Abstract
Biomolecular NMR chemical shift data are key information for the functional analysis of biomolecules and the development of new techniques for NMR studies utilizing chemical shift statistical information. Structural genomics projects are major contributors to the accumulation of protein chemical shift information. The management of the large quantities of NMR data generated by each project in a local database and the transfer of the data to the public databases are still formidable tasks because of the complicated nature of NMR data. Here we report an automated and efficient system developed for the deposition and annotation of a large number of data sets including 1H, 13C and 15N resonance assignments used for the structure determination of proteins. We have demonstrated the feasibility of our system by applying it to over 600 entries from the internal database generated by the RIKEN Structural Genomics/Proteomics Initiative (RSGI) to the public database, BioMagResBank (BMRB). We have assessed the quality of the deposited chemical shifts by comparing them with those predicted from the PDB coordinate entry for the corresponding protein. The same comparison for other matched BMRB/PDB entries deposited from 2001–2011 has been carried out and the results suggest that the RSGI entries greatly improved the quality of the BMRB database. Since the entries include chemical shifts acquired under strikingly similar experimental conditions, these NMR data can be expected to be a promising resource to improve current technologies as well as to develop new NMR methods for protein studies.
Keywords: NMR, Chemical shift, Proteomics, Database, BMRB
Introduction
The major goals of the structural genomics projects are to fill the sparse space of the protein structural universe and to increase the chance for structural biologists to encounter data for proteins whose amino acid sequence is identical or close to that of their target protein. Owing to the significant amount of effort aimed at the determination of three dimensional protein structures, the total number of entries in the Protein Data Bank (PDB) has reached more than 81,500 (in May, 2012). The structures determined by solution NMR techniques are now approaching 9,500, occupying ~12 % of the PDB overall. NMR protein studies have produced a variety of experimental parameters along with the structure determinations, among which the assigned chemical shifts are now playing the most important role in NMR protein functional analysis. These values can be applied to NMR studies of protein interactions through chemical shift perturbations, of protein dynamics using the relaxation of assigned 13C or 15N signals, of relaxation dispersion experiments and so on. The strong correlation observed between chemical shifts and protein structure has been known for many years. One can apply this knowledge to extract structural information from experimentally determined chemical shifts, and vice versa. For the purpose of collecting and archiving NMR experimental data, the BioMagResBank (BMRB) was established as the standard public database for NMR data of biomolecules (Seavey et al. 1991; Ulrich et al. 1989, 2008). The database has grown into a large repository, embracing more than 7,800 entries for chemical shifts at present (May, 2012). Utilizing the relationship between chemical shifts and structure, many software tools have been developed, such as TALOS (Cornilescu et al. 1999), SHIFTX (Neal et al. 2003), SPARTA (Shen and Bax 2007), TALOS+ (Shen et al. 2009) and SPARTA+ (Shen and Bax 2010) to determine protein torsion angle restraints for structure determination and to back calculate chemical shifts from 3D structures. Recently, these concepts in combination with structure modeling have pioneered entirely new methods to elucidate 3D structure exclusively from chemical shifts; CHESHIRE (Cavalli et al. 2007), CS-ROSETTA (Shen et al. 2008) and CS23D (Wishart et al. 2008). To further promote these technologies, the quantity and quality of the experimental data in the BMRB archive will need to continue to expand.
Chemical shift data deposition has been mandatory when NMR structure coordinates are submitted to the PDB since December 2010. This will provide chemical shift data for all future PDB entries. New versions of the ADIT-NMR deposition system (BMRB and PDBj-BMRB) and Auto-Dep (PDBe) have been developed to support the new deposition requirements. The molecular systems addressed in recent biomolecular NMR studies tend to be more complex, including oligomeric proteins, ligand binding studies, tandemly linked protein domains and so on. Several platforms for standardizing the protocols of NMR analysis can be used for tackling NMR studies of complicated systems, such as the CCPN program suite (Fogh et al. 2002, 2005, 2006; Vranken et al. 2005), SPINS (Baran et al. 2006) and KUJIRA (Kobayashi et al. 2007). There are several validation tools and web-servers for NMR data such as iCING (Doreleijers et. al. http://nmr.cmbi.ru.nl/icing/), AVS (Moseley et al. 2004; http://psvs-1_3.nesg.org/htdocs/avs.html), RPF (Huang et al. 2005; http://nmr.cabm.rutgers.edu/rpf/), PSVS (Bhattacharya et al. 2007), LACS (Wang et al. 2005; Wang and Markley 2009; http://www.bmrb.wisc.edu/software/lacs/) and PANAV (Wang et al. 2010). These platforms and tools are helpful for deposition and annotation tasks, however, automated and systematic support tools for the organizing of local NMR experimental databases are still desired. This is particularly important for large-scale projects such as structural genomics where there is a need to manage systematic workflow for large numbers of experimental and analyzed data.
In this report, we present an automated system bundled with a series of tools to organize NMR data in a local database and quickly validate the consistency between chemical shifts and spectral datasets. The system has been designed to assist the person who is responsible for deposition of NMR data to the BMRB. We demonstrate how the system facilitates depositions and annotations for a large number of BMRB entries for NMR chemical shift data that have been used for the comprehensive NMR structure analysis conducted by the RIKEN Structural Genomics/Proteomics Initiative (RSGI) (Yokoyama et al. 2000) in 2002–2007. The quality of the deposited data has been assessed by chemical shift prediction performed with SPARTA + program using released NMR structure coordinates.
Materials and methods
The program, KUJIRA, used in the data analysis, runs on Irix (Silicon Graphics Inc.) and Linux operating systems as previously described (Kobayashi et al. 2007). NMRView C-version (Johnson and Blebins 1994) was used to display spectrum contour plots and to execute Tcl/Tk scripts in the KUJIRA system. The data conversion programs, make_macro and ADD_inf (Fig. 1) are bundles of Tcl macros that run with a recent version of Tcl/Tk (8.4 or higher). BMRB Entry Support System (BESS) is a package of programs used for preparation of an entry, file conversion, and direct access to an ADIT-NMR server designed to allow a depositor to manage a BMRB deposition on a local PC (see also Fig. 1) including a function to upload the entry information to ADIT-NMR, and to merge uploaded chemical shift data into a meta file in NMR-STAR v3.1 format. Ed_BMRB is a plug-in program for KUJIRA to help the user to edit and confirm the entry information and to perform quick validation of NMR data (Figs. 2, 3).
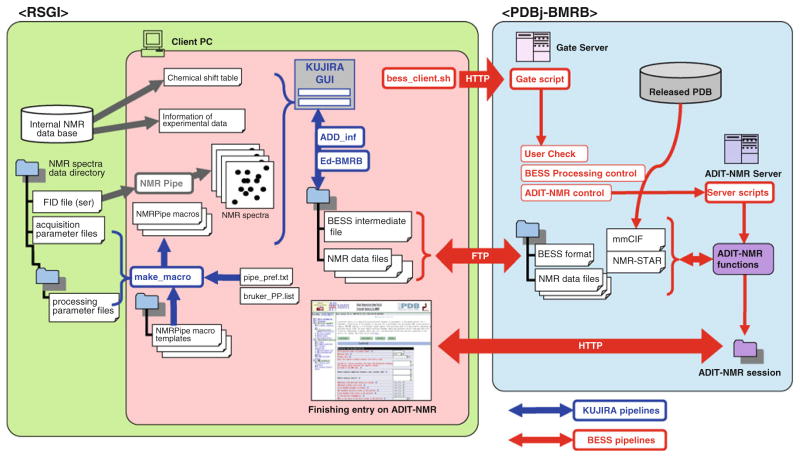
Fig. 1.
Schematic representation of the system for deposition and annotation of NMR data from RSGI to BMRB. The tools implemented in KUJIRA and BESS and related pipelines for files and tasks are colored blue and red, respectively. According to the preference settings described in the pipe_pref.txt, the program, make_macro, searches for corresponding NMR acquisition data sets in the server. The NMRPipe macro files, fid_*.com, xy_*.com and z_*.com are created for each spectrum from their corresponding template macro files by with the addition of the proper processing parameters such as phasing, extraction and zero-filling points and axis-order. The confirmation of the consensus between the NMR data and the assigned chemical shifts are carried out using the tools implemented in KUJIRA. To translate the retrieved NMR data using BESS, an entry information file is created and downloaded by bess_client.sh on the client PC using a mmCIF file derived from the corresponding released PDB entry. The program “ADD_inf” in KUJIRA fills the required information such as author information, followed by confirmation of consistency of entry information between the intermediate file and the corresponding one in the internal database on the RSGI side. The depositor can upload an entry information file and NMR data to the PDBj-BMRB side via FTP executed by “bess_client.sh”. Then the depositor can remotely execute the “Gate script” which activates “User_Check” and “BESS Processing control” followed by “ADIT-NMR control” to execute “Server scripts” on the ADIT-NMR server. These functions create a virtual ADIT-NMR session. The “bess_client.sh” can also handle HTTP protocol to call and edit the entry information on a web-browser by restarting an ADIT-NMR session
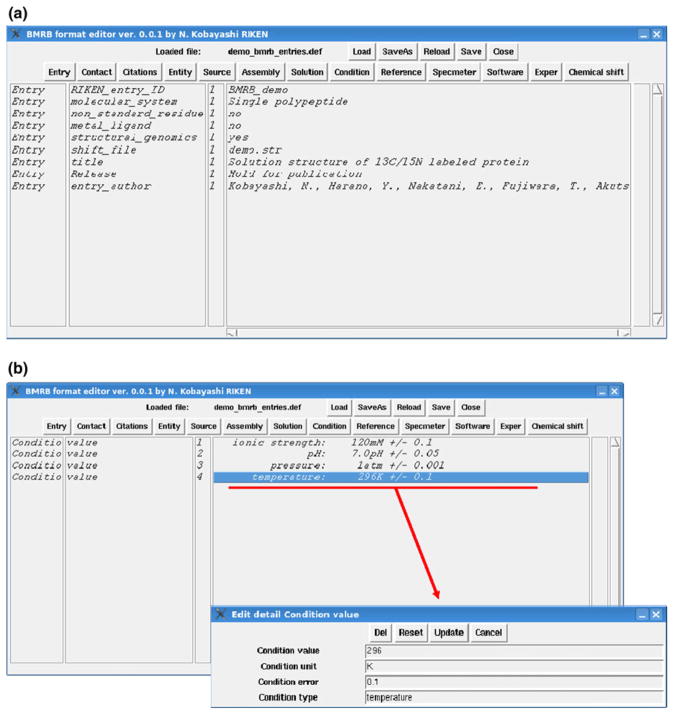
Fig. 2.
Graphical interface of the BMRB format editor implemented in KUJIRA as a plug-in module showing “Entry” (a) and “Condition” (b) items in the entry information file (*.def). The depositor can switch main items by clicking the button on the header of the window. The module has a function to save and load *.def files in the specified file format (see text). The parameters included in each item can be edited in the sub-window as shown in the subset in the panel (b) by double-clicking one of the elements in the displayed list box
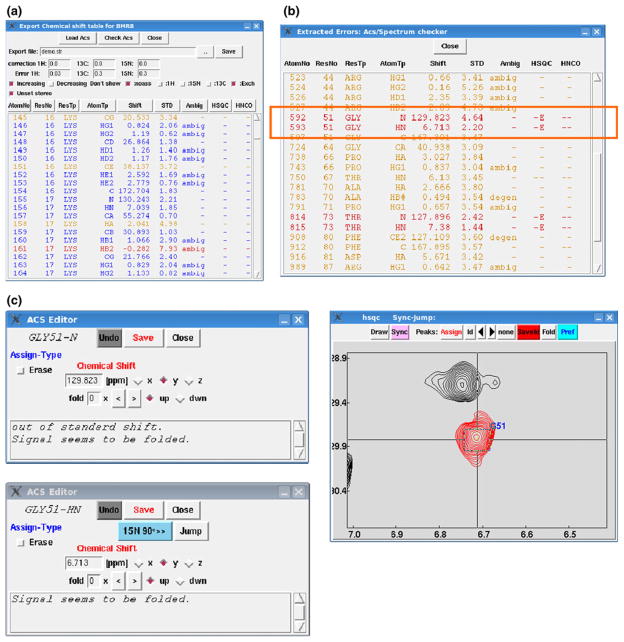
Fig. 3.
Graphical interface (a) of “Export chemical shift table for BMRB” popped up by clicking the button “Chemical shift” in the BMRB format editor shown in the figure. The editor can load chemical shifts for all atoms listed in the targeted NMR data by pressing the button “Load Acs” on the header of the GUI. The function can also indicate outliers evaluated by comparison with the standard chemical shifts listed in BMRB statistics with different colors. If the value (σ: assigned chemical shift and μ: standard value for the corresponding atom) is three or six times greater than the standard deviation listed in the statistics, the color is set at orange or red, respectively. The systematic corrections for the chemical shifts for 1H, 13C and 15N signals loaded on the listbox can be applied with the values (ppm) as specified in the “correction” entries. The error values for the signals are also specified in the entries as indicated “Error”. The check-boxes indicated as “Don’t show” for “noass”, “1H, 13C and 15N” and “Exch” are filtering the signals not assigned, assigned and exchangeable, respectively. The check-box labeled with “Unset stereo” switches all of the stereo-specific assignments for prochiral atoms treated as ambiguous ones. The buttons aligned just above the list-box are used for sorting elements displayed in the list-box, each of which corresponds to the column item. The sorting direction can be specified with check-box indicated as “Increasing” and “Decreasing”, with the manner of integer, floating point and alphabetical depending on the type of column items. b By pressing the button “Check Acs”, the sub-window pops up as shown. The function requires 2D 1H–15N, 1H–13C aliphatic and aromatic HSQC and 3D HNCO spectra. The list-box emphasizes the signals that have been considered to be outliers as mentioned above. The signals are also highlighted by red whose detected intensity is below 1.5 times the specified threshold for displaying the 2D contour plots in the corresponding position of the spectra. The lines indicated by a red box show that the 15N signal of Gly is suspected to be assigned on the aliased position inferred from the chemical shift and sign of the signal. Double-clicking the line pops up an editor sub-window for the assignment as shown in c with assessed comments. The sub-window for 1HN signal has a Jump button to display the corresponding region of the spectrum
Preparation of NMR data sets
In the present system, the NMR data deposition begins with the preparation of complete NMR data sets. NMRPipe (Delaglio et al. 1995) was commonly used for the NMR data processing for all BMRB data depositions. A tool “make_macro” has a function for retrieving sample conditions and acquisition parameters used to collect NMR experiments, for creating NMRPipe macros to convert the free induction decay (FID) data, and to execute Fourier transformation of the converted data. According to the preference file as shown in Fig. S1, “make_macro” tries to find available FIDs and parameter sets in the target directory corresponding to the requested NMR experiments. As shown in Fig. 1, “make_macro” automatically selects the appropriate template macros for the NMR experiment. The make_macro automatically fills the lines to execute the NMRPipe commands with the processing parameters and schemes as specified in the PP_list.txt and pref.txt files. In the case where the template macro is not available for the entry data, the processing is performed manually using the macro generator program implemented in NMRPipe. The current version of make_macro only supports NMR experiment data acquired on Bruker spectrometers. To complete the preparation, the make_macro runs the NMRPipe macros one by one according to the schedule as described in the preference file, followed by moving the processed spectrum data to the destination directory.
Preparation of BESS intermediate file for deposition and annotation of NMR data
The BESS intermediate file shown in Fig. 1 mediates the transfer of NMR data from RSGI to BMRB. In the intermediate file, 22–25 of the tags corresponding to mandatory deposition data items are given shorter and more straightforward names. These tags are considered to be the minimum set for the deposition of chemical shifts studied on a protein with a single chain system under a single sample condition. The intermediate file for each targeted PDB-ID (with extension .pre), is generated by BESS including the author and experimental information derived from the corresponding mmCIF file. The intermediate file also includes the experimental conditions, protein information (name, taxonomy, sequence and so on), type of NMR experiments, and program software used for the analysis. The files are then transferred to the contact person via FTP, followed by completion of remaining entry information using the program, ADD_inf. The ADD_inf tool generates the final intermediate file (with extension .fin) that is acceptable for a plug-in tool on KUJIRA.
The plug-in tool Ed-BMRB acts as a GUI tool allowing the user to confirm and correct all of the items described in the final intermediate file with the graphical interface. The graphical interface as shown in Fig. 2a is constructed with the buttons corresponding to the mandatory items as described in the intermediate file which can allow a user to edit elemental values for each item. Ed-BMRB has a special sub-window to confirm the assigned chemical shifts archived in the internal database of KUJIRA that allows the user to visually inspect the NMR spectra as well as to correct the assignments interactively with the GUI tool (Fig. 3a). In the RSGI project, 2D 1H–15N HSQC, 2D 1H–13C HSQC and 3D HNCO spectra were mainly used to confirm whether the signals as expected from the assigned chemical shifts can be observed in the corresponding spectral positions. The program Ed-BMRB automatically simulates the signal positions expected from the type of corresponding NMR experiment and assigned chemical shifts. Similarly Ed-BMRB can control the 2D and 3D spectra to expand the desired spectral region using an intrinsic KUJIRA command, Sync-Jump. Assigned chemical shifts loaded in a listbox in a sub-window have any outliers emphasized with different color coding (orange and red) indicating chemical shifts that are significantly away from the standard value. There is another sub-window to search for chemical shift outliers by detecting the intensity on the corresponding position of the spectrum (see Fig. 3c). This sub-window function can be especially useful for detecting incorrectly assigned chemical shifts derived from signal aliasing. By appearance of a short comment in the bottom of the sub-window, the user is notified that the chemical shifts have been found to be in error and provided with an explanation. After confirmation and editing of the chemical shifts, the user can export the chemical shift table in BMRB NMR-STAR file format (ver. 2.1).
The processed final intermediate file and the exported chemical shift table in NMR-STAR file format are sent back to the BMRB side via FTP using commands of the BESS client. Another command of the BESS client can merge the two files into a single file in NMR-STAR file format that is ready for deposition in the BMRB database. At the same time, the BESS client can create a virtual session for the automated deposition input tool “ADIT-NMR”. The NMR-STAR file is then loaded to the relevant file/directory for the session to complete the session as an actual BMRB deposition.
The chemical shift data for deposition
The resonance frequency of the methyl proton signal of TSP-d5 (trimethyl-silyl-propionate-d5) in 50 mM sodium phosphate buffer (pH 7.0) at 298 K was used as the chemical shift reference (zero ppm). In each NMR spectrum, the chemical shift reference of the 1H signal was calibrated from the water signal using the previously determined frequency of the TSP proton resonance at zero ppm. The chemical shifts were then corrected with the experimental temperature using the standard temperature dependency of the water signal. For the other nuclei, 13C and 15N, the frequencies of the resonances at zero ppm were estimated from that for 1H based on their gyromagnetic ratio. The chemical shifts listed for the deposition were used for the assignment of NOE peaks derived from 3D-13C-edited and 3D-15N-edited NOESY spectra, the former includes all aliphatic and aromatic carbon signals attached to protons. The 1H signals of hydrogen atoms not covalently bound to C and N atoms and the exchangeable protons were eliminated from the list, including hydroxyl protons on threonine, serine and tyrosine, and guanidinyl protons on arginine. The error values for the determined chemical shifts were identical to the tolerance of automated NOE assignments performed by CYANA (Güntert 2003). In typical cases, the values of chemical shift tolerances were set at 0.02–0.03 ppm for 1H dimensions and 0.3–0.4 ppm for 15N and 13C nuclei for either direct or indirect dimensions. The 13C chemical shifts of sequential carbonyl groups C′(i−1) in the main chain were automatically verified with 3D-HNCO data by the function implemented in the Ed_BMRB GUI tool. Only for the carbonyl carbons whose amide signal of the subsequent residue were not assigned due to lack of a 1H–15N correlation, the intra-residual signals C′(i) were manually confirmed by the direct observation of the signals in the 3D-HN(CA)CO spectrum.
Quality assessment of the deposited chemical shift data
Of the 628 RSGI BMRB chemical shift entries, there are 560 corresponding released RSGI PDB entries. The assigned chemical shift tables for the BMRB entries with matching PDB entries were downloaded from the BMRB web-site using a standard wget command in Linux. In addition to these entries, matched PDB and BMRB entries for another unrelated 1,300 single chain proteins were also downloaded. The AVS and LACS validation results available from the BMRB web-sites also were downloaded. The solvent accessible surface area (ASA), backbone order parameters for ϕ and ψ angles (S) were calculated by the program KUJIRA. SPARTA+ was used to calculate the predicted chemical shifts from the PDB structure coordinates. The experimentally determined chemical shifts for the 13C signals were corrected with the offset error values calculated by LACS. The number of outliers was counted for each assigned chemical shift (see details described in Fig. 4).
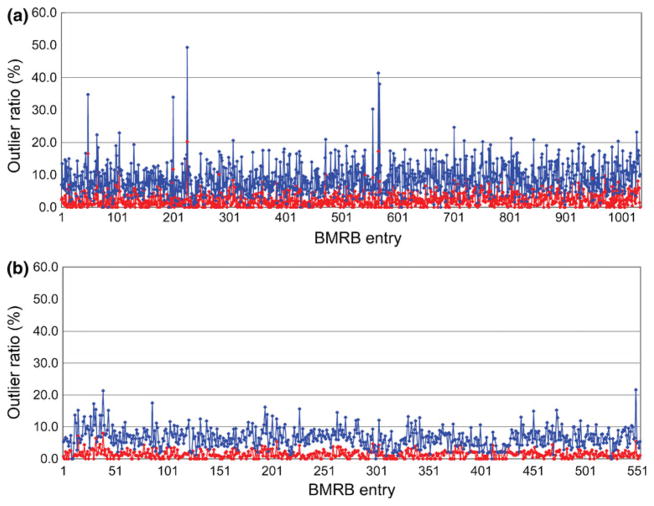
Fig. 4.
The outlier ratio plotted (a) for 1,033 BMRB entries deposited between the years 2001 to 2011 except for RSGI entries, (b) for 553 RSGI entries. The Z-value for signal i was calculated for Cα and Cβ chemical shifts by: Zi = |δexp − δpred|/Si, where δexp and δpred are experimental and predicted chemical shifts, respectively, Si prediction error for signal i. The minimum value for the prediction error was set to 0.1 ppm. The 13C offset value estimated by LACS calculation was also applied to each of the δexp values. The error ratio was then calculated with the number of assigned signals having Z-value greater than the cut-off 3.0 (red) and 2.0 (blue) and plotted. Some entries were eliminated based on the following criteria: Showing error during the calculation with KUJIRA or SPARTA+, their released BMRB data or results for LACS calculations are not available in the BMRB web-site
Results and discussions
Simplified format designed for systematic deposition and annotation of NMR data
ADIT-NMR has been established as a highly automated system for the deposition of atomic coordinates and NMR data to the PDB and BMRB public repositories, respectively. The NMR-STAR format (Ulrich et al. 2008; www.bmrb.wisc.edu/formats.html) is based on the STAR (Self Defining Text Archival and Retrieval) format developed by Hall and coworkers (Hall 1991; Hall and Spadaccini 1994; Allen et al. 1995) and is commonly used for the exchange and deposition of NMR data. The NMR-STAR format has a systematically curated data structure based on the NMR-STAR data model (http://www.bmrb.wisc.edu/formats.html). The data structure has a flexible design and accepts a wide variety of NMR parameters and experiment types, however, the save frame and loop format can be difficult to survey visually. Even in the simplest case, one may have to handle 500–700 lines (depending on the molecular system) exclusive of the chemical shift table, where at least 100–200 tag elements are commonly required. Protein NMR structural studies reported by the RSGI project were carried out by a single research group operating at a centralized research facility and the data archived in a highly regularized internal database. The protein systems studied were primarily simple single chain polymers without ligands and non-standard residues. The NMR data were derived from a limited and consistent set of NMR experiments and the experimental conditions used were standardized (details mentioned below). Because the RSGI studies were carried out on relatively simple protein systems, using a standard set of NMR experiments, and under very similar experimental conditions, we were able to further simplify the NMR-STAR data format and apply the revised format to the intermediate file used in the program BESS. As shown in Fig. S3, the intermediate file is written in a column-based text and visually straightforward. Since the information available in the PDB entry related to the target entry has been already incorporated into the file, the number of items in the intermediate file required to describe the chemical shift data is largely reduced to 22–25 in comparison with 100–200 in a normal ADIT-NMR entry. It is also remarkable that almost all of the NMR structure studies (~1,280 entries) conducted by RSGI were determined using the program KUJIRA. The NMR parameters and information including parameters for spectrum acquisition, chemical shifts, peak tables, structure coordinates were, therefore, managed in a regularized manner. Owing to the features of KUJIRA, NMR scientists can organize their experimental data as well as their author information and experimental conditions using a single analysis platform.
Quality control of analyzed chemical shifts
The accuracy of the chemical shift data deposited and archived in the BMRB NMR database is a critical issue. However, many depositors find that the effort required to validate and verify the accuracy of data outliers is a burden that slows the performance of the deposition and annotation systems. The chemical shifts deposited from RSGI to BMRB were all derived from assignments of the NOE signals observed in 15N- and 13C-edited NOESY spectra except for the carbonyl carbon signals as mentioned above. In the determination of the NMR structures corresponding to each BMRB entry, more than 95 % of the NOE peaks have been assigned by CYANA. On the basis of the KUJIRA-CYANA protocol, NOE peaks identified in the 3D 15N and 13C edited NOESY spectra have not been eliminated from the peak lists for the automated NOE assignments unless there was a specific reason, for instance, the peaks were suspected of being derived from exchangeable groups on the surface of the protein. This high completeness of the NOE assignments means that the observed NOE peaks have been well explained by the determined chemical shifts and protein structures. In other words, the reliability of the chemical shift data deposited from RSGI has been reasonably guaranteed by the structure determination. The checking function in KUJIRA for comparison with the standard chemical shifts was used effectively for detection of outliers in the assigned chemical shifts. The function generates simulated peak lists for the 2D 1H–15N spectrum and 1H–13C HSQC spectra for aliphatic and aromatic carbon signals and the 3D HNCO spectrum using the assigned chemical shifts and the information of spectrum type. The peak lists can be automatically loaded and displayed on the 2D spectrum contour plots as indicative blue boxes, allowing the user to visually inspect assignments in the chemical shift table readily as shown in Fig. 3c. As the spectral width for 13C and 15N dimensions in 2D and 3D spectra is normally narrow, many peaks outside of the spectral width appear in aliased positions, which may lead easily to mis-assignments. With the application of the function, the direct detection of spectrum intensity can also be useful to search for spuriously assigned chemical shifts as highlighted in the listbox on the “Check Acs” sub-window.
Performance of our system on deposition and annotation of the NMR data
On behalf of the RIKEN Structural Genomics/Proteomics Initiative (RSGI), the RIKEN NMR group contributed more than 1,300 structures during the 5-year structural genomics project. The project for the deposition and annotation of the chemical shift data started as a collaboration between PDBj-BMRB in Osaka University and the RIKEN NMR group beginning in 2006. As of 2009, almost all PDB entries (1,287 entries) were released, so that we were able to retrieve the author, sample and experimental information from the released files in mmCIF format which are available in the PDB archive. This information successfully reduces the labor to ensure terminological consistency between the files involved in PDB and BMRB entries. In the early years (2006–2008) of the project, the Ed_BMRB program in KUJIRA and the intermediate file formats for the client part of the program BESS were developed (Fig. S3). While the complete set of programs for BESS were used in the final year 2010, the number of yearly processed entries reached more than 300. The typical performance of the deposition system performed by a single depositor is around 30–60 min for each entry including retrieval of spectral data from the internal database in RSGI and confirmation of assigned chemical shifts directly on spectrum data sets. Although it depends on the size of protein and quality of the spectral data, the time required for the work would be more than approximately two to three times shorter than that for the depositor without the system (data not shown). The annotation work including communication with the depositors and cross-checking by a third person has been also expedited, which normally takes <1 h.
Probably the most important stages for managing public database are submission and annotation of deposited entries, as they may not be automated in order to keep quality of the deposited data. It has been a big challenge to automate and expedite this work in the database community for a long time. In contrast, the tools to confirm experimental data just before the deposition stage have been less focused. Penkett et al. have recently developed a new program suite, CcpNmr Entry Completion Interface (ECI), for simple, secure and complete deposition of NMR data (Penkett et al. 2010). The interface can directly upload and validate analyzed chemical shift data to the web based deposition site for NMR structure, AutoDep, at the European Bioinformatics Institute (EBI: http://www.ebi.ac.uk/pdbe-apps/nmr/deposition/autodep.html). The concept of their program suite is intriguingly similar to what we have reported here, which has been aimed to reduce and simplify the extra work required to deposit NMR data into the public databases. As our system has been optimized for translation of a large number of entries, we have reduced the number of deposition items in the column-based text format. This format may be widely preferred, because the data structure is amenable to standard Unix commands such as grep, sort, awk and sed as well as to a wide variety of programs that can easily handle the tabular format. However, it would not be easy to convert the files to the standard archive format such as mmCIF, NMR-STAR and XML based CCPN data model. By combining our tools with the other systems such as that of CCPN may be a promising way to enhance the efficiency of deposition work even for the NMR scientists in a small laboratory.
The impact of the high quality experimental NMR data studied under similar experimental conditions
PDB structure coordinate entries are available for all 628 RSGI NMR chemical shift data entries deposited at BMRB. With regard to the experimental conditions for the BMRB entries, one can find a high similarity among them, namely, similar temperature ranging from 298 to 303 K, pH 6.8–7.0, containing phosphate or Tris based buffers with salt concentrations in the range of 50–100 mM. Although the size of the proteins varied from 2 to 20 kDa, nearly all of the targeted proteins are monomeric singlechain proteins (as shown in Fig. S4). These facts mean that the dependencies of the NMR experimental data on the structure coordinates are strongly correlated and not influenced by the experimental conditions used. The chain lengths of the target proteins ranged from 49 to 219 amino acid residues, resulting in an average of 87 residues. The signal assignments including 1H, 13C and 15N nuclei reveal very high completeness, with average values of 94.4 and 85.7 % for main-chain and side-chain atoms, respectively (see Fig. S5). Figure 4 shows the analysis for the outlier ratio between the experimental and predicted chemical shifts. The predictions were performed using SPARTA+ and the error ratios were plotted for the 13Cα and 13Cβ signals in the ordered region (Sϕ + Sψ < 1.8). The error ratio for the RSGI entries even in the results for the Z-value cut-off 2.0 is remarkably lower than that found in the other BMRB entries. Considering the prediction power of the program SPARTA+, the deposited NMR structures and their assigned chemical shifts are well consistent and reasonably trustful in the level of the backbone conformation. These results suggest that the NMR data deposited by RSGI have contributed to the quality improvement of BioMagResBank. Future studies using the RSGI NMR data in combination with the PDB structural data will undoubtedly provide improved methods to predict chemical shifts from/to atomic coordinates as well as to develop more powerful tools to validate experimentally determined chemical shifts and NMR structures.
Supplementary Material
Acknowledgments
This work was partially supported by National Bioscience Database Center (NBDC) in Japan Science and Technology Agency (JST). We are grateful to Prof. Haruki Nakamura for intensive encouragement to us and for contribution to discussions about this study. We thank Dr. J. Doreleijers for many valuable comments, suggestions and proofreading of the manuscripts and Dr. F. Delaglio for help in establishing the macro-file library for the NMR-Pipe data process. We also thank Mr. T. Iwata for his work in preparing the web-page for downloading the BMRB related tools.
Footnotes
Electronic supplementary material The online version of this article (doi:10.1007/s10858-012-9641-6) contains supplementary material, which is available to authorized users.
Software availability
The programs and documentations for the tools used for this study are available from http://bmrbdep.protein.osakau.ac.jp/toolbox.html.
Contributor Information
Naohiro Kobayashi, Email: naohiro@protein.osaka-u.ac.jp, Institute for Protein Research, Osaka University, 3-2 Yamadaoka, Suita 565-0871, Osaka, Japan. RIKEN Systems and Structural Biology Center, 1-7-22 Suehirocho, Tsurumi, Yokohama 230-0045, Japan.
Yoko Harano, Institute for Protein Research, Osaka University, 3-2 Yamadaoka, Suita 565-0871, Osaka, Japan.
Naoya Tochio, RIKEN Systems and Structural Biology Center, 1-7-22 Suehirocho, Tsurumi, Yokohama 230-0045, Japan.
Eiichi Nakatani, Institute for Protein Research, Osaka University, 3-2 Yamadaoka, Suita 565-0871, Osaka, Japan.
Takanori Kigawa, RIKEN Systems and Structural Biology Center, 1-7-22 Suehirocho, Tsurumi, Yokohama 230-0045, Japan. Department of Computational, Intelligence and Systems Science, Interdisciplinary, Graduate School of Science and Engineering, Tokyo Institute of Technology, 4259 Nagatsuta-cho, Midori, Yokohama 226-8502, Japan.
Shigeyuki Yokoyama, RIKEN Systems and Structural Biology Center, 1-7-22 Suehirocho, Tsurumi, Yokohama 230-0045, Japan.
Steve Mading, Department of Biochemistry, University of Wisconsin-Madison, 433 Babcock Drive, Madison, WI 53706, USA.
Eldon L. Ulrich, Department of Biochemistry, University of Wisconsin-Madison, 433 Babcock Drive, Madison, WI 53706, USA
John L. Markley, Department of Biochemistry, University of Wisconsin-Madison, 433 Babcock Drive, Madison, WI 53706, USA
Hideo Akutsu, Institute for Protein Research, Osaka University, 3-2 Yamadaoka, Suita 565-0871, Osaka, Japan.
Toshimichi Fujiwara, Institute for Protein Research, Osaka University, 3-2 Yamadaoka, Suita 565-0871, Osaka, Japan.
References
- Allen FH, Barnard JM, Cook APF, Hall SR. The molecular information file (MIF): core specifications of a new standard format for chemical data. J Chem Inf Comput Sci. 1995;35:412–417. [Google Scholar]
- Baran MC, Moseley HN, Aramini JM, Bayro MJ, Monleon D, Locke JY, Montelione GT. SPINS: a laboratory information management system for organizing and archiving intermediate and final results from NMR protein structure determinations. Proteins. 2006;62:843–851. doi: 10.1002/prot.20840. [DOI] [PubMed] [Google Scholar]
- Bhattacharya A, Tejero R, Monelione GT. Evaluating protein structures determined by structural genomics consortia. Proteins. 2007;66:778–795. doi: 10.1002/prot.21165. [DOI] [PubMed] [Google Scholar]
- Cavalli A, Salvatella X, Dobson CM, Vendruscolo M. Protein structure determination from NMR chemical shifts. Proc Natl Acad Sci USA. 2007;104:9615–9620. doi: 10.1073/pnas.0610313104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornilescu G, Delaglio F, Bax A. Protein backbone angle restraints from searching a database for chemical shift and sequence homology. J Biomol NMR. 1999;13:289–302. doi: 10.1023/a:1008392405740. [DOI] [PubMed] [Google Scholar]
- Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J Biomol NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- Fogh R, Ionides J, Ulrich E, Boucher W, Vranken W, Linge JP, Habeck M, Rieping W, Bhat TN, Westbrook J, Henrick K, Gilliland G, Berman H, Thornton J, Nilges M, Markley J, Laue E. The CCPN project: an interim report on a data model for the NMR community. Nat Struct Biol. 2002;9:416–418. doi: 10.1038/nsb0602-416. [DOI] [PubMed] [Google Scholar]
- Fogh RH, Boucher W, Vranken WF, Pajon A, Stevens TJ, Bhat TN, Westbrook J, Ionides JM, Laue ED. A framework for scientific data modeling and automated software development. Bioinformatics. 2005;21:1678–1684. doi: 10.1093/bioinformatics/bti234. [DOI] [PubMed] [Google Scholar]
- Fogh RH, Vranken WF, Boucher W, Stevens TJ, Laue ED. A nomenclature and data model to describe NMR experiments. J Biomol NMR. 2006;36:147–155. doi: 10.1007/s10858-006-9076-z. [DOI] [PubMed] [Google Scholar]
- Güntert P. Automated NMR protein structure calculation. Prog NMR Spectrosc. 2003;43:105–125. [Google Scholar]
- Hall SR. The STAR file: a new format for electronic data transfer and archiving. J Chem Inf Comput Sci. 1991;31:326–333. [Google Scholar]
- Hall SR, Spadaccini N. The STAR file: detailed specifications. J Chem Inf Comput Sci. 1994;34:505–508. [Google Scholar]
- Huang YJ, Powers R, Montelione GT. Protein NMR recall, precision, and F-measure scores (RPF scores): structure quality assessment measures based on information retrieval statistics. J Am Chem Soc. 2005;127:1665–1674. doi: 10.1021/ja047109h. [DOI] [PubMed] [Google Scholar]
- Johnson BA, Blebins RA. NMRView: a computer program for the visualization and analysis of NMR data. J Biomol NMR. 1994;4:603–614. doi: 10.1007/BF00404272. [DOI] [PubMed] [Google Scholar]
- Kobayashi N, Iwahara J, Koshiba S, Tomizawa T, Tochio N, Güntert P, Kigawa T, Yokoyama S. KUJIRA, a package of integrated modules for systematic and interactive analysis of NMR data directed to high-throughput NMR structure studies. J Biomol NMR. 2007;39:31–52. doi: 10.1007/s10858-007-9175-5. [DOI] [PubMed] [Google Scholar]
- Moseley HN, Sahota G, Montelione GT. Assignment validation software suite for the evaluation and presentation of protein resonance assignment data. J Biomol NMR. 2004;28:341–355. doi: 10.1023/B:JNMR.0000015420.44364.06. [DOI] [PubMed] [Google Scholar]
- Neal S, Nip AM, Zhang H, Wishart DS. Rapid and accurate calculation of protein 1H, 13C and 15N chemical shifts. J Biomol NMR. 2003;26:215–240. doi: 10.1023/a:1023812930288. [DOI] [PubMed] [Google Scholar]
- Penkett CJ, van Ginkel G, Velankar S, Swaminathan J, Ulrich EL, Mading S, Stevens TJ, Fogh RH, Gutmanas A, Kleywegt GJ, Henrick K, Vranken WF. Straightforward and complete deposition of NMR data to the PDBe. J Biomol NMR. 2010;48:85–92. doi: 10.1007/s10858-010-9439-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seavey BR, Farr EA, Westler WM, Markley JL. A relational database for sequence-specific protein NMR data. J Biomol NMR. 1991;1:217–236. doi: 10.1007/BF01875516. [DOI] [PubMed] [Google Scholar]
- Shen Y, Bax A. Protein backbone chemical shifts predicted from searching a database for torsion angle and sequence homology. J Biomol NMR. 2007;38:289–302. doi: 10.1007/s10858-007-9166-6. [DOI] [PubMed] [Google Scholar]
- Shen Y, Bax A. SPARTA+: a modest improvement in empirical NMR chemical shift prediction by means of an artificial neural network. J Biomol NMR. 2010;48:13–22. doi: 10.1007/s10858-010-9433-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Y, Oliver L, Delaglio F, Rossi P, Aramini J, Liu G, Eletsky A, Wu Y, Singarapu KK, Lemak A, Ignatchenko A, Cheryl H, Arrowsmith CH, Szyperski T, Gaetano T, Montelione GT, Baker D, Bax A. Consistent blind protein structure generation from NMR chemical shift data. Proc Natl Acad Sci USA. 2008;105:4685–4690. doi: 10.1073/pnas.0800256105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Y, Delaglio F, Cornilescu G, Bax A. TALOS+: a hybrid method for predicting protein backbone torsion angles from NMR chemical shifts. J Biomol NMR. 2009;44:213–223. doi: 10.1007/s10858-009-9333-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulrich EL, Markley JL, Kyogoku Y. Creation of a nuclear magnetic resonance data repository and literature database. Protein Seq Data Anal. 1989;2:23–37. [PubMed] [Google Scholar]
- Ulrich EL, Akutsu H, Doreleijers JF, Harano Y, Ioannidis YE, Lin J, Livny M, Mading S, Maziuk D, Miller Z, Nakatani E, Schulte CF, Tolmie DE, Kent Wenger R, Yao H, Markley JL. BioMagResBank. Nucleic Acids Res. 2008;36:D402–D408. doi: 10.1093/nar/gkm957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vranken WF, Boucher W, Stevens TJ, Fogh RH, Pajon A, Llinas M, Ulrich EL, Markley JL, Ionides J, Laue ED. The CCPN data model for NMR spectroscopy: development of a software pipeline. Proteins. 2005;59:687–696. doi: 10.1002/prot.20449. [DOI] [PubMed] [Google Scholar]
- Wang L, Markley JL. Empirical correlation between protein backbone 15N and 13C secondary chemical shifts and its application to nitrogen chemicalshift re-referencing. J Biomol NMR. 2009;44:95–99. doi: 10.1007/s10858-009-9324-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Eghbalnia HR, Bahrami A, Markley JL. Linear analysis of carbon-13 chemical shift differences and its application to the detection and correction of errors in referencing and spin system identifications. J Biomol NMR. 2005;44:13–22. doi: 10.1007/s10858-005-1717-0. [DOI] [PubMed] [Google Scholar]
- Wang B, Wang Y, Wishart DS. A probabilistic approach for validating protein NMR chemical shift assignments. J Biomol NMR. 2010;47:85–99. doi: 10.1007/s10858-010-9407-y. [DOI] [PubMed] [Google Scholar]
- Wishart DS, Arndt D, Berjanskii M, Tang P, Zhou J, Lin G. CS23D: a web server for rapid protein structure generation using NMR chemical shifts and sequence data. Nucleic Acids Res. 2008;36:W496–W502. doi: 10.1093/nar/gkn305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokoyama S, Hirota H, Kigawa T, Yabuki T, Shirouzu M, Terada T, Ito Y, Matsuo Y, Kuroda Y, Nishimura Y, Kyogoku Y, Miki K, Masui R, Kuramitsu S. Structural genomics projects in Japan. Nat Struct Biol. 2000;7:943–945. doi: 10.1038/80712. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.