Abstract
Curcumin (diferuloylmethane), a polyphenol natural product of the plant Curcuma longa, is undergoing early clinical trials as a novel anticancer agent. However, the anticancer mechanism of curcumin remains to be elucidated. Recently, we have shown that curcumin inhibits phosphorylation of p70 S6 kinase 1 (S6K1) and eukaryotic initiation factor 4E (eIF4E) binding protein 1 (4E-BP1), two downstream effector molecules of the mammalian target of rapamycin complex 1 (mTORC1) in numerous cancer cell lines. This study was designed to elucidate the underlying mechanism. We observed that curcumin inhibited mTORC1 signaling not by inhibition of the upstream kinases, such as insulin-like growth factor 1 receptor (IGF-IR) and phosphoinositide-dependent kinase 1 (PDK1). Further, we found that curcumin inhibited mTORC1 signaling independently of protein phosphatase 2A (PP2A) or AMP-activated protein kinase AMPK-tuberous sclerosis complex (TSC). This is evidenced by the findings that curcumin was able to inhibit phosphorylation of S6K1 and 4E-BP1 in the cells pretreated with PP2A inhibitor (okadaic acid) or AMPK inhibitor (compound C), or in the cells expressing dominant-negative (dn) PP2A, shRNA to PP2A-A subunit, or dn-AMPKα. Curcumin did not alter the TSC1/2 interaction. Knockout of TSC2 did not affect curcumin inhibition of mTOR signaling. Finally, we identified that curcumin was able to dissociate raptor from mTOR, leading to inhibition of mTORC1 activity. Therefore, our data indicate that curcumin may represent a new class of mTOR inhibitor.
Introduction
The mammalian target of rapamycin (mTOR), a 289 kDa serine/threonine (Ser/Thr) protein kinase, lies downstream of insulin-like growth factor I receptor (IGF-IR) and phosphatidylinositol 3′ kinase (PI3K) and acts as a master regulator of a large array of cellular processes, including cell proliferation, growth, differentiation, survival, and motility (1). mTOR does not seem to exert its major functions as an independent entity but instead operates in cells at least as two complexes, mTOR complex 1/2 (mTORC1/2; ref. 1). mTORC1 consists of mTOR, mLST8 (also termed G-protein β-subunit–like protein, GβL, a yeast homologue of LST8), raptor (regulatory-associated protein of mTOR; refs. 2–5), and PRAS40 (proline-rich Akt substrate 40 kDa; refs. 6–8), and is rapamycin sensitive (2, 3). mTORC2 is composed of mTOR, mLST8, rictor (rapamycin-insensitive companion of mTOR; refs. 9, 11), mSin1 (mammalian stress-activated protein kinase–interacting protein 1; refs. 11–13), and protor (protein observed with rictor; ref. 14), and is rapamycin insensitive (9, 10). mTORC1 regulates phosphorylation of p70 S6 kinase 1 (S6K1) and eukaryotic initiation factor 4E (eIF4E) binding protein 1 (4E-BP1; refs. 2, 3) and functions primarily in the control of translation initiation and nutrient sensing. mTORC2 phosphorylates Akt at Ser473 (15) and regulates the actin cytoskeleton (4, 9, 10).
Recent studies have placed tuberous sclerosis complex (TSC) 1/2 as a modulator between PI3K/protein kinase B (Akt/PKB) and mTOR (16–19). The formation of TSC1/2 complex facilitates both their function and stability. The TSC1/2 complex itself undergoes extensive positive and negative regulation. In response to stimulation with growth factors or nutrients, activated Akt can phosphorylate TSC2 in vitro and in vivo at several sites, including Ser939/1130 and Thr1462 (16–19). Phosphorylation at these sites may disrupt TSC1/2 complex stability/formation and derepress mTORC1 signaling (16–19). On the other hand, in response to low level of cellular energy, AMP kinase α (AMPKα) is phosphorylated at Thr172 by the liver kinase B1 (LKB1; refs. 20–22). Activated AMPKα can phosphorylate TSC2 at Thr1227 and Ser1345, resulting in suppression of mTORC1 signaling (20–22). TSC2 has GTPase-activating protein (GAP) activity toward the Ras family small GTPase Rheb (Ras homologue enriched in brain; refs. 23–27). The Rheb/GTP complex physically binds and stimulates mTORC1 activity (28). TSC2 suppresses Rheb-mediated activation of mTORC1 by stimulating its GTPase activity, keeping Rheb in an inactive GDP-bound state (23–27). Therefore, mTORC1 seems to be directly regulated through the action of the AMPKα-TSC network.
Rapamycin inhibits mTORC1 function through association with its intracellular receptor FK-506 binding protein 12 (FKBP-12), and this complex binds to the mTORs FKBP12-rapamycin binding domain, resulting in raptor dissociation and inhibition of mTORC1 kinase activity (1). In response to stimuli, such as growth factors, nutrients (2, 3), ATP (29), and phosphatidic acid (30), mTOR is activated. Subsequently, mTORC1 phosphorylates 4E-BP1 (Thr37/46 and possibly Ser65 and Thr70; refs. 31, 32) and S6K1 (Thr389; ref. 33). mTORC1 also negatively regulates Ser/Thr protein phosphatase 2A (PP2A) activity (34). Whether mTORC1 regulates 4E-BP1 and S6K1 directly or indirectly through PP2A is controversial (1, 34). Inhibition of mTORC1 by rapamycin results in hypophosphorylation of 4E-BP1 (31, 32). Subsequently, hypophosphorylated 4E-BP1 tightly binds to eIF4E and prevents association of eIF4E with eIF4G and formation of the eIF4F initiation complex, thereby inhibiting cap-dependent translation of mRNA. In addition, inhibition of mTOR by rapamycin inactivates S6K1, blocking translation of mRNA species containing 5′ terminal oligopyrimidine tracts, although this remains controversial (1). Consequently, rapamycin inhibits cell proliferation and growth or other cellular events (1).
The polyphenol natural product curcumin (diferuloylmethane), isolated from the rhizome of the plant Curcuma longa, has garnered interest in recent years as a potential therapeutic agent for the treatment and/or prevention of various disease processes, including neurodegenerative disorders, inflammatory-related diseases, and cancer (35, 36). Various in vitro cell culture and in vivo animal studies have shown that curcumin is a potent inhibitor of almost every major stage of carcinogenesis, including transformation, initiation, promotion, invasion, angiogenesis, and metastasis (35). We are interested in investigating and identifying the anticancer mechanisms of curcumin, in the hope of uncovering its major target(s). Recently, we have shown that curcumin inhibits proliferation/growth, motility, and survival of human rhabdomyosarcoma cells (37). In an attempt to deduce the molecular mechanisms behind these effects, we studied the effect of curcumin on the mTOR signaling pathway, because as stated above, mTOR functions as a central controller of these processes. Our preliminary studies revealed that curcumin, at physiologic concentrations (2–5 μmol/L), inhibited mTORC1-mediated phosphorylation of S6K1 and 4E-BP1 in a panel of cell lines, including those derived from skeletal muscle (Rh1, Rh30), prostate (DU145), breast (MCF-7), cervical (HeLa; ref. 37), and colon (HT29; this study) cancer cells, suggesting that this effect is not cell- or cancer-type dependent. The results implicate that mTOR may in fact be the major target of curcumin. Therefore, we set out to identify the mechanisms by which curcumin inhibits mTORC1 signaling. Here, we show that curcumin inhibits mTORC1 signaling independently of the upstream kinases (IGF-IR and PDK1) and two negative regulators (PP2A and the AMPK-TSC network), and instead directly inhibits mTORC1 kinase activity by disrupting the mTOR-raptor interaction.
Materials and Methods
Materials
Curcumin (Sigma) was dissolved in 100% ethanol to prepare a 10 mmol/L stock solution and was stored at −20°C. Rapamycin (LC Laboratories) was dissolved in DMSO to prepare a 100 μg/mL stock solution and was stored at −20°C. IGF-I (PeproTech) was rehydrated in 0.1 mol/L acetic acid to prepare a 10 μg/mL stock solution and was stored at −80°C. Okadaic acid (Calbiochem) was dissolved in DMSO to prepare a 100 μmol/L stock solution and was stored at −20°C. Enhanced chemiluminescence solution was from Pierce. We used antibodies against IGF-IRβ subunit, mTOR, phospho-S6K1 (Thr389), S6K1, phospho-Akt (Thr308), Akt, Erk2, HA (Santa Cruz Biotechnology); phospho-PDK1 (Ser241), PDK1, phospho-AMPKα (Thr172), AMPK, phospho-TSC2 (Thr1462), TSC2, TSC1, phospho-Akt (S473), phospho-4E-BP1 (Thr37/46), 4E-BP1 (Thr70), phospho-Erk1/2 (Thr202/Tyr204; Cell Signaling); 4E-BP1 (Zymed); PP2A-A (Millipore), raptor, rictor (Bethyl Laboratories); mLST8 (GenWay Biotech); PP2Ac, phosphotyrosine (BD Biosciences); β-tubulin, FLAG (Sigma); goat anti-rabbit IgG-horseradish peroxidase (HRP), goat anti-mouse IgG-HRP, rabbit anti-goat IgG-HRP, and goat anti-chicken IgG-HRP (Pierce). All other chemicals were purchased from Sigma.
Cell culture and transfection
Human rhabdomyosarcoma cells (Rh1 and Rh30; gifts from Dr. Peter J. Houghton, St. Jude Children’s Research Hospital, Memphis, TN) were grown in antibiotic-free RPMI 1640 (Mediatech) supplemented with 10% fetal bovine serum (FBS; Atlanta Biologicals). Wild-type (wt) mouse embryonic fibroblasts (MEF) were kindly provided by Charles J. Sherr (Howard Hughes Medical Institute, St. Jude Children’s Research Hospital, Memphis, TN), whereas TSC2−/− MEF cells were a gift from David J. Kwiatkowski (Brigham and Women’s Hospital, Harvard Medical School, Boston, MA), respectively. HEK293, HT29, and HeLa cells were from American Type Culture Collection. HEK293 cells were grown in antibiotic-free DMEM (Mediatech) supplemented with 10% heat-inactivated FBS. HT29, HeLa, and MEF cells were grown antibiotic-free DMEM in supplemented with 10% FBS. All cell lines were grown in a humidified incubator at 37°C in an atmosphere of 5% CO2. For experiments where cells were deprived of serum, cell monolayers were washed with PBS and incubated in the serum-free DMEM. The expression vector encoding hemagglutinin (HA)–tagged dominant-negative (dn) protein phosphatase 2A catalytic subunit (PP2Ac, Leu199→Pro; dn-PP2A; ref. 38) was generously provided by Dr. Brian A. Hemmings (Friedrich Miescher Institut, Basel, Switzerland). Rh1 cells were seeded at a density of 5 × 105 per well in six-well plates and were transfected with the dn-PP2A expression plasmid by using TransIT-LT1 transfection reagent (Mirus) as directed by the manufacturer. After 48 h of transfection, cells were transferred to 100-mm dishes and were cultured for 2 wk at 37°C, 5% CO2 in growth medium containing 500 μg/mL geneticin (G418 sulfate, Mediatech). Drug-resistant cells were cloned and expanded in growth medium containing 500 μg/mL G418. Individual clones were screened for expression of the mutant proteins as well as the phosphorylation of Erk1/2 (Cell Signaling) using Western blot analysis.
Recombinant adenoviral constructs and infection of cells
Adeno-viruses encoding HA-tagged dominant-negative AMPKα1 (Ad-dn-AMPKα) was generated as described (39). Ad-dn-AMPKα and the control virus Ad-GFP encoding the green fluorescence protein (GFP) were amplified, titrated, and used as described (40).
Lentiviral shRNA cloning, production, and infection
To generate lentiviral shRNA to PP2A-A subunit, oligonucleotides containing the target sequences were synthesized, annealed, and inserted into FSIPPW lentiviral vector via the EcoR1/BamH1 restriction enzyme site. Oligonucleotides used were as follows: sense, AATTCCCGGTCAAAGAGTTCTGTGAATGCAAGAGATTCACAGAACTCTTTGACCTTTTTG; antisense, GATCCAAAAAGGTCAAAGAGTTCTGTGAATCTCTTGCATTCACAGAACTCTTTGACCGGG. Lentiviral shRNA constructs were made and used as described (40).
Western blotting
Cells were seeded in six-well plates in appropriate medium containing 10% FBS for 2 h, followed by serum starvation in serum-free DMEM for 24 h. Standard protocols involved exposure to curcumin (750 nmol/L–50 μmol/L) for 2 h before or after stimulation with 10 ng/mL IGF-I for 1 h. In the okadaic acid experiments, cells were exposed to 100 nmol/L okadaic acid for 1 h before standard treatment. Western blot analysis was performed as described (37).
Immunoprecipitation
Cells were seeded in 100-mm dishes in appropriate medium containing 10% FBS for 2 d, followed by serum starvation in serum-free DMEM for 24 h. Standard protocols involved exposure to curcumin (750 nmol/L–50 μmol/L) for 2 h before or after stimulation with 10 ng/mL IGF-I for 1 h. Cells were washed once in ice-cold 1× PBS, followed by lysis in appropriate buffer. For the mTOR coimmunoprecipitation experiments, cells were lysed in ice-cold 1× Chaps buffer [40 mmol/L HEPES (pH 7.4), 120 mmol/L NaCl, 1 mmol/L EDTA, 10 mmol/L pyrophosphate, 10 mmol/L glycerophosphate, 50 mmol/L NaF, 1.5 mmol/L Na3VO4, 0.3% Chaps, protease inhibitor cocktail (1:1,000, Sigma)]. Cell samples were subjected to freezing/thawing (dry ice/37°C water bath) six times to fully disrupt cells. After clearing, 500 μL of cell lysates were incubated overnight at 4°C with 1 μg of goat anti-mTOR antibody (Santa Cruz Biotechnology) and 30 μL of protein A/G plus agarose (Santa Cruz Biotechnology). Immunoprecipitates were washed with 1× Chaps buffer four times and twice with mTOR immunoprecipitation wash buffer [50 mmol/L HEPES (pH 7.5), 150 mmol/L NaCl]. Samples were resolved by SDS-PAGE as described above. For the TSC1/2 co-immunoprecipitation experiment, cells were lysed in ice-cold NP40 lysis buffer [10 mmol/L Tris (pH 7.5), 100 mmol/L NaCl, 1% NP40, 50 mmol/L NaF, 2 mmol/L EDTA, 1 mmol/L phenylmethanesulfonylfluoride, protease inhibitor cocktail (1:1,000, Sigma)]. After clearing, 500 μL of cell lysates were incubated overnight at 4°C with 1 μg of rabbit anti-TSC2 antibody (Cell Signaling) and 30 μL of protein A/G plus agarose (Santa Cruz Biotechnology). Immunoprecipitates were washed with NP40 lysis buffer four times. Samples were subjected to Western blotting as described above.
In vitro kinase assay
The mTORC1 was immunoprecipitated from Rh1 and HT29 cells as described above. For the mTORC1 in vitro kinase assay, immunoprecipitants were washed once in 1× Chaps buffer, once in mTORC1 kinase buffer [30 mmol/L MOPS (pH 7.5), 5 mmol/L NaF, 20 mmol/L β-glycerophosphate, 10% glycerol, 1 mmol/L DTT] containing 1 mol/L NaCl and once in mTORC1 kinase buffer. Kinase reactions were performed at 30°C for 30 min in 30 μL of mTORC1 kinase buffer containing 10 mmol/L MnCl2, 2 mmol/L ATP, and 1 μg of recombinant 4E-BP1 (Santa Cruz Biotechnology) as a substrate. For mTORC2 activity assay, 1 μg of recombinant Akt (Millipore), as a substrate, was used in the above reaction system. The reaction was stopped by the addition of 10 μL of 4× SDS sample buffer. Samples were subjected to Western blotting as described above.
Statistical analysis
Results were expressed as mean values ± SE. Statistical analysis was performed by Student’s t test (STATISTICA, Statsoft, Inc.). A level of P < 0.05 was considered to be significant.
Results
Curcumin did not affect phosphorylation of IGF-IR or PDK1
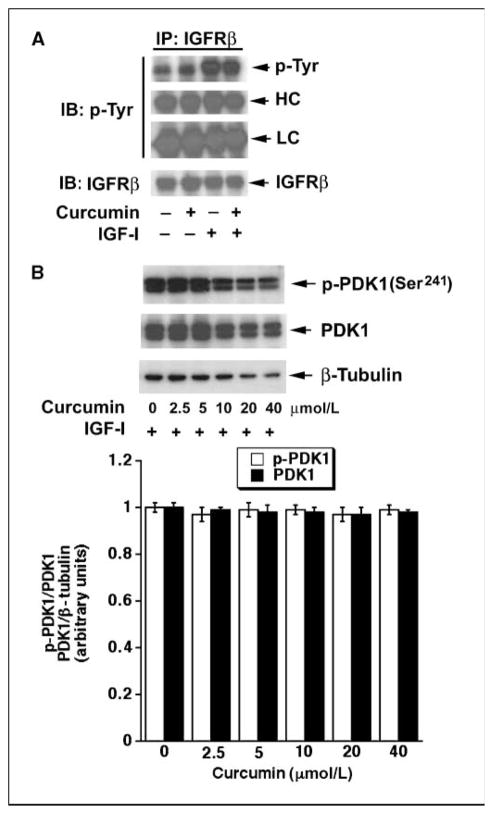
Recently, we have shown that curcumin inhibits mTORC1-mediated phosphorylation of S6K1 and 4E-BP1 and phosphorylation of mTOR at Ser2481 (autophosphorylation site; ref. 41) and Ser2448 (S6K1-mediated phosphorylation site; refs. 42, 37). Curcumin could inhibit these events by down-regulation of the activities of the upstream kinases, such as IGF-IR and PDK1. To determine whether curcumin affects IGF-IR activity, Rh1 cells were pretreated with or without curcumin (20 μmol/L) for 2 h, and then stimulated with or without IGF-I (10 ng/mL) for 1 h, followed by immunoprecipitation with antibodies to IGF-IRβ subunit (IGFRβ, p85) and immunoblotting with antibodies to phosphotyrosine (p-Tyr) and IGFRβ. As shown in Fig. 1A, IGF-I stimulated tyrosine phosphorylation of IGFRβ, but curcumin did not affect basal or IGF-I–stimulated tyrosine phosphorylation of IGFRβ, suggesting that curcumin did not affect IGFR activity.
Figure 1.
Curcumin does not affect phosphorylation of IGF-IR and PDK1. A, Rh1 cells were pretreated with or without curcumin (20 μmol/L) for 2 h and then stimulated with or without IGF-I (10 ng/mL) for 1 h, followed by immunoprecipitation with antibodies to the IGFR β-subunit (IGFRβ) plus protein A/G-agarose, and immunoblotting (IB) with antibodies to phosphotyrosine (p-Tyr) and IGFRβ, respectively. HC, heavy chain of IgG; LC, light chain of IgG. B, Rh1 cells were pretreated with curcumin (0–40 μmol/L) for 12 h and then stimulated with IGF-I (10 ng/mL) for 1 h, followed by Western blot analysis with antibodies against p-PDK1 (Ser241), PDK1, and β-tubulin (loading control), respectively. Top, representative blots; bottom, semiquantitated values of three independent experiments, performed by densitometry using NIH Image J. Statistical analysis was performed by Student’s t test. P < 0.05 was considered to be significant.
To examine whether curcumin affects PDK1 activity, Rh1 cells were pretreated with curcumin (0–40 μmol/L) for 12 h, stimulated with IGF-I (10 ng/mL) for 1 h, followed by Western blot analysis with p-PDK1 (Ser241) and PDK1, respectively. We found that treatment with curcumin at high concentrations (>10 μmol/L) for 12 h inhibited cell growth and induced cell death, as evidenced by reduction of β-tubulin (Fig. 1B) due to less cell number and floating dead cells observed under a microscope (data not shown). However, curcumin failed to alter phosphorylation of PDK1 or total PDK1 protein level, indicating that curcumin did not affect PDK1 activity. Therefore, curcumin inhibits mTOR signaling not through inhibition of the upstream kinases, such as IGF-IR or PDK1.
Curcumin inhibits mTOR-mediated phosphorylation of S6K1 and 4E-BP1 in a PP2A-independent manner
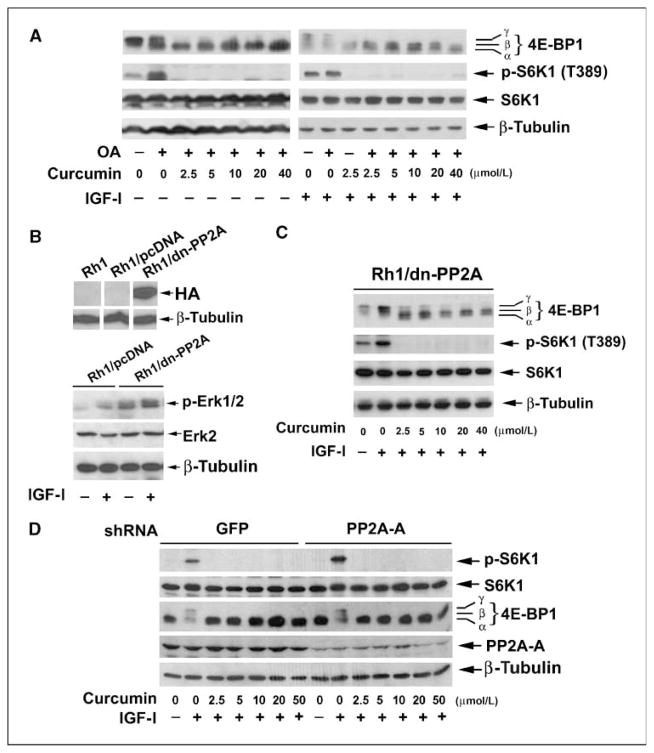
PP2A, a serine/threonine protein phosphatase, is a heterotrimeric holoenzyme composed of a catalytic subunit (PP2Ac), an A subunit (also termed PR65), and members of the B-subunit families, such as B (PR55), B′ (PR61), B″ (PR72), and B‴ (PR93/PR110; ref. 43). The phosphatase activity of PP2Ac is modulated by its association with PP2A-A and PP2A-B regulatory subunits (43). PP2A functions as a key regulator of many cellular events, such as cell proliferation, migration, and survival (43). Of interest, PP2A has been identified as the phosphatase responsible for the dephosphorylation of S6K1 and 4E-BP1, and inhibition of mTOR with rapamycin has been shown to stimulate these PP2A-mediated events (34). Therefore, we hypothesized that curcumin may inhibit mTORC1 signaling by either directly activating PP2A activity and/or disrupting mTOR-mediated inhibition of the enzyme. To this end, okadaic acid was used. Because okadaic acid selectively inhibits PP2A without inhibiting PP1 in intact cells at concentrations up to 100 nmol/L (44), serum-starved Rh30 cells were pretreated with or without okadaic acid (100 nmol/L) for 1 h, then exposed to curcumin (0–40 μmol/L) for 2 h, followed by stimulation with or without IGF-I (10 ng/mL) for 1 h. As shown in Fig. 2A, IGF-I markedly stimulated phosphorylation of S6K1 and 4E-BP1 in the absence or presence of okadaic acid. Pretreatment of cells with okadaic acid increased phosphorylation of S6K1 and 4E-BP1 but did not enhance IGF-I–stimulated phosphorylation of S6K1 and 4E-BP1. Probably, IGF-I stimulation of mTOR resulted in maximal phosphorylation of S6K1 and 4E-BP1, and addition of okadaic acid failed to further enhance their phosphorylation. Of interest, curcumin inhibited mTORC1-mediated phosphorylation of S6K1 at a concentration of 2.5 μmol/L in the absence or presence of okadaic acid in Rh30 cells (Fig. 2A). Phosphorylation of 4E-BP1 decreases its electrophoretic mobility during SDS-PAGE. Curcumin also inhibited IGF-I–stimulated phosphorylation of 4E-BP1 in Rh30 cells in the absence or presence of okadaic acid in Rh30 cells, as indicated by the decrease in the intensity of the uppermost band γ and by the increase in the higher mobility band α that corresponds to a less phosphorylated form of 4E-BP1. Similar data were observed in Rh1 and HT29 cells (data not shown). The results suggest that curcumin may exert its inhibitory effect on mTORC1 signaling independently of PP2A.
Figure 2.
Curcumin inhibits mTORC1 signaling in a PP2A-independent manner. A, serum-starved (24 h) Rh1 cells, grown in six-well plates, were treated with 100 nmol/L okadaic acid (OA) for 1 h, followed by treatment with various concentrations of curcumin for 2 h and stimulation with or without IGF-I (10 ng/mL) for 1 h. Whole-cell lysates were subjected to Western blot analysis using the indicated antibodies. B, top, Rh1 cells were stably transfected with vector alone (Rh1/pcDNA) or with a vector expressing HA-tagged dn-PP2Ac (Leu199→Pro), followed by Western blotting using the indicated antibodies. Bottom, Rh1/pcDNA and Rh1/dn-PP2A cell lines were stimulated with or without IGF-I (10 ng/mL) for 10 min, followed by Western blotting using the indicated antibodies. C, Rh1/dn-PP2A cells were exposed to curcumin (0–40 μmol/L) for 2 h followed by stimulation with IGF-I (10 ng/mL) for 1 h. Cell lysates were subjected to Western blot analysis using the indicated antibodies. D, serum-starved (24 h) HT29 cells expressing shRNAs to PP2A-A subunit and GFP (control), grown in six-well plates, were treated with curcumin (0–50 μmol/L) for 2 h, followed by stimulation with or without IGF-I (10 ng/mL) for 1 h. Whole-cell lysates were subjected to Western blot analysis using the indicated antibodies.
To confirm the above results, Rh1 clones stably expressing HA-tagged dominant negative form of PP2Ac (Leu199→Pro; dn-PP2A; ref. 40) were established (Fig. 2B, top). Because PP2A is known as a negative regulator of the extracellular regulated protein kinase 1 and 2 (Erk1/2; ref. 45), as expected, expression of dn-PP2A resulted in increase in the basal levels of phosphorylation of Erk1/2 (Fig. 2B, bottom), suggesting the dn-PP2A was functioning in the cells. However, expression of dn-PP2A did not affect the inhibitory effect of curcumin on mTORC1-mediated phosphorylation of S6K1 and 4E-BP1 in Rh1/dn-PP2A cells (Fig. 2C). We also wanted to determine the effect of curcumin on PP2Ac down-regulated cells. However, we failed to get viable PP2Ac down-regulated cells using a series of shRNAs to PP2Ac. Because the PP2A-A subunit is essential for PP2Ac activity (43), we next turned to down-regulate the PP2A-A subunit using lentiviral shRNA. As shown in Fig. 2D, down-regulation of PP2A-A by ~80% increased IGF-I–stimulated phosphorylation of S6K1 but did not affect curcumin inhibition of mTORC1 signaling in HT29 cells. Taken together, our data strongly support the notion that curcumin inhibits mTORC1 signaling in a PP2A-independent manner.
Curcumin inhibits mTOR signaling independently of the AMPK-TSC network
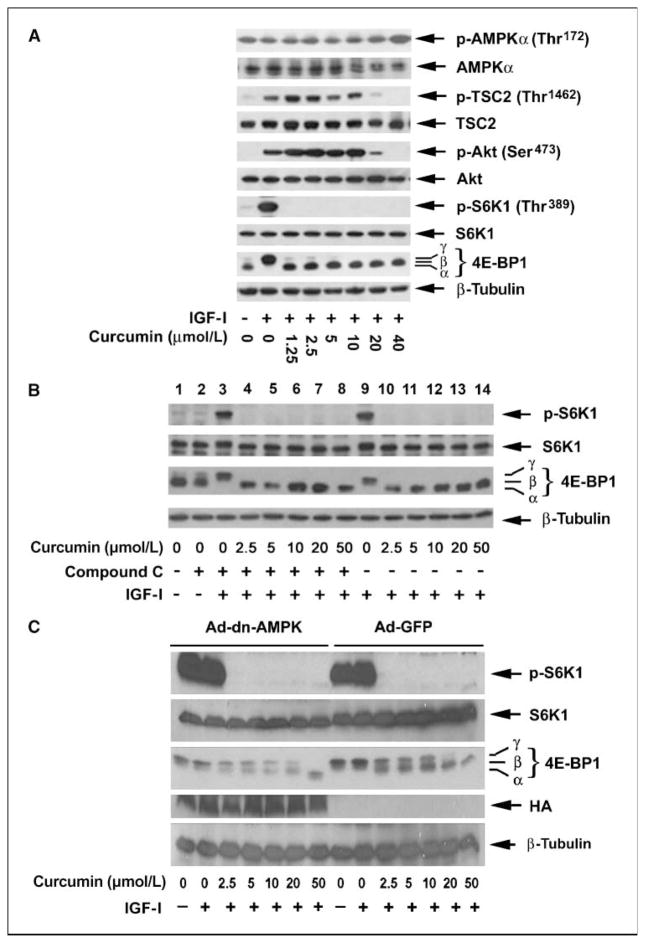
AMPK-TSC network negatively regulates mTOR signaling (1). We therefore hypothesized that curcumin might inhibit mTORC1 signaling by activation of AMPK-TSC network. In response to the low level of energy, AMPKα is activated by LKB1, leading to phosphorylation of TSC2 at Thr1227 and Ser1345, an event that serves to facilitate its inhibitory effect on mTOR signaling (1). Because no commercial antibodies are available for phospho-TSC2 at Thr1227 and Ser1345, the LKB1-mediated phospho-AMPKα at Thr172 was used as a readout of AMPKα activity toward TSC2. We observed that under serum starvation conditions, the phosphorylation level of AMPKα was robust in HT29 cells. Stimulation with IGF-I did not obviously alter the phosphorylation level of AMPKα. Addition of low concentrations of curcumin (0–20 μmol/L) did not affect AMPKα phosphorylation status either (Fig. 3A). However, treatment with curcumin at 40 μmol/L strikingly increased AMPKα phosphorylation, an event that correlates with the loss of both phosphorylation of Akt (Ser473) and TSC2 (Thr1462), but well above the initially observed inhibition of mTORC1 signaling (Fig. 3A). To address whether the inhibition of mTORC1 signaling by curcumin is directly dependent on AMPK kinase activity, we exposed Rh30 cells to the AMPK inhibitor compound C. As shown in Fig. 3B, inhibition of AMPK with compound C alone marginally increased phosphorylation of 4E-BP1 (lane 1 versus lane 2), but did not obviously enhance IGF-I–stimulated phosphorylation of S6K1 and 4E-BP1. Curcumin (2.5–50 μmol/L) potently inhibits the IGF-I–stimulated phosphorylation of S6K1 and 4E-BP1 in the absence and presence of compound C. These data suggest that curcumin inhibits mTORC1 signaling probably independently of AMPK functional kinase status.
Figure 3.
Curcumin inhibits mTORC1 signaling independently of AMPK. A, serum-starved (24 h) HT29 cells were exposed to curcumin (0–40 μmol/L) for 2 h followed by stimulation with IGF-I (10 ng/mL) for 1 h. Cell lysates were subjected to Western blot analysis using the indicated antibodies. B, serum-starved (24 h) HT29 cells, grown in six-well plates, were pretreated with compound C (10 μmol/L) for 1 h and then exposed to curcumin (0–50 μmol/L) for 2 h, followed by stimulation with IGF-I (10 ng/mL) for 1 h. Cell lysates were subjected to Western blot analysis using the indicated antibodies. C, serum-starved Rh30 cells, infected with Ad-dn-AMPKα1 and Ad-GFP (control), were exposed to curcumin (0–50 μmol/L) for 2 h, followed by stimulation with IGF-I (10 ng/mL) for 1 h. Cell lysates were subjected to Western blot analysis using the indicated antibodies.
To further confirm the above findings, Rh30 cells were infected with adenoviral vectors encoding GFP (Ad-GFP, for control) and the dominant-negative variant of AMPKα1 (Ad-dn-AMPK, Asp159-→Ala). As shown in Fig. 3C, curcumin (2.5–50 μmol/L) potently inhibited the IGF-I–stimulated phosphorylation of both S6K1 and 4E-BP1 in the cells infected with the control virus (Ad-GFP). Consistent with our findings from the compound C experiment, curcumin (2.5–50 μmol/L) also potently inhibited mTORC1 signaling in the cells expressing the dominant-negative variant of AMPKα, indicating that the effects of polyphenol on mTORC1 signaling are mediated independently of AMPK kinase activity.
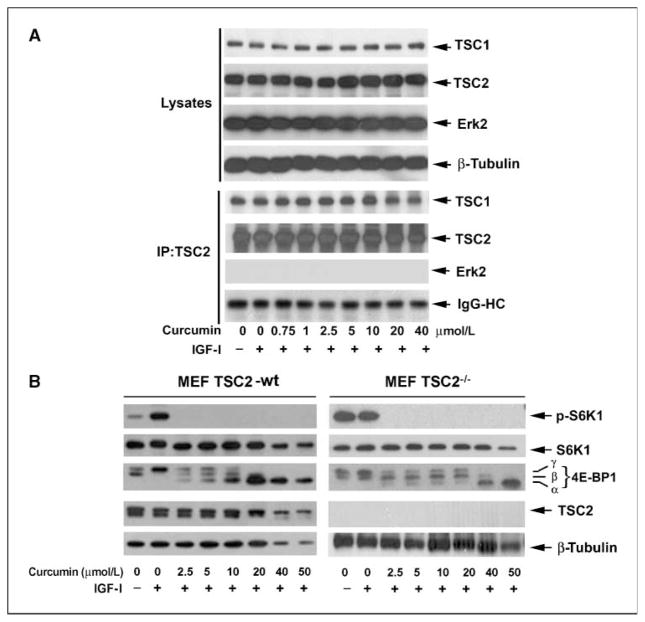
The formation of TSC1/2 complex is necessary for TSC2-mediated inhibition of the Rheb-mTORC1 axis (1). Next, we examined whether curcumin influences the TSC1/2 complex formation. As shown in Fig. 4A, curcumin did not obviously alter cellular protein expression of TSC1 and TSC2 in serum-starved Rh1 cells (top). A strong association of TSC1 with TSC2 was detected under a serum-free condition by coimmunoprecipitation. As a control, no Erk2 was found to be coimmunoprecipitated with TSC2 (bottom). Surprisingly, addition of IGF-I did not obviously affect TSC1/2 interaction although robust phosphorylation of S6K1 and 4E-BP1 was stimulated (Fig. 3A). At low concentrations (0–10 μmol/L), curcumin treatment dose-dependently inhibited mTORC1 signaling (Fig. 3A), but seemed to have no effect on TSC1/2 complex association (Fig. 4A), suggesting that curcumin inhibits mTORC1 signaling probably independently of TSC1/2. However, at higher concentrations (20–40 μmol/L), curcumin slightly destabilizes TSC1/2 interactions (Fig. 4A), which was consistent with decreased phosphorylation of Akt (Ser473) and TSC2 (Thr1462; Fig. 3A). To verify this finding, MEFs that contain a genomic knockout of TSC2 (46) were used. As shown in Fig. 4B, IGF-I stimulated phosphorylation of S6K1 and 4E-BP1 in serum-starved wt MEFs cells, but failed to enhance phosphorylation of the two proteins in serum-starved TSC2−/− cells, as knockout of TSC2 resulted in constitutive activation of mTOR signaling (46). Curcumin (2.5–50 μmol/L) inhibited the IGF-I–stimulated phosphorylation of S6K1 and 4E-BP1 in both wt and TSC2−/− MEF cells. These data suggest that curcumin is not acting as a direct activator of TSC2 function and actually can inhibit TSC function at high concentrations (>20 μmol/L). Collectively, our data clearly indicate that curcumin inhibits mTORC1 signaling independently of AMPK-TSC network.
Figure 4.
Curcumin inhibits mTORC1 signaling independently of TSC. A, Rh1 cells were exposed to curcumin (0–40 μmol/L) for 2 h, followed by stimulation with IGF-I (10 ng/mL) for 1 h. TSC2 was immunoprecipitated (IP) from the cell lysates, followed by immunoblotting with the indicated antibodies. B, MEF/TSC2−/− and wt cells were exposed to curcumin (0–50 μmol/L) for 2 h, followed by stimulation with IGF-I (10 ng/mL) for 1 h. Cell lysates were subjected to Western blot analysis using the indicated antibodies.
Curcumin inhibits the activity of mTOR
Because the upstream kinases (IGF-IR and PDK1) and two negative regulators (PP2A and AMPK-TSC network) are not involved in curcumin inhibition of mTORC1 signaling, we reasoned that curcumin might directly inhibit mTORC1 activity. For this, we first examined whether curcumin in vitro directly inhibits the mTORC1 activity. Rh1 cells were stimulated with IGF-I for 1 h. mTOR was immunoprecipitated from the cell lysates using antibodies to mTOR. After the isolated mTOR was incubated with curcumin (0–10 μmol/L) in vitro for 2 h, recombinant 4E-BP1, as a substrate, was added for in vitro kinase assay. We found that curcumin in vitro did not affect mTORC1 activity because phosphorylation of 4E-BP1 (Thr37/46) was not altered when curcumin was present (data not shown).
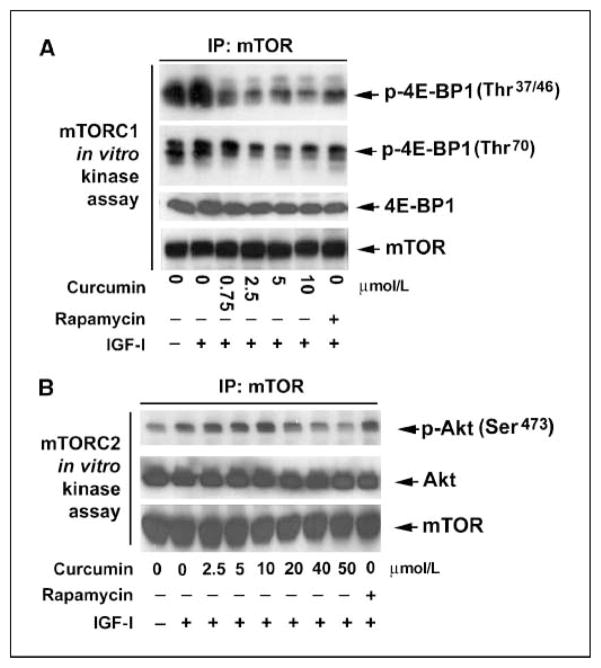
Next, we looked at the effect of curcumin on the kinase activity of mTORC1 in cultured cells (in vivo). Rh1 or HT29 cells were pretreated with curcumin (0–10 μmol/L), followed by stimulation with IGF-I (10 ng/mL) for 1 h. mTOR was immunoprecipitated from the cell lysates and the mTORC1 activity was determined by in vitro kinase assay, using recombinant 4E-BP1 as a substrate, as described above. As shown in Fig. 5A, curcumin dose-dependently inhibited the mTORC1 activity in Rh1 cells, as evidenced by the decreased phosphorylation levels of 4E-BP1 at Thr37/46 and Thr70, respectively. This inhibition was correlated well with loss of S6K1 phosphorylation at Thr389 in whole-cell lysates, as detected by Western blotting (ref. 37; Fig. 3A). Similar results were observed in HT29 cells (data not shown). Furthermore, we found that curcumin, at high concentrations (>40 μmol/L), obviously inhibited mTORC2 activity as well. This is evidenced by the findings that mTOR immunoprecipitated from the cells treated with curcumin (40–50 μmol/L) was not able to phosphorylate recombinant Akt (a substrate of mTORC2) as much as that pulled down from the control cells stimulated with IGF-I (Fig. 5B). As a control, treatment of cells with rapamycin (100 ng/mL) for 2 h inhibited mTORC1 activity (Fig. 5A) but did not affect mTORC2 activity (Fig. 5B). Therefore, our results reveal that curcumin inhibited mTORC1 activity at low concentrations (<20 μmol/L) and mTORC2 activity at high concentrations (>40 μmol/L) in cultured cells.
Figure 5.
Curcumin inhibits the kinase activity of mTOR. Serum-starved (24 h) Rh1 cells, grown in 100-mm dishes, were treated with various concentrations of curcumin or rapamycin (100 ng/mL) for 2 h, followed by stimulation with IGF-I (10 ng/mL) for 1 h. mTOR was immunoprecipitated from whole-cell lysates with antibodies to mTOR and the immunoprecipitates were used in mTORC1 in vitro kinase assay using recombinant 4E-BP1 as a substrate (A), or in mTORC2 in vitro kinase assay using recombinant Akt as a substrate (B), as described in Materials and Methods. The kinase assay products were subjected to Western blot analysis using the indicated antibodies.
Curcumin disrupts the stability of mTOR complexes
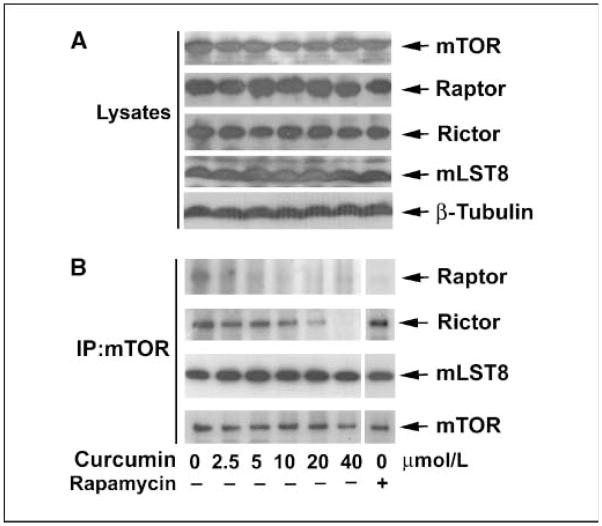
As discussed, the major components of mTORC1 are mTOR, mLST8, and raptor (2–5). Rapamycin inhibits mTORC1 activity by dissociating raptor from mTOR (2, 3). To investigate whether curcumin inhibits mTORC1 activity by affecting the association of mTOR with mLST8 and raptor, mTOR was immunoprecipitated from IGF-stimulated and curcumin-treated Rh1 cells, followed by immunoblotting with antibodies to mTOR, raptor, and mLST8, respectively. Consistent with previous findings (2, 3), rapamycin, as a positive control, did not affect binding of mLST8 or rictor to mTOR, but dissociated raptor from mTOR (Fig. 6B). Of interest, curcumin did not obviously affect cellular protein levels of mTOR, raptor, rictor, and mLST8 in the cells (Fig. 6A), but dose dependently dissociated raptor and rictor from mTOR, despite no effect on the interaction of mLST8 with mTOR (Fig. 6B). At low concentrations (2.5–5 μmol/L), curcumin almost completely abolished the association of raptor with mTOR, and at high concentrations (>40 μmol/L), curcumin also decreased the binding of rictor with mTOR (Fig. 6B). Similar data were observed in HT29 cells (data not shown). This is correlated to curcumin inhibition of mTORC1-mediated phosphorylation of S6K1 and 4E-BP1 at low concentrations (2.5–20 μmol/L) and mTORC2-mediated phosphorylation of Akt at high concentrations (>40 μmol/L).
Figure 6.
Curcumin disrupts mTOR complexes. Serum-starved (24 h) Rh1 cells, grown in 100-mm dishes, were treated with various concentrations of curcumin or rapamycin (100 ng/mL) for 2 h, followed by stimulation with IGF-I (10 ng/mL) for 1 h. mTOR was immunoprecipitated from whole-cell lysates. The cell lysates (A) and the immunoprecipitates (B) were subjected to Western blot analysis using the indicated antibodies.
Discussion
mTOR has been recognized as a key therapeutic target for the prevention and/or treatment of cancer and many other diseases because of the fact that it functions as a central hub for the regulation of numerous key cellular processes that are not only critical for normal cellular function but are also absolutely necessary for tumorigenesis (1). At the same time, a large amount of effort and energy have been allotted to the study and development of curcumin and its analogues as chemopreventative and chemotherapeutic agents (35, 36). Despite the great effect that both of these factors have exerted in the cancer research field, mTOR and curcumin have only recently begun to cross paths with each other (37, 47–49). After the publication of our initial observation that curcumin targets mTOR-mediated pathways in cancer cells (37), several recent studies have also shown that curcumin targeting of the mTOR pathway represents a potentially effective and powerful therapeutic tool for use against cancer (47–49). Curcumin induces autophagy in malignant glioma cells through inhibition of the Erk1/2 and mTOR signal transduction pathways (47, 49). Furthermore, Li and colleagues. (48) show that curcumin significantly down-regulates the expression of the p53 suppressor MDM2 in prostate cancer cells, partially through inhibition of the mTOR pathway.
To identify the mechanism by which curcumin inhibits mTOR signaling in cancer cells, first of all, we focused on the upstream kinases, because curcumin inhibits mTORC1-mediated phosphorylation of S6K1 and 4E-BP1, as well as phosphorylation of mTOR at Ser2481 (autophosphorylation site; ref. 41) and at Ser2448 (S6K1-phosphorylation site; refs. 42, 37). Our data implicate that curcumin inhibits mTOR signaling not by inhibition of any of the upstream kinases. This is supported by the findings that curcumin did not affect the kinase activity of IGF-IR or PDK1, as curcumin was not affecting their phosphorylation levels. Because PI3K, which is negatively regulated by the phosphatase and tensin homologue deleted on chromosome ten (PTEN), lies upstream of PDK1 and regulates phosphorylation of PDK1 (1), we deduce that curcumin may not affect either PTEN or PI3K activity.
Next, we considered the Ser/Thr protein phosphatase PP2A as a potential target of curcumin because PP2A has been reported as a major negative regulator of S6K1 and 4E-BP1 and itself also as a major target of mTOR (34). We hypothesized that curcumin inhibition of mTOR may suppress mTOR-mediated inhibition of PP2A activity allowing the phosphatase to target mTOR substrates, or curcumin may itself function as a direct allosteric activator of PP2A, thus directly stimulating its activity regardless of mTOR activity status. In our studies, we found that the compound inhibited mTORC1-mediated phosphorylation of S6K1 and 4E-BP1 in the absence of PP2A activity as obtained through use of a PP2A inhibitor (okadaic acid), down-regulation of PP2A-A subunit via shRNA, or overexpression of a dominant-negative form of PP2Ac. Therefore, our data suggest that curcumin inhibits mTORC1 signaling independently of PP2A.
The above findings led us to investigate whether curcumin inhibits mTOR signaling by targeting AMPK-TSC network, a major upstream negative regulator of mTORC1 signaling events and itself a major target of mTORC2-mediated regulation (1). We hypothesized that curcumin may alleviate Akt-mediated inhibition of TSC2 function, stabilize TSC1/2 interactions, or directly stimulate TSC2 GAP activity. We also hypothesized that curcumin may stimulate the kinase activity of AMPK, thereby leading to activation of TSC2 and inhibition of mTORC1 signaling. However, we found that the polyphenol did not obviously alter phosphorylation of TSC2 (Thr1462) or affect TSC1/TSC2 interaction at low concentrations (2.5–10 μmol/L) sufficient to inhibit mTORC1. Curcumin also could effectively inhibit mTORC1 signaling in wt normal MEF cells as well as cells containing a genomic knockout of TSC2. Our data further reveal that curcumin did not affect phosphorylation of AMPKα (Thr172) at low concentrations (2.5–20 μmol/L), but was able to inhibit mTORC1 signaling independently of AMPK kinase activity, as indicated by the ability of the compound to block phosphorylation of S6K1 and 4E-BP1 in the presence of an AMPK inhibitor (compound C), or a dominant-negative variant of AMPKα. To our surprise, curcumin could inhibit phosphorylation of TSC2 (Thr1462), cause a slight destabilization of TSC1/2 interaction, and stimulate phosphorylation of AMPKα at high concentrations (>40 μmol/L), through unidentified mechanism(s). Collectively, our results suggest that curcumin inhibits mTORC1 signaling independently of the AMPK-TSC network. Our data also implicate that at high concentrations (>40 μmol/L), curcumin may act as an AMPK-TSC stimulator that may be involved in the inhibition of the mTORC2-Akt pathway.
After ruling out the upstream kinases and two major negative regulators of mTOR signaling as the mediators of the effects of curcumin, we decided to investigate mTOR itself as the possible target of curcumin. Finally, we identified that curcumin blocked the kinase activity of both the mTORC1 and the mTORC2 and that these effects were the consequence of the disruption of raptor-mTOR association in the mTORC1 and the disruption of rictor-mTOR association in the mTORC2. This disruption of necessary complex associations, however, occurs at different concentrations of the compound, which is in good agreement with curcumin inhibition of mTORC1-mediated phosphorylation of S6K1 and 4E-BP1 at low concentrations (2.5–20 μmol/L) and mTORC2-mediated phosphorylation of Akt at high concentrations (>40 μmol/L). In our studies, we found that down-regulation of raptor by RNA interference inhibited mTORC1-mediated phosphorylation of S6K1 and 4E-BP1, but increased mTORC2-mediated phosphorylation of Akt in Rh30 cells. Curcumin could not inhibit phosphorylation of S6K1 and 4E-BP1 synergistically in raptor down-regulated cells, but was able to inhibit mTORC2-mediated phosphorylation of Akt.4 We also observed that overexpression of raptor was not able to prevent curcumin inhibition of mTORC1 signaling in Rh1 or Rh30 cells,4 which supports the notion that curcumin-dissociating raptor from mTOR results in inhibition of mTORC1 signaling. Clearly, more studies are needed to address how curcumin disrupts the mTOR complexes. Particularly, it is of great importance to know whether curcumin directly binds mTOR or a component of the mTOR complexes to function as a specific mTOR inhibitor. It has been reported that oxidants are involved in the activation (50) or inactivation (3) of mTOR signaling. Alternatively, curcumin may function as an antioxidant (35) to inhibit mTOR signaling. In the studies, we also observed that curcumin might not induce cell death until at higher concentrations (>10 μmol/L), which is evidenced in the findings that curcumin (>10 μmol/L) treatment for 12 h resulted in reduction of β-tubulin (Fig. 1B) due to less cell number and floating dead cells observed under a microscope (data not shown). Whether this is related to curcumin inhibition of rictor binding to mTOR as well as mTORC2-mediated Akt phosphosphorylation remains to be defined.
In summary, here we have shown that curcumin inhibits mTORC1 signaling independently of the upstream kinases (IGF-IR and PDK1) and two negative regulators (PP2A or AMPK̃TSC network) of mTOR and directly inhibits mTORC1 kinase activity by disrupting the mTOR-raptor interaction. Our findings suggest that curcumin may represent a new class of mTOR inhibitor.
Acknowledgments
Grant support: American Cancer Society Award (RSG-08-135-01-CNE; S. Huang), Feist-Weiller Cancer Research Award (S. Huang), and Start-up Fund (S. Huang) jointly from Louisiana State University Health Sciences Center in Shreveport, LA.
We thank Drs. Brian A. Hemmings, Peter J. Houghton, David J. Kwiatkowski, and Charles J. Sherr for providing cell lines and constructs.
Footnotes
Our unpublished data.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- 1.Guertin DA, Sabatini DM. Defining the role of mTOR in cancer. Cancer Cell. 2007;12:9–22. doi: 10.1016/j.ccr.2007.05.008. [DOI] [PubMed] [Google Scholar]
- 2.Hara K, Maruki Y, Long X, et al. Raptor, a binding partner of target of rapamycin (TOR), mediates TOR action. Cell. 2002;110:177–89. doi: 10.1016/s0092-8674(02)00833-4. [DOI] [PubMed] [Google Scholar]
- 3.Kim DH, Sarbassov DD, Ali SM, et al. mTOR interacts with raptor to form a nutrient-sensitive complex that signals to the cell growth machinery. Cell. 2002;110:163–75. doi: 10.1016/s0092-8674(02)00808-5. [DOI] [PubMed] [Google Scholar]
- 4.Loewith R, Jacinto E, Wullschleger S, et al. Two TOR complexes, only one of which is rapamycin sensitive, have distinct roles in cell growth control. Mol Cell. 2002;10:457–68. doi: 10.1016/s1097-2765(02)00636-6. [DOI] [PubMed] [Google Scholar]
- 5.Kim DH, Sarbassov DD, Ali SM, et al. GβL, a positive regulator of the rapamycin-sensitive pathway required for the nutrient-sensitive interaction between raptor and mTOR. Mol Cell. 2003;11:895–904. doi: 10.1016/s1097-2765(03)00114-x. [DOI] [PubMed] [Google Scholar]
- 6.Fonseca BD, Smith EM, Lee VH, Mackintosh C, Proud CG. PRAS40 is a target for mammalian target of rapamycin complex 1 and is required for signaling downstream of this complex. J Biol Chem. 2007;282:24514–24. doi: 10.1074/jbc.M704406200. [DOI] [PubMed] [Google Scholar]
- 7.Sancak Y, Thoreen CC, Peterson TR, et al. PRAS40 is an insulin-regulated inhibitor of the mTORC1 protein kinase. Mol Cell. 2007;25:903–15. doi: 10.1016/j.molcel.2007.03.003. [DOI] [PubMed] [Google Scholar]
- 8.Vander Haar E, Lee SI, Bandhakavi S, Griffin TJ, Kim DH. Insulin signalling to mTOR mediated by the Akt/PKB substrate PRAS40. Nat Cell Biol. 2007;9:316–23. doi: 10.1038/ncb1547. [DOI] [PubMed] [Google Scholar]
- 9.Jacinto E, Loewith R, Schmidt A, et al. Mammalian TOR complex 2 controls the actin cytoskeleton and is rapamycin insensitive. Nat Cell Biol. 2004;6:1122–8. doi: 10.1038/ncb1183. [DOI] [PubMed] [Google Scholar]
- 10.Sarbassov DD, Ali SM, Kim DH, et al. Rictor, a novel binding partner of mTOR, defines a rapamycin-insensitive and raptor-independent pathway that regulates the cytoskeleton. Curr Biol. 2004;14:1296–302. doi: 10.1016/j.cub.2004.06.054. [DOI] [PubMed] [Google Scholar]
- 11.Frias MA, Thoreen CC, Jaffe JD, et al. mSin1 is necessary for Akt/PKB phosphorylation, and its isoforms define three distinct mTORC2s. Curr Biol. 2006;16:1865–70. doi: 10.1016/j.cub.2006.08.001. [DOI] [PubMed] [Google Scholar]
- 12.Jacinto E, Facchinetti V, Liu D, et al. SIN1/MIP1 maintains rictor-mTOR complex integrity and regulates Akt phosphorylation and substrate specificity. Cell. 2006;127:125–37. doi: 10.1016/j.cell.2006.08.033. [DOI] [PubMed] [Google Scholar]
- 13.Yang Q, Inoki K, Ikenoue T, Guan KL. Identification of Sin1 as an essential TORC2 component required for complex formation and kinase activity. Genes Dev. 2006;20:2820–32. doi: 10.1101/gad.1461206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pearce LR, Huang X, Boudeau J, et al. Identification of Protor as a novel Rictor-binding component of mTOR complex-2. Biochem J. 2007;405:513–22. doi: 10.1042/BJ20070540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sarbassov DD, Guertin DA, Ali SM, Sabatini DM. Phosphorylation and regulation of Akt/PKB by the rictor-mTOR complex. Science. 2005;307:1098–101. doi: 10.1126/science.1106148. [DOI] [PubMed] [Google Scholar]
- 16.Gao X, Zhang Y, Arrazola P, et al. Tsc tumour suppressor proteins antagonize amino-acid-TOR signalling. Nat Cell Biol. 2002;4:699–704. doi: 10.1038/ncb847. [DOI] [PubMed] [Google Scholar]
- 17.Inoki K, Li Y, Zhu T, Wu J, Guan KL. TSC2 is phosphorylated and inhibited by Akt and suppresses mTOR signalling. Nat Cell Biol. 2002;4:648–57. doi: 10.1038/ncb839. [DOI] [PubMed] [Google Scholar]
- 18.Manning BD, Tee AR, Logsdon MN, Blenis J, Cantley LC. Identification of the tuberous sclerosis complex-2 tumor suppressor gene product tuberin as a target of the phosphoinositide 3-kinase/akt pathway. Mol Cell. 2002;10:151–62. doi: 10.1016/s1097-2765(02)00568-3. [DOI] [PubMed] [Google Scholar]
- 19.Tee AR, Fingar DC, Manning BD, Kwiatkowski DJ, Cantley LC, Blenis J. Tuberous sclerosis complex-1 and -2 gene products function together to inhibit mammalian target of rapamycin (mTOR)-mediated downstream signaling. Proc Natl Acad Sci U S A. 2002;99:13571–76. doi: 10.1073/pnas.202476899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Inoki K, Zhu T, Guan KL. TSC2 mediates cellular energy response to control cell growth and survival. Cell. 2003;115:577–90. doi: 10.1016/s0092-8674(03)00929-2. [DOI] [PubMed] [Google Scholar]
- 21.Corradetti MN, Inoki K, Bardeesy N, DePinho RA, Guan KL. Regulation of the TSC pathway by LKB1: evidence of a molecular link between tuberous sclerosis complex and Peutz-Jeghers syndrome. Genes Dev. 2004;18:1533–8. doi: 10.1101/gad.1199104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shaw RJ, Kosmatka M, Bardeesy N, et al. The tumor suppressor LKB1 kinase directly activates AMP-activated kinase and regulates apoptosis in response to energy stress. Proc Natl Acad Sci U S A. 2004;101:3329–35. doi: 10.1073/pnas.0308061100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Garami A, Zwartkruis FJ, Nobukuni T, et al. Insulin activation of Rheb, a mediator of mTOR/S6K/4E-BP signaling, is inhibited by TSC1 and 2. Mol Cell. 2003;11:1457–66. doi: 10.1016/s1097-2765(03)00220-x. [DOI] [PubMed] [Google Scholar]
- 24.Inoki K, Li Y, Xu T, Guan KL. Rheb GTPase is a direct target of TSC2 GAP activity and regulates mTOR signaling. Genes Dev. 2003;17:1829–34. doi: 10.1101/gad.1110003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tee AR, Manning BD, Roux PP, Cantley LC, Blenis J. Tuberous sclerosis complex gene products, Tuberin and Hamartin, control mTOR signaling by acting as a GTPase-activating protein complex toward Rheb. Curr Biol. 2003;13:1259–68. doi: 10.1016/s0960-9822(03)00506-2. [DOI] [PubMed] [Google Scholar]
- 26.Zhang Y, Gao X, Saucedo LJ, Ru B, Edgar BA, Pan D. Rheb is a direct target of the tuberous sclerosis tumour suppressor proteins. Nat Cell Biol. 2003;5:578–81. doi: 10.1038/ncb999. [DOI] [PubMed] [Google Scholar]
- 27.Li Y, Inoki K, Guan KL. Biochemical and functional characterizations of small GTPase Rheb and TSC2 GAP activity. Mol Cell Biol. 2004;24:7965–75. doi: 10.1128/MCB.24.18.7965-7975.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Long X, Lin Y, Ortiz-Vega S, Yonezawa K, Avruch J. Rheb binds and regulates the mTOR kinase. Curr Biol. 2005;15:702–13. doi: 10.1016/j.cub.2005.02.053. [DOI] [PubMed] [Google Scholar]
- 29.Dennis PB, Jaeschke A, Saitoh M, Fowler B, Kozma SC, Thomas G. Mammalian TOR: a homeostatic ATP sensor. Science. 2001;294:1102–5. doi: 10.1126/science.1063518. [DOI] [PubMed] [Google Scholar]
- 30.Fang Y, Vilella-Bach M, Bachmann R, Flanigan A, Chen J. Phosphatidic acid-mediated mitogenic activation of mTOR signaling. Science. 2001;294:1942–5. doi: 10.1126/science.1066015. [DOI] [PubMed] [Google Scholar]
- 31.Mothe-Satney I, Brunn GJ, McMahon LP, Capaldo CT, Abraham RT, Lawrence JC., Jr Mammalian target of rapamycin-dependent phosphorylation of PHAS-I in four (S/T)P sites detected by phospho-specific antibodies. J Biol Chem. 2000;275:33836–43. doi: 10.1074/jbc.M006005200. [DOI] [PubMed] [Google Scholar]
- 32.Gingras AC, Raught B, Gygi SP, et al. Hierarchical phosphorylation of the translation inhibitor 4E-BP1. Genes Dev. 2001;15:2852–64. doi: 10.1101/gad.912401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dennis PB, Pullen N, Kozma SC, Thomas G. The principal rapamycin-sensitive p70(s6k) phosphorylation sites, T-229 and T-389, are differentially regulated by rapamycin-insensitive kinase kinases. Mol Cell Biol. 1996;16:6242–51. doi: 10.1128/mcb.16.11.6242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Peterson RT, Desai BN, Hardwick JS, Schreiber SL. Protein phosphatase 2A interacts with the 70-kDa S6 kinase and is activated by inhibition of FKBP12-rapamycinassociated protein. Proc Natl Acad Sci U S A. 1999;96:4438–42. doi: 10.1073/pnas.96.8.4438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Aggarwal BB, Kumar A, Bharti A. Anticancer potential of curcumin: preclinical and clinical studies. Anticancer Res. 2003;23:363–98. [PubMed] [Google Scholar]
- 36.Singh S. From exotic spice to modern drug? Cell. 2007;130:765–8. doi: 10.1016/j.cell.2007.08.024. [DOI] [PubMed] [Google Scholar]
- 37.Beevers CS, Li F, Liu L, Huang S. Curcumin inhibits the mammalian target of rapamycin-mediated signaling pathways in cancer cells. Int J Cancer. 2006;119:757–64. doi: 10.1002/ijc.21932. [DOI] [PubMed] [Google Scholar]
- 38.Evans DR, Myles T, Hofsteenge J, Hemmings BA. Functional expression of human PP2Ac in yeast permits the identification of novel C-terminal and dominant-negative mutant forms. J Biol Chem. 1999;274:24038–46. doi: 10.1074/jbc.274.34.24038. [DOI] [PubMed] [Google Scholar]
- 39.Lu M, Tang Q, Olefsky JM, Mellon PL, Webster NJ. Adiponectin activates adenosine monophosphate-activated protein kinase and decreases luteinizing hormone secretion in LβT2 gonadotropes. Mol Endocrinol. 2008;22:760–71. doi: 10.1210/me.2007-0330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu L, Li F, Cardelli JA, Martin KA, Blenis J, Huang S. Rapamycin inhibits cell motility by suppression of mTOR-mediated S6K1 and 4E-BP1 pathways. Oncogene. 2006;25:7029–40. doi: 10.1038/sj.onc.1209691. [DOI] [PubMed] [Google Scholar]
- 41.Peterson RT, Beal PA, Comb MJ, Schreiber SL. FKBP12-rapamycin-associated protein (FRAP) auto-phosphorylates at serine 2481 under translationally repressive conditions. J Biol Chem. 2000;275:7416–23. doi: 10.1074/jbc.275.10.7416. [DOI] [PubMed] [Google Scholar]
- 42.Chiang GG, Abraham RT. Phosphorylation of mammalian target of rapamycin (mTOR) at Ser-2448 is mediated by p70S6 kinase. J Biol Chem. 2005;280:25485–90. doi: 10.1074/jbc.M501707200. [DOI] [PubMed] [Google Scholar]
- 43.Janssens V, Goris J, Van Hoof C. PP2A: the expected tumor suppressor. Curr Opin Genet Dev. 2005;15:34–41. doi: 10.1016/j.gde.2004.12.004. [DOI] [PubMed] [Google Scholar]
- 44.Hardie DG, Haystead TA, Sim AT. Use of okadaic acid to inhibit protein phosphatases in intact cells. Methods Enzymol. 1991;201:469–76. doi: 10.1016/0076-6879(91)01042-z. [DOI] [PubMed] [Google Scholar]
- 45.Junttila MR, Li SP, Westermarck J. Phosphatase-mediated crosstalk between MAPK signaling pathways in the regulation of cell survival. FASEB J. 2008;22:954–65. doi: 10.1096/fj.06-7859rev. [DOI] [PubMed] [Google Scholar]
- 46.Zhang H, Bajraszewski N, Wu E, et al. PDGFRs are critical for PI3K/Akt activation and negatively regulated by mTOR. J Clin Invest. 2007;117:730–8. doi: 10.1172/JCI28984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Aoki H, Takada Y, Kondo S, Sawaya R, Aggarwal BB, Kondo Y. Evidence that curcumin suppresses the growth of malignant gliomas in vitro and in vivo through induction of autophagy: role of Akt and extracellular signal-regulated kinase signaling pathways. Mol Pharmacol. 2007;72:29–39. doi: 10.1124/mol.106.033167. [DOI] [PubMed] [Google Scholar]
- 48.Li M, Zhang Z, Hill DL, Wang H, Zhang R. Curcumin, a dietary component, has anticancer, chemosensitization, and radiosensitization effects by down-regulating the MDM2 oncogene through the PI3K/mTOR/ETS2 pathway. Cancer Res. 2007;67:1988–96. doi: 10.1158/0008-5472.CAN-06-3066. [DOI] [PubMed] [Google Scholar]
- 49.Shinojima N, Yokoyama T, Kondo Y, Kondo S. Roles of the Akt/mTOR/p70S6K and ERK1/2 signaling pathways in curcumin-induced autophagy. Autophagy. 2007;3:635–7. doi: 10.4161/auto.4916. [DOI] [PubMed] [Google Scholar]
- 50.Sarbassov DD, Sabatini DM. Redox regulation of the nutrient-sensitive raptor-mTOR pathway and complex. J Biol Chem. 2005;280:39505–9. doi: 10.1074/jbc.M506096200. [DOI] [PubMed] [Google Scholar]