Abstract
This work is part of the multi-center Alzheimer's Disease Neuroimaging Initiative (ADNI), a large multi-site study of dementia, including patients having mild cognitive impairment (MCI), probable Alzheimer's disease (AD), as well as healthy elderly controls. A major portion of ADNI involves the use of [18F]-fluorodeoxyglucose (FDG) with positron emission tomography (PET). The objective of this paper is reduction of inter-scanner differences in the FDG-PET scans obtained from the 50 participating PET centers having fifteen different scanner models. In spite of a standardized imaging protocol, systematic inter-scanner variability in PET images from various sites is observed primarily due to differences in scanner resolution, reconstruction techniques, and different implementations of scatter and attenuation corrections. Two correction steps were developed by comparison of 3-D Hoffman brain phantom scans with the ‘gold standard’ digital 3-D Hoffman brain phantom: i) high frequency correction; where a smoothing kernel for each scanner model was estimated to smooth all images to a common resolution and ii) low frequency correction; where smooth affine correction factors were obtained to reduce the attenuation and scatter correction errors. For the phantom data, the high frequency correction reduced the variability by 20%-50% and the low frequency correction further reduced the differences by another 20%-25%. Correction factors obtained from phantom studies were applied to 95 scans from normal control subjects obtained from the participating sites. The high frequency correction reduced differences similar to the phantom studies. However, the low frequency correction did not further reduce differences; hence further refinement of the procedure is necessary.
Introduction
This work is part of the ongoing multi-center Alzheimer's Disease Neuroimaging Initiative (ADNI) project, a longitudinal, multi-site observational study of healthy controls, patients with mild cognitive impairment (MCI), and mild probable Alzheimer's disease (AD) patients. This five-year research project aims to study the rate of change of cognition, brain structure and function in 200 elderly controls, 400 subjects with mild cognitive impairment, and 200 with probable Alzheimer's disease. Data is being acquired longitudinally using magnetic resonance imaging (MRI), [18F]FDG PET, [11C]PiB PET, urine serum, and cerebrospinal fluid (CSF) biomarkers, as well as clinical and psychometric assessments. PET scans are being performed on half of the subjects in each of the three groups. The Division of Nuclear Medicine PET Center at the University of Michigan is the coordinating center for quality control and pre-processing of all PET studies, while several groups are responsible for analysis of the PET data.
The objective of this work is the development of a framework for reduction of inter-scanner differences in static FDG scans acquired in ADNI. The scans are being obtained from 50 participating PET centers having different hardware and software. In all there were 15 different scanner-types in this project. In spite of using a standardized imaging protocol, systematic inter-scanner variability in PET images from various sites has been observed due to differences in scanner resolution, reconstruction techniques, and different implementations of scatter and attenuation corrections on the different scanner models. It is an important step to try to minimize these differences before the data across centers is pooled for analysis.
The differences in the human PET scans can be classified into two broad categories: 1) actual inter-subject variability, which includes both anatomic and functional differences and 2) systematic differences related to scanner hardware and software. The goal of PET is to determine the functional differences between individuals or groups of individuals, and hence removal of both the anatomic differences that exist between subjects as well as the systematic differences across scanner models is of interest. While much work has been done in reducing anatomic differences across subjects by the use of standardized atlases (Mazziotta et al. 1995; Minoshima et al. 1994a) and non-linear warping techniques (Minoshima et al. 1994b), the focus of the present work is reduction of the systematic differences between the different scanner models.
The correction factors to reduce systematic inter-scanner variability were obtained from 3-D Hoffman brain phantom (Hoffman et al. 1990) scans acquired at the participating sites. The 3-D Hoffman brain phantom is a cylindrically shaped phantom that simulates the radiotracer distribution in a normal human brain for tracers aimed at measuring cerebral glucose metabolism or blood flow. The relative concentrations of radioactivity in “gray matter”, “white matter”, and all other structures are 4:1:0, respectively. The correction factors for each scanner type were obtained by comparison of the phantom scans with a ‘gold standard’ digital representation of the true Hoffman brain phantom (i.e. representing the actual radioactivity distribution).
The systematic differences in the reconstructed images across the different scanners were classified into two general types: high frequency differences, related primarily to image resolution; and low frequency differences, related to image uniformity and the more subtle aspects of image formation such as corrections for attenuation and scatter. Resolution differences are due primarily to differences in crystal sizes, and to a lesser extent due to detector material (LSO, BGO, GSO and LYSO), detector crystal axial depths, energy windows, as well as the number of rings, crystals per ring and axial field-of-view. The low frequency uniformity differences may manifest as differences in contrast (grey-to-white matter ratios) as well as superior-to-inferior, anterior-to-posterior, and/or midline-to-lateral gradients. These non-uniformities between scanners are likely to be caused primarily by disparity in the software routines that handle attenuation and scatter. The high frequency correction proposed in this work involves smoothing the data from different scanner models to a common resolution, whereas, the low frequency correction involves application of smooth affine correction factors following the high frequency correction. Both the high and low frequency correction factors were obtained by comparison of phantom scan data with the digital phantom. The phantom-based correction factors were applied to phantom scans to determine the maximum recovery possible using this approach. Subsequently, the phantom-based corrections were applied to 95 normal control scans to test their utility in human PET studies.
Methods
Hoffman brain phantom scans were obtained from all participating sites using a standard protocol. There were in all fifteen different scanner models among the participating sites (7 PET-only and 8 PET/CT scanners). The key features of the protocol include the following.
The Hoffman phantom is filled with 0.5-0.6 mCi of 18F solution and placed in the scanner.
The chest phantom is filled with 2.0-2.4 mCi of 18F solution and placed close to the Hoffman phantom to simulate the effects of out-of-field activity.
The 3-D Hoffman phantom is imaged for 30 minutes to obtain high quality images with low statistical noise contribution.
Reconstructions parameters for each scanner model were determined by the ADNI PET core and differed between vendors based on available software
The image volume is registered to the digital Hoffman brain phantom to achieve a common orientation and image grid for all scans.
Pre-processing of phantom scans
Two phantom scans were obtained for test/retest purposes at each site. All scans passing quality control tests were registered to the digital Hoffman phantom. The voxel-grid for registered phantom images of all scanner-types was 160 × 160 × 90 with a voxel-size of 1.548 mm3. The size of 1.548 mm was chosen such that the dimensions of the digital phantom best matched the physical dimensions individual layers of the 3-D Hoffman brain phantom. The registered images from each site were normalized using a mask (based on the digital Hoffman phantom) such that the mean of all voxels within the mask was unity. The normalized phantom images from different sites having the same scanner model were averaged to obtain an average image per scanner model. Let this normalized average image for scanner model n be represented as An (An ∈ Rp×q×r where p = 160 (x-dimension), q = 160 (y-dimension) and r = 90 (z-dimension)). High and low frequency correction factors were obtained by comparison of the average image An with the digital Hoffman brain phantom as described below.
High frequency correction
The high frequency correction was a simple smoothing operation to bring the images from the different scanner models to a uniform spatial resolution. The common minimum resolution was determined by estimating the resolution of each scanner model from the phantom scans. The digital Hoffman brain phantom was smoothed in all three dimensions with incremental full width half maximum (FWHM) Gaussian kernels to obtain a library of the digital phantom at various resolutions as shown below.
| - 1 |
where D is the unsmoothed digital Hoffman brain phantom, ki is the smoothing kernel with FWHM of i mm in all three dimensions, ⊗ is the convolution operator and Di is the smoothed phantom with i mm resolution. During implementation of this step, different in-plane (xy plane) and axial (z-axis) smoothing was done; but for brevity it has been represented in Equation 1 to be the same in all dimensions. The effective resolution of nth scanner model was estimated by determining the smoothed digital phantom (Di) that was closest to An in the least squares sense as shown below.
| - 2 |
where Ān and D̄i are lexicographically arranged vectors of all the voxels in the three-dimensional image volumes An and Di respectively. The coarsest resolution scanner of all the models was found to match the digital Hoffman phantom smoothed between 7 and 8 mm FWHM, both in plane and axially. Hence, the ‘target’ resolution for the average phantom image (An) for each scanner model was chosen to be the effective resolution that best matches isoptropically smoothed 8mm digital phantom, D8.
Kernels to smooth each scanner model's average phantom image to the target resolution were determined as follows. A library for each average phantom scan An was formed by smoothing it with incremental FWHM Gaussian kernels with as shown below.
| - 3 |
The FWHM of the smoothing kernel for the nth scanner model (ĵn) was selected such that the smoothed image (An, j) matched the ‘gold standard’ digital phantom smoothed to 8mm resolution (D8) in the least squares sense as shown below.
| - 4 |
where Ān, j and D̄8 are lexicographically arranged vectors of the three dimensional image volumes An, j and D8 respectively. As before, j was allowed to vary between in-plane and axial smoothing. Let the phantom image for scanner model ‘n’ after smoothing to 8 mm resolution be represented by An, ĵ. The smoothing kernel for each scanner model (kĵn) thus obtained from phantom data was then applied to every the human subject scan (In) obtained from scanner model n (In, ĵn = In ⊗ kĵn).
Low frequency correction
High frequency correction was followed by low frequency adjustment to correct for differences across scanner models that are presumed to be due primarily to small but consistent differences in the corrections for attenuation and scatter. The following linear model was used as the low frequency correction.
| - 5 |
where an and bn are the low frequency correction terms (multiplicative and additive respectively) to be determined from the high frequency corrected phantom images An, ĵn (εn is the residual term). Note that all terms in Equation 5 have the same dimensions and all operations are voxel-wise. The terms an and bn are smooth functions for nth scanner model and are designed as linear combinations of three dimensional, fifth order polynomials as shown below:
| - 6 |
where an,p and bn,p are values of the correction factors an and bn at voxel p, M is the total number of polynomial terms (M = 52 for three dimensional fifth order polynomials), αm and βm are the coefficients of the polynomial term m(1 ≤ m ≤ M), and λp,m is the value of the mth polynomial term at voxel p. Since the low frequency errors were expected to be symmetric across the midline of the brain, the non-symmetric polynomial terms (28 in number) were eliminated (M = 34). The correction terms an and bn can be expressed in the vector form as follows:
| - 7 |
where ān∈ RN×1 and b̄n∈ RN×1 vectors are the lexicographical arrangements of the three-dimensional terms an and bn (N is the number of voxels in the image volume), Λ ∈ RN×M is the polynomial matrix and ᾱn, β̄n ∈ RM×1 are the coefficients of the polynomial terms. The coefficient set (ᾱn, β̄n) for the nth scanner model is estimated by the following minimization:
| - 8 |
The low frequency correction factors can then be applied to the individual PET images that have undergone high frequency correction (In, ĵn). The application of low frequency correction for scanner model n would be as shown below.
| - 9 |
The multiplicative and additive correction factors can be considered to be terms that alter the profiles across the image volume to better match the true radioactivity distribution in order to correct for attenuation, scatter, and other sources of inconsistency between scanners.
Simulation for assessing the validity of low frequency correction factors
Simulations were performed to validate the low frequency correction methodology proposed above as well as to get an intuitive feeling for their physical interpretation. The following three scenarios of residual low frequency errors were simulated using a digital Hoffman phantom smoothed to 8 mm resolution (D8):
1. Simulation of residual attenuation
The digital Hoffman brain phantom smoothed to a uniform 8 mm resolution (D8) was forward-projected to obtain its emission sinogram (E) and transmission sinogram (T) based on ellipse attenuation using ASPIRE software (Fessler 1995). To simulate errors in attenuation correction, the residual attenuation sinogram was chosen to be the transmission scan T scaled by 0.1. The emission sinogram with residual attenuation was calculated as EA = Ee-0.1T (element-wise operations). No noise was added to the sinogram. EA was reconstructed using filtered back projection (FBP) to obtain the phantom image with residual attenuation. The proposed low frequency correction method was applied to test if it could correct for the residual attenuation.
2. Simulating residual scatter
The digital Hoffman brain phantom smoothed to 8 mm resolution (D8) was forward-projected to obtain its emission sinogram (E). The scatter sinogram was approximated by smoothing E with a two dimensional Gaussian filter (45 mm width and 15 mm standard deviation). The smoothed sinogram was scaled by 0.15 to approximate a residual scatter sinogram (S). The emission sinogram with residual scatter was obtained (ES=E+S) and reconstructed using FBP to obtain the phantom image with residual scatter. The proposed low frequency correction method was applied to test if it could correct for the scatter correction error.
3. Simulation of residual attenuation and scatter
Both scatter and attenuation were simulated in the forward projected digital Hoffman brain phantom (described in the above two simulations) and an emission sinogram with both residual attenuation and scatter was obtained (EA+S = EA + S). The resultant sinogram (EA+S) was reconstructed using FBP and the proposed low frequency correction method was used to test its ability to remove the combined residual error.
Application of correction factors to phantom and normal control data
Phantom data
As mentioned earlier, human studies vary due both to inter-subject as well as to inter-scanner differences. Since the same phantom was imaged at all participating sites, the phantom studies did not have any variability comparable to the “inter-subject” differences seen in humans. Thus, the differences in phantom scans are primarily due to scanner differences, though differences due to technical factors in performing the scan (e.g. proper mixing) could still exist. Since the correction factors were obtained from the average phantom scans themselves, application of correction factors to these same average phantom scans would give a measure of the maximum reduction in variability possible from this approach. Differences in phantom scans were calculated for three groups of images: phantom images with no post-reconstruction corrections, images after only high frequency correction and images after both high and subsequent low frequency correction. The measure of the difference between a phantom image from scanner i and those from the other scanner models was obtained using the following metric:
| - 10 |
where, Ȳi is the vector of lexicographically arranged voxel values of an image from scanner i and N (=15) is the total number of scanner models. This metric for each scanner model was expected to decrease after the high frequency correction and then further after low frequency correction.
Normal control data
For validation of the methods in human studies, the correction factors obtained from phantom scans were applied to the set of 95 normal control FDG PET scans obtained from various participating ADNI sites. ADNI subjects ranged in age from 55 to 90 years with a mean age of 75, with 58% male and 42% female. As for the phantom scans, the inter-scanner variability was calculated for three sets of human FDG scans: normal subject scans without any post-reconstruction correction, scans after high frequency correction alone, and scans after both high and low frequency corrections using the metric in equation 10.
Results
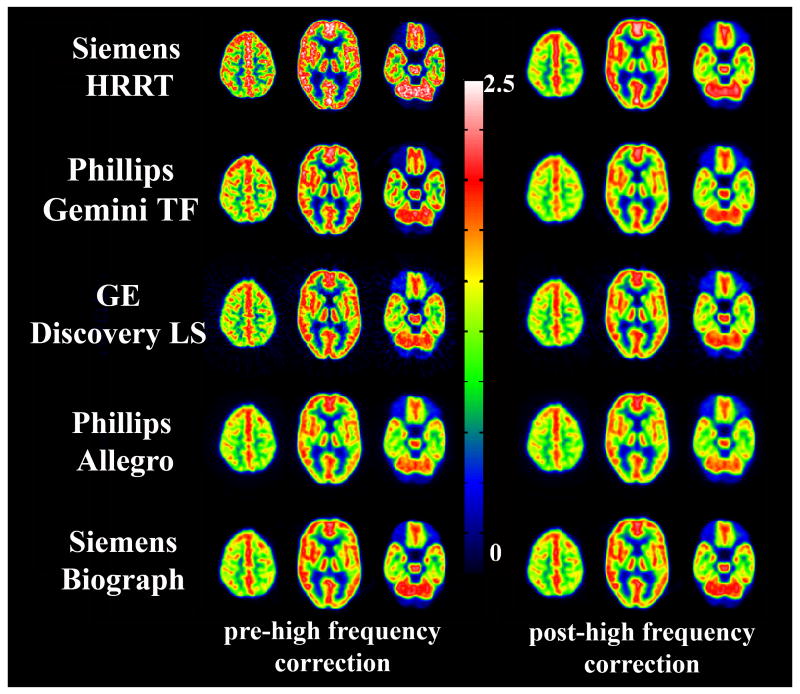
The high frequency correction factors (FWHM of the smoothing kernels) for smoothing the images from the fifteen scanner models to 8 mm resolution are listed in the Table 1. Figure 1 shows visually the reduction in resolution differences after application of the smoothing kernels to five of the 15 scanner models.
Table 1.
Scanner models and the FWHM (in mm) of the smoothing kernels to attain a resolution of 8 mm FWHM (in-plane and axial).
| Scanner Model | PET or PET/CT | FWHM in-plane (mm) | FWHM axially (mm) |
|---|---|---|---|
| Siemens HRRT | PET | 6 | 6 |
| Siemens Biograph HiRez | PET/CT | 6 | 5 |
| Phillips Gemini TF | PET/CT | ||
| Siemens HR+ | PET | 5 | 5 |
| GE Discovery RX | PET/CT | 5 | 4 |
| Phillips G-PET | PET | ||
| GE Advance | PET | 5 | 3 |
| GE Discovery LS | PET/CT | ||
| GE Discovery ST | PET/CT | 4 | 3 |
| Phillips Gemini | PET/CT | 3 | 3 |
| Phillips Gemini GXL | PET/CT | ||
| Phillips Allegro | PET | ||
| Siemens Accel | PET | 2 | 3 |
| Siemens Exact | PET | ||
| Siemens Biograph | PET/CT | ||
Figure 1.
Three levels in the Hoffman brain phantom scans for 5 different scanner models pre- and post- high frequency corrections.
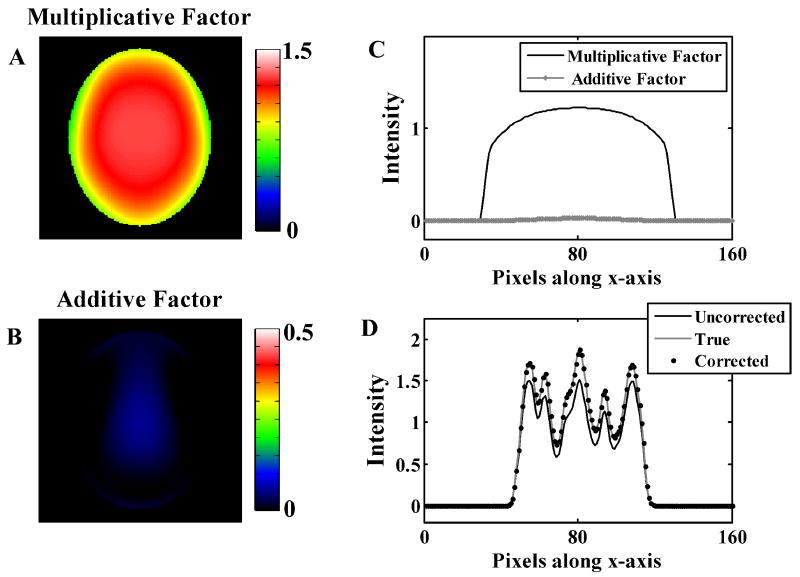
Figure 2 shows image slices of the additive and multiplicative factors obtained from the simulation study where the reconstructed image contains residual attenuation alone. The correction-factors are symmetric due to the symmetry constraint applied to the polynomial basis functions as attenuation errors are primarily multiplicative. The additive factor was very close to zero and the multiplicative factor is the major contributor to the correction, as attenuation errors are primarily multiplicative. Panel C shows the profiles of the correction factors in the x-axis (medial lateral) for fixed y (anterior posterior) and z (inferior superior) locations. The application of the correction factors removed the attenuation error as seen by the phantom image profiles in Panel D.
Figure 2.
Low frequency correction factors for simulations of images with residual attenuation error alone. Panels A and B show the multiplicative and additive correction factors. Panel C shows a sample profile through the 3-D correction factors. Panel D shows the same profile through the true, uncorrected, and corrected digital phantom images.
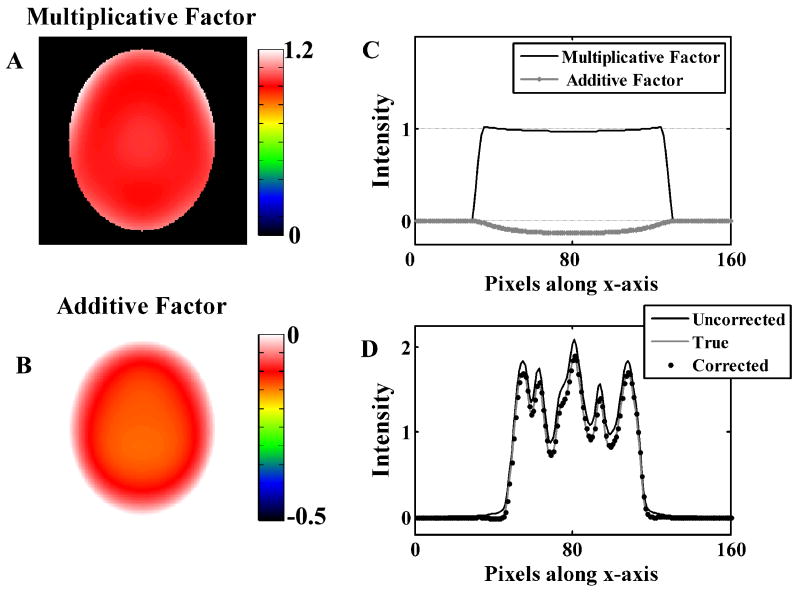
Figure 3 shows image slices of additive and multiplicative factors obtained from the simulation study where the reconstructed image contained residual scatter alone. Scatter being primarily though not entirely an additive error, the multiplicative factor was small while the additive factor was the major contributor to the correction. Panel C shows the profiles through the correction factor images. The application of the correction factors removes the scatter error as seen by the image profiles in Panel D.
Figure 3.
Low frequency correction factors for simulations of images with residual scatter error alone. Panels A and B show the multiplicative and additive correction factors. Panel C shows a sample profile through the 3-D correction factors. Panel D shows the same profile through the true, uncorrected, and corrected digital phantom images.
For the simulation case where residual scatter and attenuation were included, both additive and multiplicative factors made significant contributions to the overall correction (results not shown). As in the cases shown in Figs 2 and 3, nearly all the error was removed by the correction procedure.
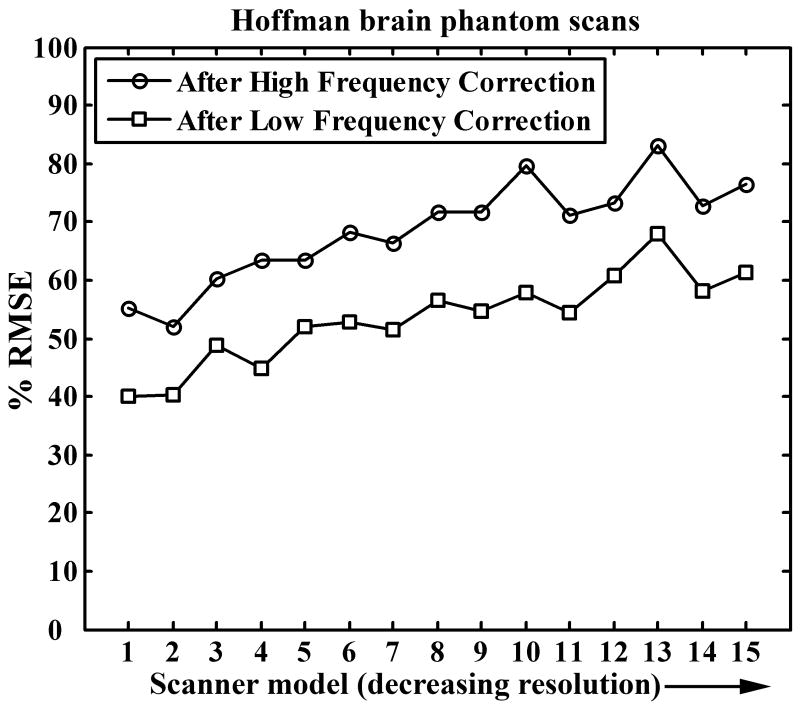
The improvement in average phantom scans by the application of the phantom-based correction factors can be seen in Figure 4. The data in Figure 4 is normalized such that the average RMSE for the group with no correction is 100% for each of the fifteen scanner models. The high frequency correction reduces the variability by 20% – 50% (higher reduction for high resolution scanners). The low frequency correction further reduced the variability further by another 20% - 25%. In spite of these two steps, 40% - 60% residual variability is seen in the phantom scans. This may be attributed to three primary causes: first, the affine low frequency correction term is a first order correction step and is not a complete model for low frequency variability. Second, a single smoothing kernel for high frequency correction was used for the entire image, which may not be optimal throughout the entire imaging volume as resolution degrades from the center of the image moving outward. Third, some of the remaining variability can be attributed to differences in phantom orientation within the scanner field-of-view, small misregistration errors, interpolation in the registration step, non-uniform mixing of 18F solution in the phantom, and other technical errors.
Figure 4.
Reduction in average between-scanner RMSE for Hoffman phantom scans. Correction factors derived from the phantom data are applied to the phantom data itself.
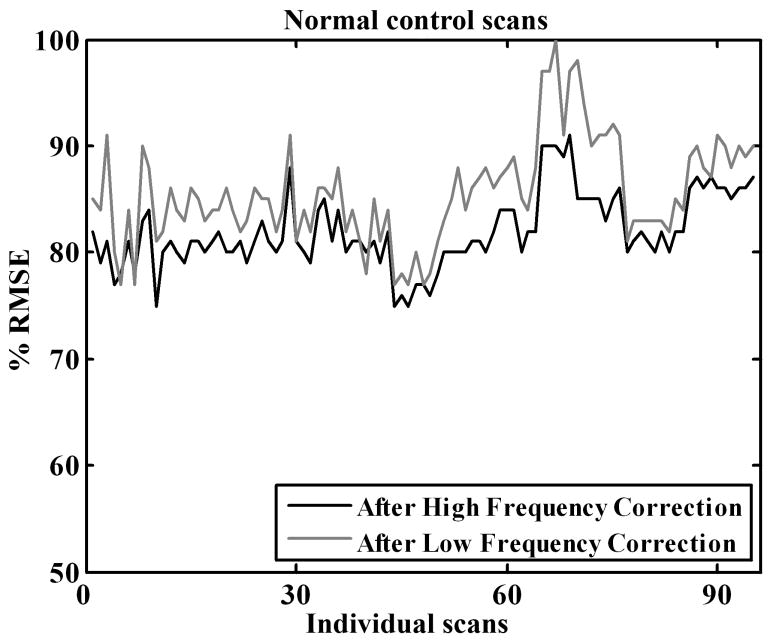
Similar to the results for phantom data, the phantom-based high frequency correction reduced the resolution variability between the normal control scans (Figure 5). However, the reduction in variability (15% – 25%) is less than that in phantom studies (Figure 4). This was expected as normal subjects, unlike phantom studies, have inter-subject variations in addition to the consistent scanner-related differences. Application of the low frequency correction, however, did not bring about a further decrease in variability thus indicating that the low frequency correction factors obtained from the phantom scans were not appropriate for the human scans.
Figure 5.
Reduction in average between-subject RMSE for normal control FDG PET scans. Correction factors derived from the phantom data are applied to normal control scans.
Discussion
This paper develops a framework for reducing the variability in PET scans obtained across different scanner models in a large multi-center study; an important step prior to pooling the data for analysis. The correction factors were derived from PET image data obtained by scanning the same object (the 3-D Hoffman brain phantom) at all 50 participating sites. Human PET scans from different centers are different not only because of the functional and anatomical differences between subjects but also due to the vendor-specific hardware and software. This work attempted to reduce these systematic vendor-specific differences by applying both high frequency and low frequency corrections.
Three-dimensional smoothing was used to minimize the high frequency resolution differences across scanner models. The coarsest effective resolution of all fifteen scanner models was found to be between 7 and 8 mm, hence 8 mm was chosen as the target resolution. This step obviously reduces the higher anatomic detail that the high-resolution scanners provide. However, making the resolution uniform between scanners was essential for the achievement of the various goals of the ADNI project. While much of the ADNI analyses are focused on changes over time, many analyses are being performed on different sub-groups of the NC, MCI and AD populations. These include separation by age, older or younger than 75 years; patients with or without positive APOE status; and gender. Analysis of such group-wise data necessitates resolution uniformity across scanners. Improving resolution of low-resolution scanner models is another option (Hom et al. 2007), though a challenging one, and would part of the future work of this effort.
The high frequency correction kernels (reported in Table 1) were found to be useful in both phantom and human control data and are being used to adjust all ADNI PET image data on a routine basis. Phantom scan-based low-frequency corrections reduced the variability in phantom scans but were found to be unsuccessful in further reducing variability in normal human FDG scans. There are two likely reasons for this result. One likely cause for the lack of success in applying Hoffman brain phantom-derived low frequency correction factors is the cylindrical shape of the Hoffman brain phantom (with no skull or neck) that is very different from the ellipsoidal shape of the human brain. Since the low frequency correction factors minimize the residual scatter and attenuation, both of which are geometry dependent phenomena, a more realistic humanoid phantom would be a better choice for obtaining low frequency correction factors. At the same time, a more realistic torso phantom should also be used to simulate the out-of-field scatter.
Furthermore, since brain sizes are different for different subjects, the extent of attenuation and scatter is also different. Thus, application of the same phantom-derived correction factors to all the human scans from a particular scanner model is not optimal. Thus, though the high frequency correction factors were found to reduce variability in the human data and are being used for all ADNI scans, more work is required for refining the approach for low frequency correction. Simulation studies will need to be performed to study the effect of brain size on the correction factors, with the goal of developing individualized correction factors.
Though this work was developed for minimizing inter-scanner PET image variability, the general techniques may be extended to multi-center studies involving other imaging modalities as well.
Acknowledgments
This work was supported by the Alzheimer's Disease Neuroimaging Initiative AG024904.
References
- Fessler JA. Technical Report 293. Comm. and Sign. Proc. Lab., Dept. of EECS, Univ. of Michigan; Ann Arbor, MI: 1995. ASPIRE 3.0 user's guide: A sparse iterative reconstruction library; pp. 48109–2122. [Google Scholar]
- Hoffman EJ, Cutler PD, Digby WM, Mazziotta JC. 3-D phantom to simulate cerebral blood flow and metabolic images for PET. IEEE Trans Nucl Sci. 1990;37:616–620. [Google Scholar]
- Hom EF, Marchis F, Lee TK, Haase S, Agard DA, Sedat JW. AIDA: an adaptive image deconvolution algorithm with application to multi-frame and three-dimensional data. J Opt Soc Am A Opt Image Sci Vis. 2007;24:1580–1600. doi: 10.1364/josaa.24.001580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazziotta JC, Toga AW, Evans A, Fox P, Lancaster J. A probabilistic atlas of the human brain: theory and rationale for its development. The International Consortium for Brain Mapping (ICBM) Neuroimage. 1995;2:89–101. doi: 10.1006/nimg.1995.1012. [DOI] [PubMed] [Google Scholar]
- Minoshima S, Koeppe RA, Frey KA, Ishihara M, Kuhl DE. Stereotactic PET atlas of the human brain: aid for visual interpretation of functional brain images. J Nucl Med. 1994a;35:949–954. [PubMed] [Google Scholar]
- Minoshima S, Koeppe RA, Frey KA, Kuhl DE. Anatomic standardization: linear scaling and nonlinear warping of functional brain images. J Nucl Med. 1994b;35:1528–1537. [PubMed] [Google Scholar]