Abstract
The development and application of methods for the laboratory evolution of biomolecules has rapidly progressed over the last few decades. Advancements in continuous microbe culturing and selection design have facilitated the development of new technologies that enable the continuous directed evolution of proteins and nucleic acids. These technologies have the potential to support the extremely rapid evolution of biomolecules with tailor-made functional properties. Continuous evolution methods must support all of the key steps of laboratory evolution—translation of genes into gene products, selection or screening, replication of genes encoding the most fit gene products, and mutation of surviving genes—in a self-sustaining manner that requires little or no researcher intervention. Continuous laboratory evolution has been historically used to study problems including antibiotic resistance, organismal adaptation, phylogenetic reconstruction, and host-pathogen interactions, with more recent applications focusing on the rapid generation of proteins and nucleic acids with useful, tailor-made properties. The advent of increasingly general methods for continuous directed evolution should enable researchers to address increasingly complex questions and to access biomolecules with more novel or even unprecedented properties.
Introduction
Initially demonstrated as a surrogate for Darwinian evolution by Spiegelman and coworkers almost five decades ago [1], continuous directed evolution has long been envisioned as a potentially highly efficient method to discover novel biomolecules with activities of interest. In Spiegelman’s seminal study, Q bacteriophage genomic RNA was amplified based on its ability to serve as a substrate for purified Q RNA-dependent RNA-replicase. After 74 serial passages of this RNA genome, a variant was produced that was both 83% smaller and could be replicated 15 times faster than the starting Q RNA genome. Importantly, this study highlighted the critical nature of selection pressure design, as the evolved Q RNA genome could be replicated at a significantly faster rate than the parental genome, but could no longer direct the synthesis of viral particles since this requirement was not implicit in the selection.
Thirty years later, Joyce and coworkers described the first system for the continuous directed evolution of catalytic function. The researchers established a self-sustaining RNA replication cycle for RNA molecules capable of catalyzing their own ligation to an RNA-DNA substrate [2]. Like the Q system, the RNA ligase ribozyme continuous evolution system relied on exogenously supplied materials (reverse transcriptase and T7 RNA polymerase). Since these early studies, a number of additional continuous directed evolution systems have been described, the majority of which are limited to either catalytic RNAs or involve the evolution of alternative RNA functions [2–7]. Additionally, these methods have been exclusively carried out in vitro, where the comparative ease of manipulation and selection stringency adjustment can be exploited compared to in vivo methods. Nevertheless, these early landmark studies vividly demonstrated the potential of continuous directed evolution and provided a foundation for subsequent advances.
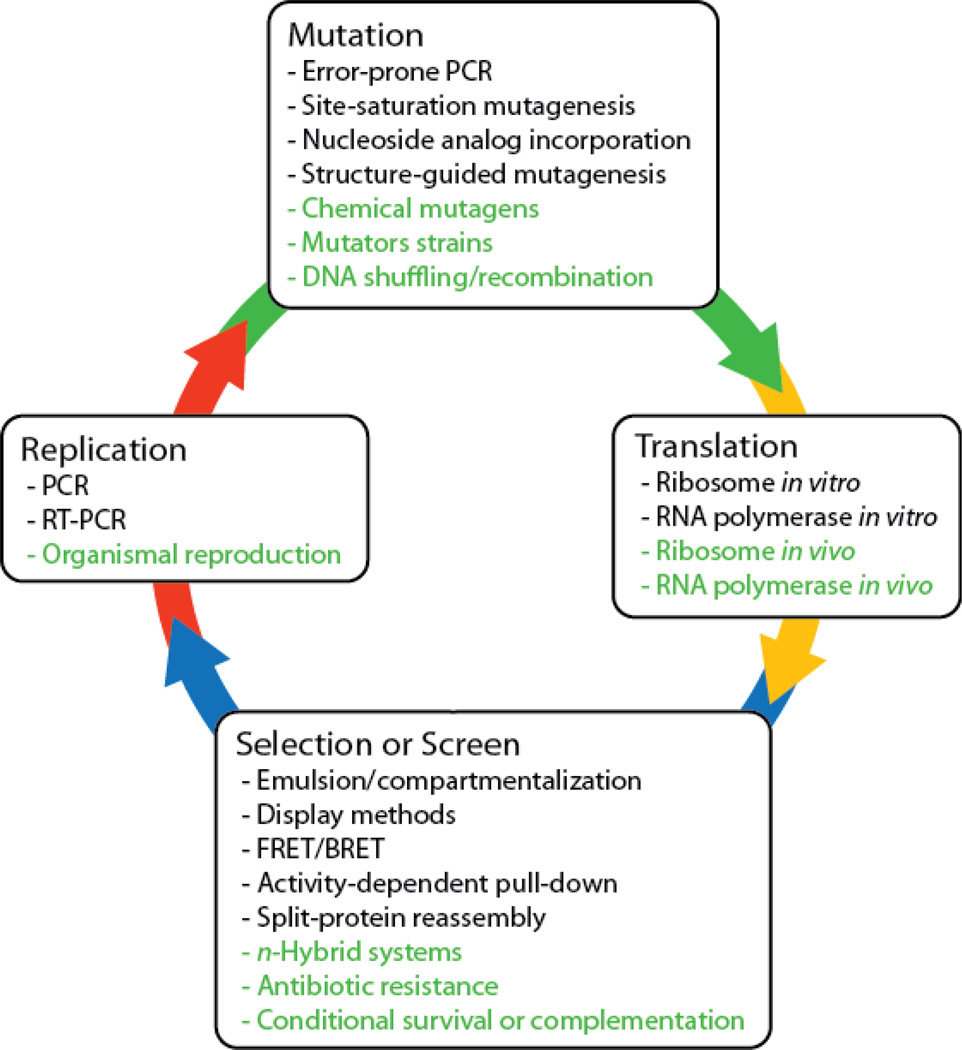
All forms of Darwinian evolution must support four fundamental processes: translation (when the evolving molecule is not identical to the information carrier), selection, replication, and mutation (Figure 1). Traditional directed evolution methods handle each of these processes discretely, frequently requiring steps in which the researcher must perform experimental manipulations. In contrast, continuous directed evolution systems must seamlessly integrate all of these processes into an uninterrupted cycle. For the purposes of this review, we define continuous directed evolution as a general method capable of evolving a specific phenotype, at the level of an organism or a set of genes, over many cycles of mutation, selection, and replication with minimal researcher intervention. A continuous evolution method therefore must support all four stages of Darwinian evolution and allow the surviving genes from one generation to spontaneously enter the translation, selection, replication, and mutation processes of the subsequent generation.
Figure 1. Schematic representation of directed evolution.
All complete directed evolution methods must provide four major components: translation, selection or screening, replication, and mutation. Historical examples of each of these components are listed. Techniques that are particularly amenable to in vivo continuous evolution are shown in green.
Many key studies on continuous evolution have relied on manual serial dilution of the evolving population as a mechanism for propagating genes and controlling selection stringency. We do not consider this technical manipulation to violate our definition, as such systems have been shown to be amenable to automation, yielding similar results [8]. While in vivo continuous evolution methods have exploited the natural occurrence of the three steps of Darwinian evolution in living organisms, these methods have also historically been constrained by modest scope, often focusing on easily selected phenotypes such as antibiotic resistance rather than traits associated with diverse applications and unmet needs. Recently, more general in vivo continuous directed evolution methods have been developed using various organisms, ranging from viruses to bacteria to higher eukaryotes. In this review, we highlight a sampling of recent developments in the field of in vivo continuous directed evolution over the past two decades and their impact, both realized and prospective, on the directed evolution community.
Viral Continuous Evolution
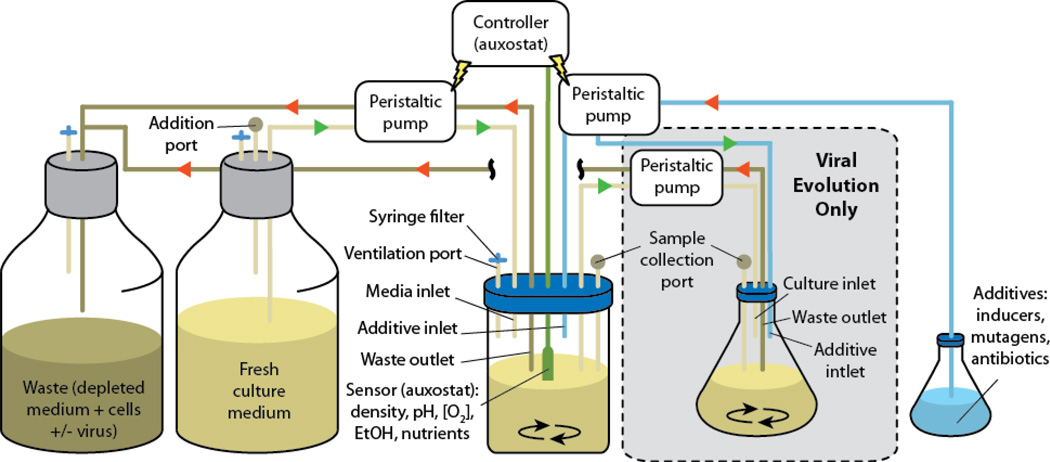
With their high mutation rates [9] and relative ease of manipulation and study, bacteriophages were used early on as model organisms for rapid directed evolution of readily selectable traits [10]. Moreover, the development of methods for continually culturing bacterial strains has greatly reduced the barrier for studying bacteriophage evolution under continuous selection pressure [10,11]. Of historical note is the early and widespread use of both chemostats, cultures maintained through the continuous inflow of fresh growth medium, and auxostats, cultures regulated by a feedback mechanism that controls the inflow of growth medium based on a monitored culture parameter (Figure 2). Both methods have proven useful in continuous culturing of bacteriophage-infectable microbes. Methods for bacterial continuous evolution will be covered in the next section.
Figure 2. Auxostats and chemostats.
Fresh culture medium is pumped into a pre-sterilized container containing the microorganism of interest. Medium with cells is pumped out of the culture container. The rates of medium input and output are held constant in a chemostat system, and the growth rate is regulated by the composition of the medium. In an auxostat system, the medium flow rates are dynamically regulated by a controller in response to measurements made in the growing culture, which correspond directly (turbidity meter) or indirectly (other sensors) to the culture density. In schemes for viral continuous evolution, the auxostat or chemostat culture is pumped into a new vessel where the virus of interest is supplied (cellstat). Both cultures can be supplemented with additives supplied through a dedicated inlet. Selection stringency can be regulated by varying flow rates, changing temperature, or adding compounds to the auxostat, chemostat or cellstat. The depleted medium, cells and virus are pumped to a waste container.
Among the earliest examples of viral continuous evolution is the 1997 work of Molineux and coworkers using X174, a bacteriophage capable of propagating using E. coli or S. typhimurium [12,13]. These studies highlighted the ability of this phage to rapidly adjust to novel environments such as higher temperatures, as well as an abnormally high degree of mutational convergence depending on selection conditions. Additionally, substantial improvements in phage fitness were observed over relatively short time frames, with an average of 1–2 mutations per 24 hours (corresponding to 0.2–0.5 % of the viral genome, and 0.003–0.005 mutations per generation per kilobase) in the absence of any added mutagens. In an expansion of this work by Bull and coworkers, X174 phage was propagated on bacterial hosts under native conditions for six months, corresponding to ~13,000 phage generations [14]. Mutations continued to accumulate at a constant rate for the majority of the experiment, suggesting a potential arms race within the propagating phage pool. The researchers speculated that this arms race was driven by the high concentration of phage with respect to host cells, resulting in high levels of co-infection and competition between phage. While the mutation rates observed under these conditions were high with respect to the size of the viral genome and sufficient for rapid phage adaptation, higher rates are typically required to access gene-specific evolution goals (1–2 mutations per round per gene) [15].
Similar phage propagation experiments were carried out by Molineux and coworkers using bacteriophage T7 to study organismal phylogenetic histories [16]. To enhance the mutation rate, T7 phage was serially passaged in the presence of the chemical mutagen MNNG (N-methyl-N’-nitro-N’-nitrosoguanidine), resulting in variants with differential restriction enzyme cleavage patterns than the wild-type phage. The correct phylogeny of the diverging phage populations could be determined using these cleavage patterns, supporting their method for phylogenetic reconstruction and suggesting maximization of parsimony as a promising method for determining ancestral characteristics. While this approach addressed the need for high mutagenesis rates during evolution, the selection was highly qualitative, requiring the accumulation of non-deleterious mutations that enabled restriction enzyme profiling. A similar strategy was later used by the same group to study the effects of successive severe population bottlenecking and expansion on viral fitness, demonstrating the remarkable plasticity of the T7 genome towards mutations and the interdependence of the accumulated mutations in specific phage linages [17]. These studies also highlight the utility of user-defined mutagenesis rates in continuous directed evolution, although chemical mutagens generally offer narrow mutational spectra and are potent carcinogens.
Recently, T7 phage was used by Barrick and coworkers to study organismal adaptation to an expanded genetic code [18]. Two versions of this phage, wild-type T7 and the 2 T7 hypermutator, were serially propagated using an E. coli strain capable of efficiently incorporating 3-iodotyrosine at amber stop codons. The use of a genetically encodable hypermutator resulted in broad mutational load to the T7 phage without the hazards typically associated with alternative chemical mutagens. Notably, two amber codon substitutions reached high frequencies in the evolving populations: Tyr88Amber in T7 RNA polymerase (T7 RNAP) and Tyr39Amber in T7 type II holin. Whereas the mutation in the RNAP was assumed to be tolerated due to a lack of interaction of this surface-exposed side chain with the remainder of the protein, the mutation in the type II holin was shown to be critical for function in the amber suppressor strain. The findings of this study suggest that an expanded genetic code may increase the evolvability of an organism.
Lambda bacteriophage has been used extensively to study host-virus interactions. Lambda phage natively recognizes LamB as the main point of entry into E. coli. Using a strictly lytic lambda phage, Lenski and coworkers studied the ability of this phage to continuously evolve the ability to use alternative outer membrane receptors as the point of entry [19]. Upon co-culturing this phage with host cells in glucose-limited conditions, the bacteria rapidly evolved resistance through downregulation of lamB expression. The mutating pool of lambda phage then evolved compensatory mutations in the LamB recognition protein J, resulting in improved fitness on LamB, as well as facilitating the use a new receptor, OmpF. In response, the E. coli host reduced fitness of the mutant lambda phage through mutations to manYZ, two proteins that form an inner membrane channel required for lambda phage entry. This coevolution highlights the utility of continuous evolution methods in revealing the dynamic interplay between host and virus evolution.
While these systems have enhanced our understanding of fundamental aspects of evolution, they generally are limited by an uncontrolled (usually low) mutagenesis rate intrinsic to the bacteriophage and host, and/or the requirement for broad organismal adaptation to an environmental challenge, rather than enabling the evolution of a specific biomolecular activity of interest. Our group has developed a bacteriophage-based continuous evolution system that attempts to address these two limitations. Phage-assisted continuous evolution (PACE) is a general system for the directed evolution of biomolecules that relies on previously discussed methods for the continued culturing of E. coli, can be used to evolve, in principle, any activity that can be coupled to gene transcription in E. coli, and allows real-time modulation of selection stringency and mutagenesis levels [20].
Briefly, PACE takes advantage of the critical role of the minor coat protein pIII from the filamentous bacteriophage M13 during both infection and progeny release [21]. Phage lacking functional pIII have virtually no ability to propagate and are rapidly lost under continuous culturing conditions. However, host cell-provided pIII restores the virulence of the phage and allows robust phage propagation in continuous culture. Accordingly, phage-borne genes encoding the ability to trigger pIII in host cells gain a fitness advantage and propagate at the expense of unfit phage. To increase the mutation rate during PACE, an inducible plasmid driving the expression of error-prone E. coli polymerase subunits results in ~100-fold increased mutagenesis with a more-uniform distribution of mutations as compared to traditional chemical mutagens [20]. Due to the continuous flow nature of this system, mutations accumulate only within the phage genome and the gene to be evolved, reducing the likelihood of “cheaters” overtaking the population during selection.
PACE was initially used to evolve novel activity in T7 RNAP [20]. Whereas T7 RNAP efficiently initiates transcription using the native T7 promoter, it lacks detectable activity on the related T3 promoter. Using a combined T7/T3 promoter as an evolutionary stepping-stone, T7 RNAP variants capable of efficiently initiating transcription from the T3 promoter were evolved within a few days. This method has since been expanded to address the effects of varying levels of mutagenesis and selection stringency on enzyme evolution [22], the path dependence of convergent protein evolution [23], the effects of neutral drift, negative selection, and stringency modulation on enzyme evolution [24], and the rapid evolution of protease enzymes from pathogenic viruses that predict how viral proteases evade clinically relevant antiviral drugs [25]. This system has been further modified to enhance its ability to serve as a basis for alternative selection schemes, and ongoing studies use PACE to mediate the continuous directed evolution of DNA-binding domains, protein-protein interactions, genome modification enzymes, proteases, and other enzymes [25].
Bacterial Continuous Evolution
Mechanisms that enable the efficient and unmonitored continuous propagation of bacteria and other microorganisms have served as a key component of many continuous evolution methods. While their utility has been extensively demonstrated for bacteriophage evolution efforts as highlighted in the previous section, these methods have also facilitated the study of continuous directed evolution of bacterial populations, especially in cases where the gene to be evolved is large or cannot be directly coupled to viral fitness. A recent demonstration assesses the evolution of population-wide antibiotic resistance. Using a continuous culture of E. coli, Collins and coworkers added increasing concentrations of the antibiotic norfloxacin to study population dynamics during increasing antibiotic selection pressure [26]. Surprisingly, the researchers found that single members of the population were, on average, less resistant to the antibiotic than the population as a whole. Additionally, resistant members were found to produce high levels of indole, a signaling molecule used by the surrounding, less resistant members to induce protective mechanisms against the antibiotic.
In a related study, Kishony and coworkers followed parallel populations of E. coli under differing antibiotic selection pressures using a bacterial culture device termed the morbidostat [27]. Unlike the previous study in which the concentration of antibiotic is adjusted in 24-hour intervals, the morbidostat continuously adjusts the concentration of the antibiotic to maintain a nearly constant selection pressure (as judged by changes in cell density, thereby making it a variation on the prototypical auxostat). The parallel populations were challenged with different antibiotics, and followed using whole-genome sequencing to assess the nature of the mutations endowing the populations with antibiotic resistance. For chloramphenicol and doxycycline, resistance proceeded through multiple pathways, ultimately resulting in similar levels of antibiotic resistance using different sets of mutations. Conversely, resistance to trimethoprim proceeded through a highly sequential series of mutations in DHFR and its associated promoter in all the parallel populations. Importantly, many resistance mutations that had been previously identified using plate-based methods were not found in this study, highlighting variable fitness costs for some mutations depending on selection conditions, and the stochasticity of evolution under selection conditions for which many potential pathways to increased fitness exist.
For some applications the basal rate of bacterial mutagenesis may not be high enough to provide access to necessary genetic changes on a practical timescale. Methods for the unbiased, yet targeted, mutagenesis of bacteria would dramatically facilitate these applications. An ideal method would require that the evolving gene(s) be replicated by an orthogonal, error-prone polymerase enzyme. One such system uses the E. coli DNA polymerase I (Pol I). Bacteria lacking Pol I are viable, suggesting that manipulation of this polymerase might be tolerated. Loeb and coworkers used previously reported mutations that increase the error rate of Pol I to continuously evolve TEM beta-lactamases capable of hydrolyzing a third-generation lactam antibiotic, aztreonam [28]. Three serial dilutions of a growing bacterial culture in increasing concentrations of aztreonam strongly enriched clinically relevant TEM mutations. The utility of this technique extends beyond the continuous directed evolution of antibiotic resistance, as it also enables the potent and targeted in vivo mutagenesis of a gene of interest. It should be noted, however, that an inverse relationship exists between the length of the gene of interest and the mutagenesis rate using this system and others that depend on Pol I mutants [28,29].
Bacterial continuous evolution has also been used to evolve phenotypes other than antibiotic resistance. The E. coli DNA polymerase III epsilon subunit DnaQ is responsible for the proofreading activity of most DNA replication. Dominant-negative variants of DnaQ provided in trans are known to have dramatic consequences on replication fidelity [30]. Integrating these principles, Ma and coworkers constructed a library of dnaQ mutants using error-prone PCR to be expressed constitutively in continuously growing bacterial cultures [31]. Selection pressures in the form of increasing concentrations of n-butanol or acetic acid were applied to the growing cultures. Plasmids carrying mutagenic dnaQ variants should be enriched at high concentrations of n-butanol or acetic acid because they endow their hosts with the ability to more quickly adapt to the strong selection pressure. Within 24 to 36 days of continuous evolution, the E. coli populations became adapted to the high solvent concentrations. As the researchers predicted, all the dnaQ variants that evolved in surviving cells were more mutagenic than the wildtype dnaQ, increased the mutation rate by up to 2,800-fold, and contained mutations known to dramatically enhance mutagenesis [30,31].
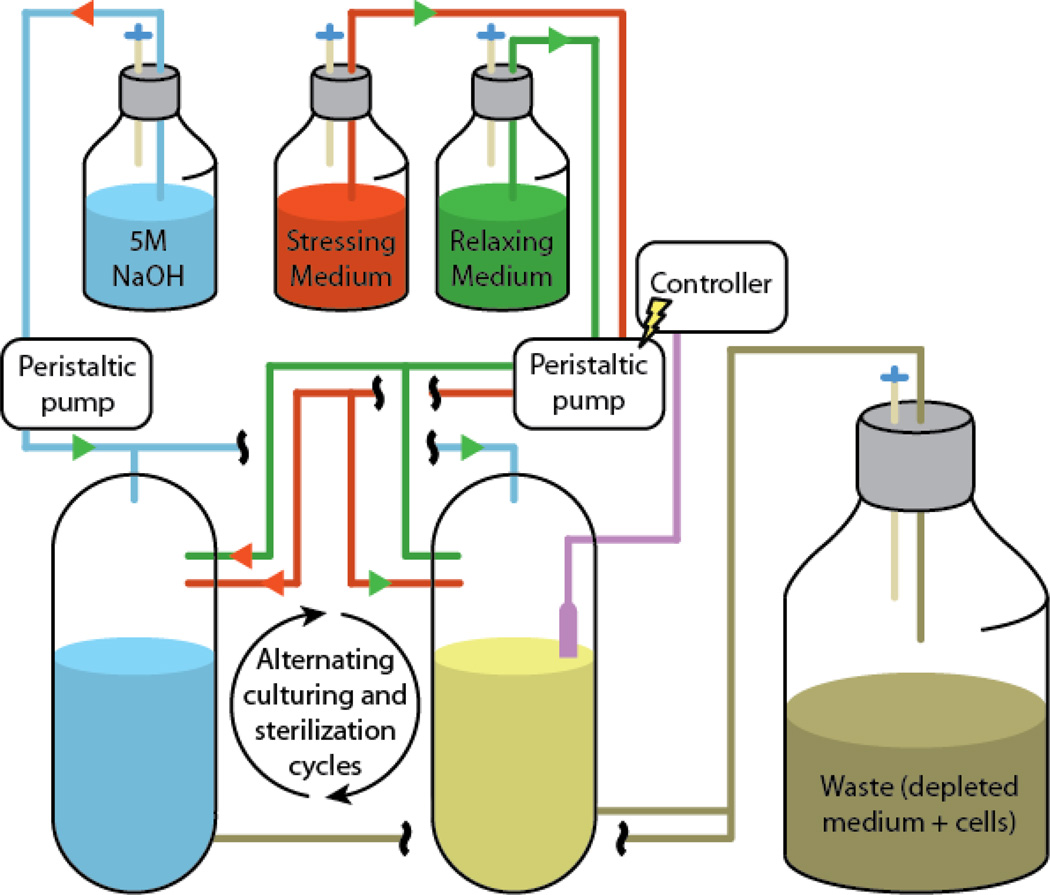
All of these examples of bacterial continuous evolution rely on the manual passaging of the bacteria being studied, or use an automated variant of a chemostat or auxostat. In either case, biofilm formation can dramatically affect the success of the experiment, as bacteria are known to attach to the culture vessel wall upon extended propagation. To ameliorate this issue, an apparatus dubbed the “GM3 cultivation device” was recently developed to allow the maintenance of steady-state microbial growth over extremely long timeframes (Figure 3). This apparatus uses two growth chambers that are cyclically used to culture the microbe of interest, with alternating rounds of chemical sterilization that ensure that members of the population do not escape selections for faster growth through biofilm formation.
Figure 3. The GM3 cultivation device.
A growing culture in the first vessel is supplemented with a defined ratio of relaxing and stressing medium. This ratio is regulated by a controller connected to a turbidity meter measuring cell density. After a defined growth time, the whole culture is transferred to the second vessel to limit selection escape through biofilm formation. Following this transfer, the first vessel is sterilized with 5 M NaOH and washed extensively to limit contamination. After another defined growth period in the second vessel, the growing culture is transferred back to the first vessel, and the cycle is repeated. The selection stringency is modulated through the ratio of the relaxing and stressing media, ultimately reaching up to 100% stressing medium. The depleted medium and cells are pumped to a waste container.
In one application of the GM3 device, Mutzel and coworkers evolved E. coli containing the large-scale substitution of chromosomal thymine with 5-chlorouracil [32]. The researchers used a thymine auxotroph grown in continuous culture on two types of media: a permissive media with all essential nutrients including thymine, and a selective media of similar composition with the exception of thymine, which was substituted with 5-chlorouracil. To slowly modulate selection stringency they mixed varying ratios of the two media, starting from 100% thymine-containing media and slowly transitioning to 100% 5-chlorouracil-containing media over six months. By the end of the experiment, strains with a significant number of mutations or chromosomal rearrangements that could grow using 5-chlorouracil were isolated. These strains were capable of incorporating 5-chlorouracil at levels up to 90% of the chromosomal DNA, with the remaining 10% attributed to thymine. After further optimization, the fraction of chromosomal thymine was reduced to less than 2% through removal of a cryptic pathway to thymine deoxynucleotide production.
In another application of the same device, Hilvert and coworkers continuously evolved an artificial chorismate mutase enzyme that had been previously designed using a nine-amino-acid code [33]. Chorismate mutase natively converts chorismate to prephenate in the shikimate pathway, a critical step in the biosynthesis of the aromatic amino acids phenylalanine and tyrosine. A strain of E. coli lacking wild-type chorismate mutase and expressing the artificial variant was continuously propagated for more than 200 days, initially in low phenylalanine-containing media and slowly transitioning to media lacking both phenylalanine and tyrosine. Bacteria carrying improved chorismate mutase mutants grew at a faster rate in the restrictive media, resulting in their enrichment over the course of the experiment. By the end of the experiment, chorismate mutase variants that expanded the nine amino acid code to ten or eleven amino acids could be isolated, and mutants generally had improved activity as compared to the parent. This study suggests that expanded genetic codes can result in improved evolvability, a similar conclusion to those reached by Barrick and coworkers using 3-iodotyrosine incorporation in T7 bacteriophage [18].
Eukaryotic Continuous Evolution
Viral and bacterial hosts are not suitable for the directed evolution of biomolecules in certain scenarios. In cases of industrial drug precursor production [34] or whole-cell biocatalysis [35], higher organisms with more extensive or relevant biosynthetic pathways may be necessary. Towards this end, continuous directed evolution techniques using eukaryotic organisms have also been developed.
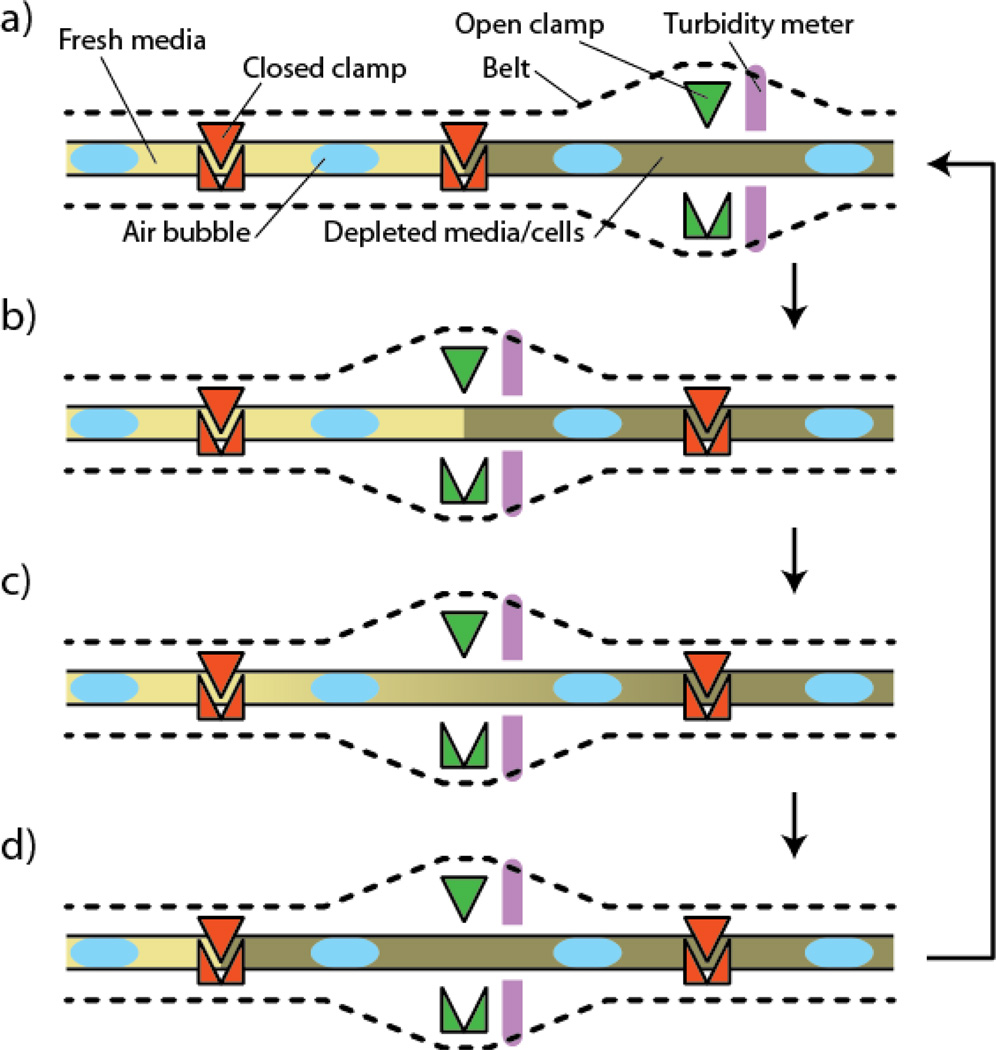
In one of the earliest examples of eukaryotic continuous evolution, the Evolugator, a proprietary continuous culturing device that reduces biofilm formation and wall growth (Figure 4), was used to evolve thermotolerant filamentous fungi. During long-term continuous culturing of filamentous fungi, wall growth can be problematic as fungal cells rapidly aggregate, leading to non-uniform culture dilution. To circumvent this issue, de Crécy-Lagard and coworkers devised an alternative method for continuous fungi culturing that dramatically ameliorates fungal wall growth [36,37]. Using this platform over the course of four months, the researchers evolved two thermotolerant variants of the entomopathogenic fungus Metarhizium anisopliae to facilitate the widespread adoption of this fungus for pest control [37]. While successful, this method for continuous directed evolution requires specialized equipment that cannot be readily generalized to alternative selection methodologies.
Figure 4. Growth chamber of the Evolugator.
a) The growing culture is shown on the right, and is completely contained within a tube clamped at regular intervals to limit unintentional mixing. The density of the culture is measured with a turbidity meter connected to a controller that dynamically regulates the in-flux of new medium. Air bubbles are included at regular intervals to increase the oxygenation level of the medium. b) Upon reaching high density, the right clamp closes, the tubing moves along the track, and the next clamp opens allowing for contact between the growing culture and the fresh medium. c) The fresh medium mixes with the growing culture through diffusion, and the culture resumes growing. d) After sufficient time, the culture again reaches high density. The middle clamp then closes, the tubing shifts on the track once again, the next clamp opens and the new medium is allowed in. This cycle is then continued for the duration of the experiment.
Cornish and coworkers recently noted that many in vivo selection methods do not take advantage of genetic recombination as a source of diversification, and reasoned that the selective induction of double-stranded breaks (DSBs) with high efficiency could dramatically increase the degree of recombination in Saccharomyces cerevisiae [38]. They developed a yeast continuous evolution system made of three parts: a plasmid that tightly controls the expression of the site-specific endonuclease I-SceI, a cassette plasmid that contains regions of homology to the target sequence flanked by I-SceI cleavage sites, and a target plasmid carrying the gene to be mutagenized. Upon induction of the endonuclease, the mutagenic cassette is liberated from its respective plasmid and goes on to mutate the region of interest through homology-directed recombination [38]. Using this system, the authors were able to evolve hisA variants capable of complementing trp1 auxotrophy in yeast in less than one month. This system is accessible and potentially powerful but requires knowledge of the relevant regions of a gene to mutate since genetic diversity is dependent on homology-directed recombination.
One of the most useful features of a continuous directed evolution system is the ability to modulate the mutagenesis of the evolving genes in a manner that does not perturb either the selection or the organism. This feature was one of the major design considerations for the development of the PACE platform [20], in which mutations accumulate only in replicating phage; the error-prone bacterial Pol I variants [28], in which mutations accumulate only in ColE1-type origin vectors; and the I-SceI-based yeast recombination system [38], in which mutations are restricted to regions highly homologous to the mutagenic cassette. All of these systems largely abrogate or avoid host strain mutagenesis. A similar approach was recently developed by Chang Liu and coworkers using a system that is completely contained in yeast. The authors used an autonomous DNA replication system from Kluveromyces lactis to enable the replication of a gene of interest in the S. cerevisiae cytosol [39]. Due to the strict requirements for the orthogonal K. lactis DNA polymerase initiation, no crosstalk was observed between the K. lactis components and the native host machinery. To make this system more amenable to directed evolution, the authors used a homology-guided approach to create error-prone DNA polymerase variants that would only replicate the orthogonal plasmid. The optimized system was able to induce ~300× higher rates of mutagenesis in the orthogonal plasmid as compared to the background S. cerevisiae mutagenesis rate, with no reduction in the genomic replication fidelity. This system represents an important methodological advance in continuous directed evolution in eukaryotes.
Conclusion
Methods for the directed evolution of biomolecules have proven to be important technologies for generating new functional proteins and nucleic acids as research tools and therapeutics, as well as for facilitating the investigation of fundamental biological questions. Some of the limitations of traditional stepwise laboratory evolution approaches can be overcome by continuous directed evolution platforms. Continuous directed evolution intrinsically enables a broader exploration of sequence space in a practical timescale compared to traditional discrete laboratory evolution methods. Indeed, in vivo continuous evolution methods can enable > 1011 protein variants to be generated and subjected to selection over > 100 generations of evolution in less than a week [20]. The efficiency of continuous evolution methods can enable long evolutionary trajectories [14,23,32,33] or access to highly evolved biomolecules with new properties that would otherwise require impractical time scales [1,2,18,24,27,31,38]. In addition, some continuous evolution platforms, including several described above, can avoid genetic bottlenecks from modest population sizes, modest screening throughput, or modest mutation rates that commonly constrain some traditional laboratory evolution methods. The minimal reliance on researcher intervention during continuous evolution can also make performing multiple parallel evolution experiments more accessible than using traditional directed evolution platforms [12–14,18–20,22–24,27,31,39].
Continuous directed evolution methods are not without their own drawbacks and should be considered a complement, rather than a replacement, to other laboratory evolution approaches. Establishing a continuous cycle of translation, mutation, selection, and replication that is general for a range of biomolecules is challenging, and therefore most continuous evolution methods perform these key steps inside cells. As a result, while traditional in vitro directed evolution techniques enable exquisite control of selection stringency and mutation rate through simple adjustments of additive concentrations, analogous perturbations are more difficult to implement in vivo and generally must be empirically determined. Because in vivo continuous evolution platforms typically require continuous culture of microorganisms, contamination either by “cheaters” (uninteresting gene variants that bypass selections) or by inoculation with undesired organisms poses additional hurdles for implementing these methods.
Finally, the development of selection methods compatible with continuous evolution is typically more difficult than the development of analogous selections or screens for traditional stepwise laboratory evolution methods. Evolved biomolecules with desired properties in a continuous evolution system must be linked to gene and organism replication, rather than to the generation of fluorescent or colorimetric signals that have proven to be robust staples of in vitro and cell-based screening. While establishing this linkage between desired biomolecule function and cell survival is relatively straightforward for genes that are natively associated with organismal fitness (such as antibiotic resistance, temperature or solvent tolerance, or improved growth rate), the design of selections for continuous evolution is more challenging for genes and gene products that do not intrinsically affect cellular survival. Fortunately, many creative strategies have been described for coupling a wide variety of molecular activities to cell survival, including protein-fragment complementation of enzymatic activities and organelle-specific localization signals, n-hybrid systems for binding activity and bond-formation catalysis [40,41], derepression-based methods for bond-cleaving activities [42], gene regulation-based selections for activities associated with transcription or translation [43], and cis-acting elements for metabolite sensing [44]. We anticipate that the expansion of such selection methods to address an increasingly broad scope of molecular problems of interest will play a key role in fully realizing the potential and defining the future impact of continuous directed evolution.
Highlights.
General and recent advances in the field of in vivo continuous evolution
Advances in the continuous culture of microbes have enabled continuous evolution
Viruses, prokaryotes, and eukaryotes have mediated in vivo continuous evolution
Continuous evolution studies have contributed insights into evolutionary biology
Acknowledgements
We thank David Thompson and Brent Dorr for critical reading of the manuscript, and David Bryson for assistance with figures. Support was generously provided by DARPA HR0011-11-2-0003, DARPA N66001-12-C-4207, and the Howard Hughes Medical Institute.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Mills DR, Peterson RL, Spiegelman S. An extracellular Darwinian experiment with a self-duplicating nucleic acid molecule. Proc Natl Acad Sci U S A. 1967;58:217–224. doi: 10.1073/pnas.58.1.217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wright MC, Joyce GF. Continuous in vitro evolution of catalytic function. Science. 1997;276:614–617. doi: 10.1126/science.276.5312.614. [DOI] [PubMed] [Google Scholar]
- 3.Breaker RR, Banerji A, Joyce GF. Continuous in vitro evolution of bacteriophage RNA polymerase promoters. Biochemistry. 1994;33:11980–11986. doi: 10.1021/bi00205a037. [DOI] [PubMed] [Google Scholar]
- 4.McGinness KE, Wright MC, Joyce GF. Continuous in vitro evolution of a ribozyme that catalyzes three successive nucleotidyl addition reactions. Chem Biol. 2002;9:585–596. doi: 10.1016/s1074-5521(02)00136-9. [DOI] [PubMed] [Google Scholar]
- 5.Kuhne H, Joyce GF. Continuous in vitro evolution of ribozymes that operate under conditions of extreme pH. J Mol Evol. 2003;57:292–298. doi: 10.1007/s00239-003-2480-z. [DOI] [PubMed] [Google Scholar]
- 6.Johns GC, Joyce GF. The promise and peril of continuous in vitro evolution. J Mol Evol. 2005;61:253–263. doi: 10.1007/s00239-004-0307-1. [DOI] [PubMed] [Google Scholar]
- 7.Voytek SB, Joyce GF. Emergence of a fast-reacting ribozyme that is capable of undergoing continuous evolution. Proc Natl Acad Sci U S A. 2007;104:15288–15293. doi: 10.1073/pnas.0707490104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Paegel BM, Joyce GF. Darwinian evolution on a chip. PLoS Biol. 2008;6:e85. doi: 10.1371/journal.pbio.0060085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Drake JW, Charlesworth B, Charlesworth D, Crow JF. Rates of spontaneous mutation. Genetics. 1998;148:1667–1686. doi: 10.1093/genetics/148.4.1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Husimi Y. Selection and evolution of bacteriophages in cellstat. Adv Biophys. 1989;25:1–43. doi: 10.1016/0065-227x(89)90003-8. [DOI] [PubMed] [Google Scholar]
- 11.Hoskisson PA, Hobbs G. Continuous culture--making a comeback? Microbiology. 2005;151:3153–3159. doi: 10.1099/mic.0.27924-0. [DOI] [PubMed] [Google Scholar]
- 12.Bull JJ, Badgett MR, Wichman HA, Huelsenbeck JP, Hillis DM, Gulati A, Ho C, Molineux IJ. Exceptional convergent evolution in a virus. Genetics. 1997;147:1497–1507. doi: 10.1093/genetics/147.4.1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wichman HA, Badgett MR, Scott LA, Boulianne CM, Bull JJ. Different trajectories of parallel evolution during viral adaptation. Science. 1999;285:422–424. doi: 10.1126/science.285.5426.422. [DOI] [PubMed] [Google Scholar]
- 14.Wichman HA, Millstein J, Bull JJ. Adaptive molecular evolution for 13,000 phage generations: a possible arms race. Genetics. 2005;170:19–31. doi: 10.1534/genetics.104.034488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Romero PA, Arnold FH. Exploring protein fitness landscapes by directed evolution. Nat Rev Mol Cell Biol. 2009;10:866–876. doi: 10.1038/nrm2805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hillis DM, Bull JJ, White ME, Badgett MR, Molineux IJ. Experimental phylogenetics: generation of a known phylogeny. Science. 1992;255:589–592. doi: 10.1126/science.1736360. [DOI] [PubMed] [Google Scholar]
- 17.Bull JJ, Badgett MR, Rokyta D, Molineux IJ. Experimental evolution yields hundreds of mutations in a functional viral genome. J Mol Evol. 2003;57:241–248. doi: 10.1007/s00239-003-2470-1. [DOI] [PubMed] [Google Scholar]
- 18. Hammerling MJ, Ellefson JW, Boutz DR, Marcotte EM, Ellington AD, Barrick JE. Bacteriophages use an expanded genetic code on evolutionary paths to higher fitness. Nat Chem Biol. 2014;10:178–180. doi: 10.1038/nchembio.1450. The authors interrogate the ability of T7 bacteriophage to continuously evolve using a host with an expanded genetic code, resulting in variants dependent on 3-iodotyrosine incorporation for optimal fitness.
- 19. Meyer JR, Dobias DT, Weitz JS, Barrick JE, Quick RT, Lenski RE. Repeatability and contingency in the evolution of a key innovation in phage lambda. Science. 2012;335:428–432. doi: 10.1126/science.1214449. The authors continuously co-evolved lambda bacteriophage and E. coli to highlight a complex evolutionary interplay between virus and host.
- 20. Esvelt KM, Carlson JC, Liu DR. A system for the continuous directed evolution of biomolecules. Nature. 2011;472:499–503. doi: 10.1038/nature09929. The authors develop a general phage-based methodology for in vivo continuous directed evolution of biomolecules and apply the system to the continuous evolution of T7 RNA polymerase.
- 21.Rakonjac J, Bennett NJ, Spagnuolo J, Gagic D, Russel M. Filamentous bacteriophage: biology, phage display and nanotechnology applications. Curr Issues Mol Biol. 2011;13:51–76. [PubMed] [Google Scholar]
- 22.Leconte AM, Dickinson BC, Yang DD, Chen IA, Allen B, Liu DR. A population-based experimental model for protein evolution: effects of mutation rate and selection stringency on evolutionary outcomes. Biochemistry. 2013;52:1490–1499. doi: 10.1021/bi3016185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dickinson BC, Leconte AM, Allen B, Esvelt KM, Liu DR. Experimental interrogation of the path dependence and stochasticity of protein evolution using phage-assisted continuous evolution. Proc Natl Acad Sci U S A. 2013;110:9007–9012. doi: 10.1073/pnas.1220670110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Carlson JC, Badran AH, Guggiana-Nilo DA, Liu DR. Negative selection and stringency modulation in phage-assisted continuous evolution. Nat Chem Biol. 2014;10:216–222. doi: 10.1038/nchembio.1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dickinson BC, Packer MS, Badran AH, Liu DR. A system for the continuous directed evolution of proteases rapidly reveals drug-resistance mutations. Submitted. 2014 doi: 10.1038/ncomms6352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lee HH, Molla MN, Cantor CR, Collins JJ. Bacterial charity work leads to population-wide resistance. Nature. 2010;467:82–85. doi: 10.1038/nature09354. ••••••••••s demonstrate dramatic variability in the resistance of single bacterial members of a growing population to an antibiotic stress, highlighting more complex mechanisms responsible for communal resistance.
- 27. Toprak E, Veres A, Michel JB, Chait R, Hartl DL, Kishony R. Evolutionary paths to antibiotic resistance under dynamically sustained drug selection. Nat Genet. 2012;44:101–105. doi: 10.1038/ng.1034. The authors contruct an auxostat variant that regularly adjusts antibiotic stress to a growing bacterial culture, and enables parallel continuous evolution experiments to study the path-dependence of antibiotic resistance.
- 28.Camps M, Naukkarinen J, Johnson BP, Loeb LA. Targeted gene evolution in Escherichia coli using a highly error-prone DNA polymerase I. Proc Natl Acad Sci U S A. 2003;100:9727–9732. doi: 10.1073/pnas.1333928100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fabret C, Poncet S, Danielsen S, Borchert TV, Ehrlich SD, Janniere L. Efficient gene targeted random mutagenesis in genetically stable Escherichia coli strains. Nucleic Acids Res. 2000;28:E95. doi: 10.1093/nar/28.21.e95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fijalkowska IJ, Schaaper RM. Mutants in the Exo I motif of Escherichia coli dnaQ: defective proofreading and inviability due to error catastrophe. Proc Natl Acad Sci U S A. 1996;93:2856–2861. doi: 10.1073/pnas.93.7.2856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Luan G, Cai Z, Li Y, Ma Y. Genome replication engineering assisted continuous evolution (GREACE) to improve microbial tolerance for biofuels production. Biotechnol Biofuels. 2013;6:137. doi: 10.1186/1754-6834-6-137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Marliere P, Patrouix J, Doring V, Herdewijn P, Tricot S, Cruveiller S, Bouzon M, Mutzel R. Chemical evolution of a bacterium's genome. Angew Chem Int Ed Engl. 2011;50:7109–7114. doi: 10.1002/anie.201100535. The authors continuously evolve E. coli to incorporate high levels of 5-chlorouracil in its genome in lieu of thymine using the GM3 device.
- 33.Muller MM, Allison JR, Hongdilokkul N, Gaillon L, Kast P, van Gunsteren WF, Marliere P, Hilvert D. Directed evolution of a model primordial enzyme provides insights into the development of the genetic code. PLoS Genet. 2013;9:e1003187. doi: 10.1371/journal.pgen.1003187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ro DK, Paradise EM, Ouellet M, Fisher KJ, Newman KL, Ndungu JM, Ho KA, Eachus RA, Ham TS, Kirby J, et al. Production of the antimalarial drug precursor artemisinic acid in engineered yeast. Nature. 2006;440:940–943. doi: 10.1038/nature04640. [DOI] [PubMed] [Google Scholar]
- 35.Ishige T, Honda K, Shimizu S. Whole organism biocatalysis. Curr Opin Chem Biol. 2005;9:174–180. doi: 10.1016/j.cbpa.2005.02.001. [DOI] [PubMed] [Google Scholar]
- 36.de Crecy E, Metzgar D, Allen C, Penicaud M, Lyons B, Hansen CJ, de Crecy-Lagard V. Development of a novel continuous culture device for experimental evolution of bacterial populations. Appl Microbiol Biotechnol. 2007;77:489–496. doi: 10.1007/s00253-007-1168-5. [DOI] [PubMed] [Google Scholar]
- 37.de Crecy E, Jaronski S, Lyons B, Lyons TJ, Keyhani NO. Directed evolution of a filamentous fungus for thermotolerance. BMC Biotechnol. 2009;9:74. doi: 10.1186/1472-6750-9-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Romanini DW, Peralta-Yahya P, Mondol V, Cornish VW. A Heritable Recombination system for synthetic Darwinian evolution in yeast. ACS Synth Biol. 2012;1:602–609. doi: 10.1021/sb3000904. •••••••hors design and apply a recombination-based method for continuous directed evolution in S. cerevisiae, rapidly discovering enzyme variants with desired properties.
- 39. Ravikumar A, Arrieta A, Liu CC. An orthogonal DNA replication system in yeast. Nat Chem Biol. 2014;10:175–177. doi: 10.1038/nchembio.1439. The authors report the use of an error-prone orthogonal DNA replication mechanism in S. cerevisiae, which could enable continuous evolution and modulated mutagenesis in vivo without interfering with host viability.
- 40.Michnick SW, Ear PH, Manderson EN, Remy I, Stefan E. Universal strategies in research and drug discovery based on protein-fragment complementation assays. Nat Rev Drug Discov. 2007;6:569–582. doi: 10.1038/nrd2311. [DOI] [PubMed] [Google Scholar]
- 41.Stynen B, Tournu H, Tavernier J, Van Dijck P. Diversity in genetic in vivo methods for protein-protein interaction studies: from the yeast two-hybrid system to the mammalian split-luciferase system. Microbiol Mol Biol Rev. 2012;76:331–382. doi: 10.1128/MMBR.05021-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.van Rossum T, Kengen SW, van der Oost J. Reporter-based screening and selection of enzymes. FEBS J. 2013;280:2979–2996. doi: 10.1111/febs.12281. [DOI] [PubMed] [Google Scholar]
- 43.Schaerli Y, Isalan M. Building synthetic gene circuits from combinatorial libraries: screening and selection strategies. Mol Biosyst. 2013;9:1559–1567. doi: 10.1039/c2mb25483b. [DOI] [PubMed] [Google Scholar]
- 44.Wang YH, Wei KY, Smolke CD. Synthetic biology: advancing the design of diverse genetic systems. Annu Rev Chem Biomol Eng. 2013;4:69–102. doi: 10.1146/annurev-chembioeng-061312-103351. [DOI] [PMC free article] [PubMed] [Google Scholar]