Abstract
Advances in mass spectrometry have transformed the scope and impact of protein characterization efforts. Identifying hundreds of proteins from rather simple biological matrices, such as yeast, was a daunting task just a few decades ago. Now, expression of more than half of the estimated ~20,000 human protein coding genes can be confirmed in record time and from minute sample quantities. Access to proteomic information at such unprecedented depths has been fueled by strides in every stage of the shotgun proteomics workflow – from sample processing to data analysis – and promises to revolutionize our understanding of the causes and consequences of proteome variation.
Introduction
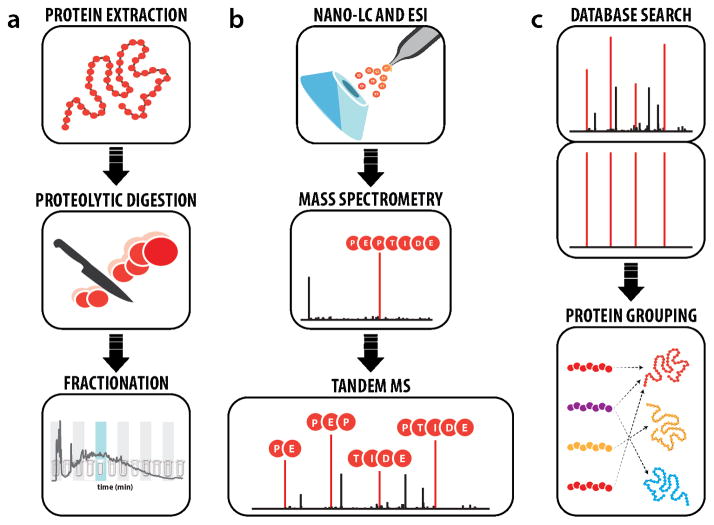
Spurred by the advent of the soft ionization methods, i.e., electrospray [1] and matrix-assisted laser desorption ionization [2], in the late 1980s, mass spectrometry (MS) has become the central method for protein analysis. Since this time, the depth and rate at which a proteome can be characterized has steadily improved so that today comprehensive analysis of most proteomes is within reach. The shotgun method, outlined in Figure 1, has proven the most useful tool for such applications. Here, proteins are extracted from lysed cells, enzymatically digested, and chromatographically separated prior to MS analysis. The MS records the masses of eluting peptide cations every second or so. In between these so-called MS1 scans the system isolates selected peptide precursors, dissociates them using collisions or chemical reactions, and records the masses of the pieces (i.e., MS2 or tandem MS). Modern MS systems can measure peptide masses accurately to thee decimal places while at the same time collecting tandem mass spectra at a blazing rate of 20 Hz. The hundreds of thousands of spectra generated from one of these experiments are then analyzed in silico using spectral matching algorithms.
Figure 1.
Workflow for “shotgun” or “bottom-up” proteomics. (a) Preparing proteomic samples for LC-MS/MS analysis requires protein extraction, proteolysis, and, optionally, peptide-level fractionation. (b) Online LC separation of complex peptide mixtures introduces analytes into the mass spectrometer for precursor and fragment ion mass analysis. (c) Tandem mass spectra are matched to theoretical spectra generated in silico to garner peptide sequences that are used for protein inference.
Mammalian proteomes are complex [3]. The human proteome contains ~20,300 protein-coding genes; however, non-synonymous single nucleotide polymorphisms (nsSNPs), alternative splicing events, and post-translational modifications (PTMs) all occur and exponentially increase the number of distinct proteoforms [4–6]. Detection of ~5,000 proteins in a proteomic experiment was a considerable achievement just a few years ago [7–9]. More recently, two groups identified over 10,000 protein groups in a single experiment. Through extensive protein and peptide fractionation (72 fractions) and digestion with multiple enzymes, Nagaraj et al. identified 10,255 protein groups from HeLa cells over 288 hours of instrument analysis [10•]. A comparison with paired RNA-Seq data revealed nearly complete overlap between the detected proteins and the expressed transcripts. In that same year, a similar strategy enabled the identification of 10,006 proteins from the U2OS cell line [11•].
A more comprehensive analysis of the human proteome can be achieved by applying similar technologies to large-scale comparisons of multiple cell lines and tissues [12,13,14•,15•]. Kim and co-workers analyzed 30 human tissues and primary cells over 2,000 LC-MS/MS experiments, resulting in the detection of 293,000 peptides with unique amino acid sequences and evidence for 17,294 gene products [16••]. Wilhelm et al. amassed a total of 16,857 LC-MS/MS experiments from human cell lines, tissues, and body fluids. These experiments produced a total of 946,000 unique peptides, which map to 18,097 protein-coding genes [17••]. Together, these two studies provide direct evidence for protein translation of over 90% of human genes (Figure 2). Despite providing the deepest coverage to date, the latter study required non-stop operation of a mass spectrometer for four straight years! New developments in mass spectrometer technology have increased the rate at which proteomes can be analyzed. Using such a device, we recently described a method that characterizes nearly every protein in yeast in just over one hour (4,000 of the 4,500 expressed yeast proteins) [18••]. In this review, we describe developments in sample preparation, MS instrumentation, and bioinformatics that have been key to obtaining comprehensive proteomic coverage. Further, we consider how access to such proteomic detail will impact genomic research.
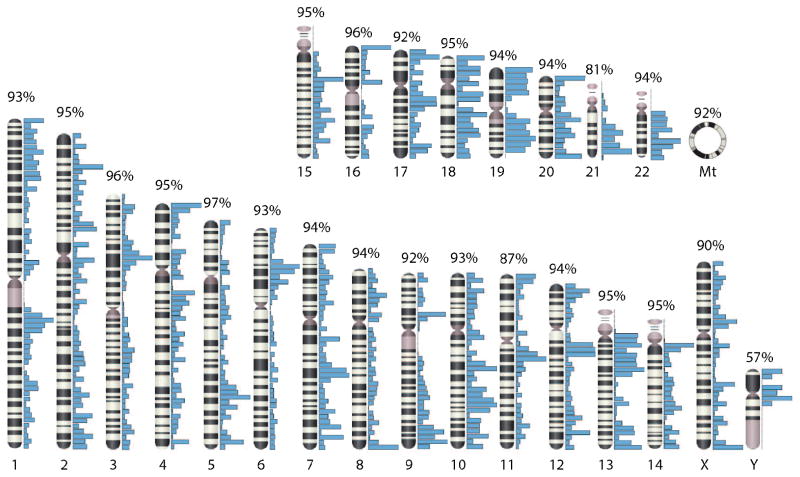
Figure 2.
Chromosomal coverage of the human proteome (reproduced with permission from ref. [17]). In one of two recent large-scale investigations of the human proteome, Wilhelm and coworkers identified 18,097 proteins, covering over 90% of all but three chromosomes (11, 21, and Y). The density of proteins covered in any particular chromosomal region is indicated by the blue bars.
Advances in proteomic sample preparation
For any proteomic method, proteins must first be liberated from their host cells, via mechanical and/or chemical disruption, often into a denaturing solution. Reduction of disulfide bonds and alkylation of cysteine residues disrupts protein structure, leaving proteins amenable to site-specific cleavage with one or more proteases. This initial step – protein extraction and solubilization – is paramount, as it dictates which proteins will be accessible for eventual MS detection. Strong detergents, such as sodium dodecyl sulfate (SDS), are exceptional denaturants, but their removal, a requirement for efficient proteolytic digestion and sensitive mass-spectrometric analysis, is challenging. Standard filtration devices can be employed as proteomic reactors (filter-aided sample preparation, FASP), allowing dissolution of proteins in high concentrations of SDS which are then depleted before digestion [19]. Alternatively, an unbiased proteomic characterization can be achieved without SDS by digesting unclarified cellular lysate, a tactic that improves coverage of proteins harbored in poorly soluble membrane and nuclear organelles [18••,20,21••].
The maximum coverage obtainable for a protein is theoretically determined by its amino acid sequence and the cleavage specificity of the chosen proteolytic enzyme, typically trypsin. A straightforward and long-recognized approach for boosting protein and proteome coverage is to digest a sample separately with multiple proteases [22]. Recently, digestion with α-lytic proteases, semi-specific enzymes that preferentially cleave after aliphatic residues, increased trypsin-only protein identifications and sequence coverage by 24% and 101%, respectively [23]. Some downsides of such multi-protease strategies include heightened sample quantity and analysis time demands. Digesting with multiple enzymes sequentially instead of in parallel, however, can afford better coverage without the extra requirements [24,25].
Even following efficient solubilization and proteolysis, many proteins are only represented following detection of a few unique peptides [6]. This mainly stems signal suppression during the electrospray ionization – that is, peptides having higher overall basicity tend to preferentially ionize rendering the more acidic peptides undetected. The most successful approach to curb this problem is to reduce the number of unique peptide sequences present in the ionization source at one time. To this end, separation chemistries and implementations thereof are central to proteomic analysis. The extreme separation resolution provided by some of these platforms, such as the automated coupling of three physicochemically orthogonal stages of chromatography [26] and high-resolution isoelectric focusing [27••], is key to achieving genome-scale coverage of the proteome.
Overall, recent developments in sample processing for shotgun proteomics have emphasized simplification and scalability [21••]. Furthermore, a robust workflow with minimal sample loss and contamination opens the door for applications with limited starting material. Using current technology, 9,500 proteins can be identified from just 100 nL of formalin-fixed and paraffin-embedded (FFPE) tissue [28•]. Although it is impossible to ionize and sequence every peptide, efficient sample preparation, coupled with advances in MS instrumentation [29–31], separation methodology [32–35], and fragmentation techniques [36–39] have vastly increased the observable portion of the human proteome.
Advances in peptide separation and MS instrumentation
Modern hybrid mass spectrometers couple highly accurate MS1 scans with ultra-fast MS/MS sequencing rates. A recent study used a linear ion trap (LIT)-Orbitrap hybrid mass spectrometer [40], which, compared to the previous generation instrument, achieves approximately twice the resolving power at the same scan speed, to analyze eleven human cell lines [14•]. Across all cell lines, 11,731 proteins were identified, with an average of 10,361 proteins identified per cell line. The number of identified protein groups is comparable to a previous study from HeLa lysate [10•], but is generated in a fraction of the time (3 versus ~ 12 days). A newly released mass spectrometer combining a quadrupole mass filter, a collision cell, a dual cell LIT and an Orbitrap mass analyzer, operates at a maximum MS/MS acquisition speed of 20 Hz [41], doubling the number of tryptic yeast peptides identified per second as compared with the Orbitrap Elite (19 versus 10 peptides/second) [18••]. Ion mobility coupled with MS has also been explored as an option for decreasing sample complexity and improving identification efficiency [42]. Traveling wave ion mobility spectrometry (TWIMS) significantly improved the duty-cycle of a time-of-flight (TOF) instrument, identifying ~7,500 protein groups from HeLa cells in one day of analysis time [31].
The sequencing speed of modern mass spectrometers is best harnessed when coupled to efficient, online peptide separation. HPLC systems operating at high pressures (>8,000 psi) [33] and longer columns packed with small particles have become standard (< 2 μm) [18••,34]. The linear relationship between the number of identified peptides and peak capacity, the number of resolvable peaks across an elution, has been demonstrated [32]. Many recent workflows have focused on optimizing chromatographic separations rather than extensive fractionation for whole proteome analysis [43]. Forgoing sample pre-fractionation in favor of long columns packed with 2 μm particles, a recent study identified 4,825 protein groups from the A375 cancer cell line over a three hour LC-MS/MS experiment [44]. A comparison of column lengths revealed that, for this particular instrument platform, a 50 cm column allowed identification of more proteins than either a 15 or 25 cm column at all gradient lengths tested, although a decrease in cumulative identified protein groups after three hours was reported. Note that the combination of long columns and small particles significantly raises column backpressure, necessitating either a UHPLC system capable of operating at pressures > 10,000 psi, or a column heater. Silica monolithic columns, which can achieve separation efficiencies similar to traditional packed columns without a substantial increase in back pressure, have also been used [35].
Advances in computational proteomics
The scale of proteomic data generated by streamlined sample processing pipelines and high-resolution mass spectrometry now approaches that of analogous genomic and transcriptomic technologies. Given that proteins more closely resemble phenotype than their encoding nucleic acid counterparts, they harbor unique biological details that can inform larger biological processes.
For organisms with sequenced genomes, peptides detected by mass spectrometry can assist in refining prediction-based gene annotations, a primary goal of the emergent proteogenomics field [45]. In addition to validating predicted genes, deep proteomic coverage can suggest novel protein-coding loci [46], N-terminal signal peptides [47], splice sites [48], and nonsynonymous variants [49]. A long-term objective of proteogenomic mapping is to associate certain variations in protein sequence with disease states. One recent study combined a customized protein database with in-depth transcriptome and proteome profiling of livers from two inbred rat strains [50•]. Interestingly, the results associated a genomic variant in the promoter region of a mis-annotated gene with the observed hypertensive phenotype of one strain, illustrating the advantages of such integrated approaches. Proteogenomic endeavors match tandem mass spectra to a database containing, ideally, all possible protein sequences encoded by an individual genome. This poses a computational challenge for large genomes with low protein-coding content, requiring extensive search-space reductions to boost sensitivity. A fresh strategy enabled unbiased proteogenomic mapping against the full human genome, along with deep proteome coverage, by blending isoelectric focusing for high-resolution peptide separation with accurate isoelectric point prediction for rational reduction of the search space [27••].
Systems-level analyses have also benefitted from the growing robustness and availability of informatics tools for label-free quantification strategies. Even highly fractionated proteomes can now be accurately compared in the absence of stable isotope labels [51]. Aided by highly accurate mass measurements, the confident transfer of peptide identifications between matching runs provides a 25–30% boost in the number of proteins quantified across multiple samples [14]. This feature makes label-free approaches very attractive for deep proteome quantification, though stable isotope labeling strategies are still more straightforward for the comparative analysis of low-abundance PTMs [52]. Furthermore, statistical analysis of signatures at the peptide-level can reveal information regarding the presence and expression patterns of one or more proteomes, an approach that will be greatly empowered by high protein sequence coverage [53].
Conclusions and outlook
Breakthroughs in every stage of the shotgun proteomics workflow have collectively ushered in a new era of proteomics, one in which identification and quantification of complete proteomes can be routinely achieved [54]. Beyond propelling basic research, this age holds great potential for personalized medicine [55]. Earlier this year, two independent efforts reported evidence of protein translation for 90–95% human genes, an impressive display of progressing technologies for proteome characterization [16••,17••]. As deep cataloguing of protein expression becomes widespread, the spotlight will shift to extensive functional mapping of proteoforms and determining how their expression is regulated by genomic elements [56]. To this end, the complementary benefits of top-down [57], targeted [58], and antibody-based [59] approaches must be harnessed and effectively integrated. Finally, in light of the surging trend of deep proteomics, it is important to remember that, for some systems, meaningful biological insight can still be drawn from moderate depths of proteome coverage [15•], which are becoming ever more accessible to proteomic researchers of all experience levels.
Recent advances in mass spectrometry have transformed the depth of coverage of the human proteome
Expression of over half of the estimated human protein coding genes can be verified in record time
Here, we highlight the impact of advances in mass spectrometry sample preparation, instrumentation, and data analysis on the characterization of the human proteome
Acknowledgments
We thank all the members of the Joshua Coon research group for insightful discussion. This work was supported by the National Institutes of Health grant R01 GM080148 to J.J.C. A.L.R. was supported by an NHGRI training grant to the Genomic Sciences Training Program (5T32HG002760). A.E.M. was supported by an NLM training grant to the Computation and Informatics in Biology and Medicine Training Program (NLM T15LM007359).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References and recommended reading
Papers of particular interest, published within the period of review, have been highlighted as:
• of special interest
•• of outstanding interest
- 1.Fenn JB, Mann M, Meng CK, Wong SF, Whitehouse CM. Electrospray ionization for mass spectrometry of large biomolecules. Science. 1989;2464:64–71. doi: 10.1126/science.2675315. [DOI] [PubMed] [Google Scholar]
- 2.Karas M, Bachmann D, Bahr U, Hillenkamp F. Matrix-assisted ultraviolet-laser desorption of nonvolatile compounds. Int J Mass Spectrom. 1987;78:53–68. [Google Scholar]
- 3.Harrow J, Frankish A, Gonzalez JM, Tapanari E, Diekhans M, Kokocinski F, Aken BL, Barrell D, Zadissa A, Searle S, Barnes I, et al. GENCODE: The reference human genome annotation for The ENCODE Project. Genome Res. 2012;22:1760–1774. doi: 10.1101/gr.135350.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.A user’s guide to the encyclopedia of DNA elements (ENCODE) PLoS Biol. 2011;9:e1001046. doi: 10.1371/journal.pbio.1001046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Abecasis GR, Altshuler D, Auton A, Brooks LD, Durbin RM, Gibbs RA, Hurles ME, McVean GA. A map of human genome variation from population-scale sequencing. Nature. 2010;467:1061–1073. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zubarev RA. The challenge of the proteome dynamic range and its implications for in-depth proteomics. Proteomics. 2013;13:723–726. doi: 10.1002/pmic.201200451. [DOI] [PubMed] [Google Scholar]
- 7.Burkard TR, Planyavsky M, Kaupe I, Breitwieser FP, Burckstummer T, Bennett KL, Superti-Furga G, Colinge J. Initial characterization of the human central proteome. BMC Syst Biol. 2011;5:17. doi: 10.1186/1752-0509-5-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wisniewski JR, Zougman A, Mann M. Combination of FASP and StageTip-based fractionation allows in-depth analysis of the hippocampal membrane proteome. J Proteome Res. 2009;8:5674–5678. doi: 10.1021/pr900748n. [DOI] [PubMed] [Google Scholar]
- 9.Hubner NC, Ren S, Mann M. Peptide separation with immobilized pI strips is an attractive alternative to in-gel protein digestion for proteome analysis. Proteomics. 2008;8:4862–4872. doi: 10.1002/pmic.200800351. [DOI] [PubMed] [Google Scholar]
- 10•.Nagaraj N, Wisniewski JR, Geiger T, Cox J, Kircher M, Kelso J, Paabo S, Mann M. Deep proteome and transcriptome mapping of a human cancer cell line. Mol Syst Biol. 2011;7:548. doi: 10.1038/msb.2011.81. This study identified >10,000 proteins from a human cell line in a single experiment, providing an estimate of the minimum number of proteins expressed in a mammalian system. Through extensive protein fractionation and digestion with trypsin, LysC and GluC, 10,300 protein groups were identified from HeLa cells. RNA-Seq data of the same cells provided evidence for less than 12,000 genes, suggesting near complete sequencing of the HeLa proteome. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11•.Beck M, Schmidt A, Malmstroem J, Claassen M, Ori A, Szymborska A, Herzog F, Rinner O, Ellenberg J, Aebersold R. The quantitative proteome of a human cell line. Mol Syst Biol. 2011;7:549. doi: 10.1038/msb.2011.82. In this report, over 10,000 proteins were identified from the U2OS cell line using a combination of peptide-level fractionation, and charge state and gas phase fractionation. Additionally, a large-scale estimation of protein concentration was performed, determining the cellular abundance for 7,300 of the detected proteins. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Phanstiel DH, Brumbaugh J, Wenger CD, Tian SL, Probasco MD, Bailey DJ, Swaney DL, Tervo MA, Bolin JM, Ruotti V, Stewart R, et al. Proteomic and phosphoproteomic comparison of human ES and iPS cells. Nat Methods. 2011;8:821–U884. doi: 10.1038/nmeth.1699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Munoz J, Low TY, Kok YJ, Chin A, Frese CK, Ding V, Choo A, Heck AJR. The quantitative proteomes of human-induced pluripotent stem cells and embryonic stem cells. Mol Syst Biol. 2011;7:550. doi: 10.1038/msb.2011.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14•.Geiger T, Wehner A, Schaab C, Cox J, Mann M. Comparative proteomic analysis of eleven common cell lines reveals ubiquitous but varying expression of most proteins. Mol Cell Proteomics. 2012;11:M111.014050. doi: 10.1074/mcp.M111.014050. This report compared the proteomes of eleven human cell lines. Over three days of analysis time, each cell line yielded an average of 10,361 protein identifications, with a total of 11,731 proteins identified. For all cells, expression values spanned a dynamic range of ~7 orders of magnitude. Although a common group of proteins was present amongst all cell lines, protein expression levels were highly variable. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15•.Gholami AM, Hahne H, Wu ZX, Auer FJ, Meng C, Wilhelm M, Kuster B. Global proteome analysis of the NCI-60 cell line panel. Cell Rep. 2013;4:609–620. doi: 10.1016/j.celrep.2013.07.018. This report provides a comprehensive resource containing quantitative proteome and kinome profiles for the 59 human cancer cell lines comprising the NCI-60 panel. Bioinformatic analysis reveals molecular signatures of cancer that are both shared across, and unique to, different cell lines. Furthermore, these expression data can be leveraged with other resources (e.g., mutational status, anticancer therapeutic response, etc.) to predict drug sensitivity or resistance. [DOI] [PubMed] [Google Scholar]
- 16••.Kim MS, Pinto SM, Getnet D, Nirujogi RS, Manda SS, Chaerkady R, Madugundu AK, Kelkar DS, Isserlin R, Jain S, Thomas JK, et al. A draft map of the human proteome. Nature. 2014;509:575–581. doi: 10.1038/nature13302. This report describes a mass spectrometry-centered map of the human proteome. A total of 30 human tissues and cell lines were investigated over 2,000 LC-MS/MS runs. Over all analyses, 293,000 unique peptides were mapped to proteins encoded by 17,294 genes. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17••.Wilhelm M, Schlegl J, Hahne H, Gholami AM, Lieberenz M, Savitski MM, Ziegler E, Butzmann L, Gessulat S, Marx H, Mathieson T, et al. Mass-spectrometry-based draft of the human proteome. Nature. 2014;509:582–587. doi: 10.1038/nature13319. In this mass spectrometry-based map of the human proteome, the authors combine data from their own lab with ~10,000 available raw files, identifying 18,097 protein-coding genes from human tissues, cell lines and body fluids. Together, both human proteome studies provide direct evidence for the actual translation of over 90% of protein-coding genes. [DOI] [PubMed] [Google Scholar]
- 18••.Hebert AS, Richards AL, Bailey DJ, Ulbrich A, Coughlin EE, Westphall MS, Coon JJ. The one hour yeast proteome. Mol Cell Proteomics. 2014;13:339–347. doi: 10.1074/mcp.M113.034769. This article describes the near complete characterization of the yeast proteome in approximately one hour of instrument analysis time. Through optimized sample preparation, chromatographic conditions, and the use of a novel Orbitrap hybrid mass spectrometer, an average of 34,255 tryptic yeast peptides were identified in 70 minutes. Up to 67 proteins were identified per minute, for a total of 4,002 proteins, or ~90% of the expressed yeast proteome. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wisniewski JR, Zougman A, Nagaraj N, Mann M. Universal sample preparation method for proteome analysis. Nat Methods. 2009;6:359–U360. doi: 10.1038/nmeth.1322. [DOI] [PubMed] [Google Scholar]
- 20.Pirmoradian M, Budamgunta H, Chingin K, Zhang B, Astorga-Wells J, Zubarev RA. Rapid and deep human proteome analysis by single-dimension shotgun proteomics. Mol Cell Proteomics. 2013;12:3330–3338. doi: 10.1074/mcp.O113.028787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21••.Kulak NA, Pichler G, Paron I, Nagaraj N, Mann M. Minimal, encapsulated proteomic-sample processing applied to copy-number estimation in eukaryotic cells. Nat Methods. 2014;11:319–U300. doi: 10.1038/nmeth.2834. This study validates an in-StageTip (iST) reactor for all steps of proteomic sample preparation, from protein extraction to peptide fractionation, and utilizes it to estimate protein copy numbers in yeast and human cells. Most importantly, iST-based methods can be automated and, thus, offer great potential for the analysis of large populations of clinical samples. [DOI] [PubMed] [Google Scholar]
- 22.Swaney DL, Wenger CD, Coon JJ. Value of using multiple proteases for large-scale mass spectrometry-based proteomics. J Proteome Res. 2010;9:1323–1329. doi: 10.1021/pr900863u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Meyer JG, Kim S, Maltby DA, Ghassemian M, Bandeira N, Komives EA. Expanding proteome coverage with orthogonal-specificity-lytic proteases. Molecular & Cellular Proteomics. 2014;13:823–835. doi: 10.1074/mcp.M113.034710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wisniewski JR, Mann M. Consecutive proteolytic digestion in an enzyme reactor increases depth of proteomic and phosphoproteomic analysis. Anal Chem. 2012;84:2631–2637. doi: 10.1021/ac300006b. [DOI] [PubMed] [Google Scholar]
- 25.Guo XF, Trudgian DC, Lemoff A, Yadavalli S, Mirzaei H. Confetti: A multiprotease map of the HeLa proteome for comprehensive proteomics. Molecular & Cellular Proteomics. 2014;13:1573–1584. doi: 10.1074/mcp.M113.035170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhou F, Lu Y, Ficarro SB, Adelmant G, Jiang W, Luckey CJ, Marto JA. Genome-scale proteome quantification by DEEP SEQ mass spectrometry. Nat Commun. 2013;4:2171. doi: 10.1038/ncomms3171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27••.Branca RM, Orre LM, Johansson HJ, Granholm V, Huss M, Perez-Bercoff A, Forshed J, Kall L, Lehtio J. HiRIEF LC-MS enables deep proteome coverage and unbiased proteogenomics. Nat Methods. 2014;11:59–62. doi: 10.1038/nmeth.2732. This report introduces a new approach for deep proteome coverage and unbiased proteogenomics. High-resolution isoelectric focusing (HiRIEF) provides a robust dimension of peptide fractionation, key for identifying >10,000 proteins from complex mammalian mixtures. Additionally, HiRIEF can be combined with accurate peptide isoelectric point (pI) prediction to extensively reduce the search space, a necessity for searching protein databases generated from six-reading-frame translations of large genomes. [DOI] [PubMed] [Google Scholar]
- 28•.Wisniewski JR, Dus K, Mann M. Proteomic workflow for analysis of archival formalin-fixed and paraffin-embedded clinical samples to a depth of 10 000 proteins. Proteom Clin Appl. 2013;7:225–233. doi: 10.1002/prca.201200046. This paper describes a streamlined workflow for deep proteomic profiling of microdissected formalin-fixed and paraffin-embedded (FFPE) clinical tissue samples. In approximately one day of MS analysis time, nearly 10,000 proteins can be identified from just 100 nL of archival FFPE samples. [DOI] [PubMed] [Google Scholar]
- 29.Michalski A, Damoc E, Hauschild JP, Lange O, Wieghaus A, Makarov A, Nagaraj N, Cox J, Mann M, Horning S. Mass spectrometry-based proteomics using Q Exactive, a high-performance benchtop quadrupole Orbitrap mass spectrometer. Mol Cell Proteomics. 2011;10:M111 011015. doi: 10.1074/mcp.M111.011015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Andrews GL, Simons BL, Young JB, Hawkridge AM, Muddiman DC. Performance characteristics of a new hybrid quadrupole time-of-flight tandem mass spectrometer (TripleTOF 5600) Anal Chem. 2011;83:5442–5446. doi: 10.1021/ac200812d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Helm D, Vissers JP, Hughes CJ, Hahne H, Ruprecht B, Pachl F, Grzyb A, Richardson K, Wildgoose J, Maier SK, Marx H, et al. Ion mobility tandem mass spectrometry enhances performance of bottom-up proteomics. Mol Cell Proteomics. 2014 doi: 10.1074/mcp.M114.041038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kocher T, Swart R, Mechtler K. Ultra-high-pressure RPLC hyphenated to an LTQ-Orbitrap Velos reveals a linear relation between peak capacity and number of identified peptides. Anal Chem. 2011;83:2699–2704. doi: 10.1021/ac103243t. [DOI] [PubMed] [Google Scholar]
- 33.Cristobal A, Hennrich ML, Giansanti P, Goerdayal SS, Heck AJR, Mohammed S. In-house construction of a UHPLC system enabling the identification of over 4000 protein groups in a single analysis. Analyst. 2012;137:3541–3548. doi: 10.1039/c2an35445d. [DOI] [PubMed] [Google Scholar]
- 34.Hsieh EJ, Bereman MS, Durand S, Valaskovic GA, MacCoss MJ. Effects of column and gradient lengths on peak capacity and peptide identification in nanoflow LC-MS/MS of complex proteomic samples. J Am Soc Mass Spectrom. 2013;24:148–153. doi: 10.1007/s13361-012-0508-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Iwasaki M, Sugiyama N, Tanaka N, Ishihama Y. Human proteome analysis by using reversed phase monolithic silica capillary columns with enhanced sensitivity. J Chromatogr A. 2012;1228:292–297. doi: 10.1016/j.chroma.2011.10.059. [DOI] [PubMed] [Google Scholar]
- 36.Syka JE, Coon JJ, Schroeder MJ, Shabanowitz J, Hunt DF. Peptide and protein sequence analysis by electron transfer dissociation mass spectrometry. Proc Natl Acad Sci U S A. 2004;101:9528–9533. doi: 10.1073/pnas.0402700101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Olsen JV, Macek B, Lange O, Makarov A, Horning S, Mann M. Higher-energy C-trap dissociation for peptide modification analysis. Nat Methods. 2007;4:709–712. doi: 10.1038/nmeth1060. [DOI] [PubMed] [Google Scholar]
- 38.Swaney DL, McAlister GC, Coon JJ. Decision tree-driven tandem mass spectrometry for shotgun proteomics. Nat Methods. 2008;5:959–964. doi: 10.1038/nmeth.1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Frese CK, Altelaar AFM, Hennrich ML, Nolting D, Zeller M, Griep-Raming J, Heck AJR, Mohammed S. Improved peptide identification by targeted fragmentation using CID, HCD and ETD on an LTQ-Orbitrap Velos. J Proteome Res. 2011;10:2377–2388. doi: 10.1021/pr1011729. [DOI] [PubMed] [Google Scholar]
- 40.Michalski A, Damoc E, Lange O, Denisov E, Nolting D, Muller M, Viner R, Schwartz J, Remes P, Belford M, Dunyach JJ, et al. Ultra high resolution linear ion trap Orbitrap mass spectrometer (Orbitrap Elite) facilitates top down LC MS/MS and versatile peptide fragmentation modes. Mol Cell Proteomics. 2012;11:O111 013698. doi: 10.1074/mcp.O111.013698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Senko MW, Remes PM, Canterbury JD, Mathur R, Song QY, Eliuk SM, Mullen C, Earley L, Hardman M, Blethrow JD, Bui H, et al. Novel parallelized quadrupole/linear ion trap/Orbitrap tribrid mass spectrometer improving proteome coverage and peptide identification rates. Anal Chem. 2013;85:11710–11714. doi: 10.1021/ac403115c. [DOI] [PubMed] [Google Scholar]
- 42.Distler U, Kuharev J, Navarro P, Levin Y, Schild H, Tenzer S. Drift time-specific collision energies enable deep-coverage data-independent acquisition proteomics. Nat Methods. 2014;11:167–170. doi: 10.1038/nmeth.2767. [DOI] [PubMed] [Google Scholar]
- 43.Thakur SS, Geiger T, Chatterjee B, Bandilla P, Frohlich F, Cox J, Mann M. Deep and highly sensitive proteome coverage by LC-MS/MS without prefractionation. Mol Cell Proteomics. 2011;10:M110 003699. doi: 10.1074/mcp.M110.003699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pirmoradian M, Budamgunta H, Chingin K, Zhang B, Astorga-Wells J, Zubarev RA. Rapid and deep human proteome analysis by single-dimension shotgun proteomics. Mol Cell Proteomics. 2013;12:3330–3338. doi: 10.1074/mcp.O113.028787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Renuse S, Chaerkady R, Pandey A. Proteogenomics. Proteomics. 2011;11:620–630. doi: 10.1002/pmic.201000615. [DOI] [PubMed] [Google Scholar]
- 46.Khatun J, Yu Y, Wrobel JA, Risk BA, Gunawardena HP, Secrest A, Spitzer WJ, Xie L, Wang L, Chen X, Giddings MC. Whole human genome proteogenomic mapping for encode cell line data: Identifying protein-coding regions. BMC Genomics. 2013;14:141. doi: 10.1186/1471-2164-14-141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hartmann EM, Armengaud J. N-terminomics and proteogenomics, getting off to a good start. Proteomics. 2014 doi: 10.1002/pmic.201400157. [DOI] [PubMed] [Google Scholar]
- 48.Sheynkman GM, Shortreed MR, Frey BL, Smith LM. Discovery and mass spectrometric analysis of novel splice-junction peptides using RNA-Seq. Mol Cell Proteomics. 2013;12:2341–2353. doi: 10.1074/mcp.O113.028142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sheynkman GM, Shortreed MR, Frey BL, Scalf M, Smith LM. Large-scale mass spectrometric detection of variant peptides resulting from nonsynonymous nucleotide differences. J Proteome Res. 2014;13:228–240. doi: 10.1021/pr4009207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50•.Low TY, van Heesch S, van den Toorn H, Giansanti P, Cristobal A, Toonen P, Schafer S, Hubner N, van Breukelen B, Mohammed S, Cuppen E, et al. Quantitative and qualitative proteome characteristics extracted from in-depth integrated genomics and proteomics analysis. Cell Rep. 2013;5:1469–1478. doi: 10.1016/j.celrep.2013.10.041. This paper integrates in-depth genomic, transcriptomic, and proteomic analyses of liver tissue from two rat strains, one of which is a widely used model for hypertension studies. The use of five orthogonal proteases and extensive pre-fractionation, as well as matching to a customized protein database, generated peptide-level evidence for over 26,000 rat liver proteins. In addition, this comprehensive strategy can serve as a tool for linking specific genomic variants to overall phenotype. [DOI] [PubMed] [Google Scholar]
- 51.Cox J, Hein MY, Luber CA, Paron I, Nagaraj N, Mann M. MaxLFQ allows accurate proteome-wide label-free quantification by delayed normalization and maximal peptide ratio extraction. Mol Cell Proteomics. 2014 doi: 10.1074/mcp.M113.031591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.RCJDS, Knittle AM, Nagaraj N, van Dinther M, Choudhary C, Ten Dijke P, Mann M, Sharma K. Time-resolved dissection of early phosphoproteome and ensuing proteome changes in response to TGF-β. Sci Signal. 2014;7:rs5. doi: 10.1126/scisignal.2004856. [DOI] [PubMed] [Google Scholar]
- 53.Webb-Robertson BJ, Matzke MM, Datta S, Payne SH, Kang J, Bramer LM, Nicora CD, Shukla AK, Metz TO, Rodland KD, Smith RD, et al. Bayesian proteoform modeling improves protein quantification of global proteomic measurements. Mol Cell Proteomics. 2014 doi: 10.1074/mcp.O113.030932. [DOI] [PubMed] [Google Scholar]
- 54.Mann M, Kulak NA, Nagaraj N, Cox J. The coming age of complete, accurate, and ubiquitous proteomes. Mol Cell. 2013;49:583–590. doi: 10.1016/j.molcel.2013.01.029. [DOI] [PubMed] [Google Scholar]
- 55.Munoz J, Heck AJ. From the human genome to the human proteome. Angew Chem Int Ed Engl. 2014 doi: 10.1002/anie.201406545. [DOI] [PubMed] [Google Scholar]
- 56.Paik YK, Hancock WS. Uniting ENCODE with genome-wide proteomics. Nat Biotechnol. 2012;30:1065–1067. doi: 10.1038/nbt.2416. [DOI] [PubMed] [Google Scholar]
- 57.Catherman AD, Durbin KR, Ahlf DR, Early BP, Fellers RT, Tran JC, Thomas PM, Kelleher NL. Large-scale top-down proteomics of the human proteome: Membrane proteins, mitochondria, and senescence. Mol Cell Proteomics. 2013;12:3465–3473. doi: 10.1074/mcp.M113.030114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Farrah T, Deutsch EW, Omenn GS, Sun Z, Watts JD, Yamamoto T, Shteynberg D, Harris MM, Moritz RL. State of the human proteome in 2013 as viewed through peptideatlas: Comparing the kidney, urine, and plasma proteomes for the biology- and disease-driven human proteome project. J Proteome Res. 2014;13:60–75. doi: 10.1021/pr4010037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ponten F, Gry M, Fagerberg L, Lundberg E, Asplund A, Berglund L, Oksvold P, Bjorling E, Hober S, Kampf C, Navani S, et al. A global view of protein expression in human cells, tissues, and organs. Mol Syst Biol. 2009;5:337. doi: 10.1038/msb.2009.93. [DOI] [PMC free article] [PubMed] [Google Scholar]