. EGFR expression was increased in M14 and M219 treated with 5-Aza compared to treatment by DMSO control for 72h.
EGFR expression is regulated by the methylation status of EGFR enhancers
Genome-wide DNA methylation and transcriptomic integrative analysis were carried out to further determine if enhanced EGFR expression in BRAFi resistant cells was regulated by changes in CpG site methylation. The data from bead chip methylation array was assessed to establish a linear regression and was used to estimate the Pearson’s correlation coefficient(r) between the methylation level and the EGFR expression level. The analysis included a 65-kb upstream region and a 180-kb downstream region, both measured from the transcription start site (TSS) of the EGFR gene (Figure 3A). The targeted genomic region analysis included the promoter, 5’UTR, 3’UTR, and gene coding regions. The promoter region of EGFR gene presents two CpG islands with respective shores and shelves that were also targeted by this analysis. One enhancer region was located upstream of the TSS, represented by probe number 4, and the other was located downstream of the TSS in the first intron of the gene, represented by probe number 51 (Figure 3B). The regions that showed significant correlation between DNA methylation and EGFR expression level were located in enhancer elements (r; P <0.05; Figure 3C). The comparative methylation analysis of parental and BRAFi resistant cells demonstrated a consistent hypomethylation in BRAFi resistant cells in CpG sites located in the upstream and downstream enhancers. EGFR downstream enhancer methylation status was verified by MSP. Supplementary Figure 12 demonstrated that methylation of EGFR downstream enhancers was much lower in M14R and M219R cells than in respective parental M14 and M219 cells. This strongly suggested that EGFR expression is regulated by methylation status of EGFR enhancers in the BRAFi resistant melanoma cells.
EGFR expression is associated with cell resistance to BRAFi
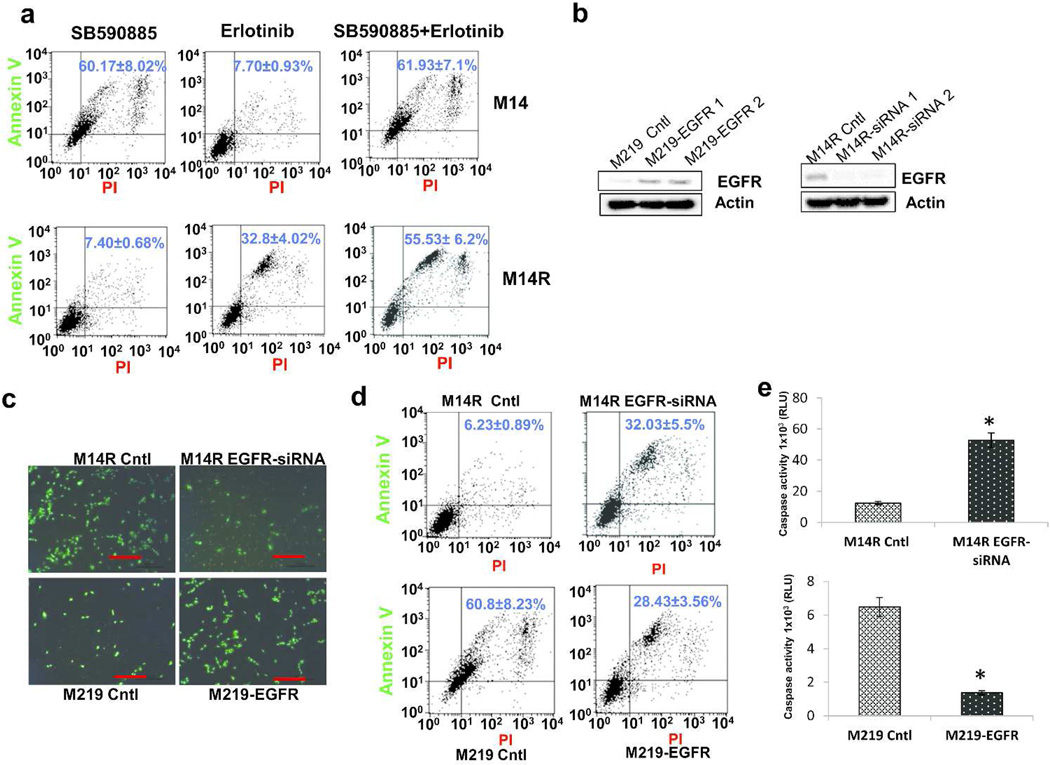
The effect of EGFR inhibitor (EGFRi) on BRAFi resistant melanoma cells was evaluated by treating M14 and M14R cells by BRAFi with or without EGFRi. M14 cells were sensitive to BRAFi but not EGFRi, whereas M14R were not sensitive to BRAFi but sensitive to EGFRi (Figure 4A, Supplementary Figures 13 and 14). Furthermore, M14R cells were more sensitive to the combination of EGFRi and BRAFi than to EGFRi alone.
Figure 4. EGFR confers resistance to BRAFi in melanoma lines.
A. Representative FACS scatter plots of treated cells stained with Annexin V(y-axis) versus propidium iodide(PI; x-axis). B. Immunoblots of EGFR expression in M219(left) and M14R(right) cells, before and after transfection with EGFR expression plasmid or EGFR small interfering RNA(siRNA). C. Fluorescence microscopy of cells treated by SB590885 and stained with PI(5ug/mL) and SYTO-13(1umol/mL). Scale bar = 100 µm. D. Representative FACS scatter plots of SB590885-treated cells that were stained with Annexin V(y-axis) versus PI(x-axis). E. Caspase3/7-Glo luminescent assay of cells treated with SB590885. Error bars, s.d.(*p <0.05).
To determine if EGFR expression confers BRAFi resistance to melanoma cells, we transfected M219 cells with an EGFR expression plasmid, and selected stable clones with high EGFR expression (Figure 4B, left). We also transfected M14R with EGFR small interfering RNA (siRNA) to reduce expression of EGFR (Figure 4B, right). M219 cells were more sensitive to BRAFi than M219-EGFR cells, and M14R-siRNA cells were more sensitive to BRAFi than M14R (Figure 4C, 4D and 4E). Taken together, these results suggested that EGFR activation was associated with BRAFi resistance in melanoma cells.
EGFR/PI3K/AKT pathway is activated in BRAFi resistant melanoma cells
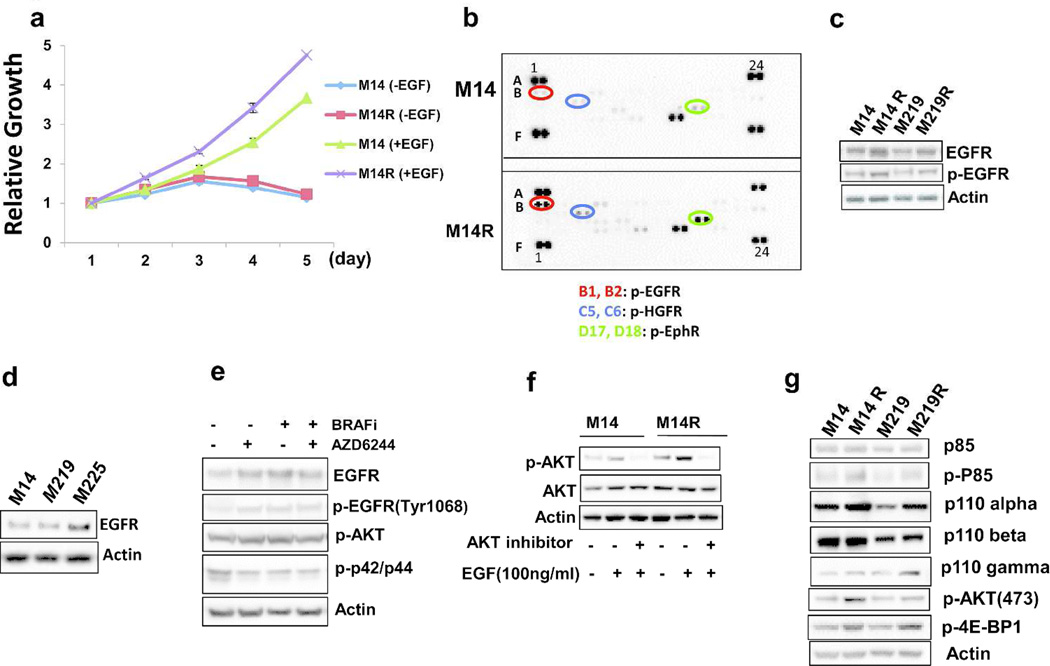
To determine if growth of BRAFi resistant cells depends on EGFR activation, parental and resistant cells were cultured in RPMI 1640 medium with or without EGF (20 ng/mL). There was no significant difference in cell growth of parental and resistant cells cultured in medium without EGF (Figure 5A). However, BRAFi resistant cells grew faster than parental cells when cultured in medium with EGF. This suggested that growth of resistant cells was more dependent on EGF than respective parent cells.
Figure 5. EGFR/PI3K/AKT pathway activation in BRAFi resistant melanoma cells.
A. Growth curve of M14 and M14R cells in RPMI1640 medium with or without EGF. B. Human phospho-RTK array assay of M14 cells (top figure) and M14R cells (bottom figure) [B1, B2(Red circle): p-EGFR; C5, C6(Blue circle):p-HGFR; D17, D18(Green circle): p-EphR]. Both pictures was exposured for 10 sec. C. Immunoblots of p-EGFR(Tyr1068) expression in parental and resistant cells D. Expression of EGFR in three BRAFmt-bearing cell lines: M14, M219 and M225. E. Immunoblots show that MEK inhibitor AZD6244 blocked MEK-mediated regulation of EGFR in BRAFmt cells. F. Immunoblots of AKT and phosphorylated-AKT expression in M14 and M14R cells exposed to EGF after a 1-hour treatment with AKT inhibitor. G. Immunoblots show expression of components on the PI3K/AKT pathway. Expression levels of p110 alpha, phosphorylated-p85(Tyr458), p-AKT(473) and phosphorylated 4E-BP1 were higher in resistant cells than in parental cells.
To further investigate EGFR activation in resistant melanoma cells, we employed Human Phosphor-RTK Array to assess phosphorylation of tyrosine kinase receptors (RTK). EGFR was more phosphorylated in resistant cells than in parental cells (Figure 5B). This suggested that overexpression of EGFR in resistant cells leads to enhanced tyrosine phosphorylation of EGFR. To further confirm that EGFR activation in cells was involved in acquired resistance to BRAFi, we performed immunoblotting to evaluate phosphorylation of EGFR at Tyr1068, which indicated kinase activation. Results demonstrated that phosphorylation of EGFR was significantly enhanced (Figure 5C).
To examine if EGFR was activated when RAS/RAf/MAPK/ERK pathway is blocked-, M225 cells (high expression of EGFR, Figure 5D) were treated with the MEK inhibitor AZD6244 or with BRAFi. After 24 hours, phosphorylated-EGFR (Tyr1068) and p-AKT were significantly increased, EGFR was also significantly increased, and p-ERK1/2 (p42/p44) was significantly decreased in BRAFi-treated cells (Figure 5E, Supplementary Figure 15). Expression of EGFR was increased in BRAFi treated cells. EGFR dimerization is formed as EGFR level increases. EGFR dimerization stimulates its intrinsic intracellular protein tyrosine kinase activity (autophosphorylation of EGFR), thereby the expression of p-EGFR increases.
These results suggested that EGFR was activated when RAS/RAF/MAPK/ERK pathway was blocked.
To determine whether activation of EGFR leads to signaling via the PI3K/AKT pathway, AKT phosphorylation was monitored after treatment of M14 and M14R cells with EGF, alone or in combination with the AKT inhibitor CCT128930(10 uM). AKT phosphorylation was significantly higher in M14R cells and was inhibited by CCT128930 (Figure 5F).
To further confirm activation of the EGFR/PI3K/AKT pathway, phosphorylation of p85, expression of PI3K components, and downstream targets of the EGFR/PI3K/AKT pathway were assessed. Expression levels of p110 alpha, phosphorylated-p85(Tyr458), p-AKT(473) and phosphorylated 4E-BP1(downstream targets of the EGFR/PI3K/AKT pathway) were higher in resistant cells than in parental cells (Figure 5G). Taken together, these results suggested that the EGFR/PI3K/AKT pathway is activated in BRAFi resistant melanoma cells.
Combination of EGFRi and BRAFi overcomes acquired resistance to BRAFi
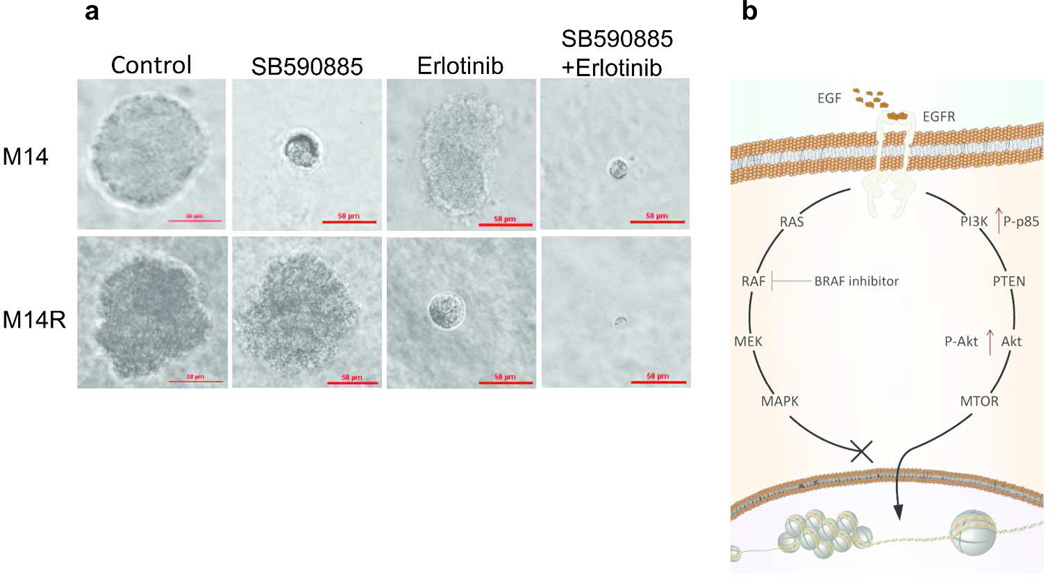
To further confirm the role of EGFR in acquired resistance to BRAFi, the effect of combining EGFRi and BRAFi on M14, M14R, M219, and M219R cells grown in a 3D matrix tumor-like microenvironment was assessed. M14R and M219R cells were more sensitive to combined inhibitors than to either single inhibitor. However, M14 and M219 cells (low EGFR expression) were sensitive to BRAFi but not to EGFRi. The photographs of the cells were taken at days 1 and 18 for comparison (Figure 6A, Supplementary Figures 16, 17 and 18). M14R and M219R cells (high EGFR expression) were sensitive to the combined inhibitors, whereas M14 and M219(low EGFR expression) were not sensitive to EGFRi. These results indicated that activated EGFR is responsive to EGFRi in BRAFi resistant melanoma cells.
Figure 6. Response of cells exposed to EGFRi and BRAFi in a 3D-tumor-like microenvironment.
A. M14 parental cells and M14R resistant cells grown in a 3D-tumor-like microenvironment were treated with DMSO, BRAFi(SB590885) and/or EGFRi(erlotinib). Scale bar = 50 µm.B. Flow diagram of EGFR/PI3K/AKT pathway activation in resistant cell lines treated by BRAFi. EGFR/PI3K/AKT pathway was activated when RAS/RAF/ MEK/ERK pathway was blocked by BRAFi.
DISCUSSION
We present evidence that BRAFi resistant melanoma cells develop an EMT phenotype, and EGFR-mediated activation of the PI3K-AKT pathway is induced as a consequence through BRAFi resistance. Enhanced expression of EGFR in BRAFi resistant cells was due to epigenetic changes in EGFR activation. This epigenetic changes led to hyperactivity of the EGFR/PI3K/AKT pathway. BRAFi resistant melanoma cells were sensitive to combined treatment with EGFRi and BRAFi. Our results suggested that epigenetic changes of EGFR led to enhanced expression and played an important role in BRAFi resistant cells. The drug combination co-targeting BRAFmt and EGFR positive melanomas may provide an effective alternative treatment strategy to improve durable clinical responses.
BRAFi resistant cells displayed a distinct EMT-like phenotype with spindle-shaped morphology as well as upregulation of EMT markers. These morphological changes of BRAFi resistant cells confirmed previous results(Nazarian et al., 2010). EMT has been associated with metastasis of lung and pancreatic cancers that are resistant to treatment with EGFR and AKT inhibitors (Byers et al., 2013; Marais et al., 2013). Our results demonstrated that migration and invasion of resistant cells were enhanced compared to parental cells. The EMT phenotype and EGFR expression are known to be closely related to cell migration and invasion (Zhang et al., 2012; Zhu et al., 2012). This may partially explain why recurrent melanomas are more aggressive in BRAFi resistant melanoma patients. We believe these resistant cells revert to an EMT-like phenotype with activation of the EGF signal pathways. Melanoma is of embryonic neuroectoderm origin, and during advanced stages may revert to its embryonic ectoderm phenotype.
The modulation of EGFR transcription is a complex mechanism which involves both genetic and epigenetic factors. Our results suggested that the hypomethylation of two different related promoters DNA elements significantly regulates EGFR expression. Remarkably, these two regions are located in the genomic enhancer elements. DNA enhancer elements interact with protein complexes which regulate the transcriptional rate of a gene or a group of genes. In breast cancer, the mechanism involving the enhancers of EGFR gene was shown to be related to anti-EGFR therapy response (Brandt et al., 2006; McInerney et al., 2001).
In a study of colon cancer, investigators found that BRAFi effect on BRAFmt tumors causes a rapid feedback activation of EGFR, which supports cell growth (Prahallad et al., 2012). EGFR-mediated reactivation of MAPK signaling allows BRAFmt colorectal cancers to resist vemurafenib treatment (Corcoran et al., 2012). Our results demonstrated that expression of EGFR and phosphorylated-EGFR was increased in BRAFi resistant melanoma cells. Expression levels of P110α and phosphorylated-p85(Tyr458) in PI3K/AKT were enhanced, while no significant enhancement on other components of the MAPK/MEK/ERK pathway was observed. We believe that the EGFR/PI3K/AKT pathway is activated when the MAPK/MEK/ERK pathway is blocked by BRAFi. EGFR IHC further confirmed that there was higher EGFR expression in resistant melanoma tumors than in BRAFi pre-treated melanoma patients tumors. EGFR-mediated reactivation of PI3k/AKT signaling provided BRAFi resistant melanoma cells with an alternative mechanism to grow and progress.
Oncogene addiction is a phenomenon in which some cancers that contain multiple genetic, epigenetic, and chromosomal abnormalities remain dependent on (addicted to) one or a few genes for both maintenance of the malignant phenotype and cell survival, especially in drug-resistant cancer cells (Kuehl and Bergsagel, 2012; Torti and Trusolino, 2011; Weinstein, 2002). Our studies indicated BRAFi resistant melanoma cells became “addicted to” EGFR oncogenic activity as do EMT cells during tumor progression. Overexpression of EGFR rendered cells resistant to BRAFi while knockdown of EGFR rendered cells sensitive to BRAFi. Expression of EGFR in BRAFi resistant tumors was increased, which also supported the findings that EGFR rendered cells sensitive to BRAFi. These findings suggested that EGFR plays an important role in BRAFi resistance. Development of resistance to tyrosine kinase inhibitors is a common problem often associated with chronic treatment with these RTK targeted anticancer drugs (Solit and Rosen, 2011). Drug combinations have recently been employed widely to treat aggressive drug resistant melanomas (Kim et al., 2011; O'Day et al., 2013). We demonstrated that growth of resistant cells was significantly inhibited by a combination of BRAFi and EGFRi, whereby growth was not affected by BRAFi alone. These findings coincided with studies described in BRAFi treatment of colon cancer (Prahallad et al., 2012). The EGFR/PI3K/AKT pathway is activated when RAS/RAF/MEK/ERK is inhibited by BRAFi. BRAFi resistant cells are epigenetically activated to overexpress EGFR and respond to EGF(Figure 6B).
This study is to demonstrate that EGFR conferred epigenetic activation upon resistant development to BRAFi in vitro and in vivo. Our MSP results showed that the methylation level of EGFR in resistant cells was much lower than that in parental cells. It is known that enhancer elements can play a major regulation of gene expression and not strictly the methylation status of the gene promoter region. Our results also showed that there was consistent hypomethylation of EGFR in BRAFi resistant cells in CpG sites located in the enhancer region. Taken together, our results suggested that the increase of EGFR expression in resistant cells was due to epigenetic regulation including hypomethylation of the promoter and enhancer regions. We believe that epigenetic changes of EGFR play an important role aggressive tumor growth in BRAFi resistant cutaneous melanomas.
Our study sheds light on the molecular basis of resistance and suggestive mechanisms of growth when BRAFi resistance develops. The EGFR and EGF axis activation in melanomas may be underestimated. It is known that primary melanoma metastasizes to distant organ sites that are ectoderm in origin. When invaded and damaged these organ sites release EGF for repair and regrowth. As a consequence, the melanoma cells bearing EGFR can take advantage of this microenvironment damage repair to survive and continue to grow through this process.
MATERIALS AND METHODS
Cells lines and tissues
The established human melanoma cell lines(M14, M219 and M225) that contained BRAFV600Emt were used in the studies. Tumor tissues from stage III/IV melanoma patients were obtained from Melanoma Institute Australia, Sydney and JWCI with written informed consent from each patient under approved Human Research Ethics committee protocols. Melanoma metastatic biopsies were taken from patients prior to commencing BRAFi and after surgical resection when resistance and progression occurred.
Tissue Microarray(TMA)
TMA were developed as previously described, and were well clinically annotated with >5 yrs follow-up (Camp et al., 2008; Nguyen et al., 2010). AJCC Stage IV melanoma TMA included 268 distant organ metastases and 39 autologous stage III/LN metastases, as well as 29 normal tissues from each respective organ(cancer-free).
BRAFi resistant BRAFmt melanoma lines
To obtain melanoma resistant cells, BRAFmt melanoma from established cell lines M14, M219 and M225 of early passages were cultured with gradually increasing concentration(0.1µM, 0.25µM, 0.5µM, 0.75µM, 1µM) of BRAFi(SB590885 in DMSO). SB590885, a potent and highly selective inhibitor of BRAF kinase (Villanueva et al., 2010) was purchased from SYMANSIS NZ Ltd(Timaru, New Zealand). Resistant melanoma cells were maintained in the continuous treatment of 1µM of SB590885 for three months. These BRAFi resistant melanoma cell lines were designated as M14R, M219R, and M225R. The resistant cells were maintained in the BRAFi drug treatment after 3 months. All the experiments were performed while cells were maintained in the presence of the BRAFi drug.
RNA extraction and array-based gene expression analysis
Total RNA was extracted from melanoma-related specimens using the RNeasy Mini Kit(QIAGEN). RNA samples were analyzed by GeneChip® Human Gene 1.0 ST Array(Affymetrix) at the USC/CHLA Genome Core Laboratory. Data was analyzed in Expression Console 1.1 software(Affymetrix) using the core transcript set and employing the robust multi-array average algorithm(RMA) for background correction and normalization of data.
RNA deep sequencing
Samples of high quality RNA(RIN ≥ 8.0) were used to create mRNA libraries using the Illumina TruSeq RNA Sample Preparation Kit v2. The mRNA libraries were then sequenced on the Illumina HiSeq 2500 high-throughput mode using TruSeq® SBS v3–HS 200 cycle kit according to standard procedures generating over 35 million 100-base pair paired-end reads per sample(Illumina). Base calling and demultiplexing were processed using CASAVA v1.8(Illumina), alignment was performed using TopHat2 (Kim et al., 2013), and expression values were generated using Cufflinks (Roberts et al., 2011).
Infinium HumanMethylation450K Beadchip analysis
DNA was extracted from cell lines with the QIAamp DNA mini kit(QIAGEN, Valencia, CA). Extracted DNA(1µg) was first treated with sodium bisulfite and then recovered using the Zymo EZ DNA methylation kit(Zymo Research, Irvine, CA), quantified and assessed for quality according to the manufacturer’s specifications (Campan et al., 2009). The Illumina Human Methylation450K BeadChip was used to assay the methylation status of >450K CpG sites across the genome (Marzese et al., 2014). Data were processed at the USC High Performance Computing Center(HPCC) using a dedicated, Linux-based, high performance computational cluster. The methylation level was reported as a beta-value(β=intensity of the methylated allele/intensity of the unmethylated allele + intensity of the methylated allele), which was calculated using the signal intensity value for each CpG site.
Biostatistical Analysis
The results are presented as mean ± SD of samples measured in triplicate, and repeated three times. Student's t-test was used to calculate differences between the various study groups. Significant difference was considered at p<0.05.
Additional materials and methods information is available in Supplemental Material.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by National Institute of Health; National Cancer Institute(grant number PO1 CA029605 Project II and Core C to(DH)), grant number 1R01CA167967-01A1(DH) and the Dr. Miriam and Sheldon G. Adelson Medical Research Foundation(D.H.).
ABBREVIATIONS
- Ab
antibody
- BRAFi
BRAF inhibitors
- BRAFmt
BRAF V600 mutation
- EGFRi
EGFR inhibitor
- EMT
Epithelial to Mesenchymal Transition
- IHC
immunohistochemistry
- M
methylated
- qMSP
methyl-specific PCR
- PBST
phosphate buffered saline tween-20
- PDGFR β
platelet-derived growth factor receptor β
- PI
propidium iodide
- RMA
robust multi-array average algorithm
- TMA
tissue microarray
- TSS
transcription start site
- RTK
receptor tyrosine kinase
- U
unmethylated
Footnotes
DATA ACCESS
The study data are deposited in NCBI’s Sequence Read Archive(SRA) databank(http://www.ncbi.nlm.nih.gov/sra/) and Accession file SRP022029.
CONFLICT OF INTEREST
All authors disclose no conflict of interest.
REFERENCES
- Brandt B, Meyer-Staeckling S, Schmidt H, et al. Mechanisms of egfr gene transcription modulation: relationship to cancer risk and therapy response. Clin Cancer Res. 2006;12:7252–7260. doi: 10.1158/1078-0432.CCR-06-0626. [DOI] [PubMed] [Google Scholar]
- Byers LA, Diao L, Wang J, et al. An epithelial-mesenchymal transition gene signature predicts resistance to EGFR and PI3K inhibitors and identifies Axl as a therapeutic target for overcoming EGFR inhibitor resistance. Clin Cancer Res. 2013;19:279–290. doi: 10.1158/1078-0432.CCR-12-1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai J, Wu J, Zhang H, et al. miR-186 downregulation correlates with poor survival in lung adenocarcinoma, where it interferes with cell-cycle regulation. Cancer Res. 2013;73:756–766. doi: 10.1158/0008-5472.CAN-12-2651. [DOI] [PubMed] [Google Scholar]
- Camp RL, Neumeister V, Rimm DL. A decade of tissue microarrays: progress in the discovery and validation of cancer biomarkers. J Clin Oncol. 2008;26:5630–5637. doi: 10.1200/JCO.2008.17.3567. [DOI] [PubMed] [Google Scholar]
- Campan M, Weisenberger DJ, Trinh B, et al. MethyLight. Methods Mol Bio. 2009;507:325–337. doi: 10.1007/978-1-59745-522-0_23. [DOI] [PubMed] [Google Scholar]
- Chapman PB, Hauschild A, Robert C, et al. Improved survival with vemurafenib in melanoma with BRAF V600E mutation. N Engl J Med. 2011;364:2507–2516. doi: 10.1056/NEJMoa1103782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi YJ, Li X, Hydbring P, et al. The requirement for cyclin D function in tumor maintenance. Cancer Cell. 2012;22:438–451. doi: 10.1016/j.ccr.2012.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corcoran RB, Ebi H, Turke AB, et al. EGFR-mediated re-activation of MAPK signaling contributes to insensitivity of BRAF mutant colorectal cancers to RAF inhibition with vemurafenib. Cancer Discov. 2012;2:227–235. doi: 10.1158/2159-8290.CD-11-0341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craene BD, Berx G. Regulatory networks defining EMT during cancer initiation and progression. Nat Rev Cancer. 2013;13:97–110. doi: 10.1038/nrc3447. [DOI] [PubMed] [Google Scholar]
- Davies H, Bignell GR, Cox C, et al. Mutations of the BRAF gene in human cancer. Nature. 2002;417:949–954. doi: 10.1038/nature00766. [DOI] [PubMed] [Google Scholar]
- Falchook GS, Long GV, Kurzrock R, et al. Dabrafenib in patients with melanoma, untreated brain metastases, and other solid tumours: a phase 1 dose-escalation trial. Lancet. 2012;379:1893–1901. doi: 10.1016/S0140-6736(12)60398-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flaherty KT, Infante JR, Daud A, et al. Combined BRAF and MEK inhibition in melanoma with BRAF V600 mutations. N Engl J Med. 2012;367:1694–1703. doi: 10.1056/NEJMoa1210093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flaherty KT, Puzanov I, Kim KB, et al. Inhibition of mutated, activated BRAF in metastatic melanoma. N Engl J Med. 2010;363:809–819. doi: 10.1056/NEJMoa1002011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girotti MR, Pedersen M, Sanchez-Laorden B, et al. Inhibiting EGF receptor or SRC family kinase signaling overcomes BRAF inhibitor resistance in melanoma. Cancer Discov. 2013;3:158–167. doi: 10.1158/2159-8290.CD-12-0386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gopal YN, Deng W, Woodman SE, et al. Basal and treatment-induced activation of AKT mediates resistance to cell death by AZD6244(ARRY-142886) in Braf-mutant human cutaneous melanoma cells. Cancer Res. 2010;70:8736–8747. doi: 10.1158/0008-5472.CAN-10-0902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauschild A, Grob JJ, Demidov LV, et al. Dabrafenib in BRAF-mutated metastatic melanoma: a multicentre, open-label, phase 3 randomised controlled trial. Lancet. 2012;380:358–365. doi: 10.1016/S0140-6736(12)60868-X. [DOI] [PubMed] [Google Scholar]
- Jakob JA, Bassett RL, Jr, Ng CS, et al. NRAS mutation status is an independent prognostic factor in metastatic melanoma. Cancer. 2012;118:4014–4023. doi: 10.1002/cncr.26724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johannessen CM, Boehm JS, Kim SY, et al. COT drives resistance to RAF inhibition through MAP kinase pathway reactivation. Nature. 2010;468:968–972. doi: 10.1038/nature09627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Pertea G, Trapnell C, et al. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013;14:R36. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim KB, Sosman JA, Fruehauf JP, et al. BEAM: a randomized phase II study evaluating the activity of bevacizumab in combination with carboplatin plus paclitaxel in patients with previously untreated advanced melanoma. J Clin Oncol. 2011;30:34–41. doi: 10.1200/JCO.2011.34.6270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kudo-Saito C, Shirako H, Takeuchi T, et al. Cancer metastasis is accelerated through immunosuppression during Snail-induced EMT of cancer cells. Cancer Cell. 2009;15:195–206. doi: 10.1016/j.ccr.2009.01.023. [DOI] [PubMed] [Google Scholar]
- Kuehl WM, Bergsagel PL. MYC addiction: a potential therapeutic target in MM. Blood. 2012;120:2351–2352. doi: 10.1182/blood-2012-08-445262. [DOI] [PubMed] [Google Scholar]
- Lito P, Pratilas CA, Joseph EW, et al. Relief of profound feedback inhibition of mitogenic signaling by RAF inhibitors attenuates their activity in BRAFV600E melanomas. Cancer Cell. 2012;22:668–682. doi: 10.1016/j.ccr.2012.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long GV, Menzies AM, Nagrial AM, et al. Prognostic and clinicopathologic associations of oncogenic BRAF in metastatic melanoma. J Clin Oncol. 2011;29:1239–1246. doi: 10.1200/JCO.2010.32.4327. [DOI] [PubMed] [Google Scholar]
- Long GV, Trefzer U, Davies MA, et al. Dabrafenib in patients with Val600Glu or Val600Lys BRAF-mutant melanoma metastatic to the brain(BREAK-MB): a multicentre, open-label, phase 2 trial. Lancet Oncol. 2012;13:1087–1095. doi: 10.1016/S1470-2045(12)70431-X. [DOI] [PubMed] [Google Scholar]
- Marais R, Sellers W, Livingston D, et al. Twenty-fourth annual Pezcoller symposium: Molecular basis for resistance to targeted agents. Cancer Res. 2013;73:1046–1049. doi: 10.1158/0008-5472.CAN-12-3236. [DOI] [PubMed] [Google Scholar]
- Marzese DM, Scolyer RA, Huynh JL, et al. Epigenome-wide DNA methylation landscape of melanoma progression to brain metastasis reveals aberrations on homeobox D cluster associated with prognosis. Hum Mol Genet. 2014;23:226–238. doi: 10.1093/hmg/ddt420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McInerney JM, Wilson MA, Strand KJ, et al. A strong intronic enhancer element of the EGFR gene is preferentially active in high EGFR expressing breast cancer cells. J Cell Biochem. 2001;80:538–549. doi: 10.1002/1097-4644(20010315)80:4<538::aid-jcb1008>3.0.co;2-2. [DOI] [PubMed] [Google Scholar]
- Montagut C, Sharma SV, Shioda T, et al. Elevated CRAF as a potential mechanism of acquired resistance to BRAF inhibition in melanoma. Cancer Res. 2008;68:4853–4861. doi: 10.1158/0008-5472.CAN-07-6787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nazarian R, Shi H, Wang Q, et al. Melanomas acquire resistance to B-RAF(V600E) inhibition by RTK or N-RAS upregulation. Nature. 2010;468:973–977. doi: 10.1038/nature09626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen T, Kuo C, Nicholl MB, et al. Downregulation of microRNA-29c is associated with hypermethylation of tumor-related genes and disease outcome in cutaneous melanoma. Epigenetics. 2010;6:388–394. doi: 10.4161/epi.6.3.14056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Day SJ, Eggermont AM, Chiarion-Sileni V, et al. Final Results of Phase III SYMMETRY Study: Randomized, Double-Blind Trial of Elesclomol Plus Paclitaxel Versus Paclitaxel Alone As Treatment for Chemotherapy-Naive Patients With Advanced Melanoma. J Clin Oncol. 2013;31:1211–1218. doi: 10.1200/JCO.2012.44.5585. [DOI] [PubMed] [Google Scholar]
- Paraiso KH, Xiang Y, Rebecca VW, et al. PTEN loss confers BRAF inhibitor resistance to melanoma cells through the suppression of BIM expression. Cancer Res. 2011;71:2750–2760. doi: 10.1158/0008-5472.CAN-10-2954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poulikakos PI, Persaud Y, Janakiraman M, et al. RAF inhibitor resistance is mediated by dimerization of aberrantly spliced BRAF(V600E) Nature. 2011;480:387–390. doi: 10.1038/nature10662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prahallad A, Sun C, Huang S, et al. Unresponsiveness of colon cancer to BRAF(V600E) inhibition through feedback activation of EGFR. Nature. 2012;483:100–103. doi: 10.1038/nature10868. [DOI] [PubMed] [Google Scholar]
- Ray PS, Wang J, Qu Y, et al. FOXC1 is a potential prognostic biomarker with functional significance in basal-like breast cancer. Cancer Res. 2010;70:3870–3876. doi: 10.1158/0008-5472.CAN-09-4120. [DOI] [PubMed] [Google Scholar]
- Roberts A, Trapnell C, Donaghey J, et al. Improving RNA-Seq expression estimates by correcting for fragment bias. Genome Biol. 2011;12:R22. doi: 10.1186/gb-2011-12-3-r22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi H, Kong X, Ribas A, et al. Combinatorial treatments that overcome PDGFRbeta-driven resistance of melanoma cells to V600EB-RAF inhibition. Cancer Res. 2011;71:5067–5074. doi: 10.1158/0008-5472.CAN-11-0140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi H, Moriceau G, Kong X, et al. Melanoma whole-exome sequencing identifies(V600E)B-RAF amplification-mediated acquired B-RAF inhibitor resistance. Nat Commun. 2012;3:724. doi: 10.1038/ncomms1727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegel R, Naishadham D, Jemal A. Cancer statistics, 2012. CA Cancer J Clin. 2012;62:10–29. doi: 10.3322/caac.20138. [DOI] [PubMed] [Google Scholar]
- Smalley KS, Lioni M, Dalla Palma M, et al. Increased cyclin D1 expression can mediate BRAF inhibitor resistance in BRAF V600E-mutated melanomas. Mol Cancer Ther. 2008;7:2876–2883. doi: 10.1158/1535-7163.MCT-08-0431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solit DB, Rosen N. Resistance to BRAF inhibition in melanomas. N Engl J Med. 2011;364:772–774. doi: 10.1056/NEJMcibr1013704. [DOI] [PubMed] [Google Scholar]
- Torti D, Trusolino L. Oncogene addiction as a foundational rationale for targeted anticancer therapy: promises and perils. EMBO Mol Med. 2011;3:623–636. doi: 10.1002/emmm.201100176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villanueva J, Vultur A, Lee JT, et al. Acquired resistance to BRAF inhibitors mediated by a RAF kinase switch in melanoma can be overcome by cotargeting MEK and IGF-1R/PI3K. Cancer Cell. 2010;18:683–695. doi: 10.1016/j.ccr.2010.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinstein IB. Cancer. Addiction to oncogenes--the Achilles heal of cancer. Science. 2002;297:63–64. doi: 10.1126/science.1073096. [DOI] [PubMed] [Google Scholar]
- Weiss MB, Abel EV, Mayberry MM, et al. TWIST1 Is an ERK1/2 Effector That Promotes Invasion and Regulates MMP-1 Expression in Human Melanoma Cells. Cancer Res. 2012;72:6382–6392. doi: 10.1158/0008-5472.CAN-12-1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whittaker S, Kirk R, Hayward R, et al. Gatekeeper mutations mediate resistance to BRAF-targeted therapies. Sci Transl Med. 2010;2:35ra41. doi: 10.1126/scitranslmed.3000758. [DOI] [PubMed] [Google Scholar]
- Wilson TR, Fridlyand J, Yan Y, et al. Widespread potential for growth-factor-driven resistance to anticancer kinase inhibitors. Nature. 2012;487:505–509. doi: 10.1038/nature11249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadav V, Zhang X, Liu J, et al. Reactivation of mitogen-activated protein kinase(MAPK) pathway by FGF receptor 3(FGFR3)/Ras mediates resistance to vemurafenib in human B-RAF V600E mutant melanoma. J Biol Chem. 2012;287:28087–28098. doi: 10.1074/jbc.M112.377218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Liang Q, Lei Y, et al. SOX4 induces epithelial-mesenchymal transition and contributes to breast cancer progression. Cancer Res. 2012;72:4597–4608. doi: 10.1158/0008-5472.CAN-12-1045. [DOI] [PubMed] [Google Scholar]
- Zhu W, Phan QT, Boontheung P, et al. EGFR and HER2 receptor kinase signaling mediate epithelial cell invasion by Candida albicans during oropharyngeal infection. Proc Natl Acad Sci U S A. 2012;109:14194–14199. doi: 10.1073/pnas.1117676109. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.