Abstract
Enterovirus D68 (EV-D68) is a member of Picornaviridae and is a causative agent of recent outbreaks in the USA of respiratory illness in children. We report here the crystal structures of EV-D68 and its complex with pleconaril, a capsid binding compound that had been developed as an anti-rhinovirus drug. The hydrophobic drug binding pocket in viral protein 1 contained density that is consistent with a fatty acid of about 10 carbon atoms. This density could be displaced by pleconaril. We also showed that pleconaril inhibits EV-D68 at a half maximal effective concentration (EC50) of 430 nM and might, therefore, be a possible drug candidate to alleviate EV-D68 outbreaks.
Picornaviruses constitute a large family of small, icosahedral viruses with a single, positive-stranded RNA genome and an external diameter of about 300Å. The enterovirus (EV) genus includes medically-important human pathogens, such as human rhinoviruses (HRV), polioviruses (PV) and coxsackieviruses (CV) (table S1) (1, 2). Many of these viruses have been well characterized structurally and functionally (3–10). However, the species EV-D remains poorly characterized.
An upsurge of EV-D68 cases in the past few years has shown clusters of infections worldwide (11). In August 2014 an outbreak of mild to severe respiratory illnesses occurred among thousands of young children in the United States of which 1116 cases have been confirmed to be caused by EV-D68. This virus has also been associated with occasional neurological infections (12). Although EV-D68 has emerged as a considerable global public health threat, there is no available vaccine or effective antiviral treatment.
The capsids of EVs consist of 60 copies of each of four different viral proteins, VP1, VP2, VP3 and VP4 (Fig. 1A). Of these VP1 (about 300 amino acids), VP2 (about 260 amino acids) and VP3 (about 240 amino acids) each have a “jelly roll” fold arranged in the capsid with pseudo T=3 icosahedral symmetry, where T represents the triangulation number (13). Their organization in the capsid is similar to that of the T=3 RNA plant viruses except that the three subunits related by quasi-3-fold symmetry have different amino acid sequences in picornaviruses (6, 10). Each roughly 70 amino acid long VP4 molecule forms an extended peptide on the internal surface of the capsid shell. The jelly roll fold consists of two anti-parallel β-sheets which face each other to form a β-barrel with a hydrophobic interior (Fig. 1B).
Fig. 1. The structure of EV-D68.

(A) Diagrammatic representation of the virus. VP1, VP2 and VP3 are colored blue, green, and red respectively. Each pentagonal icosahedral vertex is surrounded by a “canyon” colored black. The epitopes for neutralizing antibodies for the homologous HRV14 are labelled as NIm-I, NIm-II and NIm-III. The NIm-IA and NIm-IB sites are disordered in EV-D68. (B) The VP1 jelly roll in EV-D68 shown as a ribbon diagram. If the β-strands along the polypeptide are identified sequentially as A, B, C,….I, then one of the sheets is composed of the anti-parallel strands BIDG and the other by the anti-parallel strands CHEF.(C) Surface features of EV-D68 compared to HRV16. The canyon of EV-D68 is shallower and narrower than the canyon of HRV16.
Enteroviruses have a deep surface depression (“canyon”) circulating around each of the twelve pentameric vertices (Fig. 1A). The canyon was predicted to be the site of receptor binding because the amino acids outside the canyon that form the external surface of the virus were more exposed and were shown to be accessible to neutralizing antibodies (Fig. 1A) (10, 14). The virus thus could remain faithful to a specific receptor molecule that binds into the canyon while evading the host’s immune system (10, 15). The prediction that the canyon would be the site of binding to cellular receptor molecules was subsequently confirmed for numerous different enteroviruses (16, 17). All of the receptor molecules were found to have an immunoglobulin-like structure.
A variety of small ~350 Da hydrophobic molecules that are inhibitors of enterovirus infection were shown to bind into the hydrophobic pocket in the center of the VP1 jelly roll (18) (Fig. 1B). They stabilize the virus (19) by filling the pocket with a well-fitting hydrophobic molecule, thereby inhibiting uncoating of the virus and the release of the genome into the infected cell. These compounds also prevent attachment to cells by altering the surface features of the canyon floor where the virus attaches to a cellular receptor (17, 19). The floor of the canyon is formed primarily by the GH loop of VP1, the connecting residues between β-strands G and H (18).
Most infectious enteroviruses contain a small molecule or “pocket factor”, probably a fatty acid in the VP1 binding pocket (7, 18, 20). Like the capsid binding compounds, the pocket factor presumably stabilizes the virus by filling the VP1 hydrophobic pocket. Thus the virus is stabilized while being transmitted to a new host. However, when a receptor molecule binds to the floor of the canyon it depresses the floor (that also forms the roof of the VP1 binding pocket) which then squeezes the binding pocket, likely expelling the pocket factor (17). Thus attachment of the virus to a cell surface initiates uncoating, causing the loss of the genome into the cell’s cytosol.
In most enteroviruses that have been investigated these capsid binding antiviral compounds displace the pocket factor (e,g. polioviruses, coxsackieviruses A or B, and many HRVs). The rhinoviruses HRV14 (18) and HRV3 (21) do not contain a pocket factor and the GH loop is displaced relative to its position in other enteroviruses that contain a pocket factor, reducing the volume of the VP1 pocket. Either neither HRV14 nor HRV3 bind a pocket factor in vivo or it was lost during the purification procedure. Much effort was made between 1985 and 2000 to design a compound that fits into the VP1 pocket and would inhibit the maximum number of rhinoviruses (22). The final optimal structure was pleconaril (fig. S1) which not only had good efficacy but was also stable enough to maintain good bioavailability in clinical tests (23). However, pleconaril was not licensed primarily because it put women using birth control drugs at risk of conception.
The EV-D68 prototype strain Fermon CA62-1 was propagated in human rhabdomyosarcoma cells at 33°C, which had previously been shown to be the optimal growth temperature for EV-D68, indicating that EV-D68 behaves much like members of the RV-A and RV-B species that are responsible for common colds (24). The virus was purified and crystalized as described in Materials and Methods. The cubic looking crystals had a diameter between 0.1mm and 0.2mm. After soaking in glycerol, they were flash frozen in liquid nitrogen. X-ray diffraction data were collected at sector 14 of the Advanced Photon Source. The data extended to 2.0Å resolution and was processed with HKL2000 (25). The crystal symmetry was I222 with two particles per unit cell, implying that the particles were located on a 222 symmetry position. A rotation function (26) differentiated between the two orthogonal possible orientations of the icosahedron. Initial phases were calculated based on the structure of HRV2 (27) (PDB accession number 1FPN) after removing the pocket factor. The phases were then extended in small steps to 2.0Å resolution using 15-fold averaging and solvent flattening. A model of the structure was built using the program Coot (28) and refined with the program CNS (29). The final value of Rwork was 27.5%. Rfree is essentially the same as Rwork in the presence of high non-crystallographic symmetry redundancy for a randomly selected set of reflections as is the case here (table S2).
Comparison of the amino acid sequences of EV-D68 with other enteroviruses shows that VP3 has a short, C-terminal α-helix not present in other enteroviruses (fig. S2). The EV-D68 electron density map showed that this helix decorates the north side of the canyon in the neighboring, 5-fold related, icosahedral asymmetric unit. As a result the canyon is narrower than in other enteroviruses and might, therefore, not be large enough to accommodate immunoglobulin-like receptors (Fig. 1C).
The BC and DE loops of VP1 are structurally the most variable among known picornaviruses. The EV-D68 VP1 has two disordered regions corresponding to residues 80–86 (EV-D68 numbering) in the BC loop and 129–136 in the DE loop, both of which are near fivefold axes. These regions harbor the neutralizing immunogen sites, NIm-IA and NIm-IB, on HRV14, respectively (10) (Fig. 1A). Thus the flexible immunogenic regions around the five-fold axes might be an alternative mechanism for evading host humoral immune responses.
The electron density map of EV-D68 showed density in the VP1 pocket (Fig. 2A). The height of this pocket factor density was about 3 standard deviations above the mean of the non-crystallographic symmetry averaged map, as compared with about 5 standard deviations of most of the main chain density. As in other enteroviruses that have a pocket factor, the conformation of the GH loop of VP1, which defines the interface between the VP1 pocket and the floor of the canyon, is pushed into the canyon relative to the empty pocket in HRV14 (10, 18) or HRV3 (21).
Fig. 2. The pocket factor.
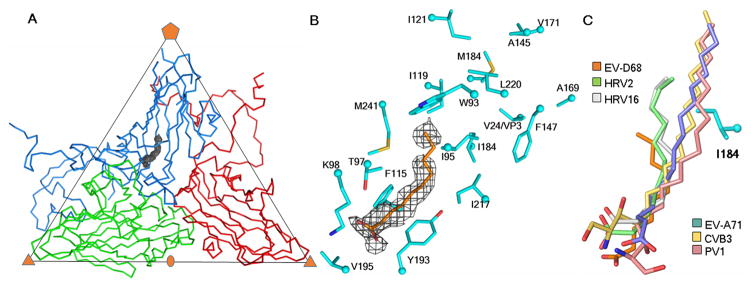
(A) One icosahedral asymmetric unit of the EV-D68 structure, showing the Cα atom backbone for VP1, VP2, VP3 in marine blue, green and red, respectively. The pocket factor electron density outline is shown in grey. (B) Enlargement of the pocket factor density with a fitted putative C10 fatty acid. Shown also are the amino acids lining the pocket. (C) Comparison of the putative pocket factor structures in six known EV structures. Shown also is the residue I184 in EV-D68 that alters the direction of the shorter pocket factors in some of the viruses as compared to others with longer pocket factors and a smaller residue at the equivalent position of 184. Atoms at the head of the pocket factors and of the VP1 residue side chains are colored red (oxygen), dark blue (nitrogen) and yellow (sulfur).
The length of the pocket factor density in the EV-D68 map corresponded to a fatty acid with an aliphatic chain of about 10 carbon atoms (Fig. 2B). Similarly, 12 carbon atom long pocket factors were observed for HRV16 (9) and HRV2 (27). In contrast, the well-formed pocket factors in poliovirus 1 (6, 7), coxsackievirus B3 (5) and EV-A71 (3) corresponded to longer fatty acids with 18, 16 and 18 carbon atoms, respectively. Furthermore, the orientation of the pocket factor tails in EV-D68, HRV16 and HRV2 are slightly different than that of the long pocket factors of EV-A71, poliovirus 1 and coxsackievirus B3 (Fig. 2C). The orientation is controlled in part by VP1 residue 184, which is Leu or Ile for the viruses with small, short pocket factors and Val for viruses with longer pocket factors (fig. S2). The larger Leu and Ile residues might push the pocket factor sideways. The similarity between the properties of the pocket factor in EV-D68 and that in the HRVs might partially explain why these viruses are less stable.
The anti-EV-D68 activity of two capsid binding compounds, pirodavir and BTA-188 (fig. S1), that had significant anti-rhinovirus activity were compared with pleconaril using plaque reduction assays in HeLa cells (Fig. 3, A and B). The half maximal effective concentration, EC50, value of these two compounds were found to be comparable to previous results using cytopathic effect inhibition assays against EV-D68 (30). However, pleconaril was found to be more potent against EV-D68 than pirodavir and BTA-188 (Fig. 3A). The inhibitory effect of pleconaril is similar against EV-D68, HRV16 and HRV14 but better than against EV-A71. It is, therefore, noteworthy that pleconaril was an effective inhibitor in extensive clinical tests for treatment of common colds (23, 31). Furthermore, fluorescence-based thermal stability assays indicated that when EV-D68 was incubated with either 10 μg/ml or 50 μg/ml pleconaril, 4°C higher temperatures were required to release the RNA genome than when no pleconaril was present (Fig. 3C and Materials and Methods in Supplementary Materials). Thus pleconaril stabilizes EV-D68 capsids, preventing the virus from uncoating during viral entry.
Fig. 3. Anti-enterovirus activity of three capsid binding compounds.
(A) List of EC50 values. Each experiment was performed at least three times. (B) Plot of % plaque reduction of pleconaril as a function of the log of its concentration. Error bars indicate standard deviations. (C) Release of EV-D68 genome upon increase in temperature as monitored by Sybr green II. The experiments were done in triplicate. For each data point, the ratio between mean fluorescence intensity and standard deviation is at least 7.6. Shown are curves fitted with a sigmoidal function for the native virus and for the virus after incubation with pleconaril.
The structure of EV-D68 was also determined to 2.3Å resolution when co-crystalized with pleconaril (Fig. S3). The crystallographic procedure was the same as for the native structure determination (Materials and Methods in Supplementary Materials). The electron density of the ligand inside the hydrophobic pocket was of the same height but much longer than that of the pocket factor in the native EV-D68 structure (Fig. 4A) and could be easily fitted with the structure of pleconaril (Fig. 4B). This demonstrated that the native pocket factor was replaced by pleconaril. However, the density of the ligand and of the pocket factor are slightly lower than that of the polypeptide main chain. In part this is to be expected because the ligand or fatty acid pocket factor are composed primarily of only carbon atoms as opposed to the heavier combination of carbon, nitrogen plus oxygen atoms of the main chain. In part the slightly lower density of the ligand might be due to some variations of conformation within the VP1 binding pocket.
Fig. 4. Structure of pleconaril bound into the VP1 pocket of EV-D68.
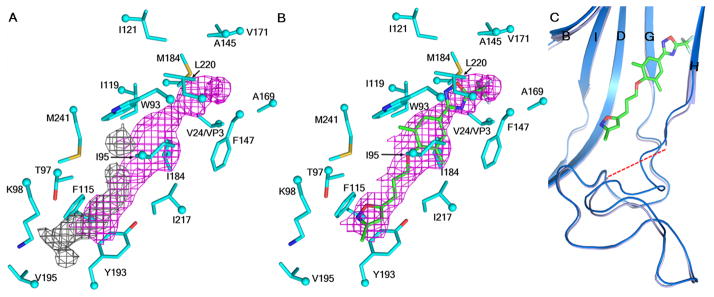
(A) Pocket factor density (grey) compared to the pleconaril density (magenta). (B) Pleconaril (green) fitted to density in the structure of the complex. (C) Conformational change of the VP1 GH loop as a consequence of the presence of pleconaril. The native and complex structures are shown in marine blue and baby blue, respectively. Oxygen, nitrogen, sulfur and fluorine atoms are shown in red, dark blue, yellow and light green, respectively.
Part of the VP1 GH loop (residues 212–215) that forms the entrance to the VP1 pocket had become less ordered. The Cα atom of residue 211 had moved 1.2 Å towards the inside of the pocket relative to that of the native structure, possibly blocking the entrance to the pocket once pleconaril had entered (Fig. 4C). Thus the dynamics of GH loop might be an important consideration for future structure-based design of EV-D68 capsid-binding inhibitors.
A comparison of the EV-D68-pleconaril, HRV14-pleconaril and HRV16-pleconaril structures showed a similar binding mode for pleconaril in the VP1 pocket of these three viruses (Fig. S4 & Table S3). This may explain why pleconaril is similarly effective against these three enteroviruses. To investigate why pleconaril is more effective against EV-D68 than pirodavir or BTA-188, we performed in silico docking experiments. The presence of trifluoromethyl substituted oxadiazole moiety in pleconaril, rather than a more hydrophilic group in either pirodavir (ethyl carboxylate group) or BTA-188 (O-ethyloxime group) at structurally equivalent positions (Fig. S5), likely contributes to more favorable interactions of pleconaril with the hydrophobic residues deep inside the VP1 pocket of EV-D68.
Our results show that the structure of EV-D68 has considerable similarities to the well-studied HRVs for which pleconaril was specifically designed. We also show that pleconaril replaces the pocket factor and is a potent inhibitor of EV-D68, with an EC50 value of 430nM. The size and location of the pocket factor lodged in the VP1 pocket is similar to that found in other HRVs and different to the pocket factors found in poliovirus 1 and EV-A71. This correlates with the observation that pleconaril is far more active when the natural pocket factor is short as in the HRVs and in EV-D68. Furthermore, sequence alignment of 188 EV-D68 strains found between 1962 and 2013 indicates that residues in VP1 which interact with pleconaril, as identified from the complex structure, are completely conserved with one exception. Therefore, pleconaril is likely to inhibit not only the prototype strain examined here, but also many other strains. In view of the previous extensive clinical trials that have established its safety, pleconaril would be a possible drug candidate to alleviate EV-D68 outbreaks.
Supplementary Material
Acknowledgments
We thank M. Steven Oberste of the Centers for Disease Control and Prevention for supplying us with the prototype strain of EV-D68. We thank Mark A. McKinlay of the Task Force for Global Health for helpful discussion and suggestions. We thank Sheryl Kelly for help with the manuscript preparation. We thank Vukica Srajer, Robert Henning and the other staff of the Advanced Photon Source BioCARS beamline 14 for help with X-ray diffraction data collection. Use of BioCARS sector 14 was supported by the National Institutes of Health, National Center for Research Resources (NIH/NCRR) grant RR007707. Use of the Advanced Photon Source was supported by the U.S. Department of Energy, Office of Science, Office of Basic Energy Sciences, under contract DE-AC02-006CH11357. This study was supported by National Institutes of Health grant award AI11219 to MGR. Coordinates for native EV-D68 and EV-D68-pleconaril structures were deposited with the Protein Data Bank with accession numbers 4WM8 and 4WM7, respectively.
References and Notes
- 1.Adams MJ, King AM, Carstens EB. Ratification vote on taxonomic proposals to the International Committee on Taxonomy of Viruses (2013) Arch Virol. 2013;158:2023–2030. doi: 10.1007/s00705-013-1688-5. [DOI] [PubMed] [Google Scholar]
- 2.Knowles NJ, et al. In: Virus Taxonomy: Classification and Nomenclature of Viruses: Ninth Report of the International Committee on Taxonomy of Viruses. King AMQ, Adams MJ, Carstens EB, Lefkowitz EJ, editors. Elsevier; San Diego: 2012. pp. 855–880. [Google Scholar]
- 3.Plevka P, Perera R, Cardosa J, Kuhn RJ, Rossmann MG. Crystal structure of human enterovirus 71. Science. 2012;336:1274. doi: 10.1126/science.1218713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang X, et al. A sensor-adaptor mechanism for enterovirus uncoating from structures of EV71. Nat Struct Mol Biol. 2012;19:424–429. doi: 10.1038/nsmb.2255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Muckelbauer JK, et al. The structure of coxsackievirus B3 at 3.5 Å resolution. Structure. 1995;3:653–667. doi: 10.1016/s0969-2126(01)00201-5. [DOI] [PubMed] [Google Scholar]
- 6.Hogle JM, Chow M, Filman DJ. Three-dimensional structure of poliovirus at 2.9 Å resolution. Science. 1985;229:1358–1365. doi: 10.1126/science.2994218. [DOI] [PubMed] [Google Scholar]
- 7.Filman DJ, et al. Structural factors that control conformational transitions and serotype specificity in type 3 poliovirus. EMBO J. 1989;8:1567–1579. doi: 10.1002/j.1460-2075.1989.tb03541.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lentz KN, et al. Structure of poliovirus type 2 Lansing complexed with antiviral agent SCH48973: comparison of the structural and biological properties of three poliovirus serotypes. Structure. 1997;5:961–978. doi: 10.1016/s0969-2126(97)00249-9. [DOI] [PubMed] [Google Scholar]
- 9.Oliveira MA, et al. The structure of human rhinovirus 16. Structure. 1993;1:51–68. doi: 10.1016/0969-2126(93)90008-5. [DOI] [PubMed] [Google Scholar]
- 10.Rossmann MG, et al. Structure of a human common cold virus and functional relationship to other picornaviruses. Nature. 1985;317:145–153. doi: 10.1038/317145a0. [DOI] [PubMed] [Google Scholar]
- 11.Tokarz R, et al. Worldwide emergence of multiple clades of enterovirus 68. J Gen Virol. 2012;93:1952–1958. doi: 10.1099/vir.0.043935-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kreuter JD, et al. A fatal central nervous system enterovirus 68 infection. Arch Pathol Lab Med. 2011;135:793–796. doi: 10.5858/2010-0174-CR.1. [DOI] [PubMed] [Google Scholar]
- 13.Caspar DL, Klug A. Physical principles in the construction of regular viruses. Cold Spring Harb Symp Quant Biol. 1962;27:1–24. doi: 10.1101/sqb.1962.027.001.005. [DOI] [PubMed] [Google Scholar]
- 14.Sherry B, Mosser AG, Colonno RJ, Rueckert RR. Use of monoclonal antibodies to identify four neutralization immunogens on a common cold picornavirus, human rhinovirus 14. J Virol. 1986;57:246–257. doi: 10.1128/jvi.57.1.246-257.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rossmann MG. The canyon hypothesis. Hiding the host cell receptor attachment site on a viral surface from immune surveillance. J Biol Chem. 1989;264:14587–14590. [PubMed] [Google Scholar]
- 16.Olson NH, et al. Structure of a human rhinovirus complexed with its receptor molecule. Proc Natl Acad Sci USA. 1993;90:507–511. doi: 10.1073/pnas.90.2.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rossmann MG, He Y, Kuhn RJ. Picornavirus-receptor interactions. Trends Microbiol. 2002;10:324–331. doi: 10.1016/s0966-842x(02)02383-1. [DOI] [PubMed] [Google Scholar]
- 18.Smith TJ, et al. The site of attachment in human rhinovirus 14 for antiviral agents that inhibit uncoating. Science. 1986;233:1286–1293. doi: 10.1126/science.3018924. [DOI] [PubMed] [Google Scholar]
- 19.Rossmann MG. Viral cell recognition and entry. Prot Sci. 1994;3:1712–1725. doi: 10.1002/pro.5560031010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Smyth M, Pettitt T, Symonds A, Martin J. Identification of the pocket factors in a picornavirus. Arch Virol. 2003;148:1225–1233. doi: 10.1007/s00705-002-0974-4. [DOI] [PubMed] [Google Scholar]
- 21.Zhao R, et al. Human rhinovirus 3 at 3.0 Å resolution. Structure. 1996;4:1205–1220. doi: 10.1016/s0969-2126(96)00128-1. [DOI] [PubMed] [Google Scholar]
- 22.Rogers JM, Diana GD, McKinlay MA. Pleconaril. A broad spectrum antipicornaviral agent. Adv Exp Med Biol. 1999;458:69–76. [PubMed] [Google Scholar]
- 23.Hayden FG, et al. Efficacy and safety of oral pleconaril for treatment of colds due to picornaviruses in adults: results of 2 double-blind, randomized, placebo-controlled trials. Clin Infect Dis. 2003;36:1523–1532. doi: 10.1086/375069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Oberste MS, et al. Enterovirus 68 is associated with respiratory illness and shares biological features with both the enteroviruses and the rhinoviruses. J Gen Virol. 2004;85:2577–2584. doi: 10.1099/vir.0.79925-0. [DOI] [PubMed] [Google Scholar]
- 25.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Method Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 26.Tong L, Rossmann MG. Rotation function calculations with GLRF program. Method Enzymol. 1997;276:594–611. [PubMed] [Google Scholar]
- 27.Verdaguer N, Blaas D, Fita I. Structure of human rhinovirus serotype 2 (HRV2) J Mol Biol. 2000;300:1179–1194. doi: 10.1006/jmbi.2000.3943. [DOI] [PubMed] [Google Scholar]
- 28.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brünger AT, et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 30.Barnard DL, et al. In vitro activity of expanded-spectrum pyridazinyl oxime ethers related to pirodavir: novel capsid-binding inhibitors with potent antipicornavirus activity. Antimicrob Agents Chemother. 2004;48:1766–1772. doi: 10.1128/AAC.48.5.1766-1772.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pevear DC, et al. Relationship of pleconaril susceptibility and clinical outcomes in treatment of common colds caused by rhinoviruses. Antimicrob Agents Chemother. 2005;49:4492–4499. doi: 10.1128/AAC.49.11.4492-4499.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ledford RM, et al. VP1 sequencing of all human rhinovirus serotypes: insights into genus phylogeny and susceptibility to antiviral capsid-binding compounds. J Virol. 2004;78:3663–3674. doi: 10.1128/JVI.78.7.3663-3674.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tijsma A, et al. The capsid binder vapendavir and novel protease inhibitor SG85 inhibit enterovirus 71 replication. Antimicrob Agents Chemother. 2014 doi: 10.1128/AAC.03328-14. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.



