Abstract
Cytosine DNA methylation is an epigenetic modification in eukaryotes that maintains genome integrity and regulates gene expression. The DNA methylation patterns in plants are more complex than those in animals, and plants and animals have common as well as distinct pathways in regulating DNA methylation. Recent studies involving genetic, molecular, biochemical and genomic approaches have greatly expanded our knowledge of DNA methylation in plants. The roles of many proteins as well as non-coding RNAs in DNA methylation have been uncovered.
Keywords: siRNA, non-coding RNA, DNA methylation, Pol IV, Pol V, argonaute
DNA methylation is a type of epigenetic modification that occurs at the bases of the DNA strands. In both animals and plants, DNA methylation is mostly found at the 5th carbon of the cytosine pyrimidine ring. Methylation on DNA may alter the transcriptional activity of associated genes or maintain genome integrity by repressing transposable elements. The methylation status at some loci may respond to external or internal signals, however, as a type of epigenetic modification, the overall DNA methylation status is heritable. In animals, almost all methylated cytosines occur in the CG sequence context, although non-CG methylation has been found in embryonic stem cells and neurons [1, 2]. The mammalian DNA methyltransferase 3 (DNMT3) family functions in the establishment of DNA methylation during the development of germ cells [3, 4]. Thereafter, DNA methyltransferase 1 (DNMT1) maintains DNA methylation during DNA replication [4]. In contrast to DNA methylation in animals, plant cytosine methylation has been found at all cytosine sequence contexts—CG, CHG and CHH (H=A, T or C). In plants, de novo DNA methylation is mediated by the RNA-directed DNA methylation (RdDM) pathway. After DNA replication, multiple DNA methyltransferases are employed to maintain cytosine methylation at different sequence contexts. In this review, we focus on the progress of plant DNA methylation studies in recent years. We first discuss the DNA methylation landscapes in plants. Then we describe the mechanisms underlying de novo DNA methylation, maintenance of DNA methylation in different sequence contexts and DNA demethylation in plants. We mainly focus on the functions of proteins and non-coding RNAs involved in these processes in the model plant Arabidopsis. For easy reference, Arabidopsis genes that function in DNA methylation are listed in Table 1. Furthermore, the transgenerational inheritance and variation of DNA methylation in plants are also discussed.
Table 1.
| Gene | Genome Identifier | Gene product | Function | Suggested Reference |
|---|---|---|---|---|
| DRM2 | AT5G14620 | DNA methyltransferase | De novo methylation and maintainance of CHH methylation through RdDM | [40] |
| DRM1 | AT5G15380 | DNA methyltransferase | A homolog of DRM2 | [40] |
| MET1 | AT5G49160 | DNA methyltransferase | Maintains CG methylation | [43, 44] |
| CMT3 | AT1G69770 | DNA methyltransferase | Maintains CHG methylation | [50, 51] |
| CMT2 | AT4G19020 | DNA methyltransferase | Maintains DNA methylation at heterochromatin | [44, 58] |
| NRPD1 | AT1G63020 | Largest subunit of Pol IV | Generates 24-nt siRNA precursors in RdDM | [15, 16] |
| NRPE1 | AT2G40030 | Largest subunit of Pol V | Generates scaffold transcripts in RdDM | [11, 85, 86] |
| NRPB2 | AT4G21710 | Second largest subunit of Pol II | Generaes scaffold transcripts at some RdDM target loci | [27] |
| CLSY1 | AT3G42670 | SNF2-like chromatin remodelling protein | Interacts with Pol IV, functions in 24-nt siRNA precursor biogenesis | [18] |
| DRD1 | AT2G16390 | SNF2-like chromatin remodelling protein | Part of the DDR complex, functions in the recruitment of Pol V to RdDM target loci | [28, 29] |
| DDM1 | AT5G66750 | SNF2-like chromatin remodelling protein | Functions in the maintenance of DNA methylation at heterochromatin | [58, 87] |
| SUVH2 | AT2G33290 | SRA domain methylcytosine-binding protein | Functions in the recruitment of Pol V to RdDM loci | [33, 88] |
| SUVH9 | AT4G13460 | SRA domain methylcytosine-binding protein | A homolog of SUVH2 | [33, 88] |
| VIM1 | AT1G57820 | SRA domain methylcytosine-binding protein | Functions in the maintenance of CG methylation | [45, 46] |
| VIM2 | AT1G66050 | SRA domain methylcytosine-binding protein | A homolog of VIM1 | [46] |
| VIM3 | AT5G39550 | SRA domain methylcytosine-binding protein | A homolog of VIM1 | [46] |
| RDM4 | AT2G30280 | IWR (Interacts with Pol II)-type transcription factor | Interacts with Pol IV, functions in 24-nt siRNA precursor biogenesis | [19, 20] |
| RDR2 | AT4G11130 | RNA dependent RNA polymerase | Interacts with Pol IV, functions in 24-nt siRNA precursor biogenesis | [89, 90] |
| SHH1 | AT1G15215 | SAWADEE domain and homeodomain containing protein | Interacts with Pol IV, functions in 24-nt siRNA precursor biogenesis | [17, 21] |
| DCL3 | AT3G43920 | Dicer-like protein | Generates 24-nt siRNA duplexes in RdDM | [89] |
| AGO4 | AT2G27040 | Argonaute protein | Binds to 24-nt siRNAs, functions in RdDM | [25, 42, 91] |
| HEN1 | AT4G20910 | Small RNA methyltransferase | Methylates siRNA duplexes | [24] |
| DMS3 | AT3G49250 | Structural maintenance of chromosomes (SMC) hinge domain-containing protein | Part of the DDR complex, functions in the recruitment of Pol V to RdDM target loci | [29, 31] |
| RDM1 | AT3G22680 | Contains a domain of unknown function | Part of the DDR complex, functions in the recruitment of Pol V to RdDM target loci | [28–30] |
| STA1 | AT4G03430 | PRP6-like splicing factor | Functions in scaffold transcript biogenesis | [35] |
| KTF1 | AT5G04290 | Transcription elongation factor SPT5-like protein | Functions downstream of scaffold transcripts in RdDM | [36, 37] |
| IDN2 | AT3G48670 | dsRNA-binding protein | Functions downstream of scaffold transcripts in RdDM | [38, 92] |
| KYP | AT5G13960 | Histone H3K9 methytransferase | Functions in the maintenance of CHG methylation through CMT3 | [93] |
| SUVH5 | AT2G35160 | Histone H3K9 methytransferase | A homolog of KYP | [94, 95] |
| SUVH6 | AT2G22740 | Histone H3K9 methytransferase | A homolog of KYP | [54] |
| IBM1 | AT3G07610 | Histone demethylase | Functions in preventing ectopic DNA methylation at gene bodies | [55, 56] |
| ROS1 | AT2G36490 | DNA glycosylase | Demethylates DNA | [60, 62] |
| DML2 | AT3G10010 | DNA glycosylase | A homolog of ROS1 | [59, 96] |
| DML3 | AT4G34060 | DNA glycosylase | A homolog of ROS1 | [59, 96] |
| DME | AT5G04560 | DNA glycosylase | A homolog of ROS1 that is active in central cells | [69–71] |
| ROS4 | AT3G14980 | MBD domain-containing Histone H3 acetyltransferase | Functions in DNA demethylation at some ROS1 target loci | [61] |
| ROS3 | AT5G58130 | RNA-binding protein | Functions in DNA demethylation | [66] |
| ZDP | AT3G14890 | Phosphoesterase | Functions downstream of ROS1 in DNA demethylation | [63] |
| ATXRCC1 | AT1G80420 | DNA repair protein | Functions in DNA demethylation | [65] |
The DNA methylation landscape in plants
By combining bisulfite conversion and high-throughput sequencing, genome-wide DNA methylation has been profiled in the model plant Arabidopsis [5, 6]. The DNA methylation profiling using five-week-old Arabidopsis plants indicates that cytosine methylation levels in CG, CHG and CHH sequence contexts are 24%, 6.7% and 1.7% respectively [5]. For cytosine sites in Arabidopsis, most CG sites are either unmethylated or methylated at the 80%–100% level [5, 6], suggesting that CG methylation is precisely maintained after DNA replication. In contrast, methylated CHG sites have a wide range of methylation levels, and most methylated CHH sites have a low methylation level [5, 6], suggesting that differentially methylated CHG and CHH loci exist in different cells. In consistency with its function in transposon silencing, DNA methylation is highly enriched at pericentromeric regions where transposon and repeat frequencies are high [5, 6]. At transposons or other repeats, cytosines in all three types of sequence contexts could be methylated; whereas at the bodies of some protein-coding genes, CG methylation exists without repressing the expression of the gene [5]. Although the role CG methylation at gene bodies is not well understood, a study using different Arabidopsis accessions has shown positive correlation between gene body CG methylation levels and gene expression levels [7]. Similar to Arabidopsis, the DNA methylation profiles from soybean [8] and rice [9] also show similar distribution patterns of CG, CHG and CHH methylation in transposable elements and gene bodies.
De novo methylation and the RNA-directed DNA Methylation pathway
The establishment of DNA methylation in plants is mediated by the RNA-directed DNA Methylation (RdDM) pathway. In 1994, Wassenegger et al. first reported that the potato spindle tuber viroid (PSTVd) transgene in tobacco was methylated when the viroid RNA-RNA replication occurred, which indicated that DNA methylation was directed by homologous RNA [10]. It is now clear that the RNAs that direct DNA methylation are 24-nt (nucleotide) small interfering RNAs (siRNAs). In addition to the 24-nt siRNAs, longer non-coding RNAs specifically referred to as the scaffold RNAs also play a very important role in guiding the methyltransferase to target loci [11]. During RdDM, these two types of non-coding RNAs as well as several proteins are involved. The 24-nt siRNAs are generated by DICER-LIKE 3 (DCL3) from long double-stranded RNA (dsRNA) precursors, and then the small RNAs are loaded into AGO4 [12]. The small RNA-AGO4 complex is recruited to the RdDM target loci by the homologous nascent scaffold RNA through sequence complementarity between the siRNA and the scaffold RNA, and following this interaction, the DNA methyltransferase DOMAINS REARRANGED METHYLTRANSFERASE 2 (DRM2) is recruited to the target loci [12] (Figure 1).
Figure 1. The RNA-directed DNA methylation pathway (RdDM) in Arabidopsis.
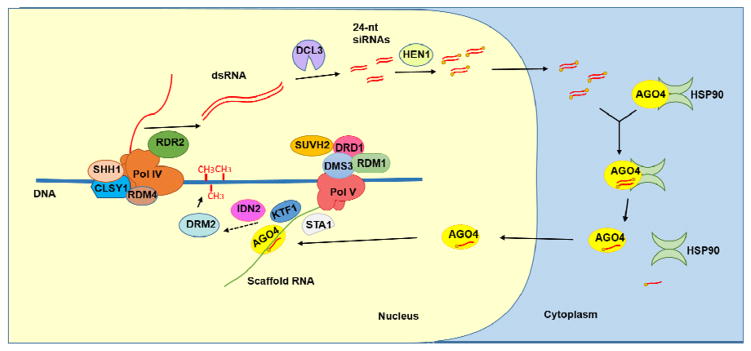
At the early stage of RdDM, RNA Polymerase IV (Pol IV) is proposed to generate a single-stranded non-coding RNA, which is converted into double-stranded RNA (dsRNA) by RNA-DEPENDENT RNA POLYMARASE 2 (RDR2). SAWADEE HOMEODOMAIN HOMOLOG 1 (SHH1), CLASSY1 (CLSY1), RNA-DIRECTED DNA METHYLATION 4 (RDM4) and RDR2 were found to be in the same complex as Pol IV and may assist the recruitment of, or transcription by, Pol IV. 24-nt siRNA duplexes are produced from dsRNA precursors by DICER-LIKE 3 (DCL3) and are methylated at their 3′ ends by HEN1. The 24-nt siRNAs are exported into the cytoplasm to be loaded into AGONAUTE4 (AGO4). The molecular chaperon HSP90 facilitates the release of the siRNA passenger strand. The AGO4 protein loaded with an siRNA is translocated into the nucleus. RNA POLYMERASE V (Pol V) generates a non-coding scaffold RNA at RdDM loci. SU(VAR)3-9 HOMOLOG 2 (SUVH2) or its homolog SUVH9 contributes to the recruitment of Pol V to methylated DNA. A protein complex containing at least three proteins, DEFECTIVE IN RNA-DIRECTED DNA METHYLATION 1 (DRD1), DEFECTIVE IN MERISTEM SILENCING 3 (DMS3) and RNA-DIRECTED DNA METHYALTION 1 (RDM1), is also required to recruit Pol V to target loci. A splicing factor STABILIZED 1 (STA1) is required for the production of scaffold RNAs, although its mechanism of action is unclear. The scaffold RNA is able to recruit the silencing effector AGO4 as well as KOW DOMAIN-CONTAINING TRANSCRIPTION FACTOR 1 (KTF1), which may help to recruit downstream factors of RdDM. A dsRNA binding protein INVOLVED IN DE NOVO 2 (IDN2) is proposed to recognize the complex of the siRNA and the scaffold RNA. These events further recruit DOMAINS REARRANGED METHYLTRANSFERASE 2 (DRM2) to the RdDM loci through an unknown mechanism.
The 24-nt siRNAs could be produced from both exogenous loci and endogenous loci. Exogenous RNAs such as transcripts derived from viruses or transgenes can be processed into siRNAs by the plant RdDM pathway to silence these foreign sequences at the transcriptional level [10, 13]. In addition, lots of endogenous loci such as transposable elements and other repeats also give rise to siRNAs, which mediate the maintenance of CHH DNA methylation through the RdDM pathway after DNA replication[14]. When the 24-nt siRNAs are produced from endogenous loci that are already methylated, the transcription of the target loci depends on the plant-specific RNA polymerase Pol IV. When the NRPD1 gene encoding the largest subunit of Pol IV is mutated, the production of almost all heterochromatic 24-nt siRNAs is abolished [14–16]. By affinity purification of NRPD1, Law et al. [17] identified several proteins that are present in the Pol IV complex, including CLASSY1 (CLSY1), RNA-DIRECTED DNA METHYLATION 4 (RDM4), SAWADEE HOMEODOMAIN HOMOLOG 1 (SHH1) and RNA-DEPENDENT RNA POLYMERASE 2 (RDR2). CLSY1 is an SNF2 domain-containing protein that is required for the accumulation of heterochromatic siRNAs and is likely to mediate the interaction between Pol IV and RDR2 [18]. RDM4 is an IWR (Interacts with Pol II)-type transcription factor that is also required for Pol II and Pol V transcription [19, 20]. The specificity of Pol IV recruitment to some target RdDM loci may be conferred by SHH1, whose SAWADEE domain is capable of recognizing histone H3 tails with unmethylated lysine (K)4 and methylated K9 [21]. RDR2 is proposed to convert the Pol IV transcripts into dsRNAs for Dicer processing. In vitro assays suggested that the activity of RDR2 in dsRNA synthesis requires Pol IV [22]. The production of 24-nt siRNAs from a dsRNA procursor requires the Dicer protein DCL3, which colocalizes with RDR2 in the nucleolus [23]. Like miRNAs in plants, the 24nt-siRNAs are methylated by the small RNA methyltransferase HEN1 at their 3′ ends to be stabilized [24]. In recent years, it was found that that the loading of 24-nt siRNA into AGO4 takes place in the cytoplasm and the maturation of the AGO4/siRNA complex requires the catalytic activity of AGO4 and the molecular chaperon HSP90 [25]. However, how siRNAs are exported to the cytoplasm and recruited to AGO4 is still unknown. After siRNA binding, AGO4 is translocated into the nucleus [25] and recruited to RdDM target loci by scaffold RNAs.
Scaffold RNAs are long nascent transcripts generated from RdDM target loci. These transcripts have triphosphates or 7meG caps at their 5′ ends and are not 3′ polyadenylated [11]. Given that scaffold RNAs physically interact with AGO4 [26], scaffold RNAs have been hypothesized to be bridges that link siRNAs to their target loci through sequence complementarity. The biogenesis of scaffold RNAs is independent of siRNA biogenesis. The scaffold RNAs are mainly produced by another plant-specific RNA polymerase, Pol V [11], and Pol II also contributes to the accumulation of scaffold RNAs at some loci [27]. In addition to the RNA polymerase, a putative chromatin-remodeling complex is also required for the biogenesis of scaffold RNAs. This complex is composed of at least three proteins: a SWI2/SNF2-like chromatin-remodeling protein DEFECTIVE IN RNA-DIRECTED DNA METHYLATION 1 (DRD1), a structural maintenance of chromosomes (SMC) hinge domain-containing protein DEFECTIVE IN MERISTEM SILENCING 3 (DMS3) and a protein without known functional domains named RNA-DIRECTED DNA METHYALTION 1 (RDM1) [28–31]. This complex is therefore termed the “DDR complex” [29]. These three proteins were biochemically co-purified [29] and genetic analysis indicates that all three genes are required for the association of Pol V with certain chomatin loci, mainly gene promoters and some evolutionarily young transposable elements [32]. A chromatin feature that attracts Pol V is the pre-existing DNA methylation, as the occupancy of NRPE1 (the largest subunit of Pol V) was lost at normal RdDM target loci and redistributed to other loci in the met1 mutant defective in the maintenance of CG methylation [33]. SU(VAR)3-9 HOMOLOG 2 (SUVH2) and its homolog SUVH9, which bind methylated DNA through the SET- and RING finger-associated (SRA) domain, recruit Pol V to methylated DNA [33, 34]. Recently, a PRP6-like splicing factor STABILIZED 1 (STA1) has been reported to be involved in the production of Pol V-dependent scaffold RNAs [35]. STA1 does not affect the splicing of most known RdDM genes and colocalizes with AGO4 in the Cajal body [35]. However, how STA1 functions in RdDM remains elusive.
The recruitment of AGO4-siRNA to RdDM target loci by Pol V-dependent scaffold transcripts initiates DNA methylation at these loci. The KOW DOMAIN-CONTAINING TRANSCRIPTION FACTOR 1 (KTF1) protein is also recruited to target loci in a Pol V-dependent manner and is proposed to function in the recruitment of chromatin modifiers together with AGO4. [36, 37]. Furthermore, a dsRNA-binding protein INVOLVED IN DE NOVO 2 (IDN2) acts downstream of scaffold RNA transcription and may function in nucleosome remodeling at target loci in RdDM [38, 39]. The final key step of RdDM is the recruitment of methyltransferase DRM2 or its homolog DRM1 [40]. However, a direct link between Pol V-dependent transcripts and the recruitment of DRM2 is still missing. Although there has been no evidence of the downstream effectors of RdDM being involved in the production of siRNAs, it should be noted that some of them such as AGO4, Pol V and DRM2 are required in the accumulation of siRNAs at a subset of RdDM loci [21, 41]. Considering that mutations in NRPE1 (encoding the largest subunit of Pol V) do not affect the accumulation of the AGO4 protein whereas those in NRPD1 and RDR2 do [41], the effect of Pol V and DRM2 on siRNA accumulation is likely due to feedback regulation from loss of DNA methylation. AGO4 could contribute to siRNA accumulation at multiple levels. First, the slicer activity of AGO4 is required for the maturation of the AGO4-siRNA complex [25], in which AGO4 may stabilize siRNAs by protecting them from nucleases. Second, AGO4 may promote the production of secondary siRNAs triggered by the slicing of scaffold RNAs by primary siRNAs. It has been found that the siRNA accumulation level at some RdDM target loci requires the slicing activity of AGO4 [41, 42], raising the possibility that secondary siRNAs are produced from these loci. Yet the cleavage products of scaffold transcripts by AGO4 in vivo have not been reported. Finally, the reduced siRNA accumulation at some loci in ago4 mutants could also result from the feedback regulation from reduced DNA methylation.
Maintenance of DNA methylation
After DNA replication, only the template strand is methylated. DNA methyltransferases in plants deposit methyl groups onto the nascent strand in order to maintain the DNA methylation patterns through cell division. It has been well characterized in Arabidopsis that the maintenance of DNA methylation in different cytosine sequence contexts is implemented by different methyltransferases through distinct pathways (Figure 2).
Figure 2. Maintenance of DNA methylation in different sequence contexts in Arabidopsis.
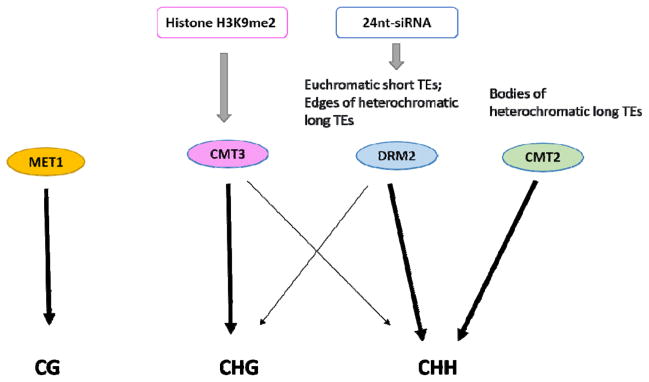
The maintenance of cytosine methylation in the CG sequence context requires DNA METHYLTRANSFERASE 1 (MET1). CHG methylation is mainly maintained by CHROMOMETHYLASE 3 (CMT3), which is guided by the histone H3K9me2 mark. DOMAINS REARRANGED METHYLTRANSFERASE 2 (DRM2) mediates the maintenance of CHH methylation at euchromatic short TEs or the edges of heterochromatic long TEs through the RNA-directed DNA methylation (RdDM) pathway. At the bodies of heterochromatic long TEs where the RdDM machineries cannot access, cytosine methylation in the CHH sequence context is maintained by CHROMOMETHYLASE 2 (CMT2). Crosstalks also exist between different DNA methylation pathways. Maintenance of CHH methylation at some loci requires CMT3 while maintenance of CHG methylation at some loci requires the RdDM pathway.
The maintenance of CG methylation is mediated primarily by DNA METHYLTRANSFERASE 1 (MET1)—the homolog of the mammalian DNMT1 [43, 44]. As a type of symmetric DNA methylation, methylated CG sites become hemi-methylated after DNA replication and MET1 is recruited to restore the target loci to a fully methylated state. In addition to MET1, the maintenance of CG methylation also requires three VARIATION IN METHYLATION family proteins: VIM1, VIM2 and VIM3. These proteins contain a plant homeodomain (PHD) domain, a SRA domain and two RING domains [45, 46]. From in vitro assays, it has been shown that VIM1 is a methylcytosine-binding protein [45] that preferentially binds hemi-methylated CG sites [47]. In the vim1 vim2 vim3 triple mutant, a global loss of DNA methylation in the CG sequence context that strongly resembled the methylation profile in met1 mutants was observed [48], suggesting that these VIM proteins may function upstream of MET1. Introduction of a wild-type MET1 into the met1 mutant restored CG methylation at transposons. In contrast, DNA methylation at gene bodies could not be restored when MET1 was introduced back into met1 mutants [48]. Since non-CG methylation maintained by other methyltransferases is preserved at transposons but not gene bodies in the met1 mutant, these different behaviors of MET1 target loci during re-methylation suggested that maintenance of DNA methylation by MET1 depends on pre-existing DNA methylation. Recently, Ruscio et al. [49] reported that in human cells, a non-coding transcript generated from the gene locus CEBPA associated with DNMT1 and prevented DNA methylation at this locus. Genome-wide profiling of DNMT1-associated RNAs revealed that the DNMT1-RNA interaction occurred at thousands of loci [49]. It remains to be tested whether Arabidopsis MET1 is regulated by non-coding RNAs in a similar manner.
The maintenance of CHG methylation mostly depends on a plant-specific DNA methyltransferase named CHROMOMETHYLASE 3 (CMT3), which is guided by the dimethylated histone H3K9 (H3K9me2) mark [50, 51]. The H3K9me2 modification in the Arabidopsis genome is distributed at loci of transposable elements and other repeats of mainly pericentromeric regions as well as some euchromatic regions [52]. The maintenance of histone H3K9me2 is mediated by the histone methyltransferase KRYPTONITE (KYP)/SUVH4 and its homologs SU(VAR)3-9 HOMOLOG 5 (SUVH5) and SUVH6 in Arabidopsis. In the kyp single mutant or the kyp suvh5 suvh6 triple mutant, not only was the H3K9me2 level reduced [53], DNA methylation in the CHG sequence context at those loci was also decreased [48]. Du et al. [50] showed that CMT3 could bind to the methylated histone H3 tails through its Chromo and BAH domains and therefore be recruited to the H3K9me2 sites. Interestingly, the histone methyltransferase KYP could preferentially bind to methylated cytosines through its SRA domain, and loss of DNA methylation also resulted in decreased H3K9me2 accumulation [54]. These lines of evidence suggest that a self-reinforcing loop exists between DNA methylation and histone H3K9me2 to silence transposable elements and repeats. Since the SRA domain of the histone H3K9 methyltransferases binds to methylated cytosines in all sequence contexts [54], a histone demethylase INCREASED IN BONSAI METHYLATION 1 (IBM1) is required to prevent ectopic H3K9me2 and DNA methylation at genes [48, 55, 56]. Besides the CMT3/SUVH pathway, the DNA methyltransferase DRM2 also contributes to the maintenance of DNA methylation in the CHG sequence context at some loci through the RdDM pathway [48]. So far, no RNA was found to directly function in the CMT3/SUVH pathway. However, by investigating a natural inverted repeat locus formed by two PHOSPHORIBOSYLANTHRANILATE ISOMERASE (PAI) genes, Enke et al. [57] found that although non-CG DNA methylation at this locus was only dependent on the CMT3/SUVH pathway and was independent of DRM2, the maintenance of non-CG DNA methylation and H3K9me2 was compromised in the dcl2 dcl3 dcl4 triple mutant, indicating that small RNAs function in the maintenance of non-CG methylation in a DRM2-independent mechanism and prevent the loss of H3K9me2 during Pol II transcription. Further mechanistic studies will be required to provide insights to the role of small RNAs in this process.
Cytosine methylation in the CHH sequence context is asymmetric and methylation at a CHH locus will be lost in one daughter DNA molecule after replication. The previously introduced RdDM pathway is responsible for the maintenance of CHH methylation at target loci, mainly small TEs at euchromatic arms or the edges of long TEs. The histone remodeler in the RdDM pathway, DRD1, may have lower efficiency in remodeling nucleosomes at highly condensed heterochromatic regions [58] and therefore these regions cannot be accessed by the RdDM machinery. At those heterochromatic loci, another SWI2/SNF2 type chromatin remodeling protein DECREASED DNA METHYLATION 1 (DDM1) functions in the remodeling of nucleosomes, allowing methyltransferases to access the DNA [58]. At these loci, the maintenance of DNA methylation in CG and CHG sequence contexts requires MET1 and CMT3 respectively, and CMT2, a CMT3 homolog, mediates the maintenance of cytosine methylation in the CHH sequence context [58].
Although plants have specialized pathways to maintain DNA methylation in different sequence contexts, genome methylation profiles in various methylation pathway mutants showed that crosstalks exist between different pathways in maintaining DNA methylation in different cytosine sequence contexts [48]. In met1 mutants, loss of both CG and non-CG methylation was observed at some loci, suggesting that non-CG methylation at these loci may depend on CG methylation. Mutations in the CMT3/SUVH pathway genes such as CMT3, KYP and SUVH5/6 also lead to loss of CHH methylation at some loci, while the mechanism of KYP- and SUVH5/6-mediated CHH methylation maintenance at some of these loci may be independent of CMT3 [48].
DNA Demethylation
The methylation status of DNA is dynamically regulated. At certain developmental stages or in certain cell types, DNA demethylation occurs, either through passive loss of DNA methylation after DNA replication, or active DNA demethylation by DNA glycosylases.
In somatic cells, a DNA glycosylase named REPRESSOR OF SILENCING 1 (ROS1) together with its homologs DEMETER-LIKE 2(DML2) and DML3 actively remove DNA methylation to prevent hyper-methylation at their target loci [59, 60]. So far, only a few proteins involved in demethylation have been identified in Arabidopsis. A histone acetyltransferase named REPRESSOR OF SILENCING 4 (ROS4) is known to act at a subset of ROS1 target loci [61]. In addition to the N-Acyltransferase domain, ROS4 also contains a MBD domain that could bind to methylated cytosines, and a PHD finger domain that could bind to histone proteins without H3K4 di- or trimethylation [61]. Thus ROS4 may recognize chromatin loci with certain features and provide a favorable environment for DNA glycosylases by changing histone modifications. ROS1 and other glycosylases demethylate DNA by excising methylated cytosines through their glycosylase/lyase activity [60, 62]. Following base excision, the 3′ phosphate generated by ROS1 is removed by the DNA phosphatase ZDP [63] and the gap is filled with an unmethylated cytosine by the DNA repair system [64]. The Arabidopsis homolog of animal X-RAY REPAIR CROSS COMPLEMENTING 1 (XRCC1) –ATXRCC1 may function in stimulating the activity of ROS1 and ZDP [65]. An RNA-binding protein REPRESSOR OF SILENCING 3 (ROS3) is also involved in DNA demethylation [66]. ROS3 colocalizes with ROS1 in vivo and may guide ROS1 to its target loci.
Demethylation of DNA also takes place during the development of reproductive cells. During male gametogenesis, DDM1 expression is down-regulated in the vegetative nuclei, resulting in loss of DNA methylation and activation of transposable elements [67]. 21-nt siRNAs generated from activated TEs are thought to move into the sperm nuclei to reinforce the DNA methylation at target loci [67]. Thus demethylation in the vegetative nuclei ensures trans-generational silencing of transposable elements. On the other hand, MET1 is down-regulated during female gametogenesis [68], causing passive DNA demethylation through cell cycles. In addition, another DNA glycosylase DEMETER (DME), which is homologous to ROS1, actively demethylates DNA in the central cell [69–71]. As a result, the CG methylation level is decreased in the central cell and the eventual endosperm cells. It has been suggested that the activated transposable elements in the endosperm also give rise to siRNAs that move into the embryo where they reinforce the silencing status of their homologous sequences [72, 73]. However, the proposed movement of siRNAs from non-germ line cells to germ-line cells has yet to be shown experimentally.
Transgenerational inheritance and variation of DNA methylation
The patterns of DNA methylation are not only maintained through mitosis, but also transmitted to the next generation. Studies on the epigenetic inheritance of DNA methylation using soybean recombinant inbred lines (RILs) [8] and maize RILs [74] showed that most differentially methylated regions between the parents of the RILs behaved like genetic polymorphisms and cosegregated with the genetic background. Yet, unusual epigenetic inheritance such as paramutation may also be related to DNA methylation. Paramutation refers to the interaction between two alleles in which one allele is able to induce a heritable change in the other. A classic case of paramutation is the interaction of the two alleles B-I and B′ at the maize b1 locus. When two plants carrying B-I and B′ respectively were crossed together, the highly expressed B-I allele would be converted into the silenced B′ allele [75]. A region containing seven tandem repeats upstream of b1 is required for paramutation [76] and the methylation status of this regulatory region is different between B-I and B′ [77]. Furthermore, MEDIATOR OF PARAMUTATION 1 (MOP1) encoding an RNA-dependent RNA polymerase was shown to function in paramutation [78], suggesting that non-coding RNA is involved. It has been proposed that RdDM is involved in the conversion of highly expressed alleles into silenced alleles in paramutation [77].
Like genetic variations, spontaneous changes in DNA methylation also happen during genome replication. In recent years, studies on the methylomes of Arabidopsis progenies from a common ancestor that was propagated for 30 generations revealed that although the overall DNA methylation patterns were well maintained over generations, a lot of epigenetic variations in DNA methylation were observed among descendent lines [79, 80]. Based on differentially methylated regions among different lines, protein-coding genes showed a higher frequency of epigenetic change compared to transposable elements that are targets of RdDM [79, 80]. Some DNA methylation changes at protein-coding genes created epialleles and changed gene expression levels [79]. Thus epigenetic variations also contribute to phenotypic diversity and adaptation.
Conclusions and future perspectives
DNA methylation is an important type of epigenetic modification in eukaryotes to maintain genome integrity, silence the transcription of exogenous DNA and regulate endogenous gene expression. Although plants and animals show similarities in the maintenance of CG methylation, plants have evolved distinct pathways in the maintenance of DNA methylation in CHG and CHH sequence contexts. The establishment of DNA methylation requires non-coding RNAs, especially small RNAs. It is noteworthy that piwi-interacting RNAs (piRNAs), a class of small RNAs found in animals, have been implicated in guiding DNA methylation in mice [81, 82] although the underlying mechanisms are unknown. It is possible that piRNAs cause DNA methylation in a similar manner as 24-nt siRNAs in plants, for which the underlying mechanism is well established. Furthermore, active DNA demethylation by glycosylases prevents ectopic methylation in the genome. Through the regulation of both DNA methylation and demethylation pathways, the plant genome maintains proper methylation levels and patterns through cell divisions and responds correctly to developmental or environmental signals.
During DNA methylation or demethylation, DNA methyltransferses or demethylases are precisely recruited to their target loci, due to the guidance from other components in their pathways such as modified histone proteins and non-coding RNAs. Yet many missing links remain to be established to fully understand how proteins in the DNA methylation pathways are accurately guided to and restricted within their target loci without disturbing neighboring regions. For example, what is the signal that triggers de novo methylation at some transgenes; and how methylcytosine-binding proteins distinguish RdDM target loci and CG methylation at gene bodies. In addition, the downstream events that affect transcription after DNA methylation are unknown. To further understand the molecular mechanisms underlying DNA methylation in plants, more factors need to be identified. Although many genes were identified through forward genetic screens using DNA methylation sensitive reporter lines in the last decade, this approach limited the discovery of genes that are functionally redundant or not stringently required in DNA methylation. Other methods such as biochemical or proteomic approaches may lead to the discovery more proteins in DNA methylation pathways.
The application of high-throughput sequencing in DNA methylation studies also greatly contributed to our knowledge of DNA methylation in the genomic scale. Future applications of some newly developed high-throughput sequencing approaches such as RNA-immunoprecipitation sequencing (RIP-seq) [83] or Crosslinking immunoprecipitation sequencing (CLIP-seq) [84] to profile RNAs associated with heterochromatin or DNA methylation related proteins would provide new insights into the role of non-coding RNAs in DNA methylation. Although the DNA methylome has been profiled in plants, however, the mixing of different cell types in methylome studies may mask some methylation patterns in specific cell types that are underrepresented in the samples. Future improvements of cell type-specific DNA isolation or methylome sequencing from a single cell or a few cells would help understand the spatial and temporal dynamics of DNA methylation in plants.
Acknowledgments
Research in the Chen lab is supported by grants from NIH (GM061146), USDA (2010-04209), Howard Hughes Medical Institute, and Gordon and Betty Moore Foundation (GBMF3046).
References
- 1.Lister R, Pelizzola M, Dowen RH, Hawkins RD, Hon G, Tonti-Filippini J, Nery JR, Lee L, Ye Z, Ngo QM, et al. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature. 2009;462:315–322. doi: 10.1038/nature08514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lister R, Mukamel EA, Nery JR, Urich M, Puddifoot CA, Johnson ND, Lucero J, Huang Y, Dwork AJ, Schultz MD, et al. Global epigenomic reconfiguration during mammalian brain development. Science (New York, NY) 2013;341:1237905. doi: 10.1126/science.1237905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kaneda M, Okano M, Hata K, Sado T, Tsujimoto N, Li E, Sasaki H. Essential role for de novo DNA methyltransferase Dnmt3a in paternal and maternal imprinting. Nature. 2004;429:900–903. doi: 10.1038/nature02633. [DOI] [PubMed] [Google Scholar]
- 4.Reik W. Stability and flexibility of epigenetic gene regulation in mammalian development. Nature. 2007;447:425–432. doi: 10.1038/nature05918. [DOI] [PubMed] [Google Scholar]
- 5.Cokus SJ, Feng S, Zhang X, Chen Z, Merriman B, Haudenschild CD, Pradhan S, Nelson SF, Pellegrini M, Jacobsen SE. Shotgun bisulphite sequencing of the Arabidopsis genome reveals DNA methylation patterning. Nature. 2008;452:215–219. doi: 10.1038/nature06745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lister R, O’Malley RC, Tonti-Filippini J, Gregory BD, Berry CC, Millar AH, Ecker JR. Highly integrated single-base resolution maps of the epigenome in Arabidopsis. Cell. 2008;133:523–536. doi: 10.1016/j.cell.2008.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schmitz RJ, Schultz MD, Urich MA, Nery JR, Pelizzola M, Libiger O, Alix A, McCosh RB, Chen H, Schork NJ, et al. Patterns of population epigenomic diversity. Nature. 2013;495:193–198. doi: 10.1038/nature11968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schmitz RJ, He Y, Valdes-Lopez O, Khan SM, Joshi T, Urich MA, Nery JR, Diers B, Xu D, Stacey G, et al. Epigenome-wide inheritance of cytosine methylation variants in a recombinant inbred population. Genome research. 2013;23:1663–1674. doi: 10.1101/gr.152538.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li X, Zhu J, Hu F, Ge S, Ye M, Xiang H, Zhang G, Zheng X, Zhang H, Zhang S, et al. Single-base resolution maps of cultivated and wild rice methylomes and regulatory roles of DNA methylation in plant gene expression. BMC genomics. 2012;13:300. doi: 10.1186/1471-2164-13-300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wassenegger M, Heimes S, Riedel L, Sanger HL. RNA-directed de novo methylation of genomic sequences in plants. Cell. 1994;76:567–576. doi: 10.1016/0092-8674(94)90119-8. [DOI] [PubMed] [Google Scholar]
- 11.Wierzbicki AT, Haag JR, Pikaard CS. Noncoding transcription by RNA polymerase Pol IVb/Pol V mediates transcriptional silencing of overlapping and adjacent genes. Cell. 2008;135:635–648. doi: 10.1016/j.cell.2008.09.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Law JA, Jacobsen SE. Establishing, maintaining and modifying DNA methylation patterns in plants and animals. Nature reviews Genetics. 2010;11:204–220. doi: 10.1038/nrg2719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jones L, Hamilton AJ, Voinnet O, Thomas CL, Maule AJ, Baulcombe DC. RNA-DNA interactions and DNA methylation in post-transcriptional gene silencing. The Plant cell. 1999;11:2291–2301. doi: 10.1105/tpc.11.12.2291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Onodera Y, Haag JR, Ream T, Costa Nunes P, Pontes O, Pikaard CS. Plant nuclear RNA polymerase IV mediates siRNA and DNA methylation-dependent heterochromatin formation. Cell. 2005;120:613–622. doi: 10.1016/j.cell.2005.02.007. [DOI] [PubMed] [Google Scholar]
- 15.Zhang X, Henderson IR, Lu C, Green PJ, Jacobsen SE. Role of RNA polymerase IV in plant small RNA metabolism. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:4536–4541. doi: 10.1073/pnas.0611456104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Herr AJ, Jensen MB, Dalmay T, Baulcombe DC. RNA polymerase IV directs silencing of endogenous DNA. Science (New York, NY) 2005;308:118–120. doi: 10.1126/science.1106910. [DOI] [PubMed] [Google Scholar]
- 17.Law JA, Vashisht AA, Wohlschlegel JA, Jacobsen SE. SHH1, a homeodomain protein required for DNA methylation, as well as RDR2, RDM4, and chromatin remodeling factors, associate with RNA polymerase IV. PLoS genetics. 2011;7:e1002195. doi: 10.1371/journal.pgen.1002195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Smith LM, Pontes O, Searle I, Yelina N, Yousafzai FK, Herr AJ, Pikaard CS, Baulcombe DC. An SNF2 protein associated with nuclear RNA silencing and the spread of a silencing signal between cells in Arabidopsis. The Plant cell. 2007;19:1507–1521. doi: 10.1105/tpc.107.051540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.He XJ, Hsu YF, Zhu S, Liu HL, Pontes O, Zhu J, Cui X, Wang CS, Zhu JK. A conserved transcriptional regulator is required for RNA-directed DNA methylation and plant development. Genes & development. 2009;23:2717–2722. doi: 10.1101/gad.1851809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kanno T, Bucher E, Daxinger L, Huettel B, Kreil DP, Breinig F, Lind M, Schmitt MJ, Simon SA, Gurazada SG, et al. RNA-directed DNA methylation and plant development require an IWR1-type transcription factor. EMBO reports. 2010;11:65–71. doi: 10.1038/embor.2009.246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Law JA, Du J, Hale CJ, Feng S, Krajewski K, Palanca AM, Strahl BD, Patel DJ, Jacobsen SE. Polymerase IV occupancy at RNA-directed DNA methylation sites requires SHH1. Nature. 2013;498:385–389. doi: 10.1038/nature12178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Haag JR, Ream TS, Marasco M, Nicora CD, Norbeck AD, Pasa-Tolic L, Pikaard CS. In vitro transcription activities of Pol IV, Pol V, and RDR2 reveal coupling of Pol IV and RDR2 for dsRNA synthesis in plant RNA silencing. Molecular cell. 2012;48:811–818. doi: 10.1016/j.molcel.2012.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pontes O, Li CF, Costa Nunes P, Haag J, Ream T, Vitins A, Jacobsen SE, Pikaard CS. The Arabidopsis chromatin-modifying nuclear siRNA pathway involves a nucleolar RNA processing center. Cell. 2006;126:79–92. doi: 10.1016/j.cell.2006.05.031. [DOI] [PubMed] [Google Scholar]
- 24.Li J, Yang Z, Yu B, Liu J, Chen X. Methylation protects miRNAs and siRNAs from a 3′-end uridylation activity in Arabidopsis. Current biology: CB. 2005;15:1501–1507. doi: 10.1016/j.cub.2005.07.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ye R, Wang W, Iki T, Liu C, Wu Y, Ishikawa M, Zhou X, Qi Y. Cytoplasmic assembly and selective nuclear import of Arabidopsis Argonaute4/siRNA complexes. Molecular cell. 2012;46:859–870. doi: 10.1016/j.molcel.2012.04.013. [DOI] [PubMed] [Google Scholar]
- 26.Wierzbicki AT, Ream TS, Haag JR, Pikaard CS. RNA polymerase V transcription guides ARGONAUTE4 to chromatin. Nature genetics. 2009;41:630–634. doi: 10.1038/ng.365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zheng B, Wang Z, Li S, Yu B, Liu JY, Chen X. Intergenic transcription by RNA polymerase II coordinates Pol IV and Pol V in siRNA-directed transcriptional gene silencing in Arabidopsis. Genes & development. 2009;23:2850–2860. doi: 10.1101/gad.1868009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kanno T, Mette MF, Kreil DP, Aufsatz W, Matzke M, Matzke AJ. Involvement of putative SNF2 chromatin remodeling protein DRD1 in RNA-directed DNA methylation. Current biology: CB. 2004;14:801–805. doi: 10.1016/j.cub.2004.04.037. [DOI] [PubMed] [Google Scholar]
- 29.Law JA, Ausin I, Johnson LM, Vashisht AA, Zhu JK, Wohlschlegel JA, Jacobsen SE. A protein complex required for polymerase V transcripts and RNA- directed DNA methylation in Arabidopsis. Current biology: CB. 2010;20:951–956. doi: 10.1016/j.cub.2010.03.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gao Z, Liu HL, Daxinger L, Pontes O, He X, Qian W, Lin H, Xie M, Lorkovic ZJ, Zhang S, et al. An RNA polymerase II- and AGO4-associated protein acts in RNA-directed DNA methylation. Nature. 2010;465:106–109. doi: 10.1038/nature09025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kanno T, Bucher E, Daxinger L, Huettel B, Bohmdorfer G, Gregor W, Kreil DP, Matzke M, Matzke AJ. A structural-maintenance-of-chromosomes hinge domain-containing protein is required for RNA-directed DNA methylation. Nature genetics. 2008;40:670–675. doi: 10.1038/ng.119. [DOI] [PubMed] [Google Scholar]
- 32.Zhong X, Hale CJ, Law JA, Johnson LM, Feng S, Tu A, Jacobsen SE. DDR complex facilitates global association of RNA polymerase V to promoters and evolutionarily young transposons. Nature structural & molecular biology. 2012;19:870–875. doi: 10.1038/nsmb.2354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Johnson LM, Du J, Hale CJ, Bischof S, Feng S, Chodavarapu RK, Zhong X, Marson G, Pellegrini M, Segal DJ, et al. SRA- and SET-domain-containing proteins link RNA polymerase V occupancy to DNA methylation. Nature. 2014 doi: 10.1038/nature12931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu ZW, Shao CR, Zhang CJ, Zhou JX, Zhang SW, Li L, Chen S, Huang HW, Cai T, He XJ. The SET domain proteins SUVH2 and SUVH9 are required for Pol V occupancy at RNA-directed DNA methylation loci. PLoS genetics. 2014;10:e1003948. doi: 10.1371/journal.pgen.1003948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dou K, Huang CF, Ma ZY, Zhang CJ, Zhou JX, Huang HW, Cai T, Tang K, Zhu JK, He XJ. The PRP6-like splicing factor STA1 is involved in RNA-directed DNA methylation by facilitating the production of Pol V-dependent scaffold RNAs. Nucleic acids research. 2013;41:8489–8502. doi: 10.1093/nar/gkt639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rowley MJ, Avrutsky MI, Sifuentes CJ, Pereira L, Wierzbicki AT. Independent chromatin binding of ARGONAUTE4 and SPT5L/KTF1 mediates transcriptional gene silencing. PLoS genetics. 2011;7:e1002120. doi: 10.1371/journal.pgen.1002120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.He XJ, Hsu YF, Zhu S, Wierzbicki AT, Pontes O, Pikaard CS, Liu HL, Wang CS, Jin H, Zhu JK. An effector of RNA-directed DNA methylation in arabidopsis is an ARGONAUTE 4- and RNA-binding protein. Cell. 2009;137:498–508. doi: 10.1016/j.cell.2009.04.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ausin I, Greenberg MV, Simanshu DK, Hale CJ, Vashisht AA, Simon SA, Lee TF, Feng S, Espanola SD, Meyers BC, et al. INVOLVED IN DE NOVO 2-containing complex involved in RNA-directed DNA methylation in Arabidopsis. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:8374–8381. doi: 10.1073/pnas.1206638109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhu Y, Rowley MJ, Bohmdorfer G, Wierzbicki AT. A SWI/SNF chromatin-remodeling complex acts in noncoding RNA-mediated transcriptional silencing. Molecular cell. 2013;49:298–309. doi: 10.1016/j.molcel.2012.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cao X, Jacobsen SE. Role of the arabidopsis DRM methyltransferases in de novo DNA methylation and gene silencing. Current biology: CB. 2002;12:1138–1144. doi: 10.1016/s0960-9822(02)00925-9. [DOI] [PubMed] [Google Scholar]
- 41.Havecker ER, Wallbridge LM, Hardcastle TJ, Bush MS, Kelly KA, Dunn RM, Schwach F, Doonan JH, Baulcombe DC. The Arabidopsis RNA-directed DNA methylation argonautes functionally diverge based on their expression and interaction with target loci. The Plant cell. 2010;22:321–334. doi: 10.1105/tpc.109.072199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Qi Y, He X, Wang XJ, Kohany O, Jurka J, Hannon GJ. Distinct catalytic and non-catalytic roles of ARGONAUTE4 in RNA-directed DNA methylation. Nature. 2006;443:1008–1012. doi: 10.1038/nature05198. [DOI] [PubMed] [Google Scholar]
- 43.Kankel MW, Ramsey DE, Stokes TL, Flowers SK, Haag JR, Jeddeloh JA, Riddle NC, Verbsky ML, Richards EJ. Arabidopsis MET1 cytosine methyltransferase mutants. Genetics. 2003;163:1109–1122. doi: 10.1093/genetics/163.3.1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Finnegan EJ, Kovac KA. Plant DNA methyltransferases. Plant molecular biology. 2000;43:189–201. doi: 10.1023/a:1006427226972. [DOI] [PubMed] [Google Scholar]
- 45.Woo HR, Pontes O, Pikaard CS, Richards EJ. VIM1, a methylcytosine-binding protein required for centromeric heterochromatinization. Genes & development. 2007;21:267–277. doi: 10.1101/gad.1512007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Woo HR, Dittmer TA, Richards EJ. Three SRA-domain methylcytosine-binding proteins cooperate to maintain global CpG methylation and epigenetic silencing in Arabidopsis. PLoS genetics. 2008;4:e1000156. doi: 10.1371/journal.pgen.1000156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yao Q, Song CX, He C, Kumaran D, Dunn JJ. Heterologous expression and purification of Arabidopsis thaliana VIM1 protein: in vitro evidence for its inability to recognize hydroxymethylcytosine, a rare base in Arabidopsis DNA. Protein expression and purification. 2012;83:104–111. doi: 10.1016/j.pep.2012.03.003. [DOI] [PubMed] [Google Scholar]
- 48.Stroud H, Greenberg MV, Feng S, Bernatavichute YV, Jacobsen SE. Comprehensive analysis of silencing mutants reveals complex regulation of the Arabidopsis methylome. Cell. 2013;152:352–364. doi: 10.1016/j.cell.2012.10.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Di Ruscio A, Ebralidze AK, Benoukraf T, Amabile G, Goff LA, Terragni J, Figueroa ME, De Figueiredo Pontes LL, Alberich-Jorda M, Zhang P, et al. DNMT1-interacting RNAs block gene-specific DNA methylation. Nature. 2013;503:371–376. doi: 10.1038/nature12598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Du J, Zhong X, Bernatavichute YV, Stroud H, Feng S, Caro E, Vashisht AA, Terragni J, Chin HG, Tu A, et al. Dual binding of chromomethylase domains to H3K9me2-containing nucleosomes directs DNA methylation in plants. Cell. 2012;151:167–180. doi: 10.1016/j.cell.2012.07.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lindroth AM, Cao X, Jackson JP, Zilberman D, McCallum CM, Henikoff S, Jacobsen SE. Requirement of CHROMOMETHYLASE3 for maintenance of CpXpG methylation. Science (New York, NY) 2001;292:2077–2080. doi: 10.1126/science.1059745. [DOI] [PubMed] [Google Scholar]
- 52.Bernatavichute YV, Zhang X, Cokus S, Pellegrini M, Jacobsen SE. Genome-wide association of histone H3 lysine nine methylation with CHG DNA methylation in Arabidopsis thaliana. PloS one. 2008;3:e3156. doi: 10.1371/journal.pone.0003156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Inagaki S, Miura-Kamio A, Nakamura Y, Lu F, Cui X, Cao X, Kimura H, Saze H, Kakutani T. Autocatalytic differentiation of epigenetic modifications within the Arabidopsis genome. The EMBO journal. 2010;29:3496–3506. doi: 10.1038/emboj.2010.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Johnson LM, Bostick M, Zhang X, Kraft E, Henderson I, Callis J, Jacobsen SE. The SRA methyl-cytosine-binding domain links DNA and histone methylation. Current biology: CB. 2007;17:379–384. doi: 10.1016/j.cub.2007.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Saze H, Shiraishi A, Miura A, Kakutani T. Control of genic DNA methylation by a jmjC domain-containing protein in Arabidopsis thaliana. Science (New York, NY) 2008;319:462–465. doi: 10.1126/science.1150987. [DOI] [PubMed] [Google Scholar]
- 56.Miura A, Nakamura M, Inagaki S, Kobayashi A, Saze H, Kakutani T. An Arabidopsis jmjC domain protein protects transcribed genes from DNA methylation at CHG sites. The EMBO journal. 2009;28:1078–1086. doi: 10.1038/emboj.2009.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Enke RA, Dong Z, Bender J. Small RNAs prevent transcription-coupled loss of histone H3 lysine 9 methylation in Arabidopsis thaliana. PLoS genetics. 2011;7:e1002350. doi: 10.1371/journal.pgen.1002350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zemach A, Kim MY, Hsieh PH, Coleman-Derr D, Eshed-Williams L, Thao K, Harmer SL, Zilberman D. The Arabidopsis nucleosome remodeler DDM1 allows DNA methyltransferases to access H1-containing heterochromatin. Cell. 2013;153:193–205. doi: 10.1016/j.cell.2013.02.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Penterman J, Zilberman D, Huh JH, Ballinger T, Henikoff S, Fischer RL. DNA demethylation in the Arabidopsis genome. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:6752–6757. doi: 10.1073/pnas.0701861104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gong Z, Morales-Ruiz T, Ariza RR, Roldan-Arjona T, David L, Zhu JK. ROS1, a repressor of transcriptional gene silencing in Arabidopsis, encodes a DNA glycosylase/lyase. Cell. 2002;111:803–814. doi: 10.1016/s0092-8674(02)01133-9. [DOI] [PubMed] [Google Scholar]
- 61.Qian W, Miki D, Zhang H, Liu Y, Zhang X, Tang K, Kan Y, La H, Li X, Li S, et al. A histone acetyltransferase regulates active DNA demethylation in Arabidopsis. Science (New York, NY) 2012;336:1445–1448. doi: 10.1126/science.1219416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Agius F, Kapoor A, Zhu JK. Role of the Arabidopsis DNA glycosylase/lyase ROS1 in active DNA demethylation. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:11796–11801. doi: 10.1073/pnas.0603563103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Martinez-Macias MI, Qian W, Miki D, Pontes O, Liu Y, Tang K, Liu R, Morales-Ruiz T, Ariza RR, Roldan-Arjona T, et al. A DNA 3′ phosphatase functions in active DNA demethylation in Arabidopsis. Molecular cell. 2012;45:357–370. doi: 10.1016/j.molcel.2011.11.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gehring M, Reik W, Henikoff S. DNA demethylation by DNA repair. Trends in genetics: TIG. 2009;25:82–90. doi: 10.1016/j.tig.2008.12.001. [DOI] [PubMed] [Google Scholar]
- 65.Martinez-Macias MI, Cordoba-Canero D, Ariza RR, Roldan-Arjona T. The DNA repair protein XRCC1 functions in the plant DNA demethylation pathway by stimulating cytosine methylation (5-meC) excision, gap tailoring, and DNA ligation. The Journal of biological chemistry. 2013;288:5496–5505. doi: 10.1074/jbc.M112.427617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zheng X, Pontes O, Zhu J, Miki D, Zhang F, Li WX, Iida K, Kapoor A, Pikaard CS, Zhu JK. ROS3 is an RNA-binding protein required for DNA demethylation in Arabidopsis. Nature. 2008;455:1259–1262. doi: 10.1038/nature07305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Slotkin RK, Vaughn M, Borges F, Tanurdzic M, Becker JD, Feijo JA, Martienssen RA. Epigenetic reprogramming and small RNA silencing of transposable elements in pollen. Cell. 2009;136:461–472. doi: 10.1016/j.cell.2008.12.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Jullien PE, Mosquna A, Ingouff M, Sakata T, Ohad N, Berger F. Retinoblastoma and its binding partner MSI1 control imprinting in Arabidopsis. PLoS biology. 2008;6:e194. doi: 10.1371/journal.pbio.0060194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Choi Y, Gehring M, Johnson L, Hannon M, Harada JJ, Goldberg RB, Jacobsen SE, Fischer RL. DEMETER, a DNA glycosylase domain protein, is required for endosperm gene imprinting and seed viability in arabidopsis. Cell. 2002;110:33–42. doi: 10.1016/s0092-8674(02)00807-3. [DOI] [PubMed] [Google Scholar]
- 70.Gehring M, Huh JH, Hsieh TF, Penterman J, Choi Y, Harada JJ, Goldberg RB, Fischer RL. DEMETER DNA glycosylase establishes MEDEA polycomb gene self-imprinting by allele-specific demethylation. Cell. 2006;124:495–506. doi: 10.1016/j.cell.2005.12.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ibarra CA, Feng X, Schoft VK, Hsieh TF, Uzawa R, Rodrigues JA, Zemach A, Chumak N, Machlicova A, Nishimura T, et al. Active DNA demethylation in plant companion cells reinforces transposon methylation in gametes. Science (New York, NY) 2012;337:1360–1364. doi: 10.1126/science.1224839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Mosher RA, Melnyk CW. siRNAs and DNA methylation: seedy epigenetics. Trends in plant science. 2010;15:204–210. doi: 10.1016/j.tplants.2010.01.002. [DOI] [PubMed] [Google Scholar]
- 73.Mosher RA, Melnyk CW, Kelly KA, Dunn RM, Studholme DJ, Baulcombe DC. Uniparental expression of Pol IV-dependent siRNAs in developing endosperm of Arabidopsis. Nature. 2009;460:283–286. doi: 10.1038/nature08084. [DOI] [PubMed] [Google Scholar]
- 74.Regulski M, Lu Z, Kendall J, Donoghue MT, Reinders J, Llaca V, Deschamps S, Smith A, Levy D, McCombie WR, et al. The maize methylome influences mRNA splice sites and reveals widespread paramutation-like switches guided by small RNA. Genome research. 2013;23:1651–1662. doi: 10.1101/gr.153510.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Coe EH. A REGULAR AND CONTINUING CONVERSION-TYPE PHENOMENON AT THE B LOCUS IN MAIZE. Proceedings of the National Academy of Sciences of the United States of America. 1959;45:828–832. doi: 10.1073/pnas.45.6.828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Stam M, Belele C, Dorweiler JE, Chandler VL. Differential chromatin structure within a tandem array 100 kb upstream of the maize b1 locus is associated with paramutation. Genes & development. 2002;16:1906–1918. doi: 10.1101/gad.1006702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Haring M, Bader R, Louwers M, Schwabe A, van Driel R, Stam M. The role of DNA methylation, nucleosome occupancy and histone modifications in paramutation. The Plant journal: for cell and molecular biology. 2010 doi: 10.1111/j.1365-313X.2010.04245.x. [DOI] [PubMed] [Google Scholar]
- 78.Alleman M, Sidorenko L, McGinnis K, Seshadri V, Dorweiler JE, White J, Sikkink K, Chandler VL. An RNA-dependent RNA polymerase is required for paramutation in maize. Nature. 2006;442:295–298. doi: 10.1038/nature04884. [DOI] [PubMed] [Google Scholar]
- 79.Schmitz RJ, Schultz MD, Lewsey MG, O’Malley RC, Urich MA, Libiger O, Schork NJ, Ecker JR. Transgenerational epigenetic instability is a source of novel methylation variants. Science (New York, NY) 2011;334:369–373. doi: 10.1126/science.1212959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Becker C, Hagmann J, Muller J, Koenig D, Stegle O, Borgwardt K, Weigel D. Spontaneous epigenetic variation in the Arabidopsis thaliana methylome. Nature. 2011;480:245–249. doi: 10.1038/nature10555. [DOI] [PubMed] [Google Scholar]
- 81.Watanabe T, Tomizawa S, Mitsuya K, Totoki Y, Yamamoto Y, Kuramochi-Miyagawa S, Iida N, Hoki Y, Murphy PJ, Toyoda A, et al. Role for piRNAs and noncoding RNA in de novo DNA methylation of the imprinted mouse Rasgrf1 locus. Science (New York, NY) 2011;332:848–852. doi: 10.1126/science.1203919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Aravin AA, Sachidanandam R, Bourc’his D, Schaefer C, Pezic D, Toth KF, Bestor T, Hannon GJ. A piRNA pathway primed by individual transposons is linked to de novo DNA methylation in mice. Molecular cell. 2008;31:785–799. doi: 10.1016/j.molcel.2008.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Zhao J, Ohsumi TK, Kung JT, Ogawa Y, Grau DJ, Sarma K, Song JJ, Kingston RE, Borowsky M, Lee JT. Genome-wide identification of polycomb-associated RNAs by RIP-seq. Molecular cell. 2010;40:939–953. doi: 10.1016/j.molcel.2010.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Zhang C, Darnell RB. Mapping in vivo protein-RNA interactions at single-nucleotide resolution from HITS-CLIP data. Nature biotechnology. 2011;29:607–614. doi: 10.1038/nbt.1873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kanno T, Huettel B, Mette MF, Aufsatz W, Jaligot E, Daxinger L, Kreil DP, Matzke M, Matzke AJ. Atypical RNA polymerase subunits required for RNA-directed DNA methylation. Nature genetics. 2005;37:761–765. doi: 10.1038/ng1580. [DOI] [PubMed] [Google Scholar]
- 86.Pontier D, Yahubyan G, Vega D, Bulski A, Saez-Vasquez J, Hakimi MA, Lerbs-Mache S, Colot V, Lagrange T. Reinforcement of silencing at transposons and highly repeated sequences requires the concerted action of two distinct RNA polymerases IV in Arabidopsis. Genes & development. 2005;19:2030–2040. doi: 10.1101/gad.348405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Gendrel AV, Lippman Z, Yordan C, Colot V, Martienssen RA. Dependence of heterochromatic histone H3 methylation patterns on the Arabidopsis gene DDM1. Science (New York, NY) 2002;297:1871–1873. doi: 10.1126/science.1074950. [DOI] [PubMed] [Google Scholar]
- 88.Johnson LM, Law JA, Khattar A, Henderson IR, Jacobsen SE. SRA-domain proteins required for DRM2-mediated de novo DNA methylation. PLoS genetics. 2008;4:e1000280. doi: 10.1371/journal.pgen.1000280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Kasschau KD, Fahlgren N, Chapman EJ, Sullivan CM, Cumbie JS, Givan SA, Carrington JC. Genome-wide profiling and analysis of Arabidopsis siRNAs. PLoS biology. 2007;5:e57. doi: 10.1371/journal.pbio.0050057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Xie Z, Johansen LK, Gustafson AM, Kasschau KD, Lellis AD, Zilberman D, Jacobsen SE, Carrington JC. Genetic and functional diversification of small RNA pathways in plants. PLoS biology. 2004;2:E104. doi: 10.1371/journal.pbio.0020104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Zilberman D, Cao X, Jacobsen SE. ARGONAUTE4 control of locus-specific siRNA accumulation and DNA and histone methylation. Science (New York, NY) 2003;299:716–719. doi: 10.1126/science.1079695. [DOI] [PubMed] [Google Scholar]
- 92.Ausin I, Mockler TC, Chory J, Jacobsen SE. IDN1 and IDN2 are required for de novo DNA methylation in Arabidopsis thaliana. Nature structural & molecular biology. 2009;16:1325–1327. doi: 10.1038/nsmb.1690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Jackson JP, Lindroth AM, Cao X, Jacobsen SE. Control of CpNpG DNA methylation by the KRYPTONITE histone H3 methyltransferase. Nature. 2002;416:556–560. doi: 10.1038/nature731. [DOI] [PubMed] [Google Scholar]
- 94.Ebbs ML, Bender J. Locus-specific control of DNA methylation by the Arabidopsis SUVH5 histone methyltransferase. The Plant cell. 2006;18:1166–1176. doi: 10.1105/tpc.106.041400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Rajakumara E, Law JA, Simanshu DK, Voigt P, Johnson LM, Reinberg D, Patel DJ, Jacobsen SE. A dual flip-out mechanism for 5mC recognition by the Arabidopsis SUVH5 SRA domain and its impact on DNA methylation and H3K9 dimethylation in vivo. Genes & development. 2011;25:137–152. doi: 10.1101/gad.1980311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Ortega-Galisteo AP, Morales-Ruiz T, Ariza RR, Roldan-Arjona T. Arabidopsis DEMETER-LIKE proteins DML2 and DML3 are required for appropriate distribution of DNA methylation marks. Plant molecular biology. 2008;67:671–681. doi: 10.1007/s11103-008-9346-0. [DOI] [PubMed] [Google Scholar]


