Abstract
Thousands of proteins are subjected to posttranslational modifications that can have dramatic effects on their functions. Traditional biological methods have struggled to address some of the challenges inherit in the unbiased identification of certain posttranslational modifications. As with many areas of biological discovery, the development of chemoselective and bioorthogonal reactions and chemical probes has transformed our ability to selectively label and enrich a wide variety of posttranslational modifications. Collectively, these efforts are making significant contributions to the goal of mapping the protein modification landscape.
Introduction
Posttranslational modifications (PTMs) augment the chemical diversity of proteins, which is otherwise typically limited by the naturally-occurring amino acids and the underlying genetic code. A wide array of enzymatic and chemical modifications, ranging from simple side-chain oxidations to the addition of large polysaccharides, have been shown to dramatically alter the biochemical and biophysical properties of proteins. Despite the documented importance of individual PTMs in cellular biology, the proteome-wide identification of specific PTM substrates remains challenging. PTMs can be substoichiometric, dynamically regulated and heterogeneous in their structure, making their analysis and identification particularly difficult. During the past two decades, chemical methods have significantly contributed to the identification and analysis of posttranslationally-modified proteins. In general, these techniques can be broadly classified into two categories, which both enable the installation of visualization or affinity tags:
Methods that use chemoselective reactions to exploit the unique chemical-reactivity of the posttranslational modifications themselves (Figure 1A).
Strategies to install abiotic probes (chemical reporters, Figure 1B) that can be selectively subjected to a range of bioorthogonal reactions (Figure 2).
Figure 1. Chemical Methods to tag and enrich posttranslationally modified proteins.
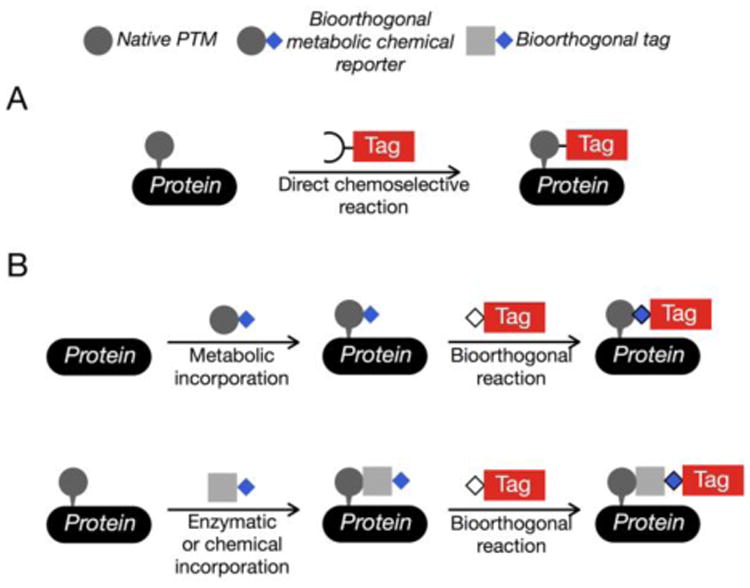
(A) Some posttranslational modifications (PTMs) can be specifically reacted with enrichment tags using chemoselective reactions. (B) Bioorthogonal reactions in combination with chemical reporters enable the installation of affinity tags. Chemical reporters can either be incorporated into PTMs using cellular metabolism or appended to existing PTMs using enzymes or selective chemical reactions.
Figure 2. Bioorthogonal reactions occur selectively between two abiotic functional-groups.
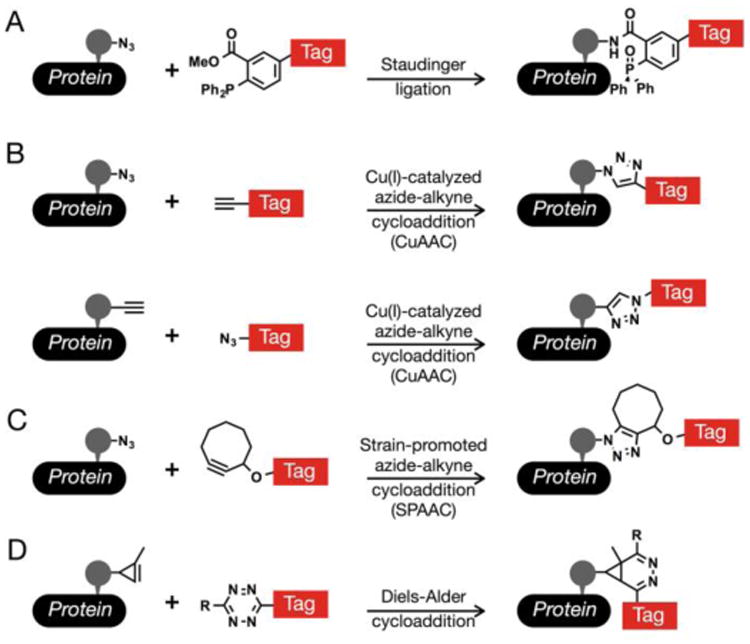
(A) The Staudinger ligation allows for the coupling of azides with triarylphosphine reagents. (B) The Cu(I)-catalyzed azide-alkyne cycloaddition (CuAAC) gives a stable triazole product from azides and alkynes. This common bioorthogonal reaction can be performed in both directions; however, azide-tags give reduced background signals. (C) Cyclooctyne reagents will react with azides in a strain-promoted azide-alkyne cycloaddition (SPAAC). (D) Tetrazines will undergo rapid inverse-demand Diels-Alder reactions with activated alkyenes, such as cyclopropenes.
In parallel, the development of more sensitive mass spectrometers and increasingly sophisticated computer-algorithms for peptide identification have enabled the routine proteomic identification of thousands of proteins from individual complex-samples. Coupled with complementary ionization chemistries and quantitative methods, mass spectrometry can offer high sensitivity, modification-site identification and the ability to quantify changes in PTM occupancy. Here, we review the currently available chemical-strategies to enrich and identify posttranslationally-modified proteins in mammalian cells and highlight their recent applications. For a more comprehensive review of the particular advantages of chemical reporters or the scope of bioorthogonal chemistries, we direct readers to excellent summaries elsewhere [1,2]. We end with an summary of future challenges and opportunities that when met will expand the utility of chemical approaches for the study of specific PTMs.
Glycosylation
There are two major types of protein glycosylation that modify large numbers of protein substrates in metazoan and plant cells [3]. Proteins localized to the secretory pathway and the cell surface, or secreted into the extracellular space, can be modified by oligosaccharide structures (e.g., N-linked and mucin O-linked glycosylation). Additionally, intracellular proteins can be substrates for the addition of the single monosaccharide N-acetyl-glucosamine, termed O-GlcNAc modification. Bioinformatic studies suggest that more than 50% of mammalian proteins are glycosylated, and these glycans play diverse cellular roles. Extracellular glycans can alter the half-life of circulating proteins and mediate cellular contacts with proteins, neighboring cells and pathogens, while intracellular glycosylation can affect protein localization and signal transduction. The inherent limitations in characterizing these diverse modifications, prompted the development of the Staudinger ligation and the first azide-bearing metabolic chemical reporter, N-azdioacetyl-mannosamine (Figure 3A), for the detection of sialic acids [4]. Together with the creation of other bioorthogonal reactions, this simple idea was extended to other azide- and aklyne-analogs of monosaccharides for the detection of mucin O-linked glycoproteins (Figure 3B), fucose-modified glycans (Figure 3C) and O-GlcNAcylated proteins (Figure 3D) [5].
Figure 3. Chemical reporters for glycosylation.
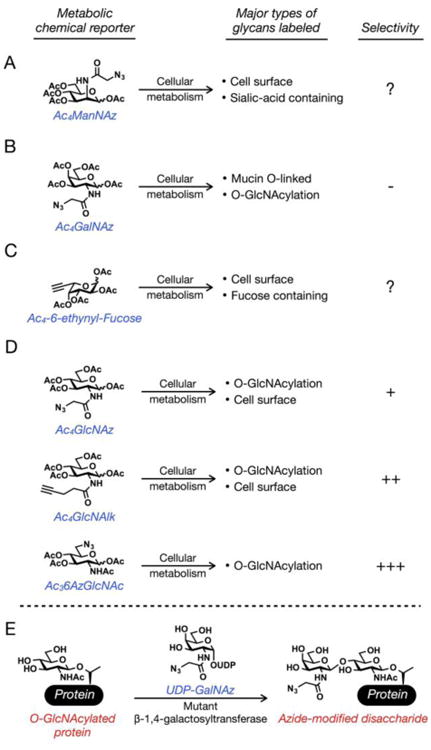
(A) Peracetylated N-azdioacetyl-mannosamine (Ac4ManNAz) labels sialic acid residues on cell-surface glycans. Its selectivity for these glycans is unknown. (B) Peracetylated N-azdioacetyl-galactosamine (Ac4GalNAz) labels both mucin O-linked glycans and O-GlcNAcylated proteins with no selectivity. (C) Peracetylated 6-ethynyl-fucose (Ac4-6-entynyl-Fucose) labels fucose residues on cell-surface glycans. Its selectivity is also unknown. (D) Peracetylated N-azidoacetyl-glucosamine (Ac4GlcNAz) and peracetylated N-pentynyl-glucosamine (Ac4GlcNAlk) label both O-GlcNAcylated proteins and cell-surface glycans with some selectivity for O-GlcNAcylated proteins. Peracetylated-6-azido-N-acetyl-glucosamine (Ac36AzGlcNAc) labels O-GlcNAcylated proteins with a high-level of selectivity. (E) O-GlcNAcylated proteins can be labeled post-lysis using a combination of UDP-GalNAz and a mutant β-1,4-galactosyltransferase (GalTY289L).
Many types of cancer display altered levels of both sialic acid and mucin O-linked glycans. Treatment of highly metastatic prostate cancer with Ac4ManNAz (Figure 3A) allowed for the identification of cell-surface proteins, the majority of which were implicated in cell motility, migration and invasion, supporting a potential role for sialic acid in metastasis [6]. In a separate study, a prostate cancer cell-line was treated with Ac4GalNAz to enable the enrichment and identification of 29 cell-surface glycoproteins, including many proteins involved in cell adhesion [7]. More recently, Ac4GalNAz (Figure 3B) was used in combination with iTRAQ (isobaric tag for relative and absolute quantitation) to enrich and compare the secreted mucin glycoproteins from two different CHO cell-lines [8], demonstrating that these chemical reporters could be used in the future to compare cellular glycoproteins from two states (e.g., cancer vs. normal tissue).
O-GlcNAc modification of serine and threonine residues is an abundant modification of proteins in the cytosol, nucleus and mitochondria. Unlike cell-surface glycosylation, O-GlcNAcylation is a dynamic modification that plays critical roles in cellular responses to changes in metabolism and stress, particularly in diseases such as cancer, diabetes and neurodegeneration [9]. The first metabolic chemical reporter of O-GlcNAcylation was peracetylated N-azidoacetyl-glucosamine (Ac4GlcNAz, Figure 3D) [10], which was recently used to identify over one-thousand potentially O-GlcNAcylated proteins from a single cell-line and confirm the modification sites on 80 of these substrates [11]. To provide improved signal-to-noise in the CuAAC bioorthogonal reaction, the alkyne-analog N-pentynyl-glucosamine (GlcNAlk, Figure 3D) has also been developed and revealed the O-GlcNAc modification of the ubiquitin ligase NEDD4 [12]. This chemical reporter was also used to identify O-GlcNAcylated proteins associated with diabetic retinopathy [13]. As an alternative to metabolic methods, a chemoenzymatic method has been developed (Figure 3E). Specifically, incubation of cell-lysates with an engineered β-1,4-galactosyltransferase and azide- or ketone-containing UDP-donor sugar results in modification of O-GlcNAc residues [14]. The resulting disaccharide can then be subjected to bioorthogonal labeling for visualization and proteomics. For example, this chemoenzymatic chemical reporter was recently used in conjunction with a photo-cleavable biotin tag to enrich and identify 274 O-GlcNAcylated proteins and 458 specific O-GlcNAc modification sites on 195 proteins [15].
Recently, research has also focused on the effects of metabolic cross-talk between glycosylation pathways on the cellular fate of different metabolic chemical reporters. For example, it has been shown that UDP-GalNAz and UDP-GlcNAz can be enzymatically interconverted in living cells [12,16], allowing the proteomic identification of O-GlcNAcylated proteins from cells treated with Ac4GalNAz [17]. While Ac4GalNAz-treatment appears to label O-GlcNAcylated proteins more robustly under some conditions [16], its lack of specificity requires cell fractionation to separate O-GlcNAcylated and mucin O-linked glycoproteins. Overcoming this limitation, treatment with peracetylated 6-azido-6-dexoy-N-acetyl-glucosamine (Ac36AzGlcNAc, Figure 3D) was recently shown to selectively label O-GlcNAcylated proteins [18]. Notably, using biochemistry and proteomics the same study demonstrated that 6AzGlcNAc is the most selective for O-GlcNAcylation, followed by GlcNAz and finally the relatively nonspecific GalNAz. These results suggest that selective metabolic chemical reporters of other types of glycoproteins could be developed in the future.
Lipidation
Lipid modification of proteins regulates membrane affinity, localization and trafficking for cell signaling [19]. The types of lipid modification are diverse and their covalent attachment to proteins has uncovered a complicated network of membranes and lipidated proteins that are central to basic cellular function and human disease. Their inherent complexity has pressured the development of specific and sensitive methods to probe their function and identify modified substrates.
Myristoylation and palmitoylation are two classes of fatty-acylation events that occur both during translation and posttranslationally. Myristoylation is the irreversible, covalent attachment of a 14-carbon fatty acid to a protein's N-terminus via an amide linkage, while palmitoylation typically involves the reversible addition of a 16-carbon fatty acid by palmitoyl transferases to cysteine residues on target proteins via a thioester linkage. Classically, fatty-acylation has been studied using metabolic radiolabeling (4H or 14C); however, radioactivity usually requires days to weeks to visualize modified proteins and offers no opportunities for their selective enrichment and identification [20]. To overcome these limitations, azide and alkyne fatty-acids have been developed [19]. Recently, Alk-12 (13-tetradecynoic acid, Figure 4A) was used in combination with a biotin-enrichment tag to identify mrysitoylated proteins in the malaria pathogen P. falciparum, including several proteins that are important for the parasite's life cycle and disease transmission [21]. In a similar fashion, 17-octadecynoic acid (Alk-16, Figure 4B) has been particularly useful for the proteomic identification and subsequent characterization of palmitoylated proteins. For example, Alk-16 enabled global proteomic profiling in a mouse dendritic cell-line and identified 150 potentially palmitoylated proteins. This included several new palmitoylation substrates, such as the innate immune effector IFITM3, which the authors demonstrated requires fatty-acylation for its anti-viral activity [22]. Subsequent proteomic-profiling of Jurkat T cells with Alk-16 and two other alkyne fatty-acids (Alk-12 and Alk-14) highlighted the selectivity of different length alkyne fatty-acids for different modifications (i.e., Alk-12 for myristoylation and Alk-16 for palmitoylation) [23]. More recently, Alk-16 was combined with stable isotope labeling in cell culture (SILAC) to perform quantitative proteomics and demonstrated that some palmitoylation events are very stable over time, while others are rapidly removed from substrate proteins [24]. As an alternative to bioorthogonal chemistry, the thioester linkage of S-palmitoylation enables the use of chemoselective reactions to perform a “biotin switch” [25]. This method, termed acyl-biotin exchange (ABE, Figure 4C), involves the alkylation of free cysteine residues with N-ethylmaleimide, followed by treatment with hydroxylamine, which cleaves any palmitate thioesters revealing sulfhydryl groups that can be selectively modified with biotinylation reagents. Most notably, this method was used for the identification of palmitoylated proteins in yeast [26] and then in neurons [27], including a palmitoylated Cdc42-isoform that plays a important role in the induction of dendritic spines [27]. Recently, this approach was also used to identify palmitoylation candidates in B lymphocytes [28] in malaria parasites [29] and in tissue from a mouse model of Huntington's disease [30].
Figure 4. Chemical methods to enrich lipidated proteins.
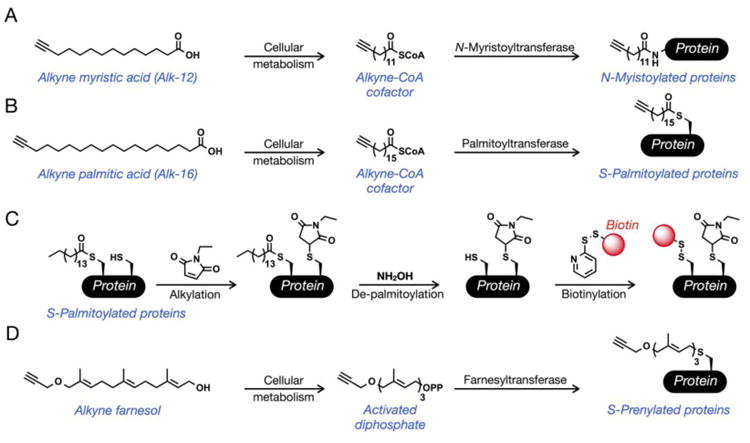
(A, B) Treatment of cells with alkyne myristic acid (Alk-12) or alkyne palmitic acid (Alk-16) results in labeling of N-myristoylated proteins or S-palmitoylated proteins, respectively. (C) Acyl-biotin exchange: S-palmitoylated proteins can be tagged post-lysis by using a combination of thiol- and thioester-selective reactions. (D) Treatment of cells with alkyne farnesol results in labeling of S-prenylated proteins.
S-Prenylation is an irreversible addition of an isoprenoid, either 15-carbon farnesyl or 20-carbon geranylgeranyl to one or two C-terminal cysteines of a protein through a thioether linkage that effects 2% of the proteome in mammals [31]. Typically, these modifications are found near the C-terminus of proteins where they are critical for localization to the cell membrane [31]. Like fatty-acids, alkyne derivatives of isoprenoids have also been created [32,33]. Recently, an alkyne-farnesol (Figure 4D) permitted a large-scale enrichment of isoprenoid-modified proteins in a macrophage cell-line that identified both known and unpredicted S-prenylated proteins [34]. Notably, these proteomic data served as a starting point to demonstrate that S-farensylation is important for the localization and antiviral activity of zinc-finger antiviral protein (ZAP).
Acetylation
Acetylation of lysine residues is a dynamic posttranslational modification closely associated with epigenetic changes to gene transcription [35], as well as the regulation of non-chromatin associated proteins [36]. Proteome-wide studies using anti-acetyl lysine antibodies have unveiled ∼4700 human acetylation sites, making the investigation of the dynamics of these modifications and the substrate specificity of acetyltransferases and deacetylases an important goal. An alkyne analog of acetate, 4-pentynoic acid, has been shown to be chemical reporter of lysine acetylation in living cells and in vitro. Treatment of a variety of cell lines with sodium 4-pentynoate resulted in metabolic labeling of a wide range of proteins, enabling the proteomic identification of both known lysine-acetylated proteins, like histones, and new potential substrates [37]. Additionally, synthetically prepared 4-pentynoyl-CoA (Figure 5A) is a substrate for purified p300 acetyltransferase, enabling the identification of transferase-specific substrates from cell lysates [38]. Acetylation can also occur on the N-terminus of proteins and serine and threonine side-chains through an ester linkage. This raises the possibility that these chemical reporters could be used to investigate these acetylation events that would not be recognized by anti-acetyl lysine antibodies.
Figure 5. Chemical reporters of protein acetylation.
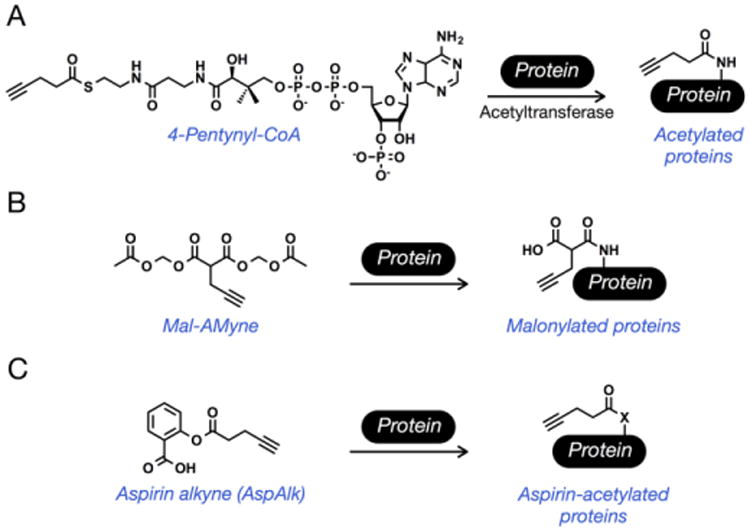
(A) The cofactor 4-pentynyl-CoA is accepted by acetyltransferases to label N-acetylated proteins in cell-lysates, while treatment with sodium pentynoate results in labeling in living cells. (B) Treatment of cells with Mal-AMyne results in labeling of malonylated proteins. (C) An alkyne aspirin-analog (AspAlk) can be used to identify aspirin-dependent protein acetylation in living cells.
Other types of lysine acetylation events, including propinylation, butyrylation, malonylation and succinylation, can also occur [39]. While it is not completely clear if these modifications are a result of enzymatic addition or simply chemical acetylation from their corresponding high-energy CoA thioesters, several of them are reversible through action of the sirtuin deacylases. Towards applying chemical reporters to the investigation of these modifications, a alkyne-analog of malonylation, 2-propargyl malonate (Mal-yne, Figure 5B) has recently been developed [40]. Notably, due to the low cell-permeability of Mal-yne, the small molecule was masked as a pro-reporter through the introduction of acetoxymethyl protecting groups that are removed by endogenous lipases in living cells. Treatment of HeLa cells with this pro-reporter (Mal-AMyne) enabled the identification of 375 potential substrates, including 14 previously confirmed malonylated proteins. Some small-molecules contain electrophilic acetates that can be chemically transferred to protein substrates. For example, aspirin (acetylsalicylic acid), a nonsteroidal anti-inflammatory drug (NSAID) commonly used for the treatment pain, is known to acetylate proteins by transfer of its acetate group to amino acid side-chains [41]. Recently, aspirin-dependent acetylation was investigated using an alkyne-aspirin chemical reporter (AspAlk, Figure 5C) that replaced the electrophilic acetate-group with 4-pentynoic acid [42]. Treatment of HCT-15 cells with AspAlk allowed for the identification of 120 potentially aspirin acetylated proteins including core histone proteins H2B and H3, implicating aspirin as a potential modulator of gene transcription.
Methylation
The reversible addition of one or more methyl groups to lysine and arginine side-chains is a critical posttranslational modification in chromatin biology and other cellular processes [43,44]. The methyl donor for the methyl transferase reaction is the cofactor S-adenosyl-L-methionine (SAM, also referred to as AdoMet). The first chemical reporters of protein methylation were two alkyne-analogs of SAM (e.g., AdoEnYn in Figure 6A) [45,46]. Notably, these two chemical reporters displayed selective turnover by different methyltransferases, raising the possibility that orthogonal chemical-reporter/transferase pairs could be developed to interrogate substrate specificity. In this regard, a series of differentially-sized azide- and alkyne-analogs of SAM have been characterized [47]. Notably, several of these compounds were not substrates for wild-type methyltransferases but were utilized by rationally-engineered methyltransferase mutants, following a “bump-hole” strategy. This enabled the selective identification of >500 methylation substrates of the closely related lysine methyltransferases EuHMT1/2 [48]. Unfortunately, the poor permeability of the SAM analogs restricted their use in live cells. To overcome this limitation, the SAM biosynthetic pathway has been recently engineered [49]. Specifically, an alkyne-analog of SAM can be generated in living cells that express a mutant SAM synthetase when they are treated with the corresponding cell-permeable alkyne-analog of methionine. Although the aforementioned methionine-based SAM reporters are functional, they are subject to degradation at physiological pH and result in nonspecific labeling. To overcome this, a more stable selenium-based SAM reporter was developed that can be utilized by both lysine and arginine methyl transferases in vitro [50]. It was subsequently demonstrated that a propargylic, selenium-containing SAM analog (ProSeAM) was compatible with native PMTs in whole-cell lysate, enabling the identification of 297 potentially methylated proteins, many of which were novel [51]. Combining all of these tools should facilitate the identification of specific methyltransferase substrates in living cells in the future.
Figure 6. Chemical reporters of other posttranslational modifications.
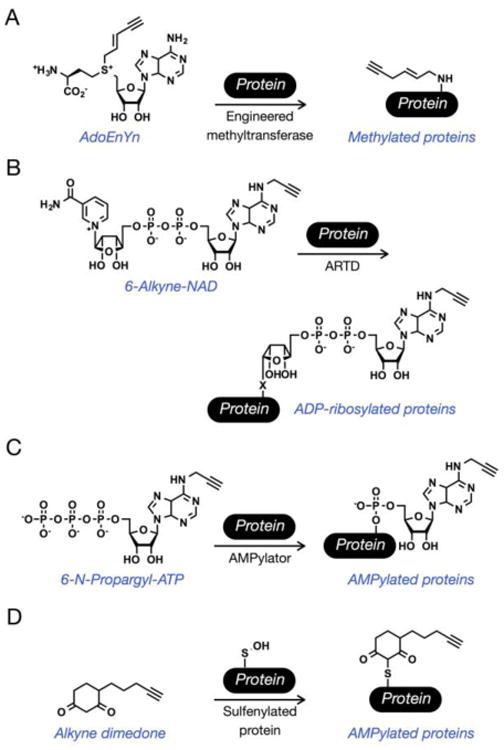
(A) AdoEnYn and other SAM analogs will be transferred by engineered methyltransferases to label N-methylated proteins. (B) The cofactor 6-alkyne-NAD is a chemical reporter for ADP-ribosylated proteins in cell lysates. (C) The ATP analog N-6-propargyl-ATP will be transferred by AMPylator enzymes to tag AMPylated proteins in cell lysates. (D) The oxidation of cysteine side-chains to sulfenic acids can be enriched by selective reaction with alkyne-dimedone.
ADP-ribosylation
ADP-ribosylation is the posttranslational addition of ADP-ribose from a nicotinamide adenine dinucleotide (NAD) donor to a range of different amino acid side-chains [52]. The modification is added by adenosine diphosphate ribosyltransferases (ARTDs) and can consist of a single ADP-ribose (mono-ADP-ribosylation) that can be subsequently extended into poly-ADP-ribosylation that can be as much as 200 units in length. Although the role of ADP-ribosylation has been characterized in DNA repair, other biological functions are relatively unknown and isolation of the ADP-ribosylated proteome would prove useful for their elucidation. Towards this goal, 6- and 8-alkyne-NAD analogs have been developed (Figure 6B) [53,54]. Incubation of cell lysates with 6-alkyne-NAD and recombinant ARTD1 (PARP1) enabled the identification of 70 potentially novel ADP-ribosylated proteins including many mitochondrial proteins [54]. Very recently, orthogonal 6-alkyne-NAD/ARTD pairs have been engineered to identify the direct substrates of specific ARTDs [55]. Using a “bump-hole” strategy, a ethyl-substituent was added to the C-5 position on the nicotinamide moiety of NAD, which could only be accepted by a corresponding mutant ARTD (K903A). Treatment of nuclear extracts with these orthogonal pairs made possible the proteomic identification of hundreds of specific substrates of either ARTD1 or ARTD2, including 42 proteins that were unique ARTD1 substrates.
AMPylation
The covalent and dynamic addition of adenylyl 5′-monophosphate through a phosphodiester bond (AMPylation) is a posttranslational modification installed by bacterial virulence factors (AMPylators) that are secreted into the mammalian host during infection where they modify target proteins to change their function in mammalian cells [56]. An alkyne-containing ATP analog, termed N6-propargyl adenosine-5′-triphosphate (N-6-propargyl-ATP, Figure 6C), was developed for the detection, enrichment and identification of AMPylated proteins [57]. N-6-propargyl-ATP is accepted by the known bacterial AMPylator VopS and was transferred on to a known target protein Cdc42 in a dose-dependent manner, and N-6-propargyl-ATP was transferred by other AMPylators to proteins in HeLa cell-lysate, suggesting that it is a general reporter. Finally, N-6-propargyl-ATP was used to identify AMPylation substrates at endogenous levels in whole-cell lysates, and therefore should be useful in characterization of AMPylated proteins in many biological systems.
Cysteine Oxidation
Oxidation of cysteine residues by reactive oxygen species (ROS) and reactive nitrogen species (RNS) is a dynamic posttranslational event that affects many protein classes and contributes to changes in signal transduction pathways and gene transcription [58]. Its importance in cellular biology has prompted the development of chemical methods to investigate the protein targets of two types of cysteine oxidation, suflenation and nitrosylation. Oxidation of the thiol group of cysteine amino acid side-chain to sulfenic acid (Cys-SOH) is a posttranslational modification caused by highly reactive ROS in a cellular environment. In fact, oxidants such as hydrogen peroxide (H2O2) have been characterized as a small signaling molecules due to their ability to oxidize cysteine side-chains and cause change in protein function. The first chemical-method for identifying sulfenylated substrates, relied on the chemoselective reduction of sulfenic acids to carry out a “biotin-switch.” Specifically, any non-modified cysteine side-chains are first alkylated, followed by transformation of suflenic acids to thiols using arsenite. Finally, reaction with biotin-maleimide enables the enrichment of previously sulfenylated proteins. This method was applied to identify novel protein targets of oxidation in rat cardiac tissue and kidney after treatment with H2O2 [59,60]. More recently, a direct method for the detection utilized an alkyne-modified dimedone (5,5-dimethyl-1,3-cyclohexanedione) probe (Figure 6D) that selectively react with sufenic acids to form stable thioethers [61]. This probe has been used to identify many proteins modified with sulfenic acid, including an oxidation event that activates the kinase EGFR [61]. S-Nitrosylation occurs when nitric oxide (NO) reacts with cysteine side-chains, and like sulfenylation, it can be detected using a “biotin-switch.” In this case, selective reduction of nitrosylation is achieved using ascorbate. The resulting free thiols can then be reacted with biotinylating reagents or directly with an affinity-resin for enrichment and proteomic identification [62,63].
Conclusions and future outlook
Chemical methods have significantly contributed to the investigation of protein modifications by enabling the isolation of specific PTMs for subsequent identification by mass spectrometry. In particular, the development of bioorthogonal reactions and chemical reporters has streamlined the application of chemistry to several different classes of PTMs but challenges and opportunities remain. Additionally, Modification of metabolites with bioorthogonal groups has the potential to affect their cellular metabolism or alter endogenous metabolic pathways. Therefore, a more detailed investigation of the metabolism and enzymatic interconversion of metabolic chemical reporters is needed to fully understand what specific types of PTM are being enriched using a particular reagent. However, it also provides unique opportunities to use changes in chemical structure to develop specific chemical reporters [18] and potentially map metabolic pathways [64]. Several cofactor-based reporters (e.g., 6-alkyne-NAD) can only be applied in cell lysates due to poor membrane permeability or chemical instability and would benefit from the continued engineering of biosynthetic pathways to enable experiments in living cells [49]. To complement chemical reporters, future work should also focus on the development of new chemoselective or enzymatic reactions that directly tag a specific PTM with bioorthogonal reactive groups or enrichment tags. Beyond identifying modified proteins, site-identification of many PTMs remains a significant challenge, since some modifications are unstable during MS/MS fragmentation and/or can be heterogeneous in their structure. This challenge needs to be met with collaborative efforts to improve and disseminate alternative ionization approaches in mass spectrometry (e.g., electron transfer dissociation) and computer algorithms to retain and then unambiguously identify PTMs. Finally, its is worth noting that the identification of any modified protein using any technique, either chemical or biological, needs to be biochemically confirmed before embarking on detailed mechanistic studies.
Highlights.
Posttranslational modifications (PTMs) increase the chemical diversity of proteins
The unbiased identification of some PTM substrates remains challenging
Several chemical methods have been developed to tackle this challenge
Acknowledgments
The Damon Runyon Cancer Research Foundation (DDR-19-12), the Concern Foundation, the Michael J. Fox Foundation, the Margaret E. Early Medical Research Trust, the National Cancer Institute of the US National Institutes of Health (CCSG P30CA014089), and Susan G. Komen for the Cure (CCR14299333) are thanked for financial support. K.N.C. is a fellow of the National Science Foundation Graduate Research Fellowship Program (DGE-0937362). We thank Prof. Howard Hang for careful reading of the manuscript and apologize to those researchers whose work we have not included due to space limitations.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Grammel M, Hang HC. chemical reporters for biological discovery. Nat Chem Biol. 2013;9:475–484. doi: 10.1038/nchembio.1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Patterson DM, Nazarova LA, Prescher JA. Finding the right (bioorthogonal) chemistry. ACS Chem Biol. 2014;9:592–605. doi: 10.1021/cb400828a. [DOI] [PubMed] [Google Scholar]
- 3.Varki A, Cummings RD, Esko JD, Freeze HH, Stanley P, Bertozzi CR, Hart GW, Etzler ME, editors. Essentials of Glycobioloy. Cold Spring Harbor Laboratory Press; 2009. [PubMed] [Google Scholar]
- **4.Saxon E, Bertozzi C. Cell surface engineering by a modified Staudinger reaction. Science. 2000;287:2007–2010. doi: 10.1126/science.287.5460.2007. The authors describe the development of the Staudinger ligation, the first truly bioorthogonal reaction, and the creation and application of the first azide-containing chemical reporter. [DOI] [PubMed] [Google Scholar]
- 5.Laughlin ST, Bertozzi CR. Imaging the glycome. Proc Natl Acad Sci USA. 2009;106:12–17. doi: 10.1073/pnas.0811481106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yang L, Nyalwidhe JO, Guo S, Drake RR, Semmes OJ. Targeted identification of metastasis-associated cell-surface sialoglycoproteins in prostate cancer. Mol Cell Proteomics. 2011;10:M110.007294. doi: 10.1074/mcp.M110.007294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hubbard SC, Boyce M, McVaugh CT, Peehl DM, Bertozzi CR. Cell surface glycoproteomic analysis of prostate cancer-derived PC-3 cells. Bioorg Med Chem Lett. 2011;21:4945–4950. doi: 10.1016/j.bmcl.2011.05.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Slade PG, Hajivandi M, Bartel CM, Gorfien SF. Identifying the CHO secretome using mucin-type O-linked glycosylation and click-chemistry. J Proteome Res. 2012;11:6175–6186. doi: 10.1021/pr300810f. [DOI] [PubMed] [Google Scholar]
- 9.Hart GW, Slawson C, Ramirez-Correa G, Lagerlof O. Cross Talk Between O-GlcNAcylation and Phosphorylation: Roles in Signaling, Transcription, and Chronic Disease. Annu Rev Biochem. 2011;80:825–858. doi: 10.1146/annurev-biochem-060608-102511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vocadlo D, Hang H, Kim E, Hanover J, Bertozzi C. A chemical approach for identifying O-GlcNAc-modified proteins in cells. Proc Natl Acad Sci USA. 2003;100:9116–9121. doi: 10.1073/pnas.1632821100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hahne H, Sobotzki N, Nyberg T, Helm D, Borodkin VS, van Aalten DMF, Agnew B, Kuster B. Proteome Wide Purification and Identification of O-GlcNAc-Modified Proteins Using Click Chemistry and Mass Spectrometry. J Proteome Res. 2013;12:927–936. doi: 10.1021/pr300967y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *12.Zaro BW, Yang YY, Hang HC, Pratt MR. Chemical reporters for fluorescent detection and identification of O-GlcNAc-modified proteins reveal glycosylation of the ubiquitin ligase NEDD4-1. Proc Natl Acad Sci USA. 2011;108:8146–8151. doi: 10.1073/pnas.1102458108. The authors demonstrate that chemical changes to metabolic chemical reporter structure can alter their specificity for different types of glycoproteins. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gurel Z, Zaro BW, Pratt MR, Sheibani N. Identification of O-GlcNAc Modification Targets in Mouse Retinal Pericytes. PLoS ONE. 2014;9:e95561. doi: 10.1371/journal.pone.0095561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **14.Clark PM, Dweck JF, Mason DE, Hart CR, Buck SB, Peters EC, Agnew BJ, Hsieh-Wilson LC. Direct In-Gel Fluorescence Detection and Cellular Imaging of O-GlcNAc-Modified Proteins. J Am Chem Soc. 2008;130:11576–11577. doi: 10.1021/ja8030467. The authors describe the development of an azide-based, chemoenzymatic method for the detection and identification of O-GlcNAcylated proteins. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Alfaro JF, Gong CX, Monroe ME, Aldrich JT, Clauss TRW, Purvine SO, Wang Z, Camp DG, Shabanowitz J, Stanley P, et al. Tandem mass spectrometry identifies many mouse brain O-GlcNAcylated proteins including EGF domain-specific O-GlcNAc transferase targets. Proc Natl Acad Sci USA. 2012;109:7280–7285. doi: 10.1073/pnas.1200425109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *16.Boyce M, Carrico IS, Ganguli AS, Yu SH, Hangauer MJ, Hubbard SC, Kohler JJ, Bertozzi CR. Metabolic cross-talk allows labeling of O-linked {beta}-N-acetylglucosamine-modified proteins via the N-acetylgalactosamine salvage pathway. Proc Natl Acad Sci USA. 2011;108:3141–3146. doi: 10.1073/pnas.1010045108. The authors directly show that some metabolic chemical reporters can be metabolically interconverted, leading to the labeling of multiple types of glycans. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Palaniappan KK, Hangauer MJ, Smith TJ, Smart BP, Pitcher AA, Cheng EH, Bertozzi CR, Boyce M. A chemical glycoproteomics platform reveals O-GlcNAcylation of mitochondrial voltage-dependent anion channel 2. CellReports. 2013;5:546–552. doi: 10.1016/j.celrep.2013.08.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **18.Chuh KN, Zaro BW, Piller F, Piller V, Pratt MR. Changes in Metabolic Chemical Reporter Structure Yield a Selective Probe of O-GlcNAc Modification. J Am Chem Soc. 2014;136:12283–12295. doi: 10.1021/ja504063c. The authors develop the first metabolic chemical reporter that is highly selective for O-GlcNAcylation and compare the proteins identified by three major azide-containing chemical reporters of glycosylation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hang HC, Wilson JP, Charron G. Bioorthogonal Chemical Reporters for Analyzing Protein Lipidation and Lipid Trafficking. Acc Chem Res. 2011;44:699–708. doi: 10.1021/ar200063v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Resh MD. Use of analogs and inhibitors to study the functional significance of protein palmitoylation. Methods. 2006;40:191–197. doi: 10.1016/j.ymeth.2006.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wright MH, Clough B, Rackham MD, Rangachari K, Brannigan JA, Grainger M, Moss DK, Bottrill AR, Heal WP, Broncel M, et al. Validation of N-myristoyltransferase as an antimalarial drug target using an integrated chemical biology approach. Nature Chemistry. 2014;6:112–121. doi: 10.1038/nchem.1830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *22.Yount JS, Moltedo B, Yang YY, Charron G, Moran TM, López CB, Hang HC. Palmitoylome profiling reveals S-palmitoylation–dependent antiviral activity of IFITM3. Nat Chem Biol. 2010;6:610–614. doi: 10.1038/nchembio.405. The authors use alkyne-palmitate reporters to identify and characterize the importance of lipidation on the antiviral activity of a membrane associated protein. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wilson JP, Raghavan AS, Yang YY, Charron G, Hang HC. Proteomic analysis of fatty-acylated proteins in mammalian cells with chemical reporters reveals S-acylation of histone H3 variants. Mol Cell Proteomics. 2011;10:M110.001198. doi: 10.1074/mcp.M110.001198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *24.Martin BR, Wang C, Adibekian A, Tully SE, Cravatt BF. Global profiling of dynamic protein palmitoylation. Nat Meth. 2011;9:84–89. doi: 10.1038/nmeth.1769. The authors combine chemical reporters with quantitative proteomics to examine the dynamics of protein palmitoylation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Drisdel RC, Green WN. Labeling and quantifying sites of protein palmitoylation. BioTechniques. 2004;36:276–285. doi: 10.2144/04362RR02. [DOI] [PubMed] [Google Scholar]
- *26.Roth AF, Wan J, Bailey AO, Sun B, Kuchar JA, Green WN, Phinney BS, Yates JR, Davis NG. Global analysis of protein palmitoylation in yeast. Cell. 2006;125:1003–1013. doi: 10.1016/j.cell.2006.03.042. This manuscript describes the first application of the acyl-biotin exchange protocol for the proteomic identification of palmitoylated proteins. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kang R, Wan J, Arstikaitis P, Takahashi H, Huang K, Bailey AO, Thompson JX, Roth AF, Drisdel RC, Mastro R, et al. Neural palmitoyl-proteomics reveals dynamic synaptic palmitoylation. Nature. 2008;456:904–909. doi: 10.1038/nature07605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ivaldi C, Martin BR, Kieffer-Jaquinod S, Chapel A, Levade T, Garin J, Journet A. Proteomic analysis of S-acylated proteins in human B cells reveals palmitoylation of the immune regulators CD20 and CD23. PLoS ONE. 2012;7:e37187. doi: 10.1371/journal.pone.0037187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jones ML, Collins MO, Goulding D, Choudhary JS, Rayner JC. Analysis of protein palmitoylation reveals a pervasive role in Plasmodium development and pathogenesis. Cell Host Microbe. 2012;12:246–258. doi: 10.1016/j.chom.2012.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wan J, Savas JN, Roth AF, Sanders SS, Singaraja RR, Hayden MR, Yates JR, Davis NG. Tracking brain palmitoylation change: predominance of glial change in a mouse model of Huntington's disease. Chem Biol. 2013;20:1421–1434. doi: 10.1016/j.chembiol.2013.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Resh MD. Trafficking and signaling by fatty-acylated and prenylated proteins. Nat Chem Biol. 2006;2:584–590. doi: 10.1038/nchembio834. [DOI] [PubMed] [Google Scholar]
- 32.DeGraw AJ, Palsuledesai C, Ochocki JD, Dozier JK, Lenevich S, Rashidian M, Distefano MD. Evaluation of alkyne-modified isoprenoids as chemical reporters of protein prenylation. Chem Biol Drug Des. 2010;76:460–471. doi: 10.1111/j.1747-0285.2010.01037.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Charron G, Tsou LK, Maguire W, Yount JS, Hang HC. Alkynyl-farnesol reporters for detection of protein S-prenylation in cells. Mol Biosyst. 2011;7:67–73. doi: 10.1039/c0mb00183j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *34.Charron G, Li MMH, MacDonald MR, Hang HC. Prenylome profiling reveals S-farnesylation is crucial for membrane targeting and antiviral activity of ZAP long-isoform. Proc Natl Acad Sci USA. 2013;110:11085–11090. doi: 10.1073/pnas.1302564110. The authors describe the application of an alkyne-farnesyl probe for the identification of prenylated proteins. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shahbazian MD, Grunstein M. Functions of site-specific histone acetylation and deacetylation. Annu Rev Biochem. 2007;76:75–100. doi: 10.1146/annurev.biochem.76.052705.162114. [DOI] [PubMed] [Google Scholar]
- 36.Yang XJ, Seto E. Lysine acetylation: codified crosstalk with other posttranslational modifications. Mol Cell. 2008;31:449–461. doi: 10.1016/j.molcel.2008.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *37.Yang YY, Ascano JM, Hang HC. Bioorthogonal Chemical Reporters for Monitoring Protein Acetylation. J Am Chem Soc. 2010;132:3640–3641. doi: 10.1021/ja908871t. The authors describe the development of a chemical reporter for protein acetylation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yu-Ying Y, Markus G, Howard HC. Identification of lysine acetyltransferase p300 substrates using 4-pentynoyl-coenzyme A and bioorthogonal proteomics. Bioorg Med Chem Lett. 2011;21:4976–4979. doi: 10.1016/j.bmcl.2011.05.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lin H, Su X, He B. Protein Lysine Acylation and Cysteine Succination by Intermediates of Energy Metabolism. ACS Chem Biol. 2012;7:947–960. doi: 10.1021/cb3001793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bao X, Zhao Q, Yang T, Fung YME, Li XD. A Chemical Probe for Lysine Malonylation. Angew Chem Int Ed. 2013;52:4883–4886. doi: 10.1002/anie.201300252. [DOI] [PubMed] [Google Scholar]
- 41.Alfonso LF, Srivenugopal KS, Bhat GJ. Does aspirin acetylate multiple cellular proteins? (Review) Mol Med Report. 2009;2:533–537. doi: 10.3892/mmr_00000132. [DOI] [PubMed] [Google Scholar]
- 42.Bateman LA, Zaro BW, Miller SM, Pratt MR. An Alkyne–Aspirin Chemical Reporter for the Detection of Aspirin-Dependent Protein Modification in Living Cells. J Am Chem Soc. 2013;135:14568–14573. doi: 10.1021/ja408322b. [DOI] [PubMed] [Google Scholar]
- 43.Greer EL, Shi Y. Histone methylation: a dynamic mark in health, disease and inheritance. Nat Rev Genet. 2012;13:343–357. doi: 10.1038/nrg3173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bedford MT, Clarke SG. Protein arginine methylation in mammals: who, what, and why. Mol Cell. 2009;33:1–13. doi: 10.1016/j.molcel.2008.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Peters W, Willnow S, Duisken M, Kleine H, Macherey T, Duncan KE, Litchfield DW, Lüscher B, Weinhold E. Enzymatic Site-Specific Functionalization of Protein Methyltransferase Substrates with Alkynes for Click Labeling. Angew Chem Int Ed Engl. 2010 doi: 10.1002/anie.201001240. [DOI] [PubMed] [Google Scholar]
- 46.Binda O, Boyce M, Rush JS, Palaniappan KK, Bertozzi CR, Gozani O. A chemical method for labeling lysine methyltransferase substrates. ChemBioChem. 2011;12:330–334. doi: 10.1002/cbic.201000433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Luo M. Current chemical biology approaches to interrogate protein methyltransferases. ACS Chem Biol. 2012;7:443–463. doi: 10.1021/cb200519y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **48.Islam K, Chen Y, Wu H, Bothwell IR, Blum GJ, Zeng H, Dong A, Zheng W, Min J, Deng H, et al. Defining efficient enzyme-cofactor pairs for bioorthogonal profiling of protein methylation. Proc Natl Acad Sci USA. 2013;110:16778–16783. doi: 10.1073/pnas.1216365110. The authors describe the creation of a chemical reporter/mutant enzyme pair for the isoform specific identification of methyltransferase substrates. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang R, Islam K, Liu Y, Zheng W, Tang H, Lailler N, Blum G, Deng H, Luo M. Profiling genome-wide chromatin methylation with engineered posttranslation apparatus within living cells. J Am Chem Soc. 2013;135:1048–1056. doi: 10.1021/ja309412s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Willnow S, Martin M, Lüscher B, Weinhold E. A selenium-based click AdoMet analogue for versatile substrate labeling with wild-type protein methyltransferases. ChemBioChem. 2012;13:1167–1173. doi: 10.1002/cbic.201100781. [DOI] [PubMed] [Google Scholar]
- 51.Bothwell IR, Islam K, Chen Y, Zheng W, Blum G, Deng H, Luo M. Se-Adenosyl- l-selenomethionine Cofactor Analogue as a Reporter of Protein Methylation. J Am Chem Soc. 2012;134:14905–14912. doi: 10.1021/ja304782r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gibson BA, Kraus WL. New insights into the molecular and cellular functions of poly(ADP-ribose) and PARPs. Nat Rev Mol Cell Biol. 2012;13:411–424. doi: 10.1038/nrm3376. [DOI] [PubMed] [Google Scholar]
- 53.Du J, Jiang H, Lin H. Investigating the ADP-ribosyltransferase activity of sirtuins with NAD analogues and 32P-NAD. Biochemistry. 2009;48:2878–2890. doi: 10.1021/bi802093g. [DOI] [PubMed] [Google Scholar]
- 54.Jiang H, Kim JH, Frizzell KM, Kraus WL, Lin H. Clickable NAD analogues for labeling substrate proteins of poly(ADP-ribose) polymerases. J Am Chem Soc. 2010;132:9363–9372. doi: 10.1021/ja101588r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Carter-O'Connell I, Jin H, Morgan RK, David LL, Cohen MS. Engineering the Substrate Specificity of ADP-Ribosyltransferases for Identifying Direct Protein Targets. J Am Chem Soc. 2014 doi: 10.1021/ja412897a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yarbrough ML, Li Y, Kinch LN, Grishin NV, Ball HL, Orth K. AMPylation of Rho GTPases by Vibrio VopS disrupts effector binding and downstream signaling. Science. 2009;323:269–272. doi: 10.1126/science.1166382. [DOI] [PubMed] [Google Scholar]
- 57.Grammel M, Luong P, Orth K, Hang HC. A Chemical Reporter for Protein AMPylation. J Am Chem Soc. 2011;133:17103–17105. doi: 10.1021/ja205137d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Antelmann H, Helmann JD. Thiol-based redox switches and gene regulation. Antioxidants & Redox Signaling. 2011;14:1049–1063. doi: 10.1089/ars.2010.3400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Saurin AT, Neubert H, Brennan JP, Eaton P. Widespread sulfenic acid formation in tissues in response to hydrogen peroxide. Proc Natl Acad Sci USA. 2004;101:17982–17987. doi: 10.1073/pnas.0404762101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tyther R, Ahmeda A, Johns E, McDonagh B, Sheehan D. Proteomic profiling of perturbed protein sulfenation in renal medulla of the spontaneously hypertensive rat. J Proteome Res. 2010;9:2678–2687. doi: 10.1021/pr1001719. [DOI] [PubMed] [Google Scholar]
- *61.Paulsen CE, Truong TH, Garcia FJ, Homann A, Gupta V, Leonard SE, Carroll KS. Peroxide-dependent sulfenylation of the EGFR catalytic site enhances kinase activity. Nat Chem Biol. 2011;8:57–64. doi: 10.1038/nchembio.736. The authors use a chemical probe of cysteine oxidation to determine that sulfenylation of in a kinase active site activates the enzyme. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Jaffrey SR, Erdjument-Bromage H, Ferris CD, Tempst P, Snyder SH. Protein S-nitrosylation: a physiological signal for neuronal nitric oxide. Nat Cell Biol. 2001;3:193–197. doi: 10.1038/35055104. [DOI] [PubMed] [Google Scholar]
- 63.Forrester MT, Thompson JW, Foster MW, Nogueira L, Moseley MA, Stamler JS. Proteomic analysis of S-nitrosylation and denitrosylation by resin-assisted capture. Nat Biotechnol. 2009;27:557–559. doi: 10.1038/nbt.1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zaro BW, Chuh KN, Pratt MR. Chemical Reporter for Visualizing Metabolic Cross-Talk between Carbohydrate Metabolism and Protein Modification. ACS Chem Biol. 2014;9:1991–1996. doi: 10.1021/cb5005564. [DOI] [PMC free article] [PubMed] [Google Scholar]


