Abstract
Botryllus schlosseri is a colonial ascidian with characteristics that make it an attractive model for studying immunology, stem cell biology, evolutionary biology, and regeneration. Transcriptome sequencing and the recent completion of a draft genome sequence for B. schlosseri have revealed a large number of genes, both with and without vertebrate homologs, but analyzing the spatial and temporal expression of these genes in situ has remained a challenge. Here we report a robust protocol for in situ hybridization that enables the simultaneous detection of multiple transcripts in whole adult B. schlosseri using Tyramide Signal Amplification in conjunction with digoxigenin- and dinitrophenol-labeled RNA probes. Using this protocol we have identified a number of genes that can serve as markers for developing and mature structures in B. schlosseri, permitting analysis of phenotypes induced in loss-of-function experiments.
Keywords: gene expression, ascidian, tyramide signal amplification, stem cells, allorecognition
Introduction
Botryllus schlosseri is a colonial marine chordate in the subphylum Tunicata, a group of invertebrates now thought to be the closest living relatives of the vertebrates (Delsuc et al., 2006). Each week, the adult body of B. schlosseri, known as a zooid, undergoes apoptosis and is replaced in an asexual budding process (Lauzon et al., 1992; Manni et al., 2007). The development of a bud in B. schlosseri requires two weeks, and as such, three simultaneous generations exist in a colony at all times (0-1 week old secondary buds, 1-2 week old primary buds, and 2-3 week old adult zooids). Up to four secondary buds may arise as thickenings of the epithelium of a primary bud, which then expand in size and pinch off to form hollow ellipsoids. Subsequent organogenesis and growth results in new bodies that are physically and genetically indistinguishable from the previous generation (Berrill, 1941).
Individuals in a B. schlosseri colony share a common extracorporeal circulatory system that terminates at the colony’s periphery in specialized structures called ampullae. When the ampullae from two colonies come into contact, a histocompatibility reaction dependent on a single locus results in one of two outcomes: (1) the vasculatures of these colonies fuse and circulating cells are freely exchanged or (2) fusion does not occur and inflammatory lesions may form between the colonies (Cima et al., 2006; De Tomaso et al., 2005; De Tomaso and Weissman, 2003; Rinkevich and Weissman, 1992b). Numerous studies have documented roles for putative receptors and ligands in the fusion/histocompatibility locus (the fuhc), and suggest that B. schlosseri allorecognition represents a protochordate “missing-self” immune response that is genetically encoded (McKitrick et al., 2011; Nyholm et al., 2006; Voskoboynik et al., 2013b).
Transfer of circulating cells between two fused B. schlosseri colonies produces a state of chimerism that may ultimately result in the complete replacement of the germline or specific somatic tissues of one colony (the “loser”) by that of the other (the “winner”) (Rinkevich and Weissman, 1992a; Sabbadin and Zaniolo, 1979; Stoner and Weissman, 1996). This phenomenon, known as parasitism, can be replicated by manually transplanting a FACS-isolated population of cells high in aldehyde dehydrogenase activity, a biomarker for stem cells in vertebrates (Kastan et al., 1990; Laird et al., 2005). Winner and loser phenotypes are hierarchical and heritable, suggesting that the ability of one colony to successfully parasitize another is likely to be genetically encoded (Stoner et al., 1999). The properties of parasitism and allorecognition make B. schlosseri an excellent model for addressing questions in the fields of immunology, stem cell biology, regeneration, and evolutionary biology.
Recent transcriptomic (Rodriguez et al., submitted) and genomic (Voskoboynik et al., 2013a) analyses of B. schlosseri have produced a treasure trove of genetic data and permitted the identification of a large number of B. schlosseri homologs of significant vertebrate genes. Notably, examination of the draft genome of B. schlosseri has revealed homologs of genes known to be critical for the development and function of the heart, eye, and immune system, some of which also play a role in human disease (Voskoboynik et al., 2013a). Given the natural suitability of B. schlosseri for addressing questions involving immunology and stem cell biology, forward and reverse genetic approaches based on this new genetic data will undoubtedly lead to important advances in these fields.
The ability to analyze spatial and temporal gene expression patterns is a critical tool required for any model organism. In this manuscript, we describe a robust in situ hybridization protocol for examining gene expression in B. schlosseri and use this protocol to analyze the expression patterns of several genes that are useful markers for developing and mature structures.
Results and Discussion
Current methods for in situ hybridization analysis in B. schlosseri are hampered by high levels of background staining in extracorporeal tissues (tunic and vasculature) and have frequently relied on sectioning to clearly reveal staining in tissues of interest (Brown et al., 2009; De Tomaso et al., 2005; Nyholm et al., 2006; Rinkevich et al., 2012; Rinkevich et al., 2013; Rosner et al., 2013). While effective for ascertaining the expression patterns of some transcripts, these methods have not been able to provide a global view of the expression domains of more than one transcript simultaneously. We sought to develop a protocol without these limitations that could be used to reproducibly analyze gene expression in whole adult B. schlosseri at all developmental stages.
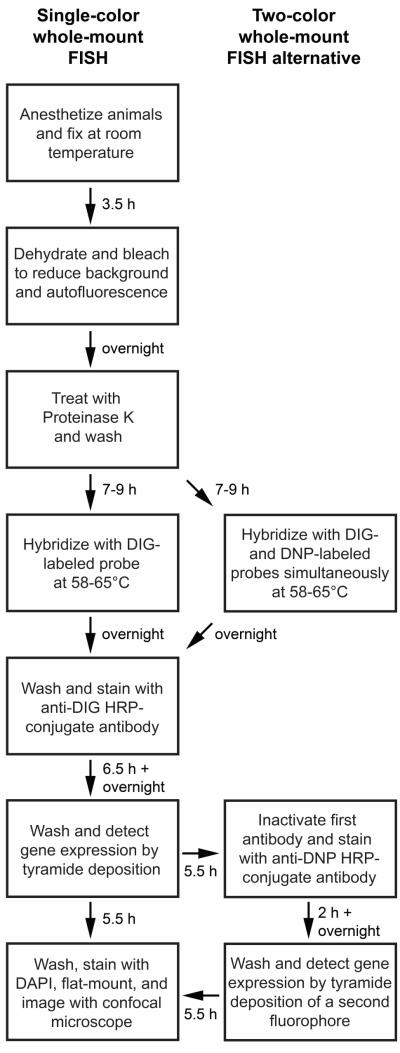
B. schlosseri are highly pigmented, and pigment cells are a major source of autofluorescent background signal in fixed samples. Different color morphs of B. schlosseri also exist, ranging from blue-black to orange-brown. We found that treatment of formaldehyde-fixed blue-black specimens with 6% hydrogen peroxide in methanol under bright light was able to nearly eliminate autofluorescence within 24 hours. Orange specimens were much more resistant to bleaching and were therefore not used in this study. Our in situ hybridization protocol utilizes several reagents designed to increase signal strength. First, we included a ribonuclease inhibitor in all solutions used prior to signal development. Second, we used Denhardt’s solution, which contains macromolecular crowding agents, in our hybridization solution since it provided a roughly two-fold increase in signal intensity (data not shown). Lastly, we used the highly sensitive Tyramide Signal Amplification method to produce strong fluorescent signals with minimal noise. Non-specific binding of antibodies to the tunic is an unavoidable problem in B. schlosseri, but detection of gene expression by fluorescent means circumvents this problem by allowing confocal imaging of samples. Furthermore, it permits the quantification of cell numbers via fluorescent nuclear counterstains. Our whole-mount fluorescent in situ hybridization (FISH) procedure, including alternative steps for detecting multiple transcripts simultaneously, is summarized in Figure 1.
Figure 1. Flowchart of single and double whole-mount fluorescent in situ hybridization (FISH) protocols.
Times listed next to arrows are the length of time needed to complete the step described in the preceding text box. A single-color FISH experiment requires approximately four days to complete, and a two-color FISH experiment requires approximately one day more.
In order to test the power of our technique and identify genes that could potentially be used as markers for different structures in B. schlosseri zooids and buds, we examined gene expression patterns in the Ciona intestinalis Adult In situ hybridization Database (http://bioinfo.s.chiba-u.jp/ciaid/) and rationalized that the B. schlosseri homologs of these genes may be expressed in similar tissues. We chose two genes for further analysis, Na+/K+ ATPase alpha 3 (atp1a3) and von Willebrand factor like 2 (vwfl2), because of their expression in the stomach and endostyle, respectively. We performed a translated BLAST (tblastn) of the Ciona protein sequences for these genes to the B. schlosseri transcriptome (Octopus Bioinformatics Server; http://octopus.obs-vlfr.fr/public/botryllus/blast_botryllus.php) and identified one putative homolog of atp1a3 (E-value = 0.0) and one putative homolog of vwfl2 (E-value = 1.0 × 10−45). We then cloned fragments of the B. schlosseri homologs of atp1a3 and vwfl2, generated antisense riboprobes, and examined their expression patterns in whole B. schlosseri using our FISH protocol. Our protocol yielded strong and specific staining patterns for both of these genes, and as we predicted, their expression domains mirrored those of their Ciona homologs. Expression of atp1a3 was present in an accessory organ to the stomach, the pyloric caecum (Colton, 1910), of both zooids (Figure 2a) and primary buds (Figure 2b), as well as tissues inside of and adjacent to the developing endostyle in primary buds (Figure 2b). Signal for vwfl2 was detected in the ventral glandular region of the endostyle of both zooids (Figure 2c) and developing primary buds (data not shown) suggesting that this region resembles Zone 2 of the Ciona endostyle both morphologically and with respect to gene expression (Sasaki et al., 2003). We also visualized both atp1a3 and vwfl2 expression simultaneously using sequential antibody detections and fluorescent depositions and did not observe any inappropriate colocalization of signal (Figure 2d), indicating that our FISH protocol can be used to reliably analyze multiple gene expression patterns simultaneously.
Figure 2. Expression patterns of markers for developing and adult structures in B. schlosseri.

(a-b) atp1a3 is expressed by cells of the pyloric caecum in the digestive system of the adult zooid (a) and the primary bud at stage C1 (b, arrow). atp1a3 expression is also present in part of the developing endostyle and adjacent tissues in primary buds at stage C1 (b, arrowheads indicate expression visible on the left side of the endostyle). (c) vwfl2 expression is present in the ventral portion of the endostyle of the zooid. (d) atp1a3 (red, arrow) and vwfl2 (green) can be simultaneously visualized in adult zooids by two-color FISH. atp1a3 expression is also visible in individual non-epithelial cells of unknown identity that are associated with the adult zooid. Scale bars indicate 50 μm.
Given the successful visualization of atp1a3 and vwfl2 expression, we attempted to identify additional markers for tissues and developing structures in B. schlosseri. By BLAST analysis, we identified homologs of Ciona fibroblast growth factor 11/12/13/14 (fgf11-14; E-value = 8.0 × 10−79), human par-6 family cell polarity regulator alpha (par6; E-value = 2.0 × 10−68), human runt-related transcription factor 1 (runx; E-value = 5.0 × 10−73) and human histone 3 (h3; E-value = 3.0 × 10−87). We detected expression of fgf11-14 in a subset of cells in the developing cerebral ganglia of stage C1 primary buds (Figure 3a) (Lauzon et al., 2002). Signal for par6 was present in all epithelial cells of the primary (Figure 3b) and secondary buds (Figure 3c). Expression of runx was restricted to the developing secondary buds. At stage A2, runx was expressed throughout the secondary bud (Figure 3d), but by stage B1, expression of runx was restricted to the junction between the secondary and primary bud (Figure 3e). Following morphogenesis of the double-vesicle, expression of runx was no longer detected in the secondary bud (Figure 3f). These data indicate that our FISH protocol is capable of visualizing developmentally relevant genes expressed by tissues in both the primary and secondary buds.
Figure 3. Expression patterns of markers for developing structures in B. schlosseri.

(a) fgf11-14 is expressed by a subset of cells in the developing cerebral ganglion (dashed line) of the primary bud at stage C1. (b-c) par6 expression is present in epithelial cells of the primary (b) and secondary (b and c) buds. Primary and secondary buds in panel b and secondary bud in panel c are demarcated by dashed lines. (d-f) runx is expressed in a stage-specific manner. At stage A2, expression of runx is present throughout the secondary bud (d, dashed lines). At stage B1, runx expression is restricted to the boundary between the primary and secondary buds (e, dashed lines). By stage C1, runx is no longer expressed by cells of the secondary bud (f). Faint staining inside the secondary buds at stages A2, B1, and C1 is indicative of trapping of reagents, and is present in a region devoid of cells (d-f). Background staining is also evident outside of the demarcated areas in panels d-f as a result of non-specific binding of reagents to the tunic and the surface of the primary and secondary buds. Solid lines in panels e and f define the portions of the secondary bud where runx expression is not detected. Scale bars indicate 50 μm.
Previous studies have detected expression of histone 3 during S-phase in mitotic cells (Gown et al., 1996; Muskhelishvili et al., 2003). We observed expression of B. schlosseri h3 in a non-ubiquitous manner in both primary and secondary buds (Figure 4a-c), suggesting that this gene may be marking those cells that are actively dividing. Interestingly, we also found expression of h3 in individual cells on the ventral side of the zooid (Figure 4d), in the vicinity of a putative hematopoietic and germline niche known as the “cell islands” (Rinkevich et al., 2013).
Figure 4. Expression pattern of marker for mitotically active cells in B. schlosseri.
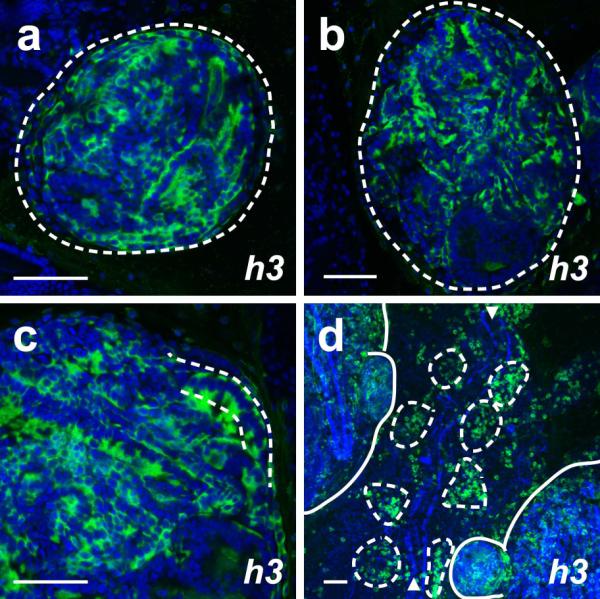
(a-c) h3 is expressed by a portion of the cells in the primary and secondary bud at stages A1 (a) and A2 (b). The majority of cells in the secondary bud at stage A2 strongly express h3 (c). Primary buds in panels a and b and the secondary bud in panel c are demarcated by dashed lines. (d) h3 expression is also present in clusters of blood cells known as “cell islands” on the ventral side of the zooid at stage C1 (surrounded by dashed lines) on both sides of the endostyle (marked by arrowheads). Other non-epithelial cells of unknown identity outside of these clusters also exhibit h3 expression. The primary and secondary buds in panel d are demarcated by solid lines. Scale bars indicate 50 μm.
Our FISH protocol also reliably detected gene expression in germline and germline support cells. The B. schlosseri vasa gene has been previously described (Brown et al., 2009; Rosner et al., 2009), and we identified a homolog of B. primigenus Piwi (piwi; E-value = 8.0 × 10−151). We found expression of these germline markers in clusters of primordial germ cells associated with the secondary and primary buds (Figure 5a-b). A novel TGFβ ligand and marker for germline support cells, tgfβ-f, was also identified during our analysis of the B. schlosseri transcriptome. Expression of tgfβ-f was present in the primitive follicle cells of juveniles (Figure 5c) as well as the follicle cells surrounding maturing oocytes and testes of primary buds (Figure 5d). Using our two-color FISH protocol, we could simultaneously visualize germline tissues and their support cells in non-fertile (Figure 5e) and fertile (Figure 5f) animals.
Figure 5. Expression patterns of markers for germline and germline-associated cells in B. schlosseri.
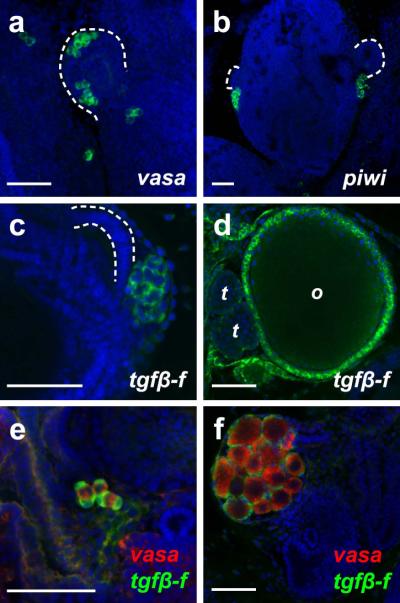
(a) vasa is expressed by clusters of germ cells associated with the secondary bud (dashed line) and just posterior to the secondary bud at stage B2. (b) At stage B1, piwi-positive germ cells are present posterior to the secondary buds (dashed lines). (c-d) tgfβ-f expression is detected in clusters of primitive follicle cells posterior to the secondary bud (dashed lines) at stage A2 in non-fertile juveniles (c) and in follicle cells surrounding the maturing oocytes (o) and testes (t) in the primary buds of fertile animals (d). (e-f) Two-color FISH can be used to simultaneously visualize vasa expression (red) and tgfβ-f expression (green) in non-fertile (e) and fertile (f) animals. Scale bars indicate 50 μm.
Taken together, these data show that our FISH protocol is capable of detecting gene expression throughout the asexual reproduction cycle in a wide array of tissues (zooid, primary bud, secondary bud, and germline). Compared to previous techniques used in B. schlosseri, this protocol provides a strong and specific signal without the need to first embed and section specimens. Combination of this technique with antisense knockdown approaches, pharmaceutical inhibitions, and microsurgical manipulations in the future will greatly increase our understanding of the molecular mechanisms underlying B. schlosseri development and physiology.
Materials and Methods
Animals
B. schlosseri colonies were raised on glass slides in a mariculture system with circulating 0.5 μm filtered seawater at 18-20 °C. Animals were fed daily with live algae. Developmental staging of animals was performed by examination of primary and secondary buds under a dissecting microscope (Lauzon et al., 2002).
Cloning of genes used for in situ hybridization analysis
B. schlosseri homologs of genes of interest were identified by tblastn searches of the B. schlosseri transcriptome (http://octopus.obs-vlfr.fr/public/botryllus/blast_botryllus.php) using human or Ciona (when available) protein sequences. If possible, primer pairs were designed to amplify a 1000 bp fragment of each transcript.
Total RNA was isolated from B. schlosseri colonies with the NucleoSpin RNA II kit (MN, 740955) and SuperScript II Reverse Transcriptase (Life Technologies, 18064-014) was used to synthesize cDNA primed by random primers (Life Technologies, 48190-011). PCR was performed with Advantage cDNA Polymerase (Clontech, 639105) and products were cloned into the pGEM-T Easy vector (Promega, A1360).
Synthesis of antisense RNA probes
Constructs were linearized and approximately 1 μg of template was used in an in vitro transcription reaction with SP6 or T7 RNA polymerase (Roche, 10810274001, 10881767001). For digoxigenin labeling of probes, DIG RNA labeling mix (Roche, 11277073910) was added to the reaction. Dinitrophenol labeling was accomplished by adding dinitrophenol-11-UTP (Perkin Elmer, NEL555001EA), UTP, CTP, GTP, and ATP (Roche, 11277057001) to the reaction at final concentrations of 0.35 mM, 0.65 mM, 1 mM, 1 mM, and 1 mM, respectively. RNases were inhibited with Protector RNase Inhibitor (Roche, 03335399001). The transcription reaction was incubated at 37 °C for 2 hours and then RNase-free DNase I (Roche, 04716728001) was added to remove the plasmid template. Probes were then precipitated twice with LiCl/ethanol to maximize removal of unincorporated labeled nucleotides (Streit and Stern, 2001).
Antisense probes used in this study include atp1a3, fgf11-14, h3, par6, piwi, runx, tgfβ-f, vasa, and vwfl2. Primer sequences used for amplifying these genes are listed in Table 1.
Table 1.
Summary of B. schlosseri sequences and primer pairs used for in situ hybridization analysis.
| Transcript name |
Accession number | Forward (F) and Reverse (R) Primers (5′ to 3′) | Size of amplified fragment (bp) |
|---|---|---|---|
| atp1a3 | KJ747407 | F: GACCCTGACCGCGAAACGTATG R: CGAGGTCCTGGCGAACACAATC |
1,051 |
| fgf11-14 | KJ747408 | F: CAGCAGCTCAAGGGTGTGAAGAC R: CCTCTTCTTCACCTCTCCCGAACC |
492 |
| h3 | KJ747409 | F: ATGGCGAGAACAAAGCAAACCGC R: CTGGCGAGCTGGATGTCCTTC |
386 |
| par6 | KJ747410 | F: TCCGTGAATAGGATCAGTGTAGGTGAT R: CTTACAAAATGGACAAGCAGTCTTTGG |
212 |
| piwi | KJ747411 | F: CGTGTGGATGGAATTGAGTGGGATATG R: CAATTTTTGTGGCGACGCTCATAAC |
865 |
| runx | KJ747412 | F: CTACCCAACACATTCATACCACGC R: CGTTAGCGGAGTCTTCTCTCTTCAC |
1,049 |
| tgfβ-f | KJ747413 | F: CATGGATTCTTGCAGGAGAG R: GTTACCGAACTTTCTGACCC |
735 |
| vasa | KJ747414 | F: AGGCACTATGATTCAGCCTGTG R: ATCATAATCACCCGTCTCGCG |
976 |
| vwfl2 | KJ747415 | F: CATCCGCTTGCAGAGACGTTG R: GGACAGACCGACTCATCACATTG |
985 |
Whole-mount in situ hybridization
B. schlosseri were anesthetized with 0.02% Tricaine (TCI, T0941) in seawater for 10 minutes, and then fixed with 4% formaldehyde in 0.5 M NaCl, 0.1 M MOPS pH 7.5 for 3 hours at room temperature with agitation. We detected an improvement in signal when an RNase inhibitor (Sigma, R7397-30ML) was added to all solutions from fixation to post-antibody washes inclusive. Fixed samples were then dehydrated with methanol and bleached overnight in 6% H2O2 in methanol under direct light. Once all pigment was eliminated, samples were rehydrated stepwise into PBST and permeabilized by treatment with 10 μg/ml proteinase K (Roche, 03115879001) in PBST for 30 minutes at room temperature. After permeabilization, a postfixation step was performed with 4% formaldehyde in PBST for 20 minutes at room temperature. Samples were then washed with PBST and incubated in hybridization buffer (65% formamide, 5X SSC, 1X Denhardt’s solution, 0.1% Tween-20, 5 mg/ml torula yeast RNA, 50 μg/ml heparin) without probe for 4-6 hours at 58-65 °C, followed by overnight incubation with digoxigenin-labeled riboprobes diluted in hybridization buffer at 58-65 °C. For simultaneous detection of two transcripts, samples were also incubated with dinitrophenol-labeled riboprobes.
Following hybridization, unbound probes were washed away by a series of low-salt washes and samples were incubated in blocking solution (PBST, 5% heat-inactivated horse serum, 2 mg/ml bovine serum albumin) for 4 hours at room temperature with agitation. HRP-conjugated anti-digoxigenin antibody (Roche, 11207733910) diluted 1:1000 in blocking solution was then added and the samples were incubated overnight at 4 °C with agitation. After incubation with antibody, the samples were washed extensively in PBST + 2 mg/ml bovine serum albumin and transferred to PBS. Digoxigenin-labeled probes were then detected by fluorophore deposition using the TSA Plus System (Perkin Elmer, NEL753001KT).
If detection of a second transcript was needed, the HRP-conjugated anti-digoxigenin antibody was inactivated by incubating samples in PBS + 2% H2O2 + 0.1% Triton-X100 for 1 hour at room temperature with agitation. Following washing in PBST, dinitrophenol-labeled probes were detected with an HRP-conjugated anti-dinitrophenol antibody (Perkin Elmer, FP1129) diluted 1:200 in blocking solution. Samples were washed extensively in PBST + 2 mg/ml bovine serum albumin to remove unbound antibody, and probes were detected with a second fluorophore deposition using the TSA Plus System. Lastly, samples were stained with DAPI (Life Technologies, D1306) and flat-mounted with VECTASHIELD (Vector Labs, H-1000). Imaging of labeled samples was performed using an Olympus FLV1000S Spectral Laser Scanning Confocal.
A detailed step-by-step protocol is provided in the Supplementary Materials.
Supplementary Material
Acknowledgements
We would like to thank Mike Caun for his expert care of the De Tomaso laboratory mariculture facility and Daryl Taketa for critical comments on this manuscript. We would also like to thank Dr. Andrew Gracey of the University of Southern California for help with the assembly of the B. schlosseri transcriptome. This research was supported by grants from the NIH to A.W.D. (AI041588, AG037966). We also acknowledge the use of the NRI-MCDB Microscopy Facility and the Spectral Laser Scanning Confocal supported by the Office of The Director, National Institutes of Health of the NIH under Award # S10OD010610. A.D.L. was supported by an NIH NRSA fellowship (F32 GM108227) and by a Tri-Counties Blood Bank Postdoctoral Fellowship.
NIH Grant Support: This work was supported by the following grants from the NIH: GM108227, AI041588, AG037966, S10OD010610.
References
- Berrill NJ. The Development of the Bud in Botryllus. Biological Bulletin. 1941;80:169–184. [Google Scholar]
- Brown FD, Tiozzo S, Roux MM, Ishizuka K, Swalla BJ, De Tomaso AW. Early lineage specification of long-lived germline precursors in the colonial ascidian Botryllus schlosseri. Development. 2009;136:3485–3494. doi: 10.1242/dev.037754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cima F, Sabbadin A, Zaniolo G, Ballarin L. Colony specificity and chemotaxis in the compound ascidian Botryllus schlosseri. Comp Biochem Physiol A Mol Integr Physiol. 2006;145:376–382. doi: 10.1016/j.cbpa.2006.07.017. [DOI] [PubMed] [Google Scholar]
- Colton HS. The “Pyloric Gland” of the Ascidian Botryllus: An Organ of Excretion? Biological Bulletin. 1910;19:35–54. [Google Scholar]
- De Tomaso AW, Nyholm SV, Palmeri KJ, Ishizuka KJ, Ludington WB, Mitchel K, Weissman IL. Isolation and characterization of a protochordate histocompatibility locus. Nature. 2005;438:454–459. doi: 10.1038/nature04150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Tomaso AW, Weissman IL. Initial characterization of a protochordate histocompatibility locus. Immunogenetics. 2003;55:480–490. doi: 10.1007/s00251-003-0612-7. [DOI] [PubMed] [Google Scholar]
- Delsuc F, Brinkmann H, Chourrout D, Philippe H. Tunicates and not cephalochordates are the closest living relatives of vertebrates. Nature. 2006;439:965–968. doi: 10.1038/nature04336. [DOI] [PubMed] [Google Scholar]
- Gown AM, Jiang JJ, Matles H, Skelly M, Goodpaster T, Cass L, Reshatof M, Spaulding D, Coltrera MD. Validation of the S-phase specificity of histone (H3) in situ hybridization in normal and malignant cells. J Histochem Cytochem. 1996;44:221–226. doi: 10.1177/44.3.8648081. [DOI] [PubMed] [Google Scholar]
- Kastan MB, Schlaffer E, Russo JE, Colvin OM, Civin CI, Hilton J. Direct demonstration of elevated aldehyde dehydrogenase in human hematopoietic progenitor cells. Blood. 1990;75:1947–1950. [PubMed] [Google Scholar]
- Laird DJ, De Tomaso AW, Weissman IL. Stem cells are units of natural selection in a colonial ascidian. Cell. 2005;123:1351–1360. doi: 10.1016/j.cell.2005.10.026. [DOI] [PubMed] [Google Scholar]
- Lauzon RJ, Ishizuka KJ, Weissman IL. A cyclical, developmentally-regulated death phenomenon in a colonial urochordate. Dev Dyn. 1992;194:71–83. doi: 10.1002/aja.1001940109. [DOI] [PubMed] [Google Scholar]
- Lauzon RJ, Ishizuka KJ, Weissman IL. Cyclical generation and degeneration of organs in a colonial urochordate involves crosstalk between old and new: a model for development and regeneration. Dev Biol. 2002;249:333–348. doi: 10.1006/dbio.2002.0772. [DOI] [PubMed] [Google Scholar]
- Manni L, Zaniolo G, Cima F, Burighel P, Ballarin L. Botryllus schlosseri: a model ascidian for the study of asexual reproduction. Dev Dyn. 2007;236:335–352. doi: 10.1002/dvdy.21037. [DOI] [PubMed] [Google Scholar]
- McKitrick TR, Muscat CC, Pierce JD, Bhattacharya D, De Tomaso AW. Allorecognition in a basal chordate consists of independent activating and inhibitory pathways. Immunity. 2011;34:616–626. doi: 10.1016/j.immuni.2011.01.019. [DOI] [PubMed] [Google Scholar]
- Muskhelishvili L, Latendresse JR, Kodell RL, Henderson EB. Evaluation of cell proliferation in rat tissues with BrdU, PCNA, Ki-67(MIB-5) immunohistochemistry and in situ hybridization for histone mRNA. J Histochem Cytochem. 2003;51:1681–1688. doi: 10.1177/002215540305101212. [DOI] [PubMed] [Google Scholar]
- Nyholm SV, Passegue E, Ludington WB, Voskoboynik A, Mitchel K, Weissman IL, De Tomaso AW. fester, A candidate allorecognition receptor from a primitive chordate. Immunity. 2006;25:163–173. doi: 10.1016/j.immuni.2006.04.011. [DOI] [PubMed] [Google Scholar]
- Rinkevich B, Douek J, Rabinowitz C, Paz G. The candidate Fu/HC gene in Botryllusschlosseri (Urochordata) and ascidians’ historecognition--an oxymoron? Dev Comp Immunol. 2012;36:718–727. doi: 10.1016/j.dci.2011.10.015. [DOI] [PubMed] [Google Scholar]
- Rinkevich B, Weissman IL. Allogeneic resorption in colonial protochordates: consequences of nonself recognition. Dev Comp Immunol. 1992a;16:275–286. doi: 10.1016/0145-305x(92)90002-t. [DOI] [PubMed] [Google Scholar]
- Rinkevich B, Weissman IL. Incidents of rejection and indifference in Fu/HC incompatible protochordate colonies. J Exp Zool. 1992b;263:105–111. doi: 10.1002/jez.1402630111. [DOI] [PubMed] [Google Scholar]
- Rinkevich Y, Voskoboynik A, Rosner A, Rabinowitz C, Paz G, Oren M, Douek J, Alfassi G, Moiseeva E, Ishizuka KJ, Palmeri KJ, Weissman IL, Rinkevich B. Repeated, long-term cycling of putative stem cells between niches in a basal chordate. Dev Cell. 2013;24:76–88. doi: 10.1016/j.devcel.2012.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosner A, Moiseeva E, Rabinowitz C, Rinkevich B. Germ lineage properties in the urochordate Botryllus schlosseri - from markers to temporal niches. Dev Biol. 2013;384:356–374. doi: 10.1016/j.ydbio.2013.10.002. [DOI] [PubMed] [Google Scholar]
- Rosner A, Moiseeva E, Rinkevich Y, Lapidot Z, Rinkevich B. Vasa and the germ line lineage in a colonial urochordate. Dev Biol. 2009;331:113–128. doi: 10.1016/j.ydbio.2009.04.025. [DOI] [PubMed] [Google Scholar]
- Sabbadin A, Zaniolo G. Sexual Differentiation and Germ-Cell Transfer in the Colonial Ascidian Botryllus-Schlosseri. Journal of Experimental Zoology. 1979;207:289–304. [Google Scholar]
- Sasaki A, Miyamoto Y, Satou Y, Satoh N, Ogasawara M. Novel endostyle-specific genes in the ascidian Ciona intestinalis. Zoolog Sci. 2003;20:1025–1030. doi: 10.2108/zsj.20.1025. [DOI] [PubMed] [Google Scholar]
- Stoner DS, Rinkevich B, Weissman IL. Heritable germ and somatic cell lineage competitions in chimeric colonial protochordates. Proc Natl Acad Sci U S A. 1999;96:9148–9153. doi: 10.1073/pnas.96.16.9148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoner DS, Weissman IL. Somatic and germ cell parasitism in a colonial ascidian: possible role for a highly polymorphic allorecognition system. Proc Natl Acad Sci U S A. 1996;93:15254–15259. doi: 10.1073/pnas.93.26.15254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Streit A, Stern CD. Combined whole-mount in situ hybridization and immunohistochemistry in avian embryos. Methods. 2001;23:339–344. doi: 10.1006/meth.2000.1146. [DOI] [PubMed] [Google Scholar]
- Voskoboynik A, Neff NF, Sahoo D, Newman AM, Pushkarev D, Koh W, Passarelli B, Fan HC, Mantalas GL, Palmeri KJ, Ishizuka KJ, Gissi C, Griggio F, Ben-Shlomo R, Corey DM, Penland L, White RA, Weissman IL, Quake SR. The genome sequence of the colonial chordate, Botryllus schlosseri. Elife. 2013a;2:e00569. doi: 10.7554/eLife.00569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voskoboynik A, Newman AM, Corey DM, Sahoo D, Pushkarev D, Neff NF, Passarelli B, Koh W, Ishizuka KJ, Palmeri KJ, Dimov IK, Keasar C, Fan HC, Mantalas GL, Sinha R, Penland L, Quake SR, Weissman IL. Identification of a colonial chordate histocompatibility gene. Science. 2013b;341:384–387. doi: 10.1126/science.1238036. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.