Abstract
Insulin plays an extensively characterized role in the control of sugar metabolism, growth and homeostasis in a wide range of organisms. In vertebrate chordates, insulin is mainly produced by the beta cells of the endocrine pancreas, while in non-chordate animals insulin-producing cells are mainly found in the nervous system and/or scattered along the digestive tract. However, recent studies have indicated the notochord, the defining feature of the chordate phylum, as an additional site of expression of insulin-like peptides. Here we show that two of the three insulin-like genes identified in Ciona intestinalis, an invertebrate chordate with a dual life cycle, are first expressed in the developing notochord during embryogenesis and transition to distinct areas of the adult digestive tract after metamorphosis. In addition, we present data suggesting that the transcription factor Ciona Brachyury is involved in the control of notochord expression of at least one of these genes, Ciona insulin-like 2. Lastly, we review the information currently available on insulin-producing cells in ascidians and on pancreas-related transcription factors that might control their expression.
Keywords: ascidian, Brachyury, Ciona, cis-regulatory module, insulin, notochord, pancreas, transcription factor
INTRODUCTION
Insulin, insulin-like growth factors (IGFs), insulin-like (INSLs) and relaxin (RLN) genes compose an evolutionarily conserved superfamily and encode a variety of regulatory polypeptides related by shared amino acid subdomains (e.g., Conlon, 2001). Among the main conserved residues are a few cysteines that establish intra- and inter-chain disulfide bonds. Disulfide bonds are necessary for the stability and the biological activity of insulin and its related peptides (Vinther et al., 2013), and enable them to control a wide range of cellular processes, including nutrient homeostasis and regulation of cell growth and survival in a variety of tissues, as well as aging (Fulzele and Clemens, 2012; Werner and Leroith, 2014; Edmondson et al., 2003; Kenyon, 2011). In widely different organisms, cells producing insulin and related peptides have been described as specialized neurons in the nervous system, both central and peripheral, or as endocrine cells that can be either disseminated in various regions of the digestive tract or concentrated in the parenchyma of the pancreas or its evolutionary equivalents. Collectively, insulin- and other hormone-producing cells belong to the neuroendocrine system (NES) and are crucial components of the brain-gut axis (Conlon et al., 1988; Konturek et al., 2004). In invertebrate protostomes, the neuronal fraction of the NES is usually predominant and most insulin-producing cells are found in the brain, as is the case in insects and molluscs (Luo et al., 2012; Smit et al., 1998). However, in echinoderms, non-chordate deuterostomes, immunolocalization experiments have shown that the gut of the pluteus larva contains cells expressing insulin-related peptides (de Pablo et al., 1988). Subsequent studies confirmed the presence of two genes of the insulin/IGF/INSL/RLN superfamily in the S. purpuratus genome, Sp-IGF1 and Sp-IGF2 (Burke et al., 2006). Related analyses in another echinoderm, the sea star Pisaster ochraceous, also pointed at the existence of cells expressing insulin-related genes in the developing gut (Wilson and Falkmer, 1965). In chordates, the main group of deuterostomes, insulin-producing cells increase in number in various areas of the digestive tracts, and ultimately, in vertebrates, are found grouped in the islets of Langerhans of the pancreas alongside other endocrine cell types (Madsen, 2007). Interestingly, in addition to being expressed in cells of the NES, in a few chordates insulin and insulin-like genes have been found in the notochord. This is the case for two of the three insulin-like genes in a urochordate, the ascidian Ciona intestinalis. Below we summarize the early reports of insulin-producing cells in the ascidian digestive organs, we describe in detail the expression of the three Ciona insulin-like genes before and after metamorphosis, and suggest the possible involvement of the transcription factor Brachyury in the regulation of the expression of one of these genes in the developing notochord. We conclude by reviewing the current knowledge on the expression of pancreas-related transcription factors and co-factors in Ciona.
LARVAL AND POST-METAMORPHOSIS ENDODERMAL DERIVATIVES, DIGESTIVE STRUCTURES AND INSULIN-PRODUCING CELLS IN CIONA
Cell fate and function of endoderm precursors in ascidians
The organization of the larval body plan of solitary ascidians is remarkably similar to a simplified version of the embryos of higher chordates. The roughly 2,600 cells that are found in a larva of the ascidian genera Halocynthia or Ciona are organized into a few main tissues, including epidermis, notochord, muscle, endoderm, nervous tissue and sensory organs, and in the mesenchyme and its specialized regions, namely trunk lateral cells and trunk ventral cells (Passamaneck and Di Gregorio, 2005; Munro et al., 2006). In particular, the notochord invariantly contains 40 cells, is overlaid by a dorsal nerve cord composed of ~400 cells, and is flanked on each side by muscle cells, amounting to a total of 36 cells in Ciona and 40 cells in Halocynthia (Satoh, 2014). Ventral to the notochord, and occupying a large part of the larval trunk, which is also regarded as the ascidian “head”, are ~500 endodermal cells, which are topologically subdivided into trunk endoderm and endodermal strand and have been compared to a primordial non-functional pharynx and to the hypochord of fish and amphibians, respectively (Lofberg and Collazo, 1997). Interestingly, the endodermal cells of the larvae of many ascidian species do not form a functional digestive tract, although they express evolutionarily conserved markers of endoderm differentiation, such as alkaline phosphatase (Whittaker, 1977; Nishida and Kumano, 1997), and orthologs of the essential endodermal transcription factors TTF-1 and Foxa2 (Ristoratore et al., 1999; Di Gregorio et al., 2001). Thus, differently from larvae of non-chordate marine invertebrates such as the sea urchin pluteus, which is able to feed on algae and diatoms and can survive up to several weeks without metamorphosing (Hinegardner, 1969), most ascidian larvae are unable to feed themselves and are usually able to swim for only a day or two before settling to undergo metamorphosis (Satoh, 2014). During early embryonic development, between the 32-cell and the 64-cell stage, the endodermal precursors, mostly vegetal blastomeres, become fate-restricted. The endoderm fate determination is coordinated by maternal determinants that are pre-localized in the egg cytoplasm (Nishida, 1996). As a consequence, presumptive endodermal blastomeres are capable of developing into endoderm even when isolated from the remainder of the embryo (Reverberi and Minganti, 1946). A key trigger of the early determination of the ascidian endoderm is the transactivator beta-catenin, the main mediator of the canonical Wnt signaling pathway (Imai et al., 2000). The nuclear translocation of beta-catenin in the vegetally-located endodermal precursors is also required to initiate the induction of the notochord cell fate in adjacent blastomeres (Nakatani and Nishida, 1994). The consequent segregation of the three germ layers, ectoderm, mesoderm and endoderm, unravels as the effect of a combinatorial code of nuclear beta-catenin activation; blastomeres with no nuclear beta-catenin activity will become part of the ectoderm, blastomeres that maintain stable high levels of nuclear beta-catenin activity will give rise to endoderm, and cells that change their nuclear levels of beta-catenin from high to low will form the notochord (Hudson et al., 2013). In addition, high levels of activation of the FGF signaling pathway are required for the endoderm induction in A-line cells (Shi and Levine, 2008; Lemaire, 2009). Ci-FoxA-a, which encodes an ortholog of the Foxa2 transcription factor, is one of the first genes expressed in early endodermal precursors (Di Gregorio et al., 2001) under the regulation of beta-catenin; morpholino-mediated ablation of its expression results in loss of endodermal markers (Imai et al., 2006). Ci-FoxA-a in turn regulates expression of two additional transcription factors, Ci-Otx and Ci-Lhx3; the latter activates genes required for endoderm differentiation in the larva, and along with Ci-FoxA-a activates Ci-Otx, which is believed to be required for the activation of Ci-GATA-a and other genes that control differentiation of the adult endoderm (Imai et al., 2006). After the specification of the presumptive endoderm is finalized, endodermal precursors divide at a steady rate until their final number of ~500 is reached, around the tailbud stages. Endodermal cells remain yolky and maintain an undifferentiated appearance until metamorphosis (Hirano and Nishida, 2000).
Metamorphosis and formation of digestive organs
At the time of metamorphosis extensive changes begin for the body plan as a whole, and for the endodermal cells as well, as the organogenesis of the juvenile digestive tract commences and the animal begins to feed. Metamorphosis is a complex, multi-step process and its timing is tightly regulated (Nakayama-Ishimura et al., 2009). At metamorphosis, the main chordate features of the larval body plan, i.e. the notochord, dorsal neural tube and ventral endoderm, are either lost or extensively rearranged, and are replaced by other chordate hallmarks, such as the endostyle, which is regarded as a primordial thyroid gland (Roche et al., 1962; Ristoratore et al., 1999; Sasaki et al., 2003), and pharyngeal gill slits (Tanaka et al., 1996). In addition, a rudimentary heart and different kinds of blood cells begin to develop, as well as the gonad and its annexes, oviduct and spermiduct (Satoh, 2014). Neither the notochord nor the tail muscles contribute to the formation of the adult body, and both these tissues and the tail are largely reabsorbed. As the tail degenerates and the rotation of the body axis is completed the notochord and muscle cells move to the trunk, and eventually their remnants are found near the holdfast (Sato et al., 1997). Remarkably, however, most cells of the larval nervous system, with the exception of those located in the nerve cord, become part of the adult CNS, which is composed of neural ganglion and neural gland, and are incorporated into structures that develop from the neural gland, including a ciliated duct and funnel, and a dorsal strand (Manni et al., 2005). In particular, some of the ependymal (non-nervous) cells of the larva can transdifferentiate into neurons in the adult, thus acting as stem-like cells (Horie et al., 2011). On the surface of the embryo, epidermal cells begin to express the cellulose synthase gene Ci-CesA beginning at the neurula stage, and at metamorphosis these cells become part of the cellulose-based tunic (Nakashima et al., 2004; Sasakura et al., 2005), along with part of the mesenchymal cells (Hirano and Nishida, 1997). Trunk ventral cells divide after metamorphosis to generate the heart (Davidson and Levine, 2003; Satou et al., 2004) and the atrial siphon muscles (Stolfi et al., 2010). Trunk lateral cells originate blood cells, part of the body wall muscle and the cells that surround the gill slits (Hirano and Nishida, 1997), and act as neural crest-like migratory cells (Jeffery et al., 2008). In the case of the endoderm, it is noteworthy that two cells of the endodermal strand, expressing the conserved marker vasa (CiVH), migrate from the tail to the gonad anlage to originate the primordial germ cells in the adult (Takamura et al., 2002). The remaining cells of the endodermal strand and their descendants give rise to the intestine, while the trunk endoderm folds into tubular structures that will develop into esophagus, stomach and endostyle (Nakazawa et al., 2013). Therefore, altogether the larval endoderm participates in the formation of a number of adult structures, including endostyle, pharyngeal band, dorsal tubercle, peribranchial epithelium, as well as the digestive organs: the pharynx, also indicated as branchial (or pharyingeal) sac or branchial basket, esophagus, stomach, intestine and pyloric gland (Chiba et al., 2004). A small hepatopancreas has been described in Halocynthia (Hirano and Nishida, 2000) and its existence has been more recently implied in Ciona (Hammond et al., 2005).
Detection of peptide- and insulin-producing cells in Ciona: early studies
In adult ascidians, sea water incoming from the oral siphon is conveyed to the pharynx, where it is moved through the gill slits, also called stigmata, by the ciliary activity of the cells of the pharyngeal wall. As the water moves through the slits, it oxygenates the blood by coming in close contact with small blood vessels that run inside the thin pharyngeal wall (Satoh, 2014). At the same time, food particles contained in the sea water, such as algal fragments, are trapped by the abundant mucus secreted by the endostyle and moved to the esophagus by the cilia of the cells bordering the gill slits. The fully developed esophagus is about one centimeter in length and is lined by a columnar epithelium rich in mucus and ciliated cells. In the esophagus, the food particles are conveyed towards the stomach, while they begin to be compacted into thin, mucus-laden strings, or “ropes”, that in transparent ascidians can be seen transiting through the digestive organ and being expelled from the atrial siphon (Yonge, 1925; Millar, 1953). For this reason, the esophagus was believed to mainly serve as a conduit and a compactor for food particles, until insulin-immunoreactive cells were described in the esophageal epithelium in Styela clava, along with a second type of predicted endocrine cells (Bevis and Thorndyke, 1978). These observations suggested that this part of the digestive tract might also serve a secretory function. This hypothesis is corroborated by our recent results, presented below.
After moving through the esophagus, the food reaches the stomach, a sac-like ovoidal organ containing in its cavity about 40 ridges that increase its inner surface, similar in function to the rugae found in the stomach of vertebrates (Millar, 1953). Three main cell types have been described in the ascidian stomach: vacuolated cells, which are believed to be equivalent to the absorptive cells of the vertebrate intestine, ciliated mucous cells, and secreting cells, which have been tentatively indicated in the past as the site of accumulation of digestive enzymes (Thorndyke, 1977) and were recently proven to express a trypsin-type protease and a chymotrypsin-like protease (gene models: KH.L97.6 and KH.S425.3, respectively; Dehal et al., 2002; Satou et al., 2008; Ogasawara et al., 2002; http://bioinfo.s.chiba-u.jp/ciaid/). An early comparative study on the physiology of digestion tested the ability of extracts prepared from the Ciona gut to digest different carbohydrates, and it revealed that the only carbohydrates that were processed were starch, glycogen and sucrose (Yonge, 1925). In support of these results, later studies showed the presence of alpha-amylases in the gills of Ciona intestinalis and other ascidians (Fiala-Medioni and Pequignat, 1975), and currently at least one gene encoding a putative alpha-amylase has been annotated in the Ciona genome (gene model: KH.C5.659). Additional genes encoding predicted enzymes involved in carbohydrate metabolism have been found in Ciona, including glucosidases, maltase-glycoamylases and sucrase-isomaltase; in particular, expressed sequence tags (ESTs) counts indicate expression of a glucosidase (gene model: KH.L170.27) gene in the pyloric gland of the adult ascidian (Satou et al., 2002, 2005; http://ghost.zool.kyoto-u.ac.jp/reference_kh.html). Extensive cytochemical and immunohistological studies in Ciona have revealed the presence of several gastroenteric and neurohormonal peptides in the gut and in the nervous system of adult ascidians. Positive immunolocalization signals were obtained in both the neural complex and the digestive tract of Ciona for bombesin (whose mammalian homologs are gastrin-releasing peptide and neuromedin B), pancreatic polypeptide, substance P, somatostatin, secretin and neurotensin, while beta-endorphin, prolactin, and vasoactive intestinal peptide and motilin appeared restricted to the nervous system; serotonin was originally reported only in the digestive tract (Fritsch et al., 1980), but more recent studies have shown its presence in the visceral ganglion of metamorphosed Ciona larvae (De Bernardi et al., 2006). Together, these early immunolocalization experiments allowed the creation of a precise map that pinpoints the location of cells that are immunoreactive for each peptide in both gut and neural complex (Fritsch et al., 1982). Recent post-genomic studies have confirmed some of these early finding by identifying two Ciona orthologs of neurotensin, Ci-ntlp-A and Ci-ntlp-B, which are both expressed in the neural complex; Ci-ntlp-B is also expressed in the digestive tract (Kawada et al., 2011). A hybrid of gastrin and cholecystokinin, called cionin, has also been identified in the neural ganglion of Ciona (Johnsen and Rehfeld, 1990). However, genes for somatostatin, pancreatic polypeptide, ghrelin, amylin and glucagon seem to be missing from the Ciona genome, although in the latter case members of the glucagon superfamily have been identified in the solitary ascidian Chelyosoma productum (McRory and Sherwood, 1997a). Collectively, these findings fueled the hypothesis that Ciona and other ascidians occupy an intermediate position between invertebrates, which predominantly rely on neurosecretion to regulate homeostasis, and vertebrates, which perform these functions through endocrine cells that are either scattered along the intestine or aggregated into large glands. In agreement with this scenario, reactivity to ox and chicken insulin was detected in extracts prepared from both the stomach and the intestine of Ciona (Davidson et al., 1971) and subsequent studies reported immunoreactivity to members of the insulin superfamily in cells of the digestive tract of this species (Reinecke et al., 1993, 1999). In addition, insulin-immunoreactive cells and a network of insulin-immunoreactive fibers were reported in the Ciona neural ganglion (O’Neil et al., 1986). Within the digestive tract, double immunofluorescence experiments showed that polyclonal antisera against human IGF1 and human insulin both reacted with epithelial cells of the gastrointestinal mucosa, showing overlap in ~95% of the cells, while no cells that were exclusively reactive to the insulin antiserum were found (Reinecke et al., 1993). Similar overlap was seen in the neural ganglion and nerve fibers, although insulin reactivity was preferentially observed in the fibers rather than in the neuronal bodies (Reinecke et al., 1993). Follow-up studies that compared the distribution of IGF1 and insulin immunoreactivity to that for relaxin, another member of the insulin/IGF/INSL/RLN superfamily, revealed the partial coexistence of reactivity to IGF1 and relaxin in cells of the Ciona ovary, whereas no insulin-reactive cells were detected in this tissue. Conversely, no positive signal for relaxin was detected in the intestine (Reinecke et al., 1999). These results resemble those obtained in the amphioxus Branchiostoma lanceolatum, where cells positive to both IGF1 and insulin antisera were detected in the gastrointestinal tract as well as in the CNS (Reinecke et al., 1999).
INSULIN-LIKE GENES IN CIONA INTESTINALIS
Genomic organization
Insulin is synthesized as a multi-domain pre-pro-peptide in which the A- and B- chains are connected by a C-peptide, and an N-terminal signal peptide is present (Conlon, 2001). Post-translational processing by a signal peptidase removes the signal peptide and converts pre-proinsulin into proinsulin as it moves to the endoplasmic reticulum; subsequently, endoproteolytical convertases cleave the intervening C-peptide, leaving the A and B chains linked by disulfide bonds in the mature insulin chain. Instead, IGFs retain the C-peptide and contain additional domains, namely chains D and E (Reinecke and Collet, 1998). Invertebrate insulin-like genes constitute large and divergent families that share the basic insulin structure and contain a number of members that vary among different species; for example, seven insulin-like genes have been described in Drosophila (Nässel, 2002) and three in Hydra (Fujisawa and Hayakawa, 2012). After the first assembly of the Ciona intestinalis genome was completed (Dehal et al., 2002), an insulin-like gene was identified and named Ci-insulin (Satou et al., 2003); however, reports from another ascidian, Chelyosoma productum, in which two insulin-like genes had been found (McRory and Sherwood, 1997b), prompted further searches for members of the insulin/IGF/INSL/RLN superfamily in Ciona. Gene clustering and phylogenetic analyses revealed two additional insulin-like genes in Ciona intestinalis, which were named Ci-INS-L1 (the former Ci-insulin), Ci-INS-L2 and Ci-INS-L3 (Olinski et al., 2006a, 2006b). All three genes have been mapped to chromosome 2q, one of the largest Ciona chromosomes (Shoguchi et al., 2006). Ci-INS-L2 (gene model: KH.C2.313) and Ci-INS-L3 (gene model: KH.C2.1012) are tightly clustered in tandem orientation within ~7.7 kb, while Ci-INS-L1 (gene model: KH.C2.75) is found ~4.4 Mb away (Fig. 1a). Both genomic regions have been shown to display conserved synteny with related gene clusters in human and other vertebrate chromosomes (Olinski et al., 2006a). Ci-INS-L1 is an ortholog of human INSLs and RLNs, and a cleavage site for a signal peptide, along with a putative processing site, were predicted within its putative amino acid sequence (Olinski et al., 2006b). A similar structure was predicted for the product of Ci-INS-L3, which is expected to retain the C-peptide and mature into a single-chained IGF-like peptide. Instead, the predicted Ci-INS-L2 peptide contains, in addition to a signal peptide cleavage sequence, two processing sites flanking the C-peptide and a suboptimal target sequence for proprotein convertase, which together make Ci-INS-L2 more related to insulin genes (Olinski et al., 2006b).
Figure 1. Genomic organization and embryonic expression of Ciona insulin-like genes.
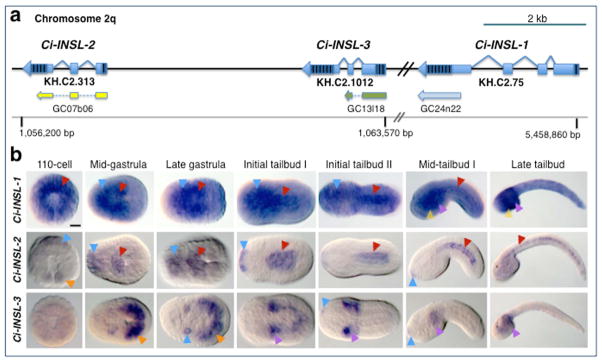
a: Schematic representation of the regions of Ciona intestinalis chromosome 02q that contain the genomic loci of Ci-INSL-2, Ci-INSL-3 and Ci-INSL-1. Genes are represented as light-blue bars (exons) and angled lines (introns); striped areas indicate untranslated regions. Current gene model identifiers are indicated underneath each gene. The ESTs used for in situ hybridizations are depicted below each gene model in yellow, green, and light blue, respectively, along with the coordinates that allow their identification within the Ciona intestinalis Gene Collection (Satou et al., 2002). A grey horizontal line symbolizes the chromosomal DNA and reports the location of essential mapping landmarks in base pairs (bp). b: Microphotographs of Ciona intestinalis whole-mount embryos at the stages indicated on top of each column, hybridized in situ with digoxygenin-labeled antisense RNA probes synthesized from the EST clones shown in a, essentially as previously described (Oda-Ishii and Di Gregorio, 2007). Gene names are indicated on the left side of each row. 110-cell embryos are shown in vegetal views, the remaining embryos are shown with anterior to the left. Arrowheads point at representative regions of larger structures, and are color-coded as follows: red, notochord; blue, nervous system and primordium of the adhesive organ, which represents the anterior-most structure in tailbud embryos; orange, muscle precursors; purple, mesenchyme; yellow, endoderm. Scale bar: ~25 microns.
Expression patterns of the Ciona insulin-like genes
Whole-mount in situ hybridization (WMISH) data had indicated that Ci-INS-L1 is expressed in numerous territories, and in particular in endodermal strand, notochord, nerve cord and peripheral nervous system (Satou et al., 2001). Expression of Ci-INS-L2 had been reported in muscle, notochord, epidermis, endoderm, nerve cord, mesenchyme, presumptive palps and other territories at the initial and early tailbud stages, however no expression was detected in the notochord of middle and late tailbuds (Satou et al., 2001), and in larvae expression Ci-INS-L2 had only been observed in the epidermis (Kusakabe et al., 2002). No in situ expression data were available for Ci-INS-L3. The lack of expression data for Ci-INS-L3 and our laboratory’s long-standing interest in notochord gene expression prompted us to further investigate the expression of this gene family, in both the pre- and post-metamorphosis body plan. Our results confirmed the widespread early expression of Ci-INS-L1 and highlighted the strong signal in notochord, trunk endoderm and mesenchyme cells throughout the tailbud stages, as well as the down-regulation of notochord expression in late tailbuds (Fig. 1b, top row). In the case of Ci-INS-L2, after detecting a transient signal in muscle precursors at the 110-cell stage, we observed sustained expression in notochord cells and their progenitors beginning at gastrulation, accompanied by transient expression in the primordium of the adhesive organ (Fig. 1b, middle row). We also observed expression of Ci-INS-L3 in muscle, mesenchyme and neural precursors during gastrulation, and, during the tailbud stages, in mesenchyme cells and a few areas of the presumptive adhesive organ (Fig. 1b, bottom row). Occasionally, what seemed a very faint, transient signal was visible in the notochord at the tailbud stage when relaxed hybridization conditions were used (data not shown).
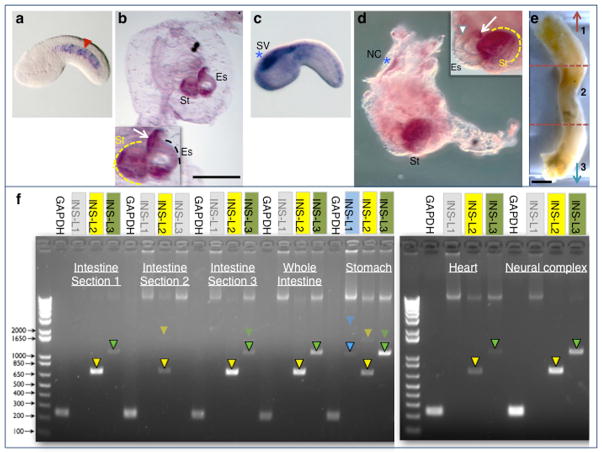
On the basis of its closer similarity to insulin genes, Ci-INS-L2 was selected for further in situ hybridization studies in juveniles. Using the same probe employed for the in situs on embryos, we detected expression of Ci-INS-L2 in esophagus and stomach of early juveniles (Fig. 2a,b; inset in b). The expression of Ci-INS-L2 in the stomach partially overlaps with the expression of Ci-Na+/K+-transporting ATPase alpha3-1 (aka ATP1A1/2/3/4, gene model: KH.L157.5), which we used as a positive control for the in situs on juveniles (Fig. 2c,d) on the basis of its strong expression in the stomach (Ogasawara et al., 2002) and for its reported presence in pancreatic beta cells of vertebrates, where it is believed that inhibition of this ion pump by glucose triggers insulin secretion (Owada et al., 1999). In juveniles, we observed expression of Ci-Na+/K+-transporting ATPase alpha3-1 in the neural complex (CNS), which is formed in part by cells that originally belonged to the sensory vesicle (see above; blue asterisks in Fig. 2c,d), in most of the surface of the stomach, and in a small part of the esophagus (Fig. 2d, and inset).
Figure 2. Expression of Ciona insulin-like genes after metamorphosis.
a: Mid-tailbud I Ciona whole-mount embryo hybridized in situ with the Ci-INSL-2 digoxygenin-labeled antisense RNA probe. Red arrowhead: notochord, showing expression. b: Whole-mount Ciona stage 4 juvenile (Chiba et al., 2004) hybridized in situ with the Ci-INSL-2 digoxygenin-labeled antisense RNA probe, as in a. Expression is seen in esophagus (Es) and stomach (St) (both magnified in inset). The stomach is partially encircled by a yellow dashed line; the esophagus is contoured by a dashed black line. c: Mid-tailbud I Ciona whole-mount embryo hybridized in situ with the Ci-Na+/K+-transporting ATPase alpha3-1 (aka ATP1A1/2/3/4, gene model: KH.L157.5) digoxygenin-labeled antisense RNA probe. Blue asterisk: sensory vesicle (SV), showing expression. d: Whole-mount Ciona juvenile, around stage 6 (Chiba et al., 2004), hybridized in situ with the Ci-Na+/K+-transporting ATPase alpha3-1 probe. Expression is seen in part of the stomach and possibly in part of the rim of the esophagus (inset), as well as in the neural complex (NC). The blue asterisks in c and d refer to the persistence of cells of the sensory vesicle in the adult neural complex. Inset shows a higher magnification view of the esophagus and stomach, as seen on a different plane of focus. The white arrow points at the rim of the esophagus; the white arrowhead highlights the ventral groove of the esophagus, an anatomical point of reference. The stomach is partially encircled by a yellow dashed line. e: Dissected intestine of an adult Ciona, which was used for the detection of the expression of Ci-INSL-1, Ci-INSL-2 and Ci-INSL-3 by RT-PCR. The intestine was arbitrarily subdivided into three sections of similar length, indicated as 1-3 and separated by dashed lines. The red arrow points towards the stomach, the teal arrow towards the anus. Scale bars: in b, 200 microns; in e, 2 millimiters. f: Gel electrophoresis of RT-PCR products obtained from RNAs extracted from different regions of the intestine and the stomach of an adult Ciona individual, and from hearts and neural complexes pooled from multiple animals. Gel lanes are labeled on top with the names of the genes that were monitored. Gene names were colored in transparent grey to symbolize lack of expression. Unlabeled lanes contain a molecular weight marker; the sizes of its most informative bands are indicated on the left side. Expression of the housekeeping gene GAPDH (Glyceraldehyde 3-phosphate dehydrogenase) was monitored in each experiment as a positive control. Expected sizes of PCR products: GAPDH, 250 bp; INS-L1, 1,074 bp for spliced transcript (blue arrowhead) and ~2.2 kb for genomic (faded blue arrowhead); INS-L2, 710 bp for spliced transcript (yellow arrowheads) and ~1.55 kb for genomic (faded yellow arrowheads); INS-L3, 1,069 bp, for spliced transcript (green arrowheads) and ~1.47 kb for genomic (faded green arrowheads). Conditions for RT-PCR: RNA was extracted by dissecting adult animals and by processing organ tissues as recommended by the Qiagen RNeasy kit (QIAGEN). DNAse treatment was used to minimize genomic contamination, according to the manufacturer’s instructions. cDNA synthesis was then conducted for each sample using the designed primers and standard RT-PCR protocol with Superscript III reverse transcriptase (Invitrogen), according to the manufacturer’s instructions. PCR conditions included an initial denaturation step at 94°C for 2.5 minutes, and 27 cycles as follows: 94°C for 15 seconds, 60°C for 30 seconds, 72°C for 30 seconds. After a last step at 72°C for 10 minutes, samples were cooled at 16°C and then stored at −20°C. cDNA products were subsequently used to amplify specific regions of the GAPDH control and of each Ci-INSL gene. Forward and reverse primers for GAPDH and each Ci-INSL gene were designed in the first and in the last exons.
The sequences of the primers were:
GAPDH.F 5′-GCAGGAGCTGGCATTGCAC-3′ and GAPDH.R 5′-ACAGTAAGATTGCATGCAACAG-3′ for GAPDH; IL-1.V1.F 5′-TCTTTTATAGGACACAGCGCATTA-3′ and IL-1.V2.R 5′-TAAACGACCATCACAGAACAATG-3′ for Ci-INSL-1; IL-2.V1.F 5′-CAGCTTTGTAACAGGACAGTGAAT-3′ and IL-2.Int.R1 5′-TGTGCAAAGCTGTGTTTTCCAAGC-3′ for Ci-INSL-2; IL-3.V1.F 5′-AGATAAAGGGATTAGGCAGAAGTG-3′ and IL-3.V2.R 5′-GCTGTCGTACTGGGGTGTAAATA-3′ for Ci-INSL-3.
For the PCR amplification of GAPDH and Ci-INS-L genes, an initial 2.5-minute cycle at 94°C preceded 30 cycles as follows: 94°C for 30 seconds, 55°C for 30 seconds and 68°C for 3 minutes. After a last step at 68°C for 30 minutes, samples were cooled at 16°C for at least 15 minutes before gel electrophoresis.
As a next step, expression of all three genes was analyzed in juveniles and adults by RT-PCR. All three genes were found to be expressed in juveniles (data not shown). The results of representative RT-PCR experiments on adults are shown in Fig. 2f. Transcripts for Ci-INS-L2 were present, although at variable levels, in all the adult tissues that were analyzed (Fig. 2f); interestingly, expression of this gene varied in different tracts of the adult intestine (Fig. 2e, f). Differently from what was reported in early immunofluorescence studies (Reinecke et al., 1993), Ci-INSL-2 is the only one of the three genes that we could detect in section 2 of the adult intestine (Fig. 2f). Expression of Ci-INS-L3 peaked in stomach and neural complex, but was not detected in the middle section of the intestine (Fig. 2e, f). Ci-INS-L1 was only detected in the stomach, at seemingly low levels (Fig. 2f).
Other components of the insulin pathway in Ciona
The Ciona genome contains an insulin receptor gene, Ci-insrr (gene model: KH.L8.8), which was reported as weakly and transiently expressed in the notochord of initial tailbud embryos and seemed to switch from the notochord to the nervous system at later stages (Satou et al., 2001, 2003). Together with our results on the notochord expression of Ci-INS-L2, these data suggest that, at least during the initial tailbud stage, Ciona insulin and/or IGF-like proteins might work in an autocrine fashion. It is known that upon binding its receptor, insulin induces, through its receptor tyrosine kinase, the phosphorylation of insulin receptor substrate proteins (IRSs); in turn, IRSs can associate with the phosphatidylinositol 3-kinase (PI 3-kinase) to activate it, as well as with several other substrates, thus triggering a variety of cascading intracellular signals (White, 1997). The Ciona genome contains a single irs gene, annotated as Ci-irs1/2/4, whose expression is still unknown.
In addition to genes directly related to insulin and its receptors, recent genome annotations indicate that Ciona possesses three insulin-like growth factor binding protein (igfbp) genes, igfbp1/2, igfbp1/3/4/5/6 and igfbp2/3/4/5 (http://www.aniseed.cnrs.fr), whose expression is still uncharacterized. On the other hand, it is noteworthy that in addition to seemingly lacking evident glucagon orthologs, Ciona appears to be devoid of adipokinetic hormone, which in Drosophila and other organisms is structurally and functionally related to glucagon. Together, these observations suggest the hypothesis that insulin-like genes in Ciona might predominantly regulate growth and longevity, and play a rather minor role in glucose metabolism.
EVOLUTIONARILY CONSERVED MARKERS OF PANCREAS DEVELOPMENT AND ACTIVATORS OF INSULIN GENE EXPRESSION ACROSS CHORDATES
The regulation of Ci-INSL-2 expression after metamorphosis remains to be investigated. This provides us with the opportunity to concisely review the current knowledge gathered in Ciona on the main evolutionarily conserved transcription factors required for pancreas formation, endocrine pancreas specification, and insulin secretion, and to hypothesize possible regulatory mechanisms. A list of the main transcription factors that control pancreas development and insulin expression across chordates is provided in Table 1, and some of the most relevant information on candidate activators of insulin/IGF gene expression in Ciona is illustrated in Fig. 4. As we mentioned above, Ci-FoxA-a is one of the earliest transcription factors expressed in all endoderm precursors during embryogenesis (Di Gregorio et al., 2001); however, in juveniles this gene is exclusively detected in the endostyle (Hiruta et al., 2005); while no expression has been reported in other endodermal derivatives, EST counts indicate that some transcripts are found in the neural complex (http://www.aniseed.cnrs.fr) (Table 1). These observations suggest that even though Ci-FoxA-a plays a conserved role in endoderm differentiation in ascidian embryos, it may not be involved in the direct control of insulin/IGF gene expression, at least in juveniles. If this hypothesis were confirmed, it would pinpoint an important difference between Ciona and vertebrates, where Foxa2, to which Ci-FoxA-a is closely related, controls expression of pancreatic duodenal homeobox 1 (Pdx1) in pancreatic beta-cells (Lee et al., 2002).
Table 1.
Transcription factors and co-factors involved in pancreas formation and/or insulin gene expression, and their Ciona orthologs.
| Vertebrate transcription factor/cofactor, name and family | Function in endoderm and/or pancreas | References | Cionaintestinalis ortholog | Ciona gene model | Expression in Ciona embryos | Expression in Ciona aftermetamorphosis |
|---|---|---|---|---|---|---|
| Sox17 (SRY-box, HMG) | Endoderm formation and specification; organ lineage segregation | Spence et al., 2009 | Ci-SoxF, Ci-Sox15/17/18 | KH.C7.407 | Epidermis, mesenchyme (Imai et al., 2004) | Stomach, other regions of the digestive tract (Ogasawara et al., 2002) |
| HNF-3β/Foxa2 (Forkhead box) | Endoderm specification, gut formation | Dufort et al., 1998 | Ci-fkh/HNF-3β/FoxA-a | KH.C11.313 | Notochord, CNS, endoderm (Di Gregorio et al., 2001) | Endostyle (Hiruta et al., 2005) |
| Ipf1/Pdx1 aka Idx1, Xlox (ParaHoxhomeodomain) | Pancreas formation, endocrine cells differentiation, insulin expression | Stoffers et al., 1997; Malecki et al., 2006 | Ci-Ipf1 | KH.C14.554 | Ubiquitous in early embryos (Imai et al., 2004); CNS, mesenchyme, adhesive organ in larvae (Corrado et al., 2001) | Not analyzed |
| Neurogenin-3/HLH-4 (bHLH) | Specification of all endocrine cells | Gradwohl et al., 2000 | Ci-neurogenin | KH.C6.129 | Neural precursors, tail epidermis, tail neurons, visceral ganglion, nerve cord (Imai et al., 2004, 2009) | Ectoderm of the atria (Mazet et al., 2005) |
| HNF-6/OC-1 (Cut homeodomain) | Endocrine cells differentiation | Jacquemin et al., 2000 | Ci-Onecut | KH.C6.185 | Sensory vesicle, visceral ganglion, nerve cord (Moret et al., 2005; Imai et al., 2009) | Not analyzed |
| Mnx/Hlxb9/HB9 (homeodomain) | Development of dorsal pancreatic lobe; beta-cells formation | Harrison et al., 1999 | Ci-Mnx/Ci-HB9 | KH.L128.12 | Notochord andneural precursors, muscle, visceral ganglion, tail neurons (Hudson et al., 2003) | Endostyle, stomach (Ogasawara et al., 2002) |
| Islet-1 (LIM-homeodomain) | Formation of pancreatic mesenchyme, dorsal exocrine cells differentiation, development of all endocrine cells | Ahlgren et al., 1997 | Ci-isl1 | KH.L152.2 | Nerve cord, visceral ganglion, primordium of adhesive organ, notochord, sensory organs (Giuliano et al., 1998). Trunk ventral cells and their descendants (Stolfi et al., 2010) | Epidermis (Ogasawara et al., 2002) Atrial siphon muscle precursors, heart(Wang et al., 2013) |
| Ldb1 (co-factor) (LIM-domain binding protein) | Co-regulator of Islet-1 target genes, including insulin, MafA, Arx | Hunter et al., 2013 | Ci-Ldb1/2 | KH.C14.544 | Not analyzed | Not analyzed |
| Sox9 (SRY-box, HMG) | Maintenance of pancreatic progenitors | Seymour et al., 2007 | Ci-SoxE, Ci-Sox10/15/8/9 | KH.L154.42 | Whole embryo, stronger in nervous system and mesenchyme (Imai et al., 2004) | Pyloric gland (Wada et al., 2010) |
| Hex (homeodomain) | Specification of ventral pancreas | Bort et al., 2004 | Ci-Hex | KH.L171.10 | Mesenchyme, trunk lateral cells (Imai et al., 2006) | Not detected http://bioinfo.s.chibau.jp/ciaid/ |
| Hes-1 (bHLH) | Endocrine cells differentiation | Jensen et al., 2000 | Ci-Hes, Ci-HER, Ci-E(spl)/hairy-c | KH.L34.9 | Whole embryo (Imai et al., 2004) | Not analyzed |
| Beta2/NeuroD1 (bHLH) | Islets maturation, control of beta-cell number | Naya et al., 1997 | Ci-atonal, Ci-ath5, Ci-HAND1/2 | KH.C8.175 | Epidermal neurons (Imai et al., 2004) | Not analyzed |
| Pax6 (Paired-box) | Differentiation of alpha-and beta-cells | Sander et al., 1997 | Ci-Pax6 | KH.C9.68 | Neural precursors, nerve cord, sensory vesicle, visceral ganglion (Imai et al., 2004, 2009; Irvine et al., 2008) | Not analyzed |
| Nkx6.1 (NK homeodomain) | Control of islet size and beta-cell number | Taylor et al., 2013 | Ci-Nkx6.1, NK6-1/2/3 | KH.C5.502 | Epidermis, visceral ganglion, nerve cord (Imai et al., 2009) | Not analyzed |
| HNF-3α/Foxa1 (Forkhead box) | Glucose homeostasis, beta-cell and pancreatic islet function | Shih et al., 1999 | Ci-FoxA-b/FoxA1/2/3 | KH.C11.718 | Whole embryo, stronger in nervous system and mesenchyme (Imai et al., 2004) | Not detected (Ogasawara et al., 2002) |
| Arx (Aristalesshomeo domain) | Acquisition and maintenance of alpha-cells identity | Wilcox et al., 2013 | Ci-Arx, Ci-aristaless | KH.L119.26 | Not analyzed | Not analyzed |
| Ptf1a-p48 (bHLH) | Regulation of exocrine cells differentiation | Krapp et al., 1998 | Ci-Ptf1a | KH.C3.967 | Not analyzed | Not analyzed |
| MafA (basicLeucine zipper) | Control of insulin transcription | Zhang et al., 2005 | Ci-Maf-like, Ci-L-maf | KH.C12.675 | Maternal ubiquitous signal (Imai et al., 2004) | Endostyle, esophagus, stomach, intestine, branchial pharynx, body wall muscle, neural complex, epidermis, heart, hemocytes (Ogasawara et al., 2002) |
| Mist1 (bHLH) | Maintenance of exocrine pancreas organization, acinar cell identity | Pin et al., 2001 | Ci-Mist, Ci-bHLHA15 | KH.C3.308 | Mesenchyme (Imai et al., 2004) | Stomach and intestine http://bioinfo.s.chibau.jp/ciaid/ |
| GATA-4 (Zinc-finger) | Activation of glucagon expression | Ritz-Laser et al., 2005 | Ci-GATAa, Ci-GATA4/5/6 | KH.L20.1 | Trunk endoderm, trunk ventral cells (Hudson et al., 2006); in larvae, it switches to sensory vesicle, mesenchyme, trunk lateral cells (D’Ambrosio et al., 2003) | Stomach and part of the intestine http://bioinfo.s.chibau.jp/ciaid/ Heart (Wang et al., 2013) |
| GATA-6 (Zinc-finger) | Acinar differentiation, maintenance of exocrine pancreas | Martinelli et al., 2013 | Same as above | Same as above | Same as above | Same as above |
| Prox1 (homeodomain) | Pancreas morphogenesis and exocrine pancreas homeostasis | Westmoreland et al., 2012 | Ci-Prox1 | KH.C4.430 | Whole embryo (Imai et al., 2004) | Not analyzed |
| Same as above | Same as above | Same as above | Ci-Prox1, Ci-prox, Ci-Prox-a | KH.C4.500 | Not analyzed | Not analyzed |
| Rfx6 (winged-helix) | Development and maintenance of beta-cells phenotype | Taleb and Polychronakos, 2011 | Ci-Rfx4/6, Ci-Rfx4b | KH.S600.1 | Not analyzed | Not analyzed |
| Cdx2/3, Cdx4 (ParaHoxhomeod omain) | Cdx2/3: Regulation of glucagon expression Cdx4: Control of number and movements of beta-cells |
Cdx2/3: Laser et al., 1996 Cdx4: Kinkel et al., 2008 |
Ci-Cdx, Ci-Cdx1/2/4 | KH.C14.408 | Nerve cord precursors, secondary muscle lineage, tail epidermis, trunk lateral cells, endodermal strand (Satou et al., 2001; Imai et al., 2006) | Seemingly small areas of stomach and intestine http://bioinfo.s.chibau.jp/ciaid/ |
| Lmx-1 (homeodomain) | Activation of insulin expression | German et al., 1992 | Ci-Lmx-like, Ci-Lmx1A | KH.C9.485 | Notochord, sensory vesicle (Jose’-Edwards et al., 2011) | Not analyzed |
| Lrh-1/NR5A2 (nuclear receptor) | Protection of beta-cells against apoptosis | Baquie’ et al., 2011 | Ci-Lrh-1, Ci-NR5A1/2/5, Ci-FTZF | KH.C6.10 | Not expressed (Imai et al., 2004) | Not analyzed |
| HNF-1A/HNF-1alpha (homeodomain) | Control of insulin secretion | Pontoglio et al., 1998 | Ci-HNF-1A | KH.C12.461 | Ubiquitous, likely maternal (Imai et al., 2004) | Endostyle http://bioinfo.s.chibau.jp/ciaid/ |
| HNF-4A/HNF-4alpha/NR2A1 (nuclear receptor) | Control of the expression of beta-cell genes involved in insulin secretory response to glucose andnutrients | Wang et al., 2000 | Ci-HNF-4A | KH.C3.84 | Ubiquitous, likely maternal; trunk endoderm (transient); a pair of tail epidermal neurons (Imai et al., 2004) | Not analyzed |
| Sox4 (SRY-box, HMG) | Insulin secretion, endocrine pancreas development, alpha-cells differentiation | Goldsworthy et al., 2008; Mavropoulos et al., 2005 | Ci-SoxC, Ci-Sox11/12/4 | KH.C7.523 | Neural precursors, trunk endoderm, CNS, adhesive organ, tail epidermis, epidermal neurons (Imai et al., 2004) | Stomach, part of the intestine, endostyle, branchial pharynx, neural complex http://bioinfo.s.chibau.jp/ciaid/ |
| Glis3 (Zinc-finger, Krüppel-like) | Direct and indirect activator of insulin expression | Yang et al., 2009 | Ci-Glis1/3, Ci-ZF137 | KH.C5.53 | Not expressed until the late tailbud stage; in late tailbuds, expressed in a small area of the anterior sensory vesicle (Miwata et al., 2006) | Not analyzed |
| Insm1 (Zinc-finger) | Transcriptional repression of BETA2/NeuroD1; development of beta-cells and intestinal endocrine cells | Liu et al., 2006 Gierl et al., 2006 | Ci-Insm1b, Ci-ZF041 | KH.C4.378 | Narrow area in the nerve cord encompassing two symmetric small groups of cells (Miwata et al., 2006) | Not analyzed |
| Grg3/Tle3 (co-factor) | Stimulation of beta-cell differentiation | Metzger et al., 2012 | Ci-Groucho, Ci-Tle1/2/3/4 | KH.L96.11 | Whole embryo, later mesenchyme (Imai et al., 2004) | Endostyle, branchial pharynx, body wall muscle, possibly part of the intestine (Ogasawara et al., 2002) |
| FoxO1 (Forkhead box) | Regulation of beta-cell proliferation, positive regulator of Beta2/NeuroD1 and MafA | Buteau and Accili, 2007 | Ci-FoxO, Ci-FoxO1/3/4/6 | KH.L20.26 | Epidermis of late tailbuds (Imai et al., 2004) | Neural complex, possibly some faint signal in stomach and intestine http://bioinfo.s.chibau.jp/ciaid/ |
| CREB (Leucine zipper) | Regulation of insulin, cyclinD1, cyclinA2 and other targets in response to different stimuli | Dalle et al., 2011 | Ci-CREB1, Ci-ATF1, Ci-CREB/ATFc | KH.C1.26 | Transient notochord, endoderm, epidermis, nervous system, mesenchyme (Imai et al., 2004) | Endostyle, possibly part of the stomach http://bioinfo.s.chibau.jp/ciaid/ |
Abbreviations: SRY, sex determining region Y-box; HMG, high mobility group; bHLH, basic helix-loop-helix. Prox1 appears twice because Ciona possesses two orthologs of this gene.
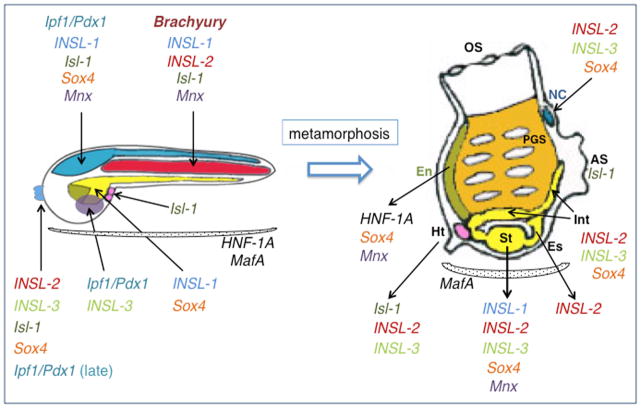
Figure 4. Graphic summary of the pre- and post-metamorphosis expression territories of Ciona orthologs of evolutionarily conserved pancreatic transcription factors.
Simplified drawings of a Ciona tailbud embryo (left) and a Ciona juvenile (right), not drawn to scale. For simplicity, the only tissues depicted in the tailbud are the notochord (red), CNS and adhesive organ (blue), trunk endoderm and endodermal strand (yellow), presumptive endostyle (green), trunk ventral cells (pink), mesenchyme (purple). In the juvenile, the only organs depicted are the endostyle (green), pharynx (orange) and pharyngeal gill slits, heart (pink), neural complex (blue), esophagus, stomach and intestine (all yellow). Gene names are abbreviated as in Table 1. Black arrows connect the genes to the territories where their expression has been reported (Table 1 and references therein). For the sake of completeness, we have tentatively included in this figure the expression patterns of Ci-INSL-1 and Ci-INSL-3; however, these patterns were detected in adults and might not be fully preserved in juveniles. Dotted semi-circles neighboring gene names symbolize ubiquitous or semi-ubiquitous expression of that particular gene. Abbreviations: OS, oral siphon; NC, neural complex; AS, atrial siphon; Int, intestine; Es, esophagus; St, stomach; Ht, heart; En, endostyle; PGS, pharyngeal gill slits.
In turn, Pdx1, aka insulin promoter factor 1 (Ipf1), a ParaHox homeodomain transcription factor, is a well-characterized and conserved activator of insulin expression in vertebrates (e.g., Madsen et al., 1997). This transcription factor, which is required for the formation of the mature pancreas (Stoffers et al., 1997), directly binds a CTAATG site in the proximal promoter region of the human insulin gene; a point mutation that changes this sequence to CTgATG reduces binding of IPF1/PDX1 in vitro and has been suggested to be a predisposing factor to type 2 diabetes (Malecki et al., 2006). Ciona possesses one ortholog of Ipf1, among numerous other homeobox genes (Di Gregorio et al., 1995). This gene is expressed weakly and ubiquitously in early embryos (Imai et al., 2004) and its product was detected via antibody staining in various regions of the CNS, in mesenchyme and in the adhesive organ of larvae (Corrado et al., 2001) (Fig. 4); the expression in the Ciona CNS is consistent with the reported expression of the orthologous gene in the developing rat CNS (Perez-Villamil et al., 1999). However, expression of Ciona Ipf1 in either juveniles or adults is yet to be analyzed, and the available EST counts do not indicate expression in any of the adult tissues that were sampled.
In rat, the insulin 1 promoter is bound by two homeodomain proteins, cdx-3 and lmx-1; both proteins can activate insulin transcription in transgenic assays (German et al., 1992). Ciona possesses one Cdx homeobox gene, which is expressed in various embryonic tissues, including endodermal strand, and seems weakly expressed in a few regions of stomach and intestine in juveniles (Table 1). Two Lmx genes are found in Ciona, likely as a result of a lineage-specific duplication, Ci-Lmx and Ci-Lmx-like (Jose’-Edwards et al., 2011). Both genes are expressed in the sensory vesicle, although in different domains; however, Ci-Lmx-like is also expressed in notochord cells beginning at gastrulation, and persists in this tissue until the mid-tailbud stage (Jose’-Edwards et al., 2011). This suggests the possibility that Ci-Lmx-like might cooperate with Ci-Bra in activating Ci-INSL-2 and/or Ci-INSL-1.
A key regulator of the specification of all pancreas endocrine precursors, neurogenin-3 (Gradwohl et al., 2000; Rukstalis and Habener, 2009), which in Ciona is present as a single-copy gene annotated as Ci-neurogenin, is expressed in vertebrates in small regions of the nervous system and later on in the precursors of pancreatic endocrine cells; interestingly, Ci-neurogenin is also expressed in small segments of the embryonic CNS (Imai et al., 2009) (Table 1), but is restricted to the ectoderm of the atria in juveniles and seems absent from both the digestive tract and the neural complex (Mazet et al., 2005). In the embryonic nervous system, Ci-neurogenin has been shown to control expression of Ci-Onecut (Pezzotti et al., 2014); interestingly, HNF-6/OC-1, one of the mouse orthologs of Ci-Onecut, is required for the differentiation of endocrine pancreatic cells, being responsible for the proper onset of Ipf1/Pdx1 expression in endoderm, and for the maintenance of neurogenin-3 (Jacquemin et al., 2000). However, expression of Ci-Onecut after metamorphosis has not been analyzed, and the EST counts indicate little or no expression in adults. In addition to controlling expression of Ci-Onecut, Ci-neurogenin has also been found to act upstream of Ci-Mnx in the developing nervous system of Ciona (Imai et al., 2009; Hudson et al., 2003; Ogasawara et al., 2002). Ci-Mnx is considered an ortholog of Mnx/Hlxb9/HB9, another vertebrate transcription factor with a dual neural/pancreatic function (Harrison et al., 1999; William et al., 2003).
Ci-isl1, the Ciona ortholog of islet-1, the LIM-homeodomain transcription factor required for the formation of all endocrine cells of the pancreatic islets (Ahlgren et al., 1997), is initially expressed in the precursors of the adhesive organ and sensory vesicle, and is transiently expressed in the notochord in tailbud-stage embryos (Giuliano et al., 1998). However, in juveniles this gene has only been detected in the epidermis (Ogasawara et al., 2002) and its expression in fully developed adult animals is still unknown. The Ciona ortholog of Nkx6.1 (Table 1), another regulator of insulin secretion found predominantly in mature pancreatic endocrine cells (Schisler et al., 2005), is weakly expressed in the embryonic epidermis and is predominant in the visceral ganglion and lateral nerve cord (Imai et al., 2009), but has not been analyzed by in situ hybridization in either juveniles or adults, and is not detected by EST counts in adult tissues.
Orthologs of additional markers of pancreatic development that are found in the Ciona genome but whose expression is still unknown include Arx, which in mammals is required for the development of pancreatic alpha-cells (Wilcox et al., 2013). Limited information is available on the expression of the Ciona orthologs of Lrh-1 and PTF-1L, which in vertebrates cooperate in controlling exocrine pancreas-specific expression of lipases, proteases and other digestive enzymes (Holmstrom et al., 2011); Ciona Lrh-1/NR5A1/2/5 (aka FTZF) (Table 1) is not detectable during early embryogenesis (Imai et al., 2004) and Ciona PTF1A is yet to be characterized. Neither gene has been analyzed after metamorphosis.
It is noteworthy that a few relevant pancreatic transcription factors appear to be absent or highly divergent in ascidians. For example, Ciona seems to lack a bona fide ortholog of Pax4, which is required for the development of both beta- and delta-cells and in particular for the differentiation of beta-cells in mammals (Sosa-Pineda et al., 1997). In addition, despite the presence of several Nkx genes, including the early endodermal marker TTF-1/Nkx2.1 (Ristoratore et al., 1999), a clear ortholog of Nkx2.2 could not be found in Ciona (Wada et al., 2003). In mouse, Nkx2.2 is expressed predominantly in CNS, islets and intestinal endocrine cells, where it controls cell-type specification. Knock-out mice lacking Nkx2.2 are hyperglycemic due to the lack of insulin-producing beta-cells, which are replaced by ghrelin-producing epsilon-cells, and they also have a reduced number of the glucagon-producing alpha-cells (Sussel et al., 1998). The situation is similar in the case of Mafb, which in vertebrates is required for beta-cell maturation (Artner et al., 2007), as neither of the two Ciona Maf-related genes can be assigned to the Mafb class. Similarly, in the case of the Ciona Pou genes, none seems to belong to the Pou3 class, which includes Pou3f4/Brn4, a transcription factor involved in both the control of glucagon expression in alpha-cells and the acquisition of alpha-cell identity (Hussain et al., 2002).
Of note, Ci-GATAa, aka GATA4/5/6, which is first expressed in trunk endoderm and is found in the digestive organs after metamorphosis, is equally related to GATA-4 and GATA-6, which are both required for endocrine and exocrine pancreas formation, respectively (Table 1), and have been reported to play functionally redundant roles (Carrasco et al., 2012).
NOTOCHORD EXPRESSION OF INSULIN-LIKE GENES: BEYOND ASCIDIANS
After the discovery of a hybrid insulin/IGF from Branchiostoma californiensis (Chan et al., 1990), the amphioxus genome project identified five additional insulin/relaxin genes from B. floridae, as well as their putative receptors (Holland et al., 2008). Four of these genes display a clustered organization and conserved synteny with the clustered insulin-like genes in humans and in Ciona (Holland et al., 2008). Thus far, expression in notochord is yet to be reported for any of these genes. However, in vertebrates, we found reports of the notochord expression of insulin-like peptides and their receptors in animals representative of different classes. Both IGF1 and its receptor, IGF1-R, were found to be expressed in the notochord of the Chilean flounder, Paralichtys adspersus and in numerous other tissues (Escobar et al., 2011). Similar results were reported in the grass carp, Ctenopharyngodon idellus, where the igf2b gene was transcribed in notochord and brain of the embryos and in multiple tissues of the adult fish (Yuan et al., 2011). In zebrafish, four IGF genes, igf-1a, igf-1b, igf-2a and igf-2b, have been reported; of these, igf-1b is exclusively detected in the gonad and igf-2b is liver-specific (Zou et al., 2009). igf-1a and igf-2a were found in multiple tissues, including the notochord (Zou et al., 2009), although another study published in the same year described igf-2a as notochord-specific (White et al., 2009). Morpholino-mediated knockdown of igf-2a led to various defects, including notochord undulations (White et al., 2009). Over-expression of this gene by mRNA injection affected notochord formation and increased the distance between contralateral somites, thus impairing midline development; in addition, 24% of the igf-2a mRNA-injected embryos had partially or fully duplicated notochords (Zou et al., 2009).
In amphibians, expression of a member of the IGFBP superfamily, Xenopus xIGFBP-rP10, has been reported in the notochord and other tissues (Kuerner and Steinbeisser, 2006). In birds, the igf1r mRNA was found at high levels in the notochord and somites in the 20-somite stage chick embryo (Holzenberger et al., 1996), confirming earlier immunolocalization studies (Ralphs et al., 1990). In mammals, expression of both insulin and IGF1 was detected in the rat notochord by immunostaining (Korgun et al., 2003). Remarkably, in humans, both IGF1 and IGF1R have been reported in chordoma, a notochord-derived tumor (Nibu et al., 2013); interestingly, 41% of the chordomas analyzed in a subsequent study contained phosphorylated IGF1R/IR, which was absent in benign notochordal tumors and in fetal notochord (Sommer et al., 2010).
Together, these results suggest that insulin-like genes, their receptors and other members of the insulin pathway are evolutionarily conserved components of the basic notochord toolkit.
CIS-REGULATORY SEQUENCES CONTROLLING NOTOCHORD EXPRESSION OF CI-INSL-2
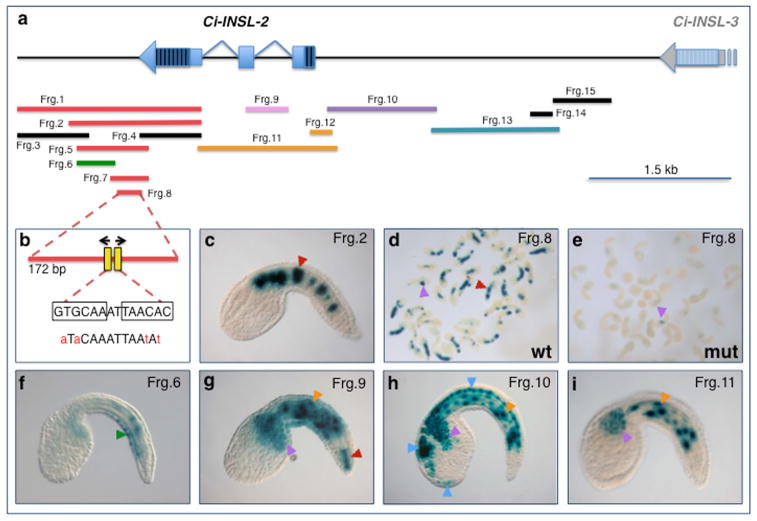
Despite the recurring presence of insulin and/or IGF gene(s) expression in the notochord in different chordates, neither the transcription factors nor the structure of the cis-regulatory sequences responsible for it are known. Therefore, we attempted the identification of cis-regulatory modules (CRMs) in the Ci-INSL-1 and Ci-INSL-2 genomic loci, and focused in particular on Ci-INSL-2 since this gene displays the most specific and sustained notochord expression among the two (Fig. 1b) and it has been described as the most closely related to vertebrate insulin genes (Olinski et al., 2006b). Tiled genomic fragments, covering approximately 6 kb, were cloned from the Ci-INSL-2 locus and tested for cis-regulatory activity in Ciona embryos (Fig. 3a and data not shown). This analysis led to the identification of a 1.645-kb genomic region active in notochord cells, located downstream of the Ci-INSL-2 transcription unit (Frg. 1, Fig. 3); this region was reduced, through serial truncations (e.g., the 1.26-kb Frg. 2, Fig. 3a,c), to a minimal 172-bp CRM (Frg. 8, Fig. 3a,b,d), which recapitulated the notochord activity of the longer fragments. Through site-directed point mutations, this minimal CRM was found to require for its activity two putative binding sites for Ciona Brachyury (Ci-Bra), a well-characterized notochord-specific transcription factor (e.g., Di Gregorio and Levine, 1999; Dunn and Di Gregorio, 2009; Katikala et al., 2013) (Fig. 3b,d,e). Moreover, testing of genomic fragments located within and upstream of the Ci-INSL-2 transcription unit revealed the existence of multiple cis-regulatory elements disseminated along this locus (Fig. 3a, f-i). In particular, additional regions able to direct notochord gene expression were uncovered, including a 418-bp sequence encompassing the first intron (Frg. 9) that directs expression in secondary notochord (the posterior-most eight notochord cells near the tip of the tail), among other tissues (Fig. 3g), and a 1.34-kb region located directly upstream of Ci-INSL-2 that directs occasional staining in primary notochord (data not shown). The latter region, along with sequences located further upstream, recapitulated the expression of Ci-INSL-2 in the primordium of the adhesive organ (Fig. 3h). In addition, one 248-bp fragment nested within the main notochord CRM was active in ventral tail epidermal neurons (Frg. 6, Fig. 3f), while the 1.34-kb region located directly upstream of Ci-INSL-2 (Frg. 10) was also active in sensory vesicle and nerve cord (Fig. 3h). A few of the fragments tested were also, or predominantly, active in muscle and mesenchyme cells (Fig. 3g-i); however, Ci-INSL-2 is not detected in larval muscle or CNS, indicating that these regulatory elements might be either inactive or repressed within their genomic context. We also tested two genomic fragments totaling ~2 kb from the Ci-INSL-1 upstream region, which were active in muscle, mesenchyme and nerve cord (data not shown). Published chromatin immunoprecipitation (ChIP) profiles (Kubo et al., 2010) indicate that the Ci-INSL-1 locus is bound in vivo by Ci-Bra, as is Ci-INSL-2, which shows high peaks of occupancy by Ci-Bra particularly in its 3′-end and downstream regions, where the notochord CRM that we have identified is found (Fig. 3). Both loci are occupied in early embryos by Ci-FoxA-a, while the Ci-INSL-3 locus is not occupied by either transcription factor (Kubo et al., 2010). These data seem consistent with the expression patterns of these three genes (Fig. 1).
Figure 3. Identification and characterization of a notochord CRM and various cis-regulatory elements in the Ci-INSL-2 genomic locus.
a: Schematic representation of the chromosomal region encompassing the Ci-INSL-2 gene and part of Ci-INSL-3. Genes are represented as light-blue bars (exons) and lines (introns); striped areas indicate untranslated regions. Part of the Ci-INSL-3 coding region is depicted for reference (grey box). Colored horizontal bars symbolize genomic fragments, abbreviated as “Frg.” followed by a number, that were individually cloned upstream of the Ci-fkh/FoxA-a basal promoter region (Oda-Ishii and Di Gregorio, 2007) and tested in vivo in Ciona embryos by electroporation (Di Gregorio and Levine, 2002). The bars are color-coded as follows: red, fragments that predominantly show activity in the notochord; pink, fragments active in secondary notochord and additional tissues; orange, fragments predominantly active in muscle; light purple, fragments active in muscle, nervous system and mesenchyme; green, fragments mainly active in epidermis; teal, fragments mainly active in muscle and nervous system; black, fragments with no detectable activity. b: schematic structure of the minimal 172-bp notochord CRM isolated from the 3′-region of Ci-INSL-2 (Frg. 8). Two putative Ci-Bra binding sites (Passamaneck et al., 2009) with inverted orientation were mutagenized as shown below the diagram; mutations are indicated in red and lower case. These mutations were sufficient to completely abolish notochord activity of the CRM. c: Representative transgenic embryo carrying one of the truncations of the notochord CRM (Frg. 2, 1.26 kb). d: Low-magnification microphotograph of a group of transgenic embryos electroporated with the wild-type (wt) 172-bp Ci-INSL-2 notochord CRM (Frg. 8). Notochord staining (red arrowhead) is seen in most embryos and is occasionally associated with mesenchyme staining (purple arrowhead). e: Low-magnification microphotograph of a group of transgenic embryos from the same clutch as the embryos shown in d, electroporated with the 172-bp Ci-INSL-2 notochord CRM carrying the mutations (mut) in the putative Ci-Bra binding sites illustrated in b and reared, fixed and stained in parallel with the embryos shown in d. Staining is only detected in mesenchyme cells (purple arrowhead). f–i: Representative transgenic embryos carrying the fragments indicated on the top right corner of each panel, photographed after X-gal staining. Arrowheads: red, notochord; green, epidermis and tail epidermal neurons; orange: muscle; purple, mesenchyme; blue, nervous system.
CONCLUDING REMARKS
BRACHYURY AND OTHER POSSIBLE ACTIVATORS OF INSULIN EXPRESSION IN ASCIDIANS
In ascidian embryos, endodermal cells induce notochord formation through the activation of the BMP and FGF signaling pathways (Darras and Nishida, 2001; Imai et al., 2002a). In particular, in Ciona, the nuclear translocation of beta-catenin activates expression of the transcription factor FoxD by binding three Tcf/LEF binding sites in its 5′-flanking region (Imai et al., 2002b). In turn, FoxD induces transcription of Ci-ZicL, a zinc-finger transcription factor that acts as a main activator of Ci-Bra expression in Aline notochord precursors (Yagi et al., 2004), possibly by working in concert with the maternally expressed Ci-p53/p73-a and Ci-p53/p73-b (Noda, 2011). Expression of Ci-Bra in B-line notochord precursors is activated by two signaling pathways, Nodal and Notch/Su(H) (Hudson and Yasuo, 2006). These pathways are complemented by the action of two FGF ligands, Ci-FGF9/16/20 and Ci-FGF8/17/18 (Yasuo and Hudson, 2007). Finally, the activation of Ci-Bra expression in notochord precursors at the 64-cell stage (Corbo et al., 1997) triggers the deployment of an extensive notochord gene battery, which is responsible for the following steps of notochord formation and includes a large number of structural genes, as well as a suite of transcription factors that likely act as Ci-Bra intermediaries (Takahashi et al., 1999; Di Gregorio and Levine, 1999; Hotta et al., 2008; Dunn and Di Gregorio, 2009; Jose’-Edwards et al., 2011, 2013; Katikala et al., 2013). These studies clearly show the causal role of the endoderm in the formation of the ascidian notochord; however, it remains unclear whether any feedback of the notochord on the endoderm is required for endoderm differentiation in ascidians. The situation is quite different in higher chordates; for example, studies in chick embryos show that during early development the notochord is in direct contact with the endoderm that will give rise to the dorsal pancreatic bud, and its removal eliminates expression of markers of pancreas development, including insulin, glucagon and carboxypeptidase A (Kim et al., 1997). Related studies in human embryos confirm that the dorsal foregut endoderm is directly adjacent to the notochord before the onset of PDX1 (Jennings et al., 2013). Together, these results indicate that the notochord plays a permissive role in the specification of pancreatic cells, and highlight the involvement of the notochord in promoting insulin expression in adjacent endodermal cells (Kim et al., 1997). Nevertheless, it remained unclear how expression of insulin genes is controlled within the notochord itself. The results that we presented here suggest that in Ciona the single-copy, notochord-specific Brachyury is involved in the control of notochord expression of at least one insulin-like gene, Ci-INSL-2. The absence of Ci-INSL-2 expression in the endoderm during embryogenesis might seem puzzling, however it is noteworthy that before metamorphosis the endoderm of Ciona and other solitary ascidians is not functional, and that in juveniles, at a time when the intestine is fully functional and the notochord is no longer present, a strong expression of Ci-INSL-2 is detected in esophagus and stomach and other regions of the digestive system. These findings raise questions on the identity of the activator(s) of Ci-INSL-2 expression in these post-metamorphosis structures, and might suggest a continued role of Ci-Bra in the activation of Ci-INSL-2 transcription after metamorphosis. Interestingly, ENU-induced Ci-Bra mutants display, in addition to the predictable disruption of notochord formation, remarkable post-metamorphosis defects in the development of the digestive organs of the juvenile (Chiba et al., 2009). It could have been simply hypothesized that these defects were due to a role of Ci-Bra in the formation of endodermal structures after metamorphosis, which would be reminiscent of the role of Brachyury orthologs in non-chordate invertebrates (Hamaguchi et al., 2012). However, the lack of Ci-Bra transcripts in hatched larvae and in juveniles (Chiba et al., 2009) makes this scenario quite unlikely. Rather, it has been proposed that the lack of functional Ci-Bra protein in Ci-Bra mutants might cause some notochord cells to stochastically transfate to endoderm, instead of undergoing apoptosis at the time of metamorphosis, with the result of generating supernumerary endodermal cells that are not properly incorporated within the developing digestive organs (Chiba et al., 2009). These findings indicate that insulin expression in the digestive tract of Ciona is controlled by transcription factor(s) other than Ci-Bra. Hence, we can use the information that is currently available on the post-metamorphosis expression of Ciona orthologs of known activators of insulin expression in other chordates, summarized in Table 1 and Fig. 4, to begin to delineate the identity of some of the putative activators of Ci-INSL-2 transcription in the digestive organs. As we discussed above, the expression of the Ciona ortholog of Ipf1/Pdx1, one of the most relevant transcription factors required for pancreas formation and an activator of insulin expression (Stoffers et al., 1997; Malecki et al., 2006) is yet to be analyzed in either juveniles or adults. Therefore, the possibility that Ciona Ipf1/Pdx1 might be activating expression of the insulin-like genes in the digestive tract remains open. Ci-isl, the Ciona ortholog of Islet-1, a transcription factor required for the development of all pancreatic endocrine cells, is detected in CNS, adhesive organ, trunk ventral cells and, transiently, in the notochord of tailbud embryos (Giuliano et al., 1998; Stolfi et al., 2010). The expression of Ci-isl in the notochord suggests that this factor might be co-regulating expression of Ci-INSL1 or Ci-INSL2 together with Ci-Bra; however, after metamorphosis its expression has only been reported in the precursors of the atrial siphon muscle and in the epidermis (Ogasawara et al., 2002; Wang et al., 2013); these results minimize the possibility that Ci-islet might be activating insulin-like expression in the digestive organs. Nevertheless, other known activators of insulin transcription, namely Ci-MafA, Ci-Mist1, Ci-GATA-4, Ci-Cdx1/2/4 and Ci-CREB (Table 1, Fig. 4), are expressed, in juveniles, in territories fully or partially overlapping with that of Ci-INSL2 expression, and might therefore contribute to the activation of Ci-INSL2 expression in the digestive tract.
EVOLUTIONARY ORIGINS OF THE ENDOCRINE PANCREAS
Lampreys and hagfish, collectively known as cyclostomes or agnathans, are considered the most primitive living vertebrates (Shimeld and Donoghue, 2012). During their larval stage, lampreys possess an “islet organ” that mainly consists of insulin-producing beta-cells accompanied by at least four different types of granular cells that might function as different types of endocrine cells (Brinn and Epple, 1976). In adult lampreys, the islet organ acquires additional cell types that are indicated as B, D, and F cells, each producing a different hormone (Youson and Al-Mahrouki, 1999), thus making this structure more closely related to the pancreas of jawed vertebrates.
The general consensus on the evolutionary origins of the pancreas is that insulin-producing beta-cells are the essential and most primitive components of its endocrine division (e.g., Madsen, 2007). This hypothesis is supported by the findings of insulin and IGF in the hepatopancreas of molluscs (Gallardo et al., 2003; Chung, 2014), and by our findings in Ciona, where the existence of glucagon, amylin, pancreatic peptide, ghrelin and somatostatin has not been confirmed. The diffused expression of Ci-INSL-2 argues against the existence of a discrete endocrine pancreatic region in Ciona. Such region is somewhat more refined in amphioxus, where the expression patterns of AmphiXlox, the ortholog of Ipf1/Pdx1, and AmphiCdx (Table 1) overlap in the posterior gut (Brooke et al., 1998), while the localization of AmphiNeurogenin and of insulin-like peptides suggest that the presumptive pancreas might lie in the mid-gut (Holland et al., 2000; Reinecke, 1981). In ascidians other than Ciona, particularly Halocynthia, a so-called hepatopancreas is anatomically distinguishable as an outpocketing of the stomach, and is believed to release digestive enzymes into the lumen of the stomach, thus acting as a primitive exocrine pancreas (Hirano and Nishida, 2000). Even though a hepatopancreas has been described in Ciona (Hammond et al., 2005), its anatomical bearings and its functional role in this species are less clear. Therefore, it would be of interest to assess the expression of genes encoding digestive enzymes and additional markers of the exocrine pancreas in other regions of the digestive tract in both juveniles and adults, in order to determine whether a primordial exocrine pancreas-like structure exists in Ciona and other ascidians. With respect to the endocrine pancreas, we could hypothesize that in Ciona the transcription factors that in vertebrates are coexpressed in pancreatic beta-cells might instead control insulin expression in different regions of the digestive system, by acting either individually or combinatorially, as opposed to being colocalized within a small organ-like structure. In conclusion, our experimental results, combined with information found in the literature that we have reviewed here, indicate that a significant fraction of the components of the pancreatic gene regulatory network, along with the molecular infrastructure required for the formation of insulin-producing cells, are present and might be operational in ascidians. Moreover, several pancreatic transcription factors are deployed in the digestive organs after metamorphosis; however, it should be remembered that some of the factors that in vertebrates control pancreatic organogenesis and differentiation might be playing derived, lineage-specific roles in ascidians.
Acknowledgments
We thank our anonymous reviewers for improving our manuscript with their excellent comments. We are grateful to Drs. Reiko Yoshida, Yasunori Sasakura and Michio Ogasawara for their kind technical advice on the in situ protocol for Ciona juveniles, to Ms. Shruti Sharma for technical help and to A. MacGyver for inspiration. This work was supported by grant NIH/NIGMS GM100466, by grant 1-FY11-468 from the March of Dimes Birth Defects Foundation, and by a grant from the Bohmfalk Trust Fund to A.D.G.
Contract Grant Sponsor: NIH/NIGMS, contract grant number: GM100466; Contract Grant Sponsor: March of Dimes Birth Defects Foundation, contract grant number: 1-FY11-468; Contract Grant Sponsor: the Bohmfalk Trust Fund.
Abbreviations
- bp
base pair(s)
- CNS
central nervous system
- kb
kilobase(s), or 1000 base pairs
- EST
Expressed Sequence Tag
- mut
mutant
- PCR
Polymerase Chain Reaction
- RT-PCR
Reverse Transcriptase Polymerase Chain Reaction
- wt
wild-type
- X-gal
5-bromo-4-chloro-3-indolyl-β-D-galactopyranoside
References
- Ahlgren U, Pfaff SL, Jessell TM, Edlund T, Edlund H. Independent requirement for ISL1 in formation of pancreatic mesenchyme and islet cells. Nature. 1997;385:257–260. doi: 10.1038/385257a0. [DOI] [PubMed] [Google Scholar]
- Artner I, Blanchi B, Raum JC, Guo M, Kaneko T, Cordes S, Sieweke M, Stein R. MafB is required for islet beta cell maturation. Proc Natl Acad Sci U S A. 2007;104:3853–3858. doi: 10.1073/pnas.0700013104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baquié M, St-Onge L, Kerr-Conte J, Cobo-Vuilleumier N, Lorenzo PI, Jimenez Moreno CM, Cederroth CR, Nef S, Borot S, Bosco D, Wang H, Marchetti P, Pattou F, Wollheim CB, Gauthier BR. The liver receptor homolog-1 (LRH-1) is expressed in human islets and protects {beta}-cells against stress-induced apoptosis. Hum Mol Genet. 2011;20:2823–2833. doi: 10.1093/hmg/ddr193. [DOI] [PubMed] [Google Scholar]
- Bevis PJ, Thorndyke MC. Endocrine cells in the oesophagus of the ascidian Styela clava, a cytochemical and immunofluorescence study. Cell Tissue Res. 1978;187:153–158. doi: 10.1007/BF00220627. [DOI] [PubMed] [Google Scholar]
- Bort R, Martinez-Barbera JP, Beddington RS, Zaret KS. Hex homeobox gene-dependent tissue positioning is required for organogenesis of the ventral pancreas. Development. 2004;131:797–806. doi: 10.1242/dev.00965. [DOI] [PubMed] [Google Scholar]
- Brinn JE, Epple A. New types of islet cells in a cyclostome, Petromyzon marinus L. Cell Tissue Res. 1976;171:317–329. doi: 10.1007/BF00224657. [DOI] [PubMed] [Google Scholar]
- Brooke NM, Garcia-Fernàndez J, Holland PW. The ParaHox gene cluster is an evolutionary sister of the Hox gene cluster. Nature. 1998;392:920–922. doi: 10.1038/31933. [DOI] [PubMed] [Google Scholar]
- Burke RD, Angerer LM, Elphick MR, Humphrey GW, Yaguchi S, Kiyama T, Liang S, Mu X, Agca C, Klein WH, Brandhorst BP, Rowe M, Wilson K, Churcher AM, Taylor JS, Chen N, Murray G, Wang D, Mellott D, Olinski R, Hallböök F, Thorndyke MC. A genomic view of the sea urchin nervous system. Dev Biol. 2006;300:434–460. doi: 10.1016/j.ydbio.2006.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buteau J, Accili D. Regulation of pancreatic beta-cell function by the forkhead protein FoxO1. Diabetes Obes Metab. 2007;9(Suppl 2):140–146. doi: 10.1111/j.1463-1326.2007.00782.x. [DOI] [PubMed] [Google Scholar]
- Carrasco M, Delgado I, Soria B, Martín F, Rojas A. GATA4 and GATA6 control mouse pancreas organogenesis. J Clin Invest. 2012;122:3504–3415. doi: 10.1172/JCI63240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan SJ, Cao QP, Steiner DF. Evolution of the insulin superfamily: cloning of a hybrid insulin/insulin-like growth factor cDNA from amphioxus. Proc Natl Acad Sci U S A. 1990;87:9319–9323. doi: 10.1073/pnas.87.23.9319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiba S, Sasaki A, Nakayama A, Takamura K, Satoh N. Development of Ciona intestinalis juveniles (through 2nd ascidian stage) Zoolog Sci. 2004;21:285–298. doi: 10.2108/zsj.21.285. [DOI] [PubMed] [Google Scholar]
- Chiba S, Jiang D, Satoh N, Smith WC. Brachyury null mutant-induced defects in juvenile ascidian endodermal organs. Development. 2009;136:35–39. doi: 10.1242/dev.030981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung JS. An insulin-like growth factor found in hepatopancreas implicates carbohydrate metabolism of the blue crab Callinectes sapidus. Gen Comp Endocrinol. 2014;199:56–64. doi: 10.1016/j.ygcen.2014.01.012. [DOI] [PubMed] [Google Scholar]
- Conlon JM, Reinecke M, Thorndyke MC, Falkmer S. Insulin and other islet hormones (somatostatin, glucagon and PP) in the neuroendocrine system of some lower vertebrates and that of invertebrates--a minireview. Horm Metab Res. 1988;20:406–410. doi: 10.1055/s-2007-1010849. [DOI] [PubMed] [Google Scholar]
- Conlon JM. Evolution of the insulin molecule: insights into structure-activity and phylogenetic relationships. Peptides. 2001;22:1183–1193. doi: 10.1016/s0196-9781(01)00423-5. [DOI] [PubMed] [Google Scholar]
- Corbo JC, Levine M, Zeller RW. Characterization of a notochord-specific enhancer from the Brachyury promoter region of the ascidian, Ciona intestinalis. Development. 1997;124:589–602. doi: 10.1242/dev.124.3.589. [DOI] [PubMed] [Google Scholar]
- Corrado M, Aniello F, Fucci L, Branno M. Ci-IPF1, the pancreatic homeodomain transcription factor, is expressed in neural cells of Ciona intestinalis larva. Mech Dev. 2001;102:271–274. doi: 10.1016/s0925-4773(01)00311-2. [DOI] [PubMed] [Google Scholar]
- Dalle S, Quoyer J, Varin E, Costes S. Roles and regulation of the transcription factor CREB in pancreatic β-cells. Curr Mol Pharmacol. 2011;4:187–195. doi: 10.2174/1874467211104030187. [DOI] [PubMed] [Google Scholar]
- Darras S, Nishida H. The BMP signaling pathway is required together with the FGF pathway for notochord induction in the ascidian embryo. Development. 2001;128:2629–2638. doi: 10.1242/dev.128.14.2629. [DOI] [PubMed] [Google Scholar]
- Davidson B, Levine M. Evolutionary origins of the vertebrate heart: Specification of the cardiac lineage in Ciona intestinalis. Proc Natl Acad Sci U S A. 2003;100:11469–11473. doi: 10.1073/pnas.1634991100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson JK, Falkmer S, Mehrotra BK, Wilson S. Insulin assays and light microscopical studies of digestive organs in protostomian and deuterostomian species and in coelenterates. Gen Comp Endocrinol. 1971;17:388–401. doi: 10.1016/0016-6480(71)90148-1. [DOI] [PubMed] [Google Scholar]
- De Bernardi F, Pennati R, Candiani S, Zega G, Groppelli S, Pestarino M. Serotonin in the morphogenesis of ascidian nervous system. Caryologia. 2006;59:375–379. [Google Scholar]
- Dehal P, Satou Y, Campbell RK, Chapman J, Degnan B, De Tomaso A, Davidson B, Di Gregorio A, Gelpke M, Goodstein DM, et al. The draft genome of Ciona intestinalis: insights into chordate and vertebrate origins. Science. 2002;298:2157–2167. doi: 10.1126/science.1080049. [DOI] [PubMed] [Google Scholar]
- de Pablo F, Chambers SA, Ota A. Insulin-related molecules and insulin effects in the sea urchin embryo. Dev Biol. 1988;130:304–310. doi: 10.1016/0012-1606(88)90436-8. [DOI] [PubMed] [Google Scholar]
- Di Gregorio A, Spagnuolo A, Ristoratore F, Pischetola M, Aniello F, Branno M, Cariello L, Di Lauro R. Cloning of ascidian homeobox genes provides evidence for a primordial chordate cluster. Gene. 1995;156:253–257. doi: 10.1016/0378-1119(95)00035-5. [DOI] [PubMed] [Google Scholar]
- Di Gregorio A, Levine M. Regulation of Ci-tropomyosin-like, a Brachyury target gene in the ascidian, Ciona intestinalis. Development. 1999;126:5599–5609. doi: 10.1242/dev.126.24.5599. [DOI] [PubMed] [Google Scholar]
- Di Gregorio A, Corbo JC, Levine M. The regulation of forkhead/HNF-3beta expression in the Ciona embryo. Dev Biol. 2001;229:31–43. doi: 10.1006/dbio.2000.9964. [DOI] [PubMed] [Google Scholar]
- Di Gregorio A, Levine M. Analyzing gene regulation in ascidian embryos: new tools for new perspectives. Differentiation. 2002;70:132–139. doi: 10.1046/j.1432-0436.2002.700402.x. [DOI] [PubMed] [Google Scholar]
- Dufort D, Schwartz L, Harpal K, Rossant J. The transcription factor HNF3beta is required in visceral endoderm for normal primitive streak morphogenesis. Development. 1998;125:3015–3025. doi: 10.1242/dev.125.16.3015. [DOI] [PubMed] [Google Scholar]
- Dunn MP, Di Gregorio A. The evolutionarily conserved leprecan gene: its regulation by Brachyury and its role in the developing Ciona notochord. Dev Biol. 2009;328:561–774. doi: 10.1016/j.ydbio.2009.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edmondson SR, Thumiger SP, Werther GA, Wraight CJ. Epidermal homeostasis: the role of the growth hormone and insulin-like growth factor systems. Endocr Rev. 2003;24:737–764. doi: 10.1210/er.2002-0021. [DOI] [PubMed] [Google Scholar]
- Escobar S, Fuentes EN, Poblete E, Valdés JA, Safian D, Reyes AE, Alvarez M, Molina A. Molecular cloning of IGF-1 and IGF-1 receptor and their expression pattern in the Chilean flounder (Paralichthys adspersus) Comp Biochem Physiol B Biochem Mol Biol. 2011;159:140–147. doi: 10.1016/j.cbpb.2011.03.003. [DOI] [PubMed] [Google Scholar]
- Fiala-Medioni A, Pequignat E. Enzyme activities in the gills of benthic filter feeders (ascidia) C R Acad Sci Hebd Seances Acad Sci D. 1975;281:1123–1126. [PubMed] [Google Scholar]
- Fritsch HA, Van Noorden S, Pearse AG. Substance P-, neurotensin- and bombesin-like immunoreactivities in the gill epithelium of Ciona intestinalis L. Cell Tissue Res. 1980;208:467–473. doi: 10.1007/BF00233878. [DOI] [PubMed] [Google Scholar]
- Fritsch HA, Van Noorden S, Pearse AG. Gastro-intestinal and neurohormonal peptides in the alimentary tract and cerebral complex of Ciona intestinalis (Ascidiaceae) Cell Tissue Res. 1982;223:369–402. doi: 10.1007/BF01258496. [DOI] [PubMed] [Google Scholar]
- Fujisawa T, Hayakawa E. Peptide signaling in Hydra. Int J Dev Biol. 2012;56:543–550. doi: 10.1387/ijdb.113477tf. [DOI] [PubMed] [Google Scholar]
- Fulzele K, Clemens TL. Novel functions for insulin in bone. Bone. 2012;50:452–456. doi: 10.1016/j.bone.2011.06.018. [DOI] [PubMed] [Google Scholar]
- Gallardo N, Carrillo O, Moltó E, Deás M, González-Suárez R, Carrascosa JM, Ros M, Andrés A. Isolation and biological characterization of a 6-kDa protein from hepatopancreas of lobster Panulirus argus with insulin-like effects. Gen Comp Endocrinol. 2003;131:284–290. doi: 10.1016/s0016-6480(03)00014-5. [DOI] [PubMed] [Google Scholar]
- German MS, Wang J, Chadwick RB, Rutter WJ. Synergistic activation of the insulin gene by a LIM-homeo domain protein and a basic helix-loop-helix protein: building a functional insulin minienhancer complex. Genes Dev. 1992;6:2165–2176. doi: 10.1101/gad.6.11.2165. [DOI] [PubMed] [Google Scholar]
- Gierl MS, Karoulias N, Wende H, Strehle M, Birchmeier C. The zinc-finger factor Insm1 (IA-1) is essential for the development of pancreatic beta cells and intestinal endocrine cells. Genes Dev. 2006;20:2465–2478. doi: 10.1101/gad.381806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giuliano P, Marino R, Pinto MR, De Santis R. Identification and developmental expression of Ci-isl, a homologue of vertebrate islet genes, in the ascidian Ciona intestinalis. Mech Dev. 1998;78:199–202. doi: 10.1016/s0925-4773(98)00143-9. [DOI] [PubMed] [Google Scholar]
- Goldsworthy M, Hugill A, Freeman H, Horner E, Shimomura K, Bogani D, Pieles G, Mijat V, Arkell R, Bhattacharya S, Ashcroft FM, Cox RD. Role of the transcription factor sox4 in insulin secretion and impaired glucose tolerance. Diabetes. 2008;57:2234–2244. doi: 10.2337/db07-0337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gradwohl G, Dierich A, LeMeur M, Guillemot F. neurogenin3 is required for the development of the four endocrine cell lineages of the pancreas. Proc Natl Acad Sci U S A. 2000;97:1607–1611. doi: 10.1073/pnas.97.4.1607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamaguchi T, Takashima S, Okamoto A, Imaoka M, Okumura T, Murakami R. Dorsoventral patterning of the Drosophila hindgut is determined by interaction of genes under the control of two independent gene regulatory systems, the dorsal and terminal systems. Mech Dev. 2012;129:236–243. doi: 10.1016/j.mod.2012.07.006. [DOI] [PubMed] [Google Scholar]
- Hammond JA, Nakao M, Smith VJ. Cloning of a glycosylphosphatidylinositol-anchored alpha-2-macroglobulin cDNA from the ascidian, Ciona intestinalis, and its possible role in immunity. Mol Immunol. 2005;42:683–694. doi: 10.1016/j.molimm.2004.09.017. [DOI] [PubMed] [Google Scholar]
- Harrison KA, Thaler J, Pfaff SL, Gu H, Kehrl JH. Pancreas dorsal lobe agenesis and abnormal islets of Langerhans in Hlxb9-deficient mice. Nat Genet. 1999;23:71–75. doi: 10.1038/12674. [DOI] [PubMed] [Google Scholar]
- Hinegardner RT. Growth and development of the laboratory cultured sea urchin. Biol Bull. 1969;137:465–475. doi: 10.2307/1540168. [DOI] [PubMed] [Google Scholar]
- Hirano T, Nishida H. Developmental fates of larval tissues after metamorphosis in ascidian Halocynthia roretzi I. Origin of mesodermal tissues of the juvenile. Dev Biol. 1997;192:199–210. doi: 10.1006/dbio.1997.8772. [DOI] [PubMed] [Google Scholar]
- Hirano T, Nishida H. Developmental fates of larval tissues after metamorphosis in the ascidian, Halocynthia roretziII. Origin of endodermal tissues of the juvenile. Dev Genes Evol. 2000;210:55–63. doi: 10.1007/s004270050011. [DOI] [PubMed] [Google Scholar]
- Hiruta J, Mazet F, Yasui K, Zhang P, Ogasawara M. Comparative expression analysis of transcription factor genes in the endostyle of invertebrate chordates. Dev Dyn. 2005;233:1031–1037. doi: 10.1002/dvdy.20401. [DOI] [PubMed] [Google Scholar]
- Holland LZ, Schubert M, Holland ND, Neuman T. Evolutionary conservation of the presumptive neural plate markers AmphiSox1/2/3 and AmphiNeurogenin in the invertebrate chordate amphioxus. Dev Biol. 2000;226:18–33. doi: 10.1006/dbio.2000.9810. [DOI] [PubMed] [Google Scholar]
- Holland LZ, Albalat R, Azumi K, Benito-Gutiérrez E, Blow MJ, Bronner-Fraser M, Brunet F, Butts T, Candiani S, Dishaw LJ, et al. The amphioxus genome illuminates vertebrate origins and cephalochordate biology. Genome Res. 2008;18:1100–1111. doi: 10.1101/gr.073676.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmstrom SR, Deering T, Swift GH, Poelwijk FJ, Mangelsdorf DJ, Kliewer SA, MacDonald RJ. LRH-1 and PTF1-L coregulate an exocrine pancreas-specific transcriptional network for digestive function. Genes Dev. 2011;25:1674–1679. doi: 10.1101/gad.16860911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holzenberger M, Lapointe F, Leibovici M, Lièvre CA. The avian IGF type 1 receptor: cDNA analysis and in situ hybridization reveal conserved sequence elements and expression patterns relevant for the development of the nervous system. Brain Res Dev Brain Res. 1996;97:76–87. doi: 10.1016/s0165-3806(96)00133-2. [DOI] [PubMed] [Google Scholar]
- Horie T, Shinki R, Ogura Y, Kusakabe TG, Satoh N, Sasakura Y. Ependymal cells of chordate larvae are stem-like cells that form the adult nervous system. Nature. 2011;469:525–528. doi: 10.1038/nature09631. [DOI] [PubMed] [Google Scholar]
- Hotta K, Takahashi H, Satoh N, Gojobori T. Brachyury-downstream gene sets in a chordate, Ciona intestinalis: integrating notochord specification, morphogenesis and chordate evolution. Evol Dev. 2008;10:37–51. doi: 10.1111/j.1525-142X.2007.00212.x. [DOI] [PubMed] [Google Scholar]
- Hudson C, Darras S, Caillol D, Yasuo H, Lemaire P. A conserved role for the MEK signalling pathway in neural tissue specification and posteriorisation in the invertebrate chordate, the ascidian Ciona intestinalis. Development. 2003;130:147–159. doi: 10.1242/dev.00200. [DOI] [PubMed] [Google Scholar]
- Hudson C, Yasuo H. A signalling relay involving Nodal and Delta ligands acts during secondary notochord induction in Ciona embryos. Development. 2006;133:2855–2864. doi: 10.1242/dev.02466. [DOI] [PubMed] [Google Scholar]
- Hudson C, Kawai N, Negishi T, Yasuo H. β-Catenin-driven binary fate specification segregates germ layers in ascidian embryos. Curr Biol. 2013;23:491–495. doi: 10.1016/j.cub.2013.02.005. [DOI] [PubMed] [Google Scholar]
- Hunter CS, Dixit S, Cohen T, Ediger B, Wilcox C, Ferreira M, Westphal H, Stein R, May CL. Islet α-, β-, and δ-cell development is controlled by the Ldb1 coregulator, acting primarily with the islet-1 transcription factor. Diabetes. 2013;62:875–886. doi: 10.2337/db12-0952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussain MA, Miller CP, Habener JF. Brn-4 transcription factor expression targeted to the early developing mouse pancreas induces ectopic glucagon gene expression in insulin-producing beta cells. J Biol Chem. 2002;277:16028–16032. doi: 10.1074/jbc.M107124200. [DOI] [PubMed] [Google Scholar]
- Imai K, Takada N, Satoh N, Satou Y. (beta)-catenin mediates the specification of endoderm cells in ascidian embryos. Development. 2000;127:3009–3020. doi: 10.1242/dev.127.14.3009. [DOI] [PubMed] [Google Scholar]
- Imai KS, Satoh N, Satou Y. Early embryonic expression of FGF4/6/9 gene and its role in the induction of mesenchyme and notochord in Ciona savignyi embryos. Development. 2002;129:1729–1738. doi: 10.1242/dev.129.7.1729. [DOI] [PubMed] [Google Scholar]
- Imai KS, Satou Y, Satoh N. Multiple functions of a Zic-like gene in the differentiation of notochord, central nervous system and muscle in Ciona savignyi embryos. Development. 2002;129:2723–2732. doi: 10.1242/dev.129.11.2723. [DOI] [PubMed] [Google Scholar]
- Imai KS, Hino K, Yagi K, Satoh N, Satou Y. Gene expression profiles of transcription factors and signaling molecules in the ascidian embryo: towards a comprehensive understanding of gene networks. Development. 2004;131:4047–4058. doi: 10.1242/dev.01270. [DOI] [PubMed] [Google Scholar]
- Imai KS, Levine M, Satoh N, Satou Y. Regulatory blueprint for a chordate embryo. Science. 2006;312:1183–1187. doi: 10.1126/science.1123404. [DOI] [PubMed] [Google Scholar]
- Imai KS, Stolfi A, Levine M, Satou Y. Gene regulatory networks underlying the compartmentalization of the Ciona central nervous system. Development. 2009;136:285–293. doi: 10.1242/dev.026419. [DOI] [PubMed] [Google Scholar]
- Jacquemin P, Durviaux SM, Jensen J, Godfraind C, Gradwohl G, Guillemot F, Madsen OD, Carmeliet P, Dewerchin M, Collen D, Rousseau GG, Lemaigre FP. Transcription factor hepatocyte nuclear factor 6 regulates pancreatic endocrine cell differentiation and controls expression of the proendocrine gene ngn3. Mol Cell Biol. 2000;20:4445–4454. doi: 10.1128/mcb.20.12.4445-4454.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeffery WR, Chiba T, Krajka FR, Deyts C, Satoh N, Joly JS. Trunk lateral cells are neural crest-like cells in the ascidian Ciona intestinalis: insights into the ancestry and evolution of the neural crest. Dev Biol. 2008;324:152–160. doi: 10.1016/j.ydbio.2008.08.022. [DOI] [PubMed] [Google Scholar]
- Jennings RE, Berry AA, Kirkwood-Wilson R, Roberts NA, Hearn T, Salisbury RJ, Blaylock J, Piper Hanley K, Hanley NA. Development of the human pancreas from foregut to endocrine commitment. Diabetes. 2013;62:3514–3522. doi: 10.2337/db12-1479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen J, Pedersen EE, Galante P, Hald J, Heller RS, Ishibashi M, Kageyama R, Guillemot F, Serup P, Madsen OD. Control of endodermal endocrine development by Hes-1. Nat Genet. 2000;24:36–44. doi: 10.1038/71657. [DOI] [PubMed] [Google Scholar]
- Johnsen AH, Rehfeld JF. Cionin: a disulfotyrosyl hybrid of cholecystokinin and gastrin from the neural ganglion of the protochordate Ciona intestinalis. J Biol Chem. 1990;265:3054–3058. [PubMed] [Google Scholar]
- José-Edwards DS, Kerner P, Kugler JE, Deng W, Jiang D, Di Gregorio A. The identification of transcription factors expressed in the notochord of Ciona intestinalis adds new potential players to the brachyury gene regulatory network. Dev Dyn. 2011;240:1793–1805. doi: 10.1002/dvdy.22656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- José-Edwards DS, Oda-Ishii I, Nibu Y, Di Gregorio A. Tbx2/3 is an essential mediator within the Brachyury gene network during Ciona notochord development. Development. 2013;140:2422–2433. doi: 10.1242/dev.094227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katikala L, Aihara H, Passamaneck YJ, Gazdoiu S, José-Edwards DS, Kugler JE, Oda-Ishii I, Imai JH, Nibu Y, Di Gregorio A. Functional Brachyury binding sites establish a temporal read-out of gene expression in the Ciona notochord. PLoS Biol. 2013;11(10):e1001697. doi: 10.1371/journal.pbio.1001697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawada T, Ogasawara M, Sekiguchi T, Aoyama M, Hotta K, Oka K, Satake H. Peptidomic analysis of the central nervous system of the protochordate, Ciona intestinalis: homologs and prototypes of vertebrate peptides and novel peptides. Endocrinology. 2011;152:2416–2427. doi: 10.1210/en.2010-1348. [DOI] [PubMed] [Google Scholar]
- Kenyon C. The first long-lived mutants: discovery of the insulin/IGF-1 pathway for ageing. Philos Trans R Soc Lond B Biol Sci. 2011;366:9–16. doi: 10.1098/rstb.2010.0276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SK, Hebrok M, Melton DA. Notochord to endoderm signaling is required for pancreas development. Development. 1997;124:4243–4252. doi: 10.1242/dev.124.21.4243. [DOI] [PubMed] [Google Scholar]
- Kinkel MD, Eames SC, Alonzo MR, Prince VE. Cdx4 is required in the endoderm to localize the pancreas and limit beta-cell number. Development. 2008;135:919–929. doi: 10.1242/dev.010660. [DOI] [PubMed] [Google Scholar]
- Konturek SJ, Konturek JW, Pawlik T, Brzozowski T. Brain-gut axis and its role in the control of food intake. J Physiol Pharmacol. 2004;55:137–154. [PubMed] [Google Scholar]
- Korgun ET, Dohr G, Desoye G, Demir R, Kayisli UA, Hahn T. Expression of insulin, insulin-like growth factor I and glucocorticoid receptor in rat uterus and embryo during decidualization, implantation and organogenesis. Reproduction. 2003;125:75–84. doi: 10.1530/rep.0.1250075. [DOI] [PubMed] [Google Scholar]
- Krapp A, Knöfler M, Ledermann B, Bürki K, Berney C, Zoerkler N, Hagenbüchle O, Wellauer PK. The bHLH protein PTF1-p48 is essential for the formation of the exocrine and the correct spatial organization of the endocrine pancreas. Genes Dev. 1998;12:3752–3763. doi: 10.1101/gad.12.23.3752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubo A, Suzuki N, Yuan X, Nakai K, Satoh N, Imai KS, Satou Y. Genomic cis-regulatory networks in the early Ciona intestinalis embryo. Development. 2010;137:1613–1623. doi: 10.1242/dev.046789. [DOI] [PubMed] [Google Scholar]
- Kuerner KM, Steinbeisser H. Expression analysis of IGFBP-rP10, IGFBP-like and Mig30 in early Xenopus development. Dev Dyn. 2006;235:2861–2867. doi: 10.1002/dvdy.20910. [DOI] [PubMed] [Google Scholar]
- Kusakabe T, Yoshida R, Kawakami I, Kusakabe R, Mochizuki Y, Yamada L, Shin-I T, Kohara Y, Satoh N, Tsuda M, Satou Y. Gene expression profiles in tadpole larvae of Ciona intestinalis. Dev Biol. 2002;242:188–203. doi: 10.1006/dbio.2002.0538. [DOI] [PubMed] [Google Scholar]
- Laser B, Meda P, Constant I, Philippe J. The caudal-related homeodomain protein Cdx-2/3 regulates glucagon gene expression in islet cells. J Biol Chem. 1996;271:28984–28994. doi: 10.1074/jbc.271.46.28984. [DOI] [PubMed] [Google Scholar]
- Lee CS, Sund NJ, Vatamaniuk MZ, Matschinsky FM, Stoffers DA, Kaestner KH. Foxa2 controls Pdx1 gene expression in pancreatic beta-cells in vivo. Diabetes. 2002;51:2546–2451. doi: 10.2337/diabetes.51.8.2546. [DOI] [PubMed] [Google Scholar]
- Lemaire P. Unfolding a chordate developmental program, one cell at a time: invariant cell lineages, short-range inductions and evolutionary plasticity in ascidians. Dev Biol. 2009;332:48–60. doi: 10.1016/j.ydbio.2009.05.540. [DOI] [PubMed] [Google Scholar]
- Liu WD, Wang HW, Muguira M, Breslin MB, Lan MS. INSM1 functions as a transcriptional repressor of the neuroD/beta2 gene through the recruitment of cyclin D1 and histone deacetylases. Biochem J. 2006;397:169–177. doi: 10.1042/BJ20051669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo J, Becnel J, Nichols CD, Nässel DR. Insulin-producing cells in the brain of adult Drosophila are regulated by the serotonin 5-HT1A receptor. Cell Mol Life Sci. 2012;69:471–484. doi: 10.1007/s00018-011-0789-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madsen OD, Jensen J, Petersen HV, Pedersen EE, Oster A, Andersen FG, Jørgensen MC, Jensen PB, Larsson LI, Serup P. Transcription factors contributing to the pancreatic beta-cell phenotype. Horm Metab Res. 1997;29:265–270. doi: 10.1055/s-2007-979035. [DOI] [PubMed] [Google Scholar]
- Madsen OD. Pancreas phylogeny and ontogeny in relation to a ‘pancreatic stem cell’. C R Biol. 2007;330:534–537. doi: 10.1016/j.crvi.2007.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malecki MT, Lebrun P, Pezzolesi M, Warram JH, Krolewski AS, Jhala US. A newly identified mutation in an IPF1 binding site of the insulin gene promoter may predispose to type 2 diabetes mellitus. Diabetologia. 2006;49:1985–1987. doi: 10.1007/s00125-006-0302-8. [DOI] [PubMed] [Google Scholar]
- Manni L, Agnoletto A, Zaniolo G, Burighel P. Stomodeal and neurohypophysial placodes in Ciona intestinalis: insights into the origin of the pituitary gland. J Exp Zool B Mol Dev Evol. 2005;304:324–339. doi: 10.1002/jez.b.21039. [DOI] [PubMed] [Google Scholar]
- Martinelli P, Cañamero M, del Pozo N, Madriles F, Zapata A, Real FX. Gata6 is required for complete acinar differentiation and maintenance of the exocrine pancreas in adult mice. Gut. 2013;62:1481–1488. doi: 10.1136/gutjnl-2012-303328. [DOI] [PubMed] [Google Scholar]
- Mavropoulos A, Devos N, Biemar F, Zecchin E, Argenton F, Edlund H, Motte P, Martial JA, Peers B. sox4b is a key player of pancreatic alpha cell differentiation in zebrafish. Dev Biol. 2005;285:211–223. doi: 10.1016/j.ydbio.2005.06.024. [DOI] [PubMed] [Google Scholar]
- Mazet F, Hutt JA, Milloz J, Millard J, Graham A, Shimeld SM. Molecular evidence from Ciona intestinalis for the evolutionary origin of vertebrate sensory placodes. Dev Biol. 2005;282:494–508. doi: 10.1016/j.ydbio.2005.02.021. [DOI] [PubMed] [Google Scholar]
- McRory J, Sherwood NM. Two protochordate genes encode pituitary adenylate cyclase-activating polypeptide and related family members. Endocrinology. 1997;138:2380–2390. doi: 10.1210/endo.138.6.5167. [DOI] [PubMed] [Google Scholar]
- McRory JE, Sherwood NM. Ancient divergence of insulin and insulin-like growth factor. DNA Cell Biol. 1997;16:939–949. doi: 10.1089/dna.1997.16.939. [DOI] [PubMed] [Google Scholar]
- Metzger DE, Gasperowicz M, Otto F, Cross JC, Gradwohl G, Zaret KS. The transcriptional co-repressor Grg3/Tle3 promotes pancreatic endocrine progenitor delamination and β-cell differentiation. Development. 2012;139:1447–1456. doi: 10.1242/dev.072892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millar RH. Ciona. Liverpool: University of Liverpool Press; 1953. [Google Scholar]
- Munro E, Robin F, Lemaire P. Cellular morphogenesis in ascidians: how to shape a simple tadpole. Curr Opin Genet Dev. 2006;16:399–405. doi: 10.1016/j.gde.2006.06.004. [DOI] [PubMed] [Google Scholar]
- Nakatani Y, Nishida H. Induction of notochord during ascidian embryogenesis. Dev Biol. 1994;166:289–299. doi: 10.1006/dbio.1994.1315. [DOI] [PubMed] [Google Scholar]
- Nakayama-Ishimura A, Chambon JP, Horie T, Satoh N, Sasakura Y. Delineating metamorphic pathways in the ascidian Ciona intestinalis. Dev Biol. 2009;326:357–367. doi: 10.1016/j.ydbio.2008.11.026. [DOI] [PubMed] [Google Scholar]
- Nakashima K, Yamada L, Satou Y, Azuma J, Satoh N. The evolutionary origin of animal cellulose synthase. Dev Genes Evol. 2004;214:81–88. doi: 10.1007/s00427-003-0379-8. [DOI] [PubMed] [Google Scholar]
- Nakazawa K, Yamazawa T, Moriyama Y, Ogura Y, Kawai N, Sasakura Y, Saiga H. Formation of the digestive tract in Ciona intestinalis includes two distinct morphogenic processes between its anterior and posterior parts. Dev Dyn. 2013;242:1172–1183. doi: 10.1002/dvdy.24009. [DOI] [PubMed] [Google Scholar]
- Nässel DR. Neuropeptides in the nervous system of Drosophila and other insects: multiple roles as neuromodulators and neurohormones. Prog Neurobiol. 2002;68:1–84. doi: 10.1016/s0301-0082(02)00057-6. [DOI] [PubMed] [Google Scholar]
- Naya FJ, Huang HP, Qiu Y, Mutoh H, DeMayo FJ, Leiter AB, Tsai MJ. Diabetes, defective pancreatic morphogenesis, and abnormal enteroendocrine differentiation in BETA2/neuroD-deficient mice. Genes Dev. 1997;11:2323–2334. doi: 10.1101/gad.11.18.2323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nibu Y, José-Edwards DS, Di Gregorio A. From notochord formation to hereditary chordoma: the many roles of Brachyury. Biomed Res Int. 2013;2013:826435. doi: 10.1155/2013/826435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishida H. Vegetal egg cytoplasm promotes gastrulation and is responsible for specification of vegetal blastomeres in embryos of the ascidian Halocynthia roretzi. Development. 1996;122:1271–1279. doi: 10.1242/dev.122.4.1271. [DOI] [PubMed] [Google Scholar]
- Nishida H, Kumano G. Analysis of the temporal expression of endoderm-specific alkaline phosphatase during development of the ascidian Halocynthia roretzi. Dev Growth Differ. 1997;39:199–205. doi: 10.1046/j.1440-169x.1997.t01-1-00008.x. [DOI] [PubMed] [Google Scholar]
- Noda T. The maternal genes Ci-p53/p73-a and Ci-p53/p73-b regulate zygotic ZicL expression and notochord differentiation in Ciona intestinalis embryos. Dev Biol. 2011;360:216–229. doi: 10.1016/j.ydbio.2011.08.029. [DOI] [PubMed] [Google Scholar]
- Oda-Ishii I, Di Gregorio A. Lineage-independent mosaic expression and regulation of the Ciona multidom gene in the ancestral notochord. Dev Dyn. 2007;236:1806–1819. doi: 10.1002/dvdy.21213. [DOI] [PubMed] [Google Scholar]
- Ogasawara M, Sasaki A, Metoki H, Shin-i T, Kohara Y, Satoh N, Satou Y. Gene expression profiles in young adult Ciona intestinalis. Dev Genes Evol. 2002;212:173–185. doi: 10.1007/s00427-002-0230-7. [DOI] [PubMed] [Google Scholar]
- Olinski RP, Dahlberg C, Thorndyke M, Hallböök F. Three insulin-relaxin-like genes in Ciona intestinalis. Peptides. 2006b;27:2535–2546. doi: 10.1016/j.peptides.2006.06.008. [DOI] [PubMed] [Google Scholar]
- Olinski RP, Lundin LG, Hallböök F. Conserved synteny between the Ciona genome and human paralogons identifies large duplication events in the molecular evolution of the insulin-relaxin gene family. Mol Biol Evol. 2006a;23:10–22. doi: 10.1093/molbev/msj002. [DOI] [PubMed] [Google Scholar]
- O’Neil GS, Falkmer S, Thorndyke MC. Insulin-like immunoreactivity in the neural ganglion of the ascidian Ciona intestinalis. Acta Zool. 1986;67:147–153. [Google Scholar]
- Owada S, Larsson O, Arkhammar P, Katz AI, Chibalin AV, Berggren PO, Bertorello AM. Glucose decreases Na+,K+-ATPase activity in pancreatic beta-cells. An effect mediated via Ca2+-independent phospholipase A2 and protein kinase C-dependent phosphorylation of the alpha-subunit. J Biol Chem. 1999;274:2000–2008. doi: 10.1074/jbc.274.4.2000. [DOI] [PubMed] [Google Scholar]
- Passamaneck YJ, Di Gregorio A. Ciona intestinalis: chordate development made simple. Dev Dyn. 2005;233:1–19. doi: 10.1002/dvdy.20300. [DOI] [PubMed] [Google Scholar]
- Passamaneck YJ, Katikala L, Perrone L, Dunn MP, Oda-Ishii I, Di Gregorio A. Direct activation of a notochord cis-regulatory module by Brachyury and FoxA in the ascidian Ciona intestinalis. Development. 2009;136:3679–3689. doi: 10.1242/dev.038141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Villamil B, Schwartz PT, Vallejo M. The pancreatic homeodomain transcription factor IDX1/IPF1 is expressed in neural cells during brain development. Endocrinology. 1999;140:3857–3860. doi: 10.1210/endo.140.8.7048. [DOI] [PubMed] [Google Scholar]
- Pezzotti MR, Locascio A, Racioppi C, Fucci L, Branno M. Auto and cross regulatory elements control Onecut expression in the ascidian nervous system. Dev Biol. 2014;390:273–287. doi: 10.1016/j.ydbio.2014.03.011. [DOI] [PubMed] [Google Scholar]
- Pin CL, Rukstalis JM, Johnson C, Konieczny SF. The bHLH transcription factor Mist1 is required to maintain exocrine pancreas cell organization and acinar cell identity. J Cell Biol. 2001;155:519–530. doi: 10.1083/jcb.200105060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pontoglio M, Sreenan S, Roe M, Pugh W, Ostrega D, Doyen A, Pick AJ, Baldwin A, Velho G, Froguel P, Levisetti M, Bonner-Weir S, Bell GI, Yaniv M, Polonsky KS. Defective insulin secretion in hepatocyte nuclear factor 1alpha-deficient mice. J Clin Invest. 1998;101:2215–2222. doi: 10.1172/JCI2548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ralphs JR, Wylie L, Hill DJ. Distribution of insulin-like growth factor peptides in the developing chick embryo. Development. 1990;109:51–58. doi: 10.1242/dev.109.1.51. [DOI] [PubMed] [Google Scholar]
- Reinecke M. Immunohistochemical localization of polypeptide hormones in endocrine cells of the digestive tract of Branchiostoma lanceolatum. Cell Tissue Res. 1981;219:445–456. doi: 10.1007/BF00209985. [DOI] [PubMed] [Google Scholar]
- Reinecke M, Betzler D, Drakenberg K, Falkmer S, Sara VR. Occurrence of members of the insulin superfamily in central nervous system and digestive tract of protochordates. Histochemistry. 1993;99:277–285. doi: 10.1007/BF00269100. [DOI] [PubMed] [Google Scholar]
- Reinecke M, Collet C. The phylogeny of the insulin-like growth factors. Int Rev Cytol. 1998;183:1–94. doi: 10.1016/s0074-7696(08)60142-4. [DOI] [PubMed] [Google Scholar]
- Reinecke M, Eppler E, David I, Georges D. Immunohistochemical evidence for the presence, localization and partial coexistence of insulin, insulin-like growth factor I and relaxin in the protochordate Ciona intestinalis. Cell Tissue Res. 1999;295:331–338. doi: 10.1007/s004410051239. [DOI] [PubMed] [Google Scholar]
- Reverberi G, Minganti A. Fenomeni di evocazione nello sviluppo dell’uovo di Ascidie. Risultati dell’indagine sperimentale sull’uovo di Ascidiella aspersa e Ascidiella malaca allo stadio di 8 blastomeri. Pubbl Stn Zool Napoli. 1946;20:199–252. [Google Scholar]
- Ristoratore F, Spagnuolo A, Aniello F, Branno M, Fabbrini F, Di Lauro R. Expression and functional analysis of Cititf1, an ascidian NK-2 class gene, suggest its role in endoderm development. Development. 1999;126:5149–5159. doi: 10.1242/dev.126.22.5149. [DOI] [PubMed] [Google Scholar]
- Ritz-Laser B, Mamin A, Brun T, Avril I, Schwitzgebel VM, Philippe J. The zinc finger-containing transcription factor Gata-4 is expressed in the developing endocrine pancreas and activates glucagon gene expression. Mol Endocrinol. 2005;19:759–770. doi: 10.1210/me.2004-0051. [DOI] [PubMed] [Google Scholar]
- Roche J, Salvatore G, Rametta G. On the presence and the biosynthesis of thyroid hormones in a tunicate, Ciona intestinalis L. Biochim Biophys Acta. 1962;63:154–165. doi: 10.1016/0006-3002(62)90348-7. [DOI] [PubMed] [Google Scholar]
- Rukstalis JM, Habener JF. Neurogenin3: a master regulator of pancreatic islet differentiation and regeneration. Islets. 2009;1:177–184. doi: 10.4161/isl.1.3.9877. [DOI] [PubMed] [Google Scholar]
- Sasaki A, Miyamoto Y, Satou Y, Satoh N, Ogasawara M. Novel endostyle-specific genes in the ascidian Ciona intestinalis. Zoolog Sci. 2003;20:1025–1030. doi: 10.2108/zsj.20.1025. [DOI] [PubMed] [Google Scholar]
- Sasakura Y, Nakashima K, Awazu S, Matsuoka T, Nakayama A, Azuma J, Satoh N. Transposon-mediated insertional mutagenesis revealed the functions of animal cellulose synthase in the ascidian Ciona intestinalis. Proc Natl Acad Sci U S A. 2005;102:15134–15139. doi: 10.1073/pnas.0503640102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato Y, Terakado K, Morisawa M. Test cell migration and tunic formation during post-hatching development of the larva of the ascidian, Ciona intestinalis. Dev Growth Differ. 1997;39:117–126. doi: 10.1046/j.1440-169x.1997.00013.x. [DOI] [PubMed] [Google Scholar]
- Satoh N. Developmental genomics of ascidians. New Jersey: Wiley-Blackwell; 2014. [Google Scholar]
- Satou Y, Takatori N, Yamada L, Mochizuki Y, Hamaguchi M, Ishikawa H, Chiba S, Imai K, Kano S, Murakami SD, Nakayama A, Nishino A, Sasakura Y, Satoh G, Shimotori T, Shin-I T, Shoguchi E, Suzuki MM, Takada N, Utsumi N, Yoshida N, Saiga H, Kohara Y, Satoh N. Gene expression profiles in Ciona intestinalis tailbud embryos. Development. 2001;128:2893–2904. doi: 10.1242/dev.128.15.2893. [DOI] [PubMed] [Google Scholar]
- Satou Y, Yamada L, Mochizuki Y, Takatori N, Kawashima T, Sasaki A, Hamaguchi M, Awazu S, Yagi K, Sasakura Y, Nakayama A, Ishikawa H, Inaba K, Satoh N. A cDNA resource from the basal chordate Ciona intestinalis. Genesis. 2002;33:153–154. doi: 10.1002/gene.10119. [DOI] [PubMed] [Google Scholar]
- Satou Y, Sasakura Y, Yamada L, Imai KS, Satoh N, Degnan B. A genomewide survey of developmentally relevant genes in Ciona intestinalis V. Genes for receptor tyrosine kinase pathway and Notch signaling pathway. Dev Genes Evol. 2003;213:254–263. doi: 10.1007/s00427-003-0317-9. [DOI] [PubMed] [Google Scholar]
- Satou Y, Imai KS, Satoh N. The ascidian Mesp gene specifies heart precursor cells. Development. 2004;131:2533–2541. doi: 10.1242/dev.01145. [DOI] [PubMed] [Google Scholar]
- Satou Y, Kawashima T, Shoguchi E, Nakayama A, Satoh N. An integrated database of the ascidian, Ciona intestinalis: towards functional genomics. Zoolog Sci. 2005;22:837–843. doi: 10.2108/zsj.22.837. [DOI] [PubMed] [Google Scholar]
- Satou Y, Mineta K, Ogasawara M, Sasakura Y, Shoguchi E, Ueno K, Yamada L, Matsumoto J, Wasserscheid J, Dewar K, Wiley GB, Macmil SL, Roe BA, Zeller RW, Hastings KE, Lemaire P, Lindquist E, Endo T, Hotta K, Inaba K. Improved genome assembly and evidence-based global gene model set for the chordate Ciona intestinalis: new insight into intron and operon populations. Genome Biol. 2008;9:R152. doi: 10.1186/gb-2008-9-10-r152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schisler JC, Jensen PB, Taylor DG, Becker TC, Knop FK, Takekawa S, German M, Weir GC, Lu D, Mirmira RG, Newgard CB. The Nkx6.1 homeodomain transcription factor suppresses glucagon expression and regulates glucose-stimulated insulin secretion in islet beta cells. Proc Natl Acad Sci U S A. 2005;102:7297–7302. doi: 10.1073/pnas.0502168102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sander M, Neubüser A, Kalamaras J, Ee HC, Martin GR, German MS. Genetic analysis reveals that PAX6 is required for normal transcription of pancreatic hormone genes and islet development. Genes Dev. 1997;11:1662–1673. doi: 10.1101/gad.11.13.1662. [DOI] [PubMed] [Google Scholar]
- Seymour PA, Freude KK, Tran MN, Mayes EE, Jensen J, Kist R, Scherer G, Sander M. SOX9 is required for maintenance of the pancreatic progenitor cell pool. Proc Natl Acad Sci U S A. 2007;104:1865–1870. doi: 10.1073/pnas.0609217104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi W, Levine M. Ephrin signaling establishes asymmetric cell fates in an endomesoderm lineage of the Ciona embryo. Development. 2008;135:931–940. doi: 10.1242/dev.011940. [DOI] [PubMed] [Google Scholar]
- Shih DQ, Navas MA, Kuwajima S, Duncan SA, Stoffel M. Impaired glucose homeostasis and neonatal mortality in hepatocyte nuclear factor 3alpha-deficient mice. Proc Natl Acad Sci U S A. 1999;96:10152–10157. doi: 10.1073/pnas.96.18.10152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimeld SM, Donoghue PC. Evolutionary crossroads in developmental biology: cyclostomes (lamprey and hagfish) Development. 2012;139:2091–2099. doi: 10.1242/dev.074716. [DOI] [PubMed] [Google Scholar]
- Shoguchi E, Kawashima T, Satou Y, Hamaguchi M, Sin-I T, Kohara Y, Putnam N, Rokhsar DS, Satoh N. Chromosomal mapping of 170 BAC clones in the ascidian Ciona intestinalis. Genome Res. 2006;16:297–303. doi: 10.1101/gr.4156606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smit AB, van Kesteren RE, Li KW, Van Minnen J, Spijker S, Van Heerikhuizen H, Geraerts WP. Towards understanding the role of insulin in the brain: lessons from insulin-related signaling systems in the invertebrate brain. Prog Neurobiol. 1998;54:35–54. doi: 10.1016/s0301-0082(97)00063-4. [DOI] [PubMed] [Google Scholar]
- Sommer J, Itani DM, Homlar KC, Keedy VL, Halpern JL, Holt GE, Schwartz HS, Coffin CM, Kelley MJ, Cates JM. Methylthioadenosine phosphorylase and activated insulin-like growth factor-1 receptor/insulin receptor: potential therapeutic targets in chordoma. J Pathol. 2010;220:608–617. doi: 10.1002/path.2679. [DOI] [PubMed] [Google Scholar]
- Sosa-Pineda B, Chowdhury K, Torres M, Oliver G, Gruss P. The Pax4 gene is essential for differentiation of insulin-producing beta cells in the mammalian pancreas. Nature. 1997;386:399–402. doi: 10.1038/386399a0. [DOI] [PubMed] [Google Scholar]
- Spence JR, Lange AW, Lin SC, Kaestner KH, Lowy AM, Kim I, Whitsett JA, Wells JM. Sox17 regulates organ lineage segregation of ventral foregut progenitor cells. Dev Cell. 2009;17:62–74. doi: 10.1016/j.devcel.2009.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoffers DA, Thomas MK, Habener JF. Homeodomain protein IDX-1: a master regulator of pancreas development and insulin gene expression. Trends Endocrinol Metab. 1997;8:145–151. doi: 10.1016/s1043-2760(97)00008-8. [DOI] [PubMed] [Google Scholar]
- Stolfi A, Gainous TB, Young JJ, Mori A, Levine M, Christiaen L. Early chordate origins of the vertebrate second heart field. Science. 2010;329:565–568. doi: 10.1126/science.1190181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sussel L, Kalamaras J, Hartigan-O’Connor DJ, Meneses JJ, Pedersen RA, Rubenstein JL, German MS. Mice lacking the homeodomain transcription factor Nkx2.2 have diabetes due to arrested differentiation of pancreatic beta cells. Development. 1998;125:2213–2221. doi: 10.1242/dev.125.12.2213. [DOI] [PubMed] [Google Scholar]
- Takahashi H, Hotta K, Erives A, Di Gregorio A, Zeller RW, Levine M, Satoh N. Brachyury downstream notochord differentiation in the ascidian embryo. Genes Dev. 1999;13:1519–1523. doi: 10.1101/gad.13.12.1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takamura K, Fujimura M, Yamaguchi Y. Primordial germ cells originate from the endodermal strand cells in the ascidian Ciona intestinalis. Dev Genes Evol. 2002;212:11–18. doi: 10.1007/s00427-001-0204-1. [DOI] [PubMed] [Google Scholar]
- Taleb N, Polychronakos C. RFX6 is needed for the development and maintenance of the β-cell phenotype. Islets. 2011;3:291–293. doi: 10.4161/isl.3.5.15944. [DOI] [PubMed] [Google Scholar]
- Tanaka KJ, Ogasawara M, Makabe KW, Satoh N. Expression of pharyngeal gill-specific genes in the ascidian Halocynthia roretzi. Dev Genes Evol. 1996;206:218–226. doi: 10.1007/s004270050047. [DOI] [PubMed] [Google Scholar]
- Taylor BL, Liu FF, Sander M. Nkx6.1 is essential for maintaining the functional state of pancreatic beta cells. Cell Rep. 2013;4:1262–1275. doi: 10.1016/j.celrep.2013.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorndyke MC. Observations on the gastric epithelium of ascidians with special reference to Styela clava. Cell Tissue Res. 1977;184:539–550. doi: 10.1007/BF00220977. [DOI] [PubMed] [Google Scholar]
- Vinther TN, Norrman M, Ribel U, Huus K, Schlein M, Steensgaard DB, Pedersen TÅ, Pettersson I, Ludvigsen S, Kjeldsen T, Jensen KJ, Hubálek F. Insulin analog with additional disulfide bond has increased stability and preserved activity. Protein Sci. 2013;22:296–305. doi: 10.1002/pro.2211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wada S, Tokuoka M, Shoguchi E, Kobayashi K, Di Gregorio A, Spagnuolo A, Branno M, Kohara Y, Rokhsar D, Levine M, Saiga H, Satoh N, Satou Y. A genomewide survey of developmentally relevant genes in Ciona intestinalis II. Genes for homeobox transcription factors. Dev Genes Evol. 2003;213:222–234. doi: 10.1007/s00427-003-0321-0. [DOI] [PubMed] [Google Scholar]
- Wada H. Origin and genetic evolution of the vertebrate skeleton. Zoolog Sci. 2010;27:119–123. doi: 10.2108/zsj.27.119. [DOI] [PubMed] [Google Scholar]
- Wang H, Maechler P, Antinozzi PA, Hagenfeldt KA, Wollheim CB. Hepatocyte nuclear factor 4alpha regulates the expression of pancreatic beta -cell genes implicated in glucose metabolism and nutrient-induced insulin secretion. J Biol Chem. 2000;275:35953–35959. doi: 10.1074/jbc.M006612200. [DOI] [PubMed] [Google Scholar]
- Wang W, Razy-Krajka F, Siu E, Ketcham A, Christiaen L. NK4 antagonizes Tbx1/10 to promote cardiac versus pharyngeal muscle fate in the ascidian second heart field. PLoS Biol. 2013;11(12):e1001725. doi: 10.1371/journal.pbio.1001725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner H, Leroith D. Insulin and insulin-like growth factor receptors in the brain: Physiological and pathological aspects. Eur Neuropsychopharmacol. 2014 Jan 31; doi: 10.1016/j.euroneuro.2014.01.020. pii: S0924-977X(14)00041-8. [DOI] [PubMed] [Google Scholar]
- Westmoreland JJ, Kilic G, Sartain C, Sirma S, Blain J, Rehg J, Harvey N, Sosa-Pineda B. Pancreas-specific deletion of Prox1 affects development and disrupts homeostasis of the exocrine pancreas. Gastroenterology. 2012;142:999–1009.e6. doi: 10.1053/j.gastro.2011.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilcox CL, Terry NA, Walp ER, Lee RA, May CL. Pancreatic β-cell specific deletion of mouse Arx leads to β-cell identity loss. PLoS One. 2013;13;8(6):e66214. doi: 10.1371/journal.pone.0066214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- William CM, Tanabe Y, Jessell TM. Regulation of motor neuron subtype identity by repressor activity of Mnx class homeodomain proteins. Development. 2003;130:1523–1536. doi: 10.1242/dev.00358. [DOI] [PubMed] [Google Scholar]
- Wilson S, Falkmer S. Starfish insulin. Can J Biochem. 1965;43:1615–1624. doi: 10.1139/o65-178. [DOI] [PubMed] [Google Scholar]
- White MF. The insulin signalling system and the IRS proteins. Diabetologia. 1997;40(Suppl 2):S2–17. doi: 10.1007/s001250051387. [DOI] [PubMed] [Google Scholar]
- White YA, Kyle JT, Wood AW. Targeted gene knockdown in zebrafish reveals distinct intraembryonic functions for insulin-like growth factor II signaling. Endocrinology. 2009;150:4366–4375. doi: 10.1210/en.2009-0356. [DOI] [PubMed] [Google Scholar]
- Whittaker JR. Segregation during cleavage of a factor determining endodermal alkaline phosphatase development in ascidian embryos. J Exp Zool. 1977;202:139–153. doi: 10.1002/jez.1402020202. [DOI] [PubMed] [Google Scholar]
- Yagi K, Satou Y, Satoh N. A zinc finger transcription factor, ZicL, is a direct activator of Brachyury in the notochord specification of Ciona intestinalis. Development. 2004;131:1279–1288. doi: 10.1242/dev.01011. [DOI] [PubMed] [Google Scholar]
- Yang Y, Chang BH, Samson SL, Li MV, Chan L. The Krüppel-like zinc finger protein Glis3 directly and indirectly activates insulin gene transcription. Nucleic Acids Res. 2009;37:2529–2538. doi: 10.1093/nar/gkp122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasuo H, Hudson C. FGF8/17/18 functions together with FGF9/16/20 during formation of the notochord in Ciona embryos. Dev Biol. 2007;302:92–103. doi: 10.1016/j.ydbio.2006.08.075. [DOI] [PubMed] [Google Scholar]
- Yonge CM. Studies on the comparative phisiology of digestion. III. Secretion, digestion, and assimilation in the gut of Ciona intestinalis. British J Exp Biol. 1925;2:373–388. [Google Scholar]
- Youson JH, Al-Mahrouki AA. Ontogenetic and phylogenetic development of the endocrine pancreas (islet organ) in fish. Gen Comp Endocrinol. 1999;116:303–335. doi: 10.1006/gcen.1999.7376. [DOI] [PubMed] [Google Scholar]
- Yuan XN, Jiang XY, Pu JW, Li ZR, Zou SM. Functional conservation and divergence of duplicated insulin-like growth factor 2 genes in grass carp (Ctenopharyngodon idellus) Gene. 2011;470:46–52. doi: 10.1016/j.gene.2010.10.003. [DOI] [PubMed] [Google Scholar]
- Zeller RW, Weldon DS, Pellatiro MA, Cone AC. Optimized green fluorescent protein variants provide improved single cell resolution of transgene expression in ascidian embryos. Dev Dyn. 2006;235:456–467. doi: 10.1002/dvdy.20644. [DOI] [PubMed] [Google Scholar]
- Zhang C, Moriguchi T, Kajihara M, Esaki R, Harada A, Shimohata H, Oishi H, Hamada M, Morito N, Hasegawa K, Kudo T, Engel JD, Yamamoto M, Takahashi S. MafA is a key regulator of glucose-stimulated insulin secretion. Mol Cell Biol. 2005;25:4969–4976. doi: 10.1128/MCB.25.12.4969-4976.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou S, Kamei H, Modi Z, Duan C. Zebrafish IGF genes: gene duplication, conservation and divergence, and novel roles in midline and notochord development. PLoS One. 2009;4(9):e7026. doi: 10.1371/journal.pone.0007026. [DOI] [PMC free article] [PubMed] [Google Scholar]