Abstract
Here we demonstrate the production of a functioning cell model by formation of giant vesicles reconstituted with the GLUT1 glucose transporter and a glucose oxidase and hydrogen peroxidase linked fluorescent reporter internally. Hence, a simplified artificial cell is formed that is able to take up glucose and process it.
Cell metabolism is highly dependent on facilitated transport of nutrients from the exterior medium to the cell interior. Internalized nutrients are metabolized by diverse enzymatic reactions to ensure growth, reproduction, maintenance of structure, and allow the cell to respond to changing environments. Facilitative glucose transporters (GLUTs) mediate the bidirectional transport of monosaccharaides across the cell membrane down its concentration gradient in an energy independent manner.1 Tumor-committed cells are known to have accelerated metabolism, resulting in high glucose requirements and increased glucose uptake.2 In addition, malignant cells display upregulation of GLUT expression.3 High levels of GLUT1 and GLUT3 mark poor prognosis and survival rate for patients,4 and inhibition of GLUT expression in leukemia, endometrial and breast cancers have an anti-proliferative effect.5 GLUT antagonists suitable for clinical development are therefore of pharmaceutical interest in antineoplastic cancer therapy.6 Despite this importance, GLUTs are an under-represented drug target,7 partly because of the general difficulty in producing and handling α-helical membrane proteins. Moreover, a general platform to study this class of proteins is required for future drug development targeting GLUTs.8
Giant vesicles offer a synthetic cell format accessible to fluorescence microscopy, where function and dynamics of individual proteins can be studied under compositionally well-defined experimental conditions.9 So far the demonstration of functional membrane protein reconstitution in cell models has mainly revolved around ion channels whose activity can be measured straightforwardly by electrophysiology10, with ligand-binding assays to prove correctly folded protein11 or with fluorescent dye translocation through nonspecific pores.12 However, being able to measure the activity of transporters of uncharged substrates is challenging due to lack of an easy accessible readout. Study of these proteins is typically performed using radioisotopic substrates and liquid scintillation counting of material from native sources,13 cultured cells14 or nanoscale liposomes serving as simplified models of biological membranes.15 While uptake studies with isotope tracers give a high signal-to-noise, this approach typically requires 106 cells per data point, which precludes investigation of individual cells in heterogeneous cell populations.16 Moreover, the destructive nature of the method is incompatible with investigating the mechanism of action for membrane protein transporters within the lipid bilayer.16
We recently reported a novel protocol for the direct incorporation of membrane proteins during giant vesicle formation using an agarose swelling method.17 Work by Koenderink and co-workers,18 and others19 have demonstrated that soluble actin filaments can be functionally reconstituted internally in giant vesicles using a similar approach. Following on from this work, we hypothesized that it might be possible to simultaneously incorporate a number of different proteins, i.e. both soluble and membrane spanning proteins, into the same giant vesicle during the giant vesicle formation procedure. The work presented here demonstrates successful reconstitution of glucose transport machinery in which the intrinsic activity of the GLUT1 glucose transporter is coupled to an interior enzymatic reaction scheme yielding a fluorescent readout that is accessible by microscopy.
To produce pure GLUT1 protein, the DNA encoding the protein was introduced into the genome of the eukaryotic host protein expression system P. pastoris. A high throughput zeocin resistance screen was developed to identify high expressers of the protein (see Supplementary Information, Figure S1). Protein was expressed in the methylotrophic P. pastoris following methanol induction. Total membrane was isolated by ultracentrifugation after harvest and cell disruption. The isolated cell membranes were washed with sodium hydroxide to remove peripheral adhering proteins. GLUT1 was solubilized from the cell membranes with n-decyl-β-D-maltopyranoside detergent and purified by anion exchange chromatography and size exclusion chromatography. This procedure yielded large amounts (mg-scale) of highly pure GLUT1 (see Supplementary Information Figure S2 and Table S1). Circular dichroism spectroscopy was applied to confirm the correct conformation of purified GLUT1 in solution (Supplementary Information, Figure S3A). Assessment of storage conditions at various temperatures furthermore showed that the protein was stable upon freezing and thawing, and for several days at 4°C (Supplementary Information, Figure S3B).
Formation of GLUT1-reconstituted giant vesicles was carried out by the hydrogel-assisted swelling procedure as previously reported.17 Giant vesicles were made of 1,2-diphytanoyl-sn-glycero-3-phosphocholine (DPhPC) lipids, which form stable lipid bilayers with low permeability of small ionic and neutral molecules.20 The entire system was swollen with buffer containing all the components of an enzymatic glucose assay. In our reaction scheme, facilitated glucose transport first leads to conversion of internalized D-glucose to D-gluconolactone and hydrogen peroxide by glucose oxidase (GOx). Subsequently, nonfluorescent Amplex Red is converted to fluorescent resorufin, in the presence of hydrogen peroxide, by horseradish peroxidase (HRP) (Figure 1A). As a proof of principle for the model cell depicted in Figure 1A, we nonspecifically labeled GLUT1 in detergent stabilized aqueous solution with ATTO390 NHS dye and separated the unreacted dye from the protein. Giant vesicles were formed from hydrogel containing the ATTO390 labeled GLUT1 and with GOx, HRP and Amplex Red included in the rehydration buffer. The giant vesicle membrane is clearly fluorescent (cyan), showing that GLUT1 is incorporated into the lipid bilayer (Figure 1B, see also Supplementary Information, Figure S4 and Figure S5). A clear red fluorescent signal caused by formation of resorufin resulted from addition of glucose (Figure 1B and Figure 2). The interior fluorescent signal is confined within the encapsulating membrane (Figure 1B), demonstrating the presence of both functionally incorporated GLUT1 in the vesicle membrane and reconstituted active enzymes internally in the same vesicle.
Figure 1.
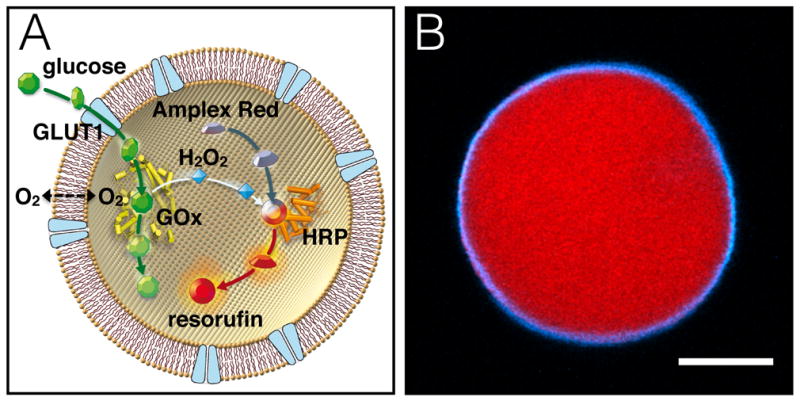
An artificial cell reconstituted with GLUT1 and an enzymatic glucose assay. (A) Illustration of reaction scheme. Facilitative glucose transport across the lipid bilayer by GLUT1 results in conversion of the internalized glucose to a fluorescent resorufin signal elicited by the combined action of GOx and HRP enzymes. (B) Fluorescent micrograph of a real model cell after addition of 4 mM glucose to the exterior medium. ATTO390-labeled GLUT1 (cyan) is clearly localized in the lipid bilayer membrane, while the interior fluorescent resorufin signal (red) is confined within the encapsulating membrane. Scale bar is 10 μm.
Figure 2.
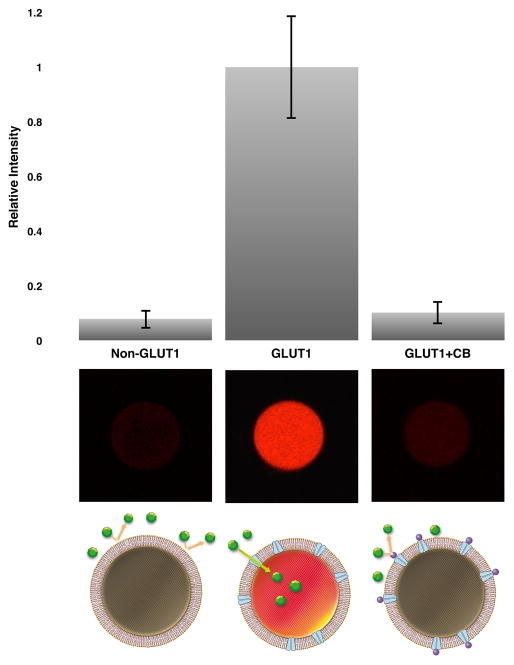
Monitoring glucose transport in giant vesicles. Endpoint measurements were performed before and 10 min after addition of 1 mM glucose. Functional reconstitution of GLUT1 and encapsulation of active enzymes elicit fluorescent resorufin after glucose addition. Non-GLUT1 containing but otherwise identical giant vesicle does not develop a prominent fluorescence signal after addition of glucose. Pre-incubation (5 min) of GLUT1-containing giant vesicle with 10 μM of CB prior to glucose addition blocks the transport activity of GLUT1. Results are n=10, error bars represent the standard deviation. Micrographs under the bar chart are representative of the visual fluorescence appearance for each experimental condition. Drawings illustrate the experimental design.
To confirm that GLUT1 is responsible for the transport of glucose across the vesicle membrane, a series of endpoint measurements were carried out in GLUT1-containing vesicles or in vesicles with no GLUT1 present (Figure 2). Measurements were carried out using a confocal microscope with image acquisition before and 10 min after addition of 1 mM glucose to the exterior medium. Non-GLUT1 vesicles served as controls and were formed from mock conditions with only buffer in the hydrogel. GLUT1-containing vesicles were formed with unlabeled protein in order to avoid potential artifacts. GLUT1-containing vesicles were measured alone or in the presence of inhibitor. Cytochalasin B (CB) is a well-characterized blocker of the GLUT1 substrate efflux site,21 and was added to GLUT1-vesicles before addition of glucose in order to inhibit glucose transport. Figure 2 show that GLUT1-swelled giant vesicles display a bright fluorescent interior 10 min after glucose addition. The averaged fluorescence intensity of GLUT1-containing giant vesicles is 12.5-fold higher than that of control vesicles. Hence, the prominent fluorescent signal in the GLUT1-swelled giant vesicles is due to functionally reconstituted GLUT1.
Addition of CB to GLUT1-vesicles caused a marked inhibition of glucose uptake, as evidenced by a 10-fold lower fluorescent signal compared to not having the inhibitor present (Figure 2). The result is in accordance with CB being a cell permeable inhibitor that readily crosses the lipid bilayer, and therefore a near complete blockage of GLUT1 activity is expected. CB did not affect the internal enzymatic reaction as measured by UV-Visible spectroscopy (Supplementary Information, Figure S6). This confirms that the diminished fluorescent signal in the presence of CB is caused by a GLUT1 specific inhibition.
To show the ability to differentiate between passive diffusion and GLUT1-mediated glucose transport we created a mixed population of giant vesicles. The lipid bilayers were labeled with fluorescent tracer dyes to identify the type of vesicle. Non-GLUT1 giant vesicles were labeled with ATTO488-DPPE fluorescent lipids (green), while the lipid bilayers of GLUT1-containing giant vesicles were labeled with ATTO390-DPPE (cyan). Figure 3 shows the clear distinction of the fluorescence intensity in a mixed giant vesicle population, demonstrating the ability to investigate individual giant vesicles in a heterogeneous population.
Figure 3.
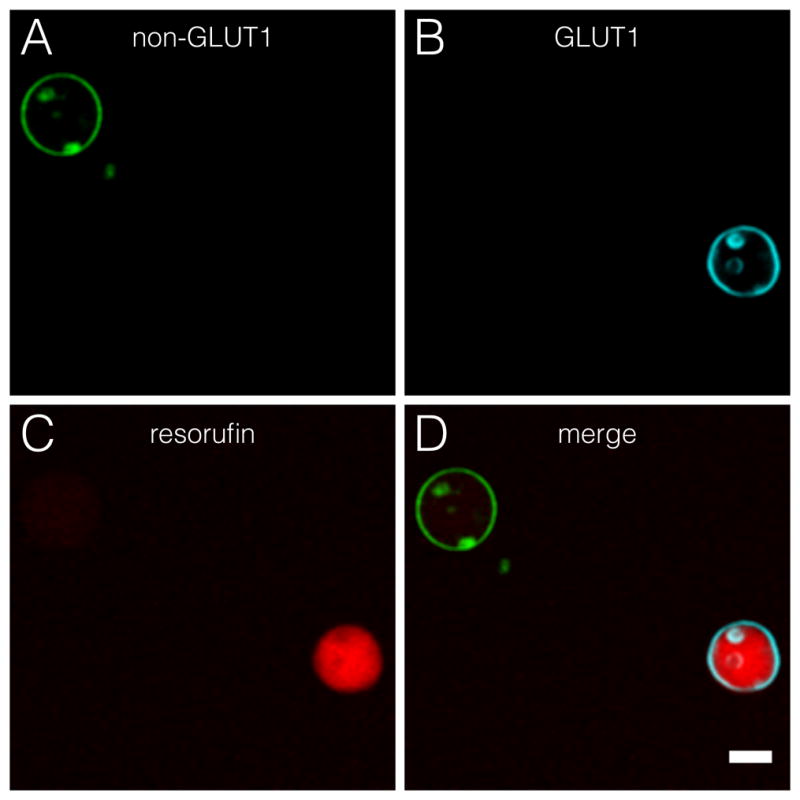
Mixed population of non-GLUT1 giant vesicles and vesicles with incorporated GLUT1 after glucose addition. Labeling with ATTO488- and ATTO390-conjugated DPPE lipids identify the type of giant vesicle. Micrograph is taken after addition of glucose. A) ATTO488-labeded lipid vesicle without GLUT1 (green). B) GLUT1-containing giant vesicle with ATTO390 lipid tracer (cyan). C) Shows that the resorufin signal is confined within the GLUT1-containing giant vesicle (red). D) Shows a merge of all micrographs. All micrographs were background normalized. Scale bar is 20 μm.
The rate of fluorescence increase, from GLUT1 containing giant vesicles, is glucose concentration dependent (Figure 4). Our results show that the rate constant is increased 1.7 times when the exterior glucose concentration is increased 4-fold, from 1 mM to 4 mM (Figure 4). GLUT1 has a Km around 1–5 mM for glucose under zero-trans uptake conditions.15c, 22 The glucose concentrations used in this study are therefore within the unsaturated range of glucose transport kinetics. GLUT-mediated glucose uptake follows Michaelis-Menten kinetics, but in our cell model the observed rate of glucose uptake is sigmoidal (Figure 4B). This is seen because the internalized glucose has to proceed through GOx and HRP catalyzed enzymatic reactions before the fluorescent signal develops (Supplementary Information, Figure S6).
Figure 4.
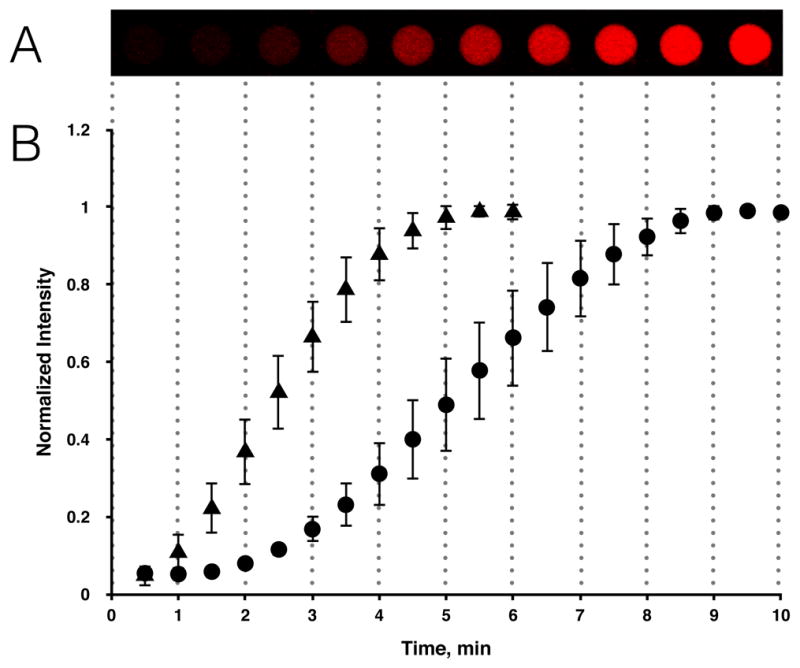
The rate of fluorescence increase in GLUT1-swelled giant vesicles exposed to different exterior glucose concentrations. (A) The image sequence shows a single giant vesicle at intervals of 1 min after addition of 1 mM glucose. (B) Plot depicting the mean pixel intensity of interior vesicle area normalized to the maximal fluorescence response observed upon saturation of resorufin for 1 mM glucose (circles, n=5) compared to the fluorescence response of 4 mM glucose (triangles, n=10). The mean linear rate constants were calculated to 0.286 min−1 for 4 mM glucose and 0.167 min−1 for 1 mM glucose, respectively. Error bars represent standard deviations.
In this study we have used the GLUT1 glucose transporter to demonstrate functional reconstitution of a polytopic intrinsic membrane protein. This twelve-membrane spanning α-helical protein belongs to the Major Facilitator Superfamily of solute carriers, which comprises the largest membrane protein class with thousands of sequenced members from organisms ranging from bacteria to mammals.23 Despite implications of members from this protein class in disorders including epilepsy, depression, osteoporosis, diabetes and cancer,8 very few approaches exist with which to study functional and regulatory properties of GLUTs and other solute carriers that transport uncharged substrates. As we demonstrate here, facilitated transport of soluble uncharged molecules, such as glucose, can be successfully coupled to multi-component enzymatic reactions inside giant vesicles with fluorescence as readout accessible by microscopy. These results strongly suggest that this approach might serve as a powerful platform for GLUT1 antagonist screening in the future.
Conclusions
In summary, this work describes the very first reconstitution of a purified glucose transporter into giant vesicles capable of reporting its function. Thus, we can finally start investigating the mechanism of action for these transporters within the lipid bilayer, using fluorescence microscopy. Besides providing a novel system where the transport and metabolism of glucose can be studied, we believe the broader implications are key for the development of future artificial cell mimics.24 Moreover, the system has central implications for future strategies aimed at preventing or modulating the activity of glucose transport, which is of specific interest for cancer research.
Supplementary Material
Acknowledgments
We thankfully acknowledge Dr. Samuel W. Cushman for extended discussions. The authors thank Troels Marstrand for his help with graphical illustrations. J.S.H. thanks the Wenner-Gren Foundations for supporting with a postdoctoral stipend and The Lars Hierta Memorial Foundation for support. K.E. thanks The Längmanska Cultural Foundation. J.R.T. and N.M. thank the National Institutes of Health (1R01GM093279) for support. The Swedish Research Council (VR-2011-2891), the Swedish Cancer Foundation and VINNOVA supported this work.
Footnotes
Electronic Supplementary Information (ESI) available: Protein expression and purification procedures, supporting methods, analysis of protein purity and folding, supporting figure S1–S6. See DOI: 10.1039/c000000x/
References
- 1.(a) Fehr M, Takanaga H, Ehrhardt DW, Frommer WB. Mol Cell Biol. 2005;25:11102. doi: 10.1128/MCB.25.24.11102-11112.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Cloherty EK, Levine KB, Carruthers A. Biochemistry. 2001;40:15549. doi: 10.1021/bi015586w. [DOI] [PubMed] [Google Scholar]
- 2.Warburg O. Science. 1956;124:269. [PubMed] [Google Scholar]
- 3.Szablewski L. Biochim Biophys Acta. 2013;1835:164. doi: 10.1016/j.bbcan.2012.12.004. [DOI] [PubMed] [Google Scholar]
- 4.(a) Krzeslak A, Wojcik-Krowiranda K, Forma E, Jozwiak P, Romanowicz H, Bienkiewicz A, Brys M. Pathol Oncol Res. 2012;18:721. doi: 10.1007/s12253-012-9500-5. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Matsushita K, Uchida K, Saigusa S, Ide S, Hashimoto K, Koike Y, Otake K, Inoue M, Tanaka K, Kusunoki M. J Pediatr Surg. 2012;47:1323. doi: 10.1016/j.jpedsurg.2011.12.007. [DOI] [PubMed] [Google Scholar]
- 5.(a) Liu T, Kishton RJ, Macintyre AN, Gerriets VA, Xiang H, Liu X, Dale Abel E, Rizzieri D, Locasale JW, Rathmell JC. Cell Death Dis. 2014;5:e1470. doi: 10.1038/cddis.2014.431. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Gautier EL, Westerterp M, Bhagwat N, Cremers S, Shih A, Abdel-Wahab O, Lutjohann D, Randolph GJ, Levine RL, Tall AR, Yvan-Charvet L. J Exp Med. 2013;210:339. doi: 10.1084/jem.20121357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhao Y, Butler EB, Tan M. Cell Death Dis. 2013;4:e532. doi: 10.1038/cddis.2013.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rask-Andersen M, Masuram S, Fredriksson R, Schioth HB. Mol Aspects Med. 2013;34:702. doi: 10.1016/j.mam.2012.07.015. [DOI] [PubMed] [Google Scholar]
- 8.Castell OK, Berridge J, Wallace MI. Angew Chem Int Ed Engl. 2012;51:3134. doi: 10.1002/anie.201107343. [DOI] [PubMed] [Google Scholar]
- 9.Hansen JS, Vararattanavech A, Vissing T, Torres J, Emneus J, Helix-Nielsen C. Chembiochem. 2011;12:2856. doi: 10.1002/cbic.201100537. [DOI] [PubMed] [Google Scholar]
- 10.(a) Holden MA, Bayley H. J Am Chem Soc. 2005;127:6502. doi: 10.1021/ja042470p. [DOI] [PubMed] [Google Scholar]; (b) Heron AJ, Thompson JR, Mason AE, Wallace MI. J Am Chem Soc. 2007;129:16042. doi: 10.1021/ja075715h. [DOI] [PubMed] [Google Scholar]; (c) Holden MA, Needham D, Bayley H. J Am Chem Soc. 2007;129:8650. doi: 10.1021/ja072292a. [DOI] [PubMed] [Google Scholar]; (d) Leptihn S, Thompson JR, Ellory JC, Tucker SJ, Wallace MI. J Am Chem Soc. 2011;133:9370. doi: 10.1021/ja200128n. [DOI] [PubMed] [Google Scholar]; (e) Bayley H, Cronin B, Heron A, Holden MA, Hwang WL, Syeda R, Thompson J, Wallace M. Mol Biosyst. 2008;4:1191. doi: 10.1039/b808893d. [DOI] [PMC free article] [PubMed] [Google Scholar]; (f) Hamada S, Tabuchi M, Toyota T, Sakurai T, Hosoi T, Nomoto T, Nakatani K, Fujinami M, Kanzaki R. Chem Commun (Camb) 2014;50:2958. doi: 10.1039/c3cc48216b. [DOI] [PubMed] [Google Scholar]
- 11.(a) Gutierrez MG, Malmstadt N. J Am Chem Soc. 2014 doi: 10.1021/ja507221m. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Estes DJ, Mayer M. Biochim Biophys Acta. 2005;1712:152. doi: 10.1016/j.bbamem.2005.03.012. [DOI] [PubMed] [Google Scholar]
- 12.(a) Dezi M, Di Cicco A, Bassereau P, Levy D. Proc Natl Acad Sci U S A. 2013;110:7276. doi: 10.1073/pnas.1303857110. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Harriss LM, Cronin B, Thompson JR, Wallace MI. J Am Chem Soc. 2011;133:14507. doi: 10.1021/ja204275t. [DOI] [PubMed] [Google Scholar]; (c) Stachowiak JC, Richmond DL, Li TH, Liu AP, Parekh SH, Fletcher DA. Proc Natl Acad Sci U S A. 2008;105:4697. doi: 10.1073/pnas.0710875105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.(a) Lowe AG, Walmsley AR. Anal Biochem. 1985;144:385. doi: 10.1016/0003-2697(85)90131-9. [DOI] [PubMed] [Google Scholar]; (b) Biochim Biophys Acta. 1986;857:146. doi: 10.1016/0005-2736(86)90342-1. [DOI] [PubMed] [Google Scholar]
- 14.(a) Harrison SA, Buxton JM, Czech MP. Proc Natl Acad Sci U S A. 1991;88:7839. doi: 10.1073/pnas.88.17.7839. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Madej MG, Sun L, Yan N, Kaback HR. Proc Natl Acad Sci U S A. 2014;111:E719. doi: 10.1073/pnas.1400336111. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Maher F, Davies-Hill TM, Simpson IA. Biochem J. 1996;315(Pt 3):827. doi: 10.1042/bj3150827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.(a) Baldwin JM, Gorga JC, Lienhard GE. J Biol Chem. 1981;256:3685. [PubMed] [Google Scholar]; (b) Sun L, Zeng X, Yan C, Sun X, Gong X, Rao Y, Yan N. Nature. 2012;490:361. doi: 10.1038/nature11524. [DOI] [PubMed] [Google Scholar]; (c) Wheeler TJ, Hinkle PC. J Biol Chem. 1981;256:8907. [PubMed] [Google Scholar]; (d) Chen CC, Kurokawa T, Shaw SY, Tillotson LG, Kalled S, Isselbacher KJ. Proc Natl Acad Sci U S A. 1986;83:2652. doi: 10.1073/pnas.83.8.2652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Barros LF, Bittner CX, Loaiza A, Ruminot I, Larenas V, Moldenhauer H, Oyarzun C, Alvarez M. J Neurochem. 2009;109(Suppl 1):94. doi: 10.1111/j.1471-4159.2009.05885.x. [DOI] [PubMed] [Google Scholar]
- 17.Hansen JS, Thompson JR, Helix-Nielsen C, Malmstadt N. J Am Chem Soc. 2013;135:17294. doi: 10.1021/ja409708e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tsai FC, Stuhrmann B, Koenderink GH. Langmuir. 2011;27:10061. doi: 10.1021/la201604z. [DOI] [PubMed] [Google Scholar]
- 19.(a) Carvalho K, Tsai FC, Lees E, Voituriez R, Koenderink GH, Sykes C. Proc Natl Acad Sci U S A. 2013;110:16456. doi: 10.1073/pnas.1221524110. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Weinberger A, Tsai FC, Koenderink GH, Schmidt TF, Itri R, Meier W, Schmatko T, Schroder A, Marques C. Biophys J. 2013;105:154. doi: 10.1016/j.bpj.2013.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.(a) Shinoda K, Shinoda W, Baba T, Mikami M. J Chem Phys. 2004;121:9648. doi: 10.1063/1.1806791. [DOI] [PubMed] [Google Scholar]; (b) Yamauchi K, Doi K, Kinoshita M, Kii F, Fukuda H. Biochim Biophys Acta. 1992;1110:171. doi: 10.1016/0005-2736(92)90355-p. [DOI] [PubMed] [Google Scholar]
- 21.Sultzman LA, Carruthers A. Biochemistry. 1999;38:6640. doi: 10.1021/bi990130o. [DOI] [PubMed] [Google Scholar]
- 22.(a) Carruthers A. Physiol Rev. 1990;70:1135. doi: 10.1152/physrev.1990.70.4.1135. [DOI] [PubMed] [Google Scholar]; (b) Kasahara T, Kasahara M. Biochem J. 1996;315(Pt 1):177. doi: 10.1042/bj3150177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.(a) Reddy VS, Shlykov MA, Castillo R, Sun EI, Saier MH., Jr FEBS J. 2012;279:2022. doi: 10.1111/j.1742-4658.2012.08588.x. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Cura AJ, Carruthers A. Compr Physiol. 2012;2:863. doi: 10.1002/cphy.c110024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.(a) Leduc PR, Wong MS, Ferreira PM, Groff RE, Haslinger K, Koonce MP, Lee WY, Love JC, McCammon JA, Monteiro-Riviere NA, Rotello VM, Rubloff GW, Westervelt R, Yoda M. Nat Nanotechnol. 2007;2:3. doi: 10.1038/nnano.2006.180. [DOI] [PubMed] [Google Scholar]; (b) Noireaux V, Libchaber A. Proc Natl Acad Sci U S A. 2004;101:17669. doi: 10.1073/pnas.0408236101. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Villar G, Graham AD, Bayley H. Science. 2013;340:48. doi: 10.1126/science.1229495. [DOI] [PMC free article] [PubMed] [Google Scholar]; (d) Walde P, Umakoshi H, Stano P, Mavelli F. Chem Commun (Camb) 2014;50:10177. doi: 10.1039/c4cc02812k. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.