Abstract
Lysobacter species are emerging as new sources of antibiotics. The regulation of these antibiotics is not well understood. Here, we identified a small molecule metabolite (LeDSF3) that regulates the biosynthesis of the antifungal antibiotic HSAF (heat-stable antifungal factor), a polycyclic tetramate macrolactam with a structure and mode of action distinct from the existing antifungal drugs. LeDSF3 was isolated from the culture broth of L. enzymogenes, and its chemical structure was established by NMR and MS. The purified compound induced green fluorescence in a reporter strain of Xanthomonas campestris, which contained gfp gene under the control of a DSF (diffusible signaling factor)-inducible promoter. Exogenous addition of LeDSF3 in L. enzymogenes cultures significantly increased the HSAF yield, the transcription of HSAF biosynthetic genes, and the antifungal activity of the organism. The LeDSF3-regulated HSAF production is dependent on the two-component regulatory system RpfC/RpfG. Moreover, LeDSF3 up-regulated the expression of the global regulator Clp (cAMP receptor-like protein). The disruption of clp led to no HSAF production. Together, the results show that LeDSF3 is a fatty acid-derived, diffusible signaling factor positively regulating HSAF biosynthesis and that the signaling is mediated by the RfpC/RpfG-Clp pathway. These findings may facilitate the antibiotic production through applied genetics and molecular biotechnology in Lysobacter, a group of ubiquitous yet underexplored microorganisms.
Keywords: Diffusible signaling factor, natural product biosynthesis, regulation, HSAF, Lysobacter enzymogenes
Introduction
Lysobacter is a genus of ubiquitous environmental bacteria that belong to the Xanthomonadaceae family within the gamma proteobacteria (Christensen and Cook 1978; Sullivan et al. 2003). The genus is emerging as a novel biocontrol agent against pathogens of crop plants (Yuen et al. 2001a; Yuen and Zhang 2001b; Zhang and Yuen 1999), including Bipolaris sorokiniana, Uromyces appendiculatus, and Rhizoctonia solani. We have been studying strains of L. enzymogenes as a new source of bioactive natural products (Li et al. 2012; Li et al. 2014; Lou et al. 2011; Lou et al. 2012; Wang et al. 2013a; Wang et al. 2013b; Yu et al. 2007; Zhang et al. 2011). We recently identified biosynthetic genes in strain OH11 for WAP-8294A, a group of cyclic lipodepsipeptides with very potent activity against methicillin-resistant Staphylococcus aureus (MRSA) (Zhang et al. 2011). We also isolated HSAF (heat-stable antifungal factor), an antifungal metabolite from strain C3 (Lou et al. 2011; Yu et al. 2007). HSAF is a polycyclic tetramate macrolactam (PTM) that has a chemical structure distinct from any existing antifungal drug or fungicide and appears to involve a novel mode of action in inhibiting hyphal growth in fungi (Li et al. 2006). The biosynthetic gene cluster for HSAF consists of a core gene (hsaf pks-nrps) encoding a hybrid polyketide synthase-nonribosomal peptide synthetase (PKS-NRPS), and tailoring genes for HSAF structural modifications (Li et al. 2014; Lou et al. 2011). Despite the research progress towards elucidating the molecular mechanism for HSAF biosynthesis, little is known about the regulation of HSAF production in L. enzymogenes.
Some endogenous or exogenous signal molecules can mediate cell-to-cell communications in microorganisms. One well-known signaling system is quorum sensing (QS) (Comella and Grossman 2005; Dusane et al. 2011; Galloway et al. 2011; Miller and Bassler 2001), which can regulate the expression of multiple genes through the accumulation and recognition of auto-induced signals, or autoinducers in the local environments. Signaling via autoinducers regulate virulence, biofilm formation and biosynthesis of secondary metabolites in microorganism. N-acyl-homoserine lactones (AHLs) are a major class of small molecule signaling factors that have been extensively studied (Case et al. 2008). Another class is the furanosyl borate diesters that exist in a number of Gram-positive and Gram-negative bacteria (Winzer et al. 2003). In L. enzymogenes, we recently found that two signaling pathways are involved in regulation of secondary metabolite biosynthesis and colony morphology (Qian et al. 2013a). One of the signaling pathways likely involves signal molecules belonging to hydroxylated benzoic acids (the so-called diffusible factors, or DFs) (He et al. 2011). The other is likely to involve analogs of the so-called diffusible signaling factor (DSF) initially reported in Xanthomonas campestris pv. campestris (Xcc) that is involved in the Rpf (regulation of pathogenicity factors) signaling system (Barber et al. 1997; Deng et al. 2011; Ryan and Dow 2011). DSF isolated from X. campestris was found to be (Z)-11-methyl-2-dodecenoic acid (Barber et al. 1997; Wang et al. 2004), and now DSF-like molecules have been reported in other microorganisms (Deng et al. 2009; Deng et al. 2010; He et al. 2010; Huang and Wong 2007; Newman et al. 2004). These signal molecules are fatty acids typically with a cis double bond at C2 position, and their chain lengths vary from 12 to 14 carbons. They regulate multiple behaviors of the cells in a density-dependent manner (quorum sensing). Some DSF contain a methyl branch, such as (Z)-11-methyl-2-dodecenoic acid from X. campestris, (2Z,5Z)-11-methyl-2,5-dodecadienoic acid from X. oryzae pv. oryzae (He et al. 2010), and 12-methyltetradecanoic acid from a citrus strain of X. fastidiosa (Simionato et al. 2007). Some DSF contain a linear fatty acid chain, such as (Z)-2-dodecenoic acid from Burkholderia cenocepacia (Boon et al. 2008) and Pseudomonas aeruginosa (Amari et al. 2013) and (Z)-2-tetradecenoic acid from a grape strain of Stenotrophomonas maltophilia (Huang and Wong 2007).
The genes in the Rpf signaling system encode enzymes for DSF biosynthesis and proteins functioning as sensor and response regulators (Barber et al. 1997; Deng et al. 2011; Ryan and Dow 2011). Among them, rpfF encodes a bifunctional 3-hydroxyacyl-ACP dehydratase and thioesterase (Bi et al. 2012), and rpfB was predicted to encode an acyl CoA synthetase (Almeida et al. 2012). The rpfC and rpfG genes encode for proteins involved in a two-component of signal transduction system, RpfC/RpfG, which serve as the sensor/response regulator of DSF (Slater et al. 2000). RpfC is a membrane-bound histidine kinase sensor protein with dual functions. Its intracellular domain is associated with RpfF, which suppresses the activity of RpfF to synthesize DSF (Cheng et al. 2010; He et al. 2006; Slater et al. 2000). When the extracellular DSF concentration reaches a threshold, RpfC undergoes autophosphorylation at its active site histidine residue. This leads to the release of RpfF, which then becomes active and in turn synthesizes more DSF (thus auto-induction of DSF). At the same time, the kinase function of RpfC phosphorylates the partner intracellular response regulator RpfG, and this activates the cyclic di-GMP phosphodiesterase activity of RpfG (Barber et al. 1997; Deng et al. 2011; Ryan and Dow 2011). Cyclic di-GMP is a second messenger involved in numerous cellular processes, including those mediated by the global regulator Clp, cAMP receptor-like protein (Chin et al. 2010). The RpfC/RpfG-Clp-mediated DSF signaling has been observed in diverse bacteria and linked to virulence, motility, biofilm dispersal, extracellular enzyme and production of extracellular polymeric substances (EPS) (Barber et al. 1997; Deng et al. 2011; Ryan and Dow 2011). In L. enzymogenes, clp has been shown to regulate the production of lytic enzymes and other antifungal factors and to be critical in biological control activity (Kobayashi et al. 2005). However, neither the direct involvement of Clp in regulating HSAF biosynthesis nor the connection between Clp and RpfC/RpfG has been demonstrated. More importantly, the actual signal molecule has not been identified from any Lysobacter species.
We have found that the genome of L. enzymogenes OH11 contains the homologous genes of the rpf cluster (Qian et al. 2013a; Qian et al. 2013b). This suggests that a DSF-like molecule is very likely produced by L. enzymogenes. The objectives of this study were to isolate and identify the DSF-like molecule from L. enzymogenes OH11 and to verify that it is involved in RpfC/RpfG-Clp regulation of HSAF biosynthesis. In this report, we describe a new DSF-like molecule (LeDSF3) from L. enzymogenes and present evidence for the involvement of LeDSF3 in regulating HSAF production through the RpfC/RpfG-Clp signaling pathway.
Materials and methods
Instruments and analytical methods
NMR spectra were recorded on a Bruker Advance 400 spectrometer at 400/100 MHz (Bruker, Fällanden, Switzerland). Mass spectra were obtained on LCQ mass spectrometers (Thermo, West Palm Beach, FL, USA). An Agilent 1120 HPLC system (Agilent, Santa Clara, CA, USA), with RF C18 columns (10.0 × 250 mm, 5 μm, for preparative HPLC; 4.6 × 150 mm, 3.5 μm, for analytic HPLC), was used in the studies. For column chromatography, a silica gel 60 (Merck, Darmstadt, Germany) column and a Sephadex LH-20 (GE healthcare, Uppsala, Sweden) column were used. TLC analyses were performed with pre-coated silica gel 60 F254 plates (Merck, Darmstadt, Germany). The fluorescent microscope pictures were obtained on an Olympus IX 81 inverted confocal microscope (Olympus, Tokyo, Japan). All general chemical reagents were purchased from Sigma-Aldrich or Fisher Scientific.
Microorganism Strains
Escherichia coli DH5α was used for the general DNA manipulation. E. coli S17-1 was used for conjugation. Lysobacter enzymogenes OH11 (CGMCC No. 1978) and its mutants including OH11ΔrpfB, OH11ΔrpfC, OH11ΔrpfF, OH11ΔrpfG, and OH11 DC were from the Liu lab (Qian et al. 2013a; Kobayashi et al. 2005). The DSF reporter strain X. campestris 8523/pKLN55 was from the Lindow lab (Newman et al. 2004), and Fusarium verticillioides A0149 (FGSC No. 7600) was used as the test organism in the antifungal assays.
Isolation of LeDSF3
L. enzymogenes OH11ΔrpfC was grown in 1/10 Tryptic Soy Broth (TSB, 100 ml 3000 shake flask, total 300 liters) at 28 °C, at 200 rpm, for 2 days. The culture broth was extracted with the same volume ethyl acetate (EtOAc) until the filtrate was colorless. The combined extract, upon evaporation, yielded the crude extract, which was partitioned between methanol and petroleum ether (250 ml each, 3 times). The petroleum ether layer was concentrated under vacuum to afford a yellow syrupy extract (1.4 g). The extract was subjected to column chromatography (60 g silica gel 60; hexane - ethyl acetate, gradient elute; 250 ml for each gradient) to afford 20 fractions, Fr. 1–20. Fraction 2 (52 mg) was subjected to preparative HPLC (RF C18 column, 10.0 × 250 mm, 5 μm; 80% acetonitrile; flow rate 3 ml/min; detection wavelength 210 nm) to afford LeDSF3 (~5 mg).
Assay for the Green Fluorescent Activity in the DSF Reporter Strain
The promoterless gfp gene fused to the promoter from the DSF-inducible engXCA gene was cloned in a plasmid conferring spectinomycin and streptomycin resistance, generating construct pKLN55 (Newman et al. 2004). The construct was mated into the rpfF mutant X. campestris 8523 to generate the DSF reporter strain X. campestris 8523/pKLN55. To detect any DSF-like molecule in L. enzymogenes, we followed the protocol described by Newman et al (Newman et al. 2004). Briefly, X. campestris 8523/pKLN55 was inoculated into liquid NYGB medium (peptone 5.0 g, yeast extract 3.0 g, glycerol 20.0 g, water 1.0 L, pH 7.2) and incubated at 30 °C for 24 h. A 5 μl of the culture was spotted onto LB plates, and the individual extracts from L. enzymogenes were added to sterile paper discs along the reporter strain. The X. campestris DSF, (Z)-11-methyl-2-dodecenoic acid (Cayman Chemical), was used as the positive control. For the test of the purified LeDSF3, 1 μg (in 5 μl DMSO) of the compound was spotted on a paper disc near the X. campestris reporter strain. The reporter strain was incubated at 28–30 °C for 48 h, and the green fluorescence was then visualized using a confocal microscopy. The excitation wavelength was 488 nm, and the emission wavelength was 509 nm.
Effect of the small molecule signal LeDSF3 on the antifungal HSAF Production
LeDSF3 (final concentration 0.2–10 μM) was added into 50 ml 1/10 TSB culture of various strains of L. enzymogenes. The cultures grew at 28 °C, 200 rpm for 2 days, and were extracted with the same volume of ethyl acetate. The organic phase was concentrated under vacuum, and the crude extract was dissolved with 0.5 ml methanol. A fraction (20 μl) of the methanol extract was injected in HPLC to analyze HSAF and analogs. For semi-quantification, the peak area of HSAF and analogs was measured to obtain the relative yield of the compounds, and each treatment was repeated at least twice. The HPLC method is shown in Table S1 in the Supplementary Material.
Test of the Antifungal Activity of L. enzymogenes strains treated with LeDSF3
Fusarium verticillioides was used as the test fungus. Various strains of L. enzymogenes and F. verticillioides were co-inoculated on 40% TSB plates, with or without LeDSF3. Half of the test plate was spread with the L. enzymogenes wild type or one of its mutants; the other half was point-inoculated with F. verticillioides mycelium. The plates were placed in a 28 °C incubator for 3 days.
RNA Extraction, Reverse-transcription PCR and Real Time-PCR
L. enzymogenes OH11 and its mutants were grown on 100 ml 1/10 strength TSB medium for 24 h. An aliquot of 3 ml cells was transferred to a sterile centrifuge tube and centrifuged for 3 min at 15,300 g. RNA was extracted from the strains using TRIZOL solution following the manufacturer’s instruction. For DNA removing and reverse transcription PCR, PrimerScript RT reagent Kit with gDNA Eraser Kit (TaKaRa biocompany) was used in this study. For real time-PCR assay, iQ SYBR green supermix kit (BIO-RAD company) was used. The primers for realtime PCR were listed in Table S2.
Results
Isolation and structural determination of the signaling molecule LeDSF3
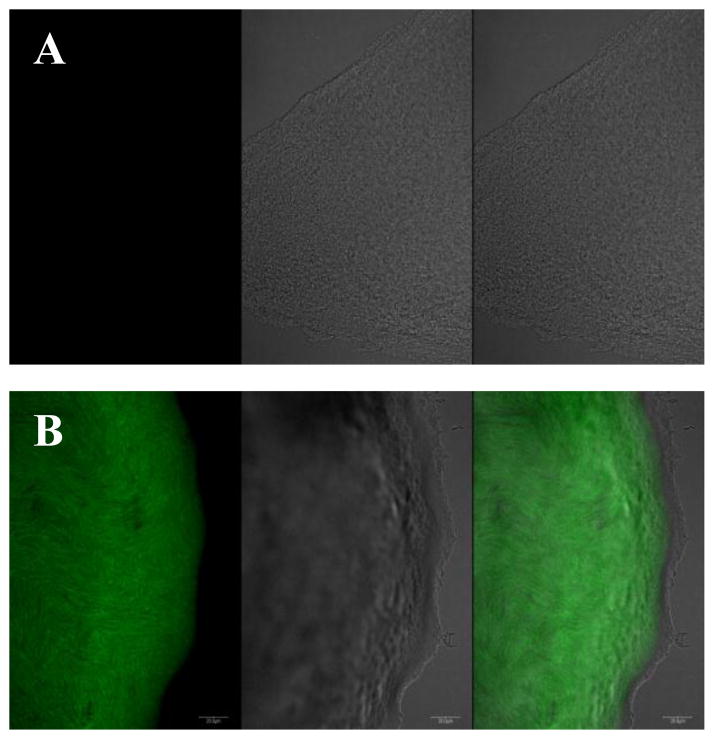
We used the DSF reporter strain X. campestris 8523/pKLN55, which contains gfp (green fluorescent protein) gene under the control of a DSF-inducible promoter (Newman et al. 2004) to isolate putative signal molecules produced by L. enzymogenes OH11. GFP expression occurs when the reporter strain is grown in the presence of a source of DSF or a DSF-like molecule. Initially, all efforts failed to isolate a sufficient amount of DSF for structural determination from fermenter broth cultures of the wild type OH11, although we could detect very weak green fluorescence using crude extract from such cultures. We subsequently shifted to the rpfC disruption mutant of OH11, with the hope that disruption of this DSF sensor/repressor would lead to a higher yield of the DSF-like compound as observed in other organisms (Chatterjee et al. 2008; Slater et al. 2000). The estimated concentration of the main DSF in the wild type and the rpfC mutant was 13 and 71 μg/L, respectively. In addition, because L. enzymogenes tends to self-lyse in broth culture when the culture is older than 4–5 days, we chose to grow the cells in flasks with 100 ml medium so that the culture could reach a relatively high cell density (OD600 nm ~1–1.5) in 2 days. With these modifications, we collected the ethyl acetate extracts from 3000 flasks of culture and followed fractions that induced GFP expression in the reporter strain (Fig. 1). The X. campestris DSF was used as a reference, which gave a retention time of 7.5 minute in HPLC. In the active fraction, three main peaks were detected by HPLC (Fig. 1). We focused on the mid-peak (LeDSF3), which had a retention time of 13.3 minute, because this peak showed the strongest activity in HSAF regulation. The purified LeDSF3 was tested for DSF-like activity using the reporter strain X. campestris 8523/pKLN55, which carries gfp gene under the control of a DSF-inducible promoter., green fluorescence was observed in the colonies (Fig. 2). Green fluorescence was observed in the colonies when 1 μg of LeDSF3 was spotted onto the solid media near the colonies of X. campestris 8523/pKLN55 (Fig. 2)
Fig. 1.
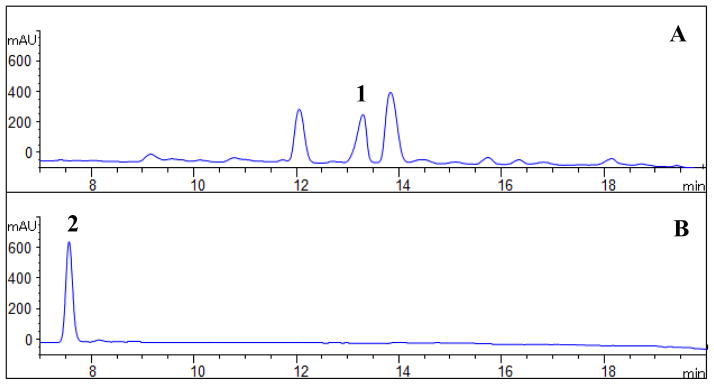
HPLC analysis of LeDSFs. A: Semi-purified extracts of LeDSF3 (1); B: X. campestris DSF (2) as reference.
Fig. 2.
DSF-like activity of LeDSF3, shown as the green fluorescence induction activity in the DSF reporter strain X. campestris 8523/pKLN55. A: untreated strain X. campestris 8523/pKLN55; B: 8523/pKLN55 treated with 1 μg LeDSF3. For each of panels, a) picture of a 8523/pKLN55 colony taken at 488 nm excitation wavelength and 509 nm emission wavelength, b) picture of the same colony taken under transmitted light, and c) merged a & b.
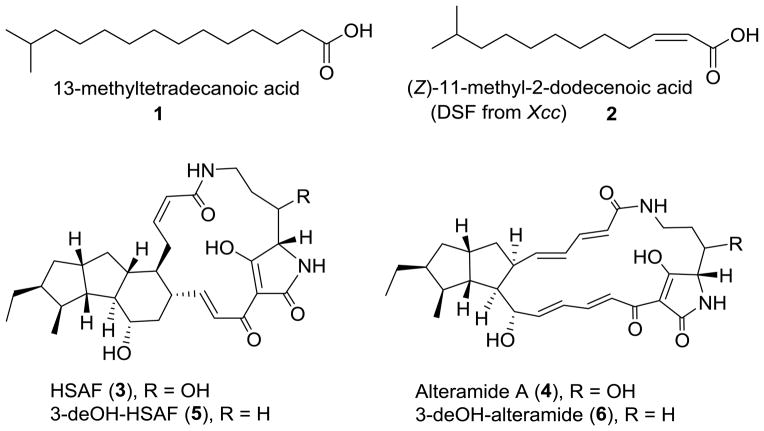
The LeDSF3 1H-NMR and the 13C NMR spectra displayed 11 CH2 (one connected with a carbonyl group), one CH, and two CH3 groups (Table S3, Fig. S1–S2). 1H-NMR signals at δH 0.88 (d, J = 6.6 Hz, 6H), 1.51–1.54 (m, 1H), and the 13C-NMR signals at δC 22.9 indicated the presence of an isopropyl group. 1H-NMR signals at δH 1.27–1.36 (m, 16 H) revealed the presence of an aliphatic carbon chain. The mass of the compound was determined by ESI-MS to be 242 Da ([M−1]− = 241; [M+1]+=243) (Fig. S3). The FT-IR absorption at 1697 cm−1 indicated the presence of C=O carbonyl group, and the FT-IR absorption from 2952 to 2871 cm−1 indicated the presence of aliphatic chain (Figure S4). These data are consistent with those reported for a chemically synthesized fatty acid (Sarpe & Kulkarni 2011). Therefore, LeDSF3 was determined to be 13-methyltetradecanoic acid (Fig. 3).
Fig. 3.
Chemical structures of LeDSF3 (1), X. campestris DSF (2), and HSAF analogs (3–6).
The small molecule signal LeDSF3 induces the antifungal HSAF production
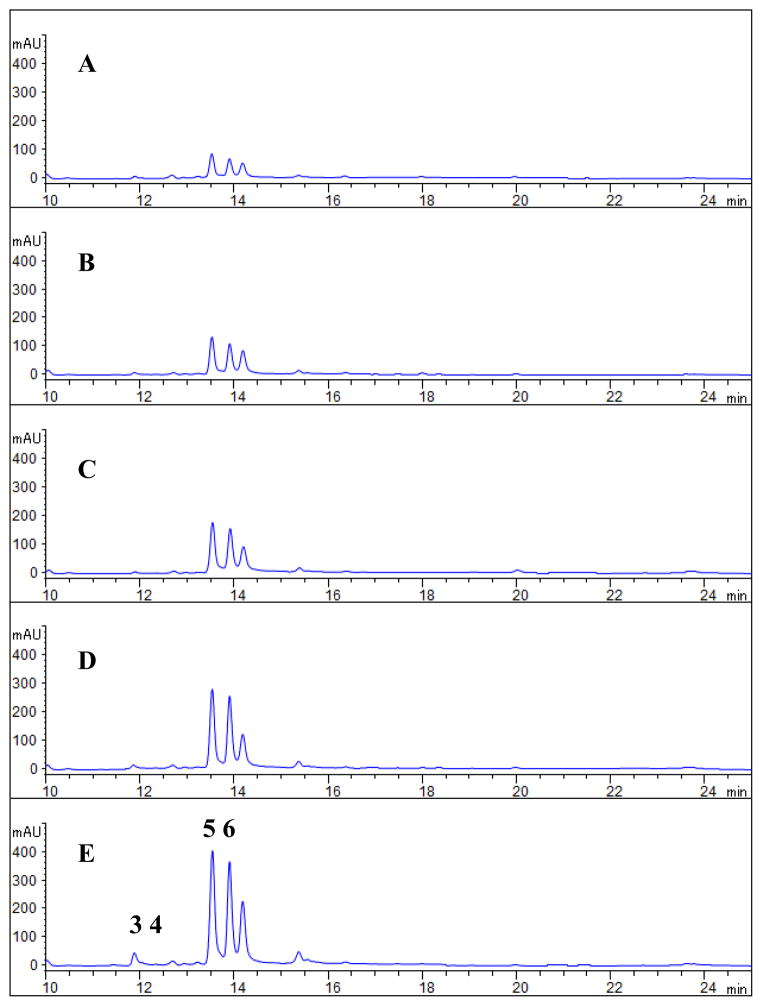
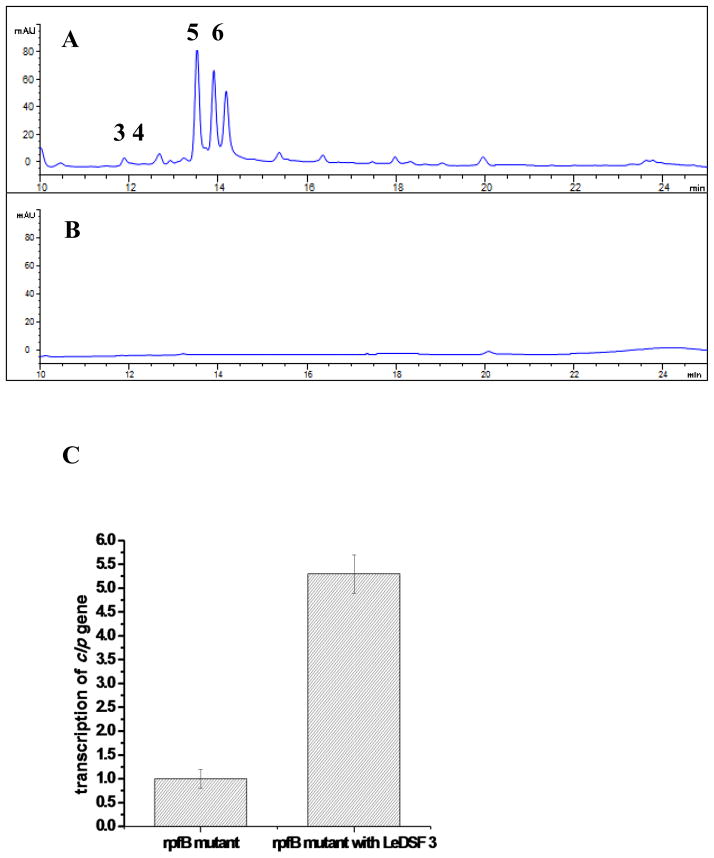
We tested the effect of LeDSF3 on the production of HSAF analogs by strain OH11. The strain mainly produced 3-deOH-HSAF (5) and 3-deOH-alteramide (6), with HSAF (3) being detected in relatively low amounts and alteramide being undetected (4) (Fig. 3 and 4). When LeDSF3 was added into the cultures, the relative amounts of the different HSAF analogs was unchanged but total HSAF production was increased in a dose-dependent manner. The yield of HSAF analogs began to increase at 0.2 μM LeDSF3 and continued to increase as the LeDSF3concentration increased (1.5–4.6 fold from 0.2 to 10 μM LeDSF3) (Fig. 4).
Fig. 4.
Induction of HSAF production by LeDSF3 in the wild type LeOH11. The L. enzymogenes OH11 cultures were grown in 10% TSB for 24 h, and HSAF was extracted from the individual cultures and analyzed by HPLC. A: No addition; B: 0.2 μM LeDSF3 added; C: 1.0 μM LeDSF3 added; D: 5.0 μM LeDSF3 added, E: 10.0 μM LeDSF3 added. For identity of the compounds, HSAF (3), alteramide A (4), 3-deOH-HSAF (5), and 3-deOH-altermide A (6).
The small molecule signal LeDSF3 regulates the antifungal HSAF biosynthesis through the two-component regulators RpfC/RpfG
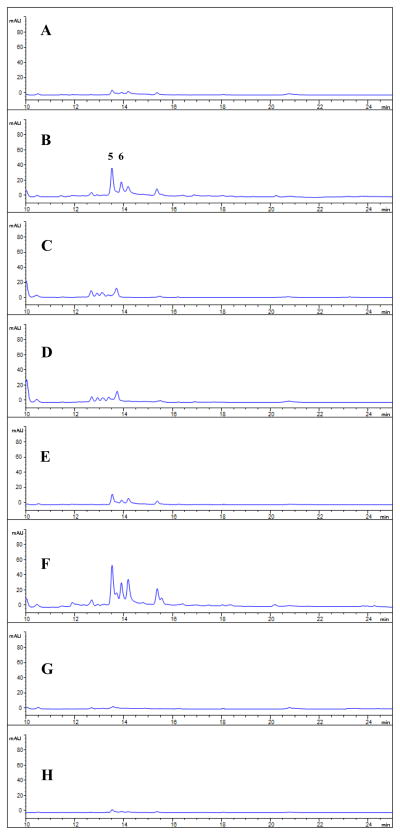
A gene cluster homologous to X. campestris rpf cluster is present in the genome of L. enzymogenes OH11 (Figure S5) (Qian et al. 2013a). The above results have demonstrated that L. enzymogenes indeed produces at least one DSF-like molecule. To test the hypothesis that LeDSF3-induced HSAF production is mediated by the rpf system, we examined the production of HSAF analogs in mutants of OH11 disrupted in individual rpf genes. As shown in Figure S6, the disruption of any of rpfB, C, F and G nearly eliminated production of HSAF analogs, verifying that the rpf signaling system is involved in the regulating HSAF production. Then, we exogenously added LeDSF3 (5 μM) to cultures of the rpf mutants. The HSAF production was partly restored in the rpfB mutant (~50%) and the rpfF mutant (~75%), but not in mutants disrupted in rpfC or rpfG mutant (Fig. 5). The results are consistent with the expected involvement of LeDSF3 as a signaling compound in the rpf system. In the rpfB and rpfF mutants, which are inactive in the DSF biosynthesis enzymes RpfB and RpfF, respectively, the two-component sensor/response regulator proteins (RpfC/RpfG) are active, whereas the entire sensor/response regulator is inactive in the rpfC and rpfG mutants. Therefore, positive response of rpfB and rpfF mutants, but not rpfC and rpfG mutants to exogenously added LeDSF3, is evidence that this compound is the signal, or one of the signals, received by RpfC.
Fig. 5.
Effect of LeDSF3 on HSAF production in various LeOH11 mutants. A: LeOH11ΔrpfB; B: ΔrpfB treated with 5 μM LeDSF3; C: ΔrpfC; D: ΔrpfC treated with 5 μM LeDSF3; E: ΔrpfF; F: ΔrpfF treated with 5 μM LeDSF3; G: ΔrpfG; H: ΔrpfG treated with 5 μM LeDSF3.
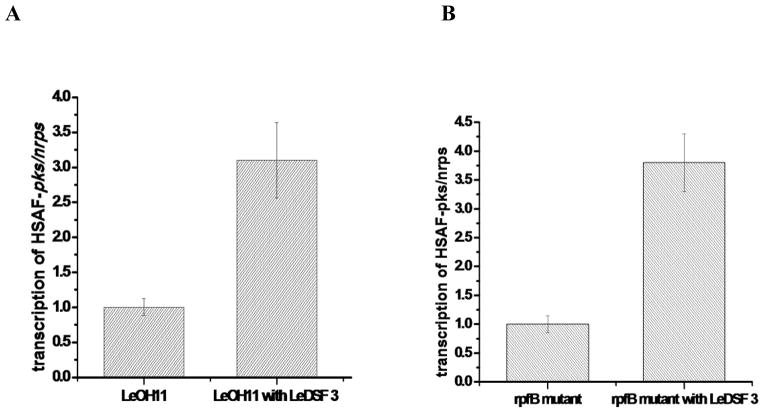
Next, we evaluated the effect of LeDSF3 on expression of HSAF pks-nrps, the key gene for HSAF biosynthesis (Li et al. 2014; Lou et al. 2011), first in the wild type OH11 and then in the rpfB mutant. In the wild type, the exogenous addition of LeDSF3 (10 μM) increased pks-nrps expression by ~ 3 fold, as determined by Q-RT-PCR (Fig. 6A). In the rpfB mutant, pks-nrps expression was elevated by ~ 4 fold by exogenous LeDSF3 (Fig. 6B). Because the rpfB mutant is expected to produce very little, if any, of its own LeDSF3, the exogenous addition of LeDSF3 would be expected to have a more significant impact on HSAF production in rpfB mutant than that in the wild type. The results from these two experiments are consistent with this model and are in agreement with the observed effects of the LeDSF3 on HSAF yield.
Fig. 6.
Effect of exogenous LeDSF3 on the transcription of HSAF pks-nrps gene in the wild type LeOH11 (A) and in rpfB mutant (B), as detected by Q-RT-PCR. The concentration of LeDSF3 was 10 μM. The relative transcription level of pks/nrps in the untreated LeOH11 was set as 1.
The small molecule signal LeDSF3 regulates the antifungal activity of L. enzymogenes through the two-component regulators RpfC/RpfG
L. enzymogenes is known for its biocontrol activity against fungal pathogens, and antifungal HSAF is known to be the main factor contributing to this antifungal activity (Li et al. 2006; Yu et al. 2007). To further understand the LeDSF signaling, we tested the activity of various rpf mutant strains, in the presence or absence of exogenous LeDSF3, against the growth of the plant pathogen Fusarium verticillioides (Fig. S7). The wild type LeOH11 exhibited a clear activity against this fungus, and this activity was markedly enhanced when LeDSF3 was added into the medium (Fig. S7A and F). Mutation of the DSF biosynthetic gene rpfB led to a decrease in the antifungal activity, which was restored by exogenous LeDSF3 (Fig. S7B and G). Mutation of the two-component sensor gene rpfC led to a complete loss of the antifungal activity, which could not be restored by exogenous LeDSF3 (Fig. S7C and H). Mutation of the other DSF biosynthetic gene rpfF led to a significant decrease in the antifungal activity, which was restored by exogenous LeDSF3 (Fig. S7D and I). Finally, mutation of the two-component response regulator gene rpfG led to a complete loss of the antifungal activity, which could not be restored by exogenous LeDSF3 (Fig. S7E and J). These results clearly support the above observed results, which showed that LeDSF3 regulates HSAF biosynthesis through the two-component regulatory system RpfC/RpfG.
The two-component regulators RpfC/RpfF and the small molecule signal LeDSF3 regulate antifungal HSAF biosynthesis through the global regulator Clp
To determine if the regulation of HSAF biosynthesis in L. enzymogenes by RpfC/RpfF and associated LeDSF3 acts through the global regulator Clp, we first analyzed the production of HSAF analogs in clp mutant. The production of HSAF analogs was completely eliminated in the clp mutant (Fig. 7A), clearly demonstrating the connection between the global regulator and HSAF biosynthesis. Next, we tested the effected of the LeDSFs on clp expression. Q-RT-PCR analysis showed that LeDSF3 increased clp expression by 5 fold in rpfB mutant (no endogenous LeDSFs) over the control (Fig. 7B). The results are consistent with the observed effect of LeDSF3 on HSAF production and pks-nrps gene expression. Together, the results showed that that the RfpC/RfpG-Clp pathway mediates LeDSF3 signaling towards antifungal HSAF biosynthesis in L. enzymogenes.
Fig. 7.
Involvement of the global regulator Clp in the RpfC/RpfG-mediated LeDSF signaling toward HSAF biosynthesis. A: HSAF production in the wild type L. enzymogenes OH11. B: HSAF production in the clp mutant. C: Effect of exogenous LeDSF3 on the transcription of clp gene in rpfB mutant, as detected by Q-RT-PCR. The concentration of LeDSF3 was 10 μM. The relative transcription level of clp gene in untreated rpfB mutant was set as 1.
Discussion
Diffusible small molecule signals are well known in antibiotic-producing bacteria, such as the Gram positive bacteria Streptomyces which often use gamma-butyrolactones as specialized regulatory small molecules for the biosynthesis of natural products (Liu et al. 2013). The so-called A-factor, a diffusible extracellular molecule [2-(6′-methylheptanoyl)-3R-hydroxymethyl-4-butanolide], triggers both aerial mycelium formation and streptomycin biosynthesis in S. griseus (Khoklov et al. 1967; Horinouchi and Beppu 1994). A-factor like signals have been found in numerous Streptomyces species, and their biosynthetic genes are commonly associated with antibiotic biosynthetic gene clusters (Liu et al. 2013). In addition to gamma-butyrolactones, small furans were found to control antibiotics in some Streptomyces, such as the epoxycyclopentenone methylenomycin A in S. coelicolor (Corre et al. 2008). Another widely studied group of small molecule signals is N-acyl homoserine lactones (AHL). These signals involve in the regulation of luminescence, antibiotic production, biofilm formation, virulence, and motility in numerous bacteria including Vibrio fischeri and Pseudomonas aeruginosa (Fuqua and Greenberg EP 2002; Schuster et al. 2013).
Lysobacter species are emerging as prolific producers for bioactive natural products (Xie et al. 2012). The biosynthetic mechanisms for these natural products have been subjected for intensive research in the past several years (Hou et al. 2011; Li et al. 2012; Li et al. 2014; Lou et al. 2011; Lou et al. 2012; Wang et al. 2013a; Wang et al. 2013b; Zhang et al. 2011). However, the molecular mechanisms by which Lysobacter control the biosynthesis of these natural products are not well understood. In this study, we report the structure of a diffusible signal molecule LeDSF3 deployed by the valuable biocontrol organism L. enzymogenes, and demonstrate that the signal molecule is essential to the up-regulation and biosynthesis of HSAF analogs, the primary antifungal antibiotics in L. enzymogenes. The chemical structure of the signal molecule was elucidated by spectroscopic analyses, which showed to be 13-methyltetradecanoic acid. Although 14-carbon DSF had been isolated from other organisms (Deng et al. 2011), LeDSF3 is a new structure. In addition to LeDSF3, several fatty acid-derived molecules were produced by L. enzymogenes (Fig. 1A). The main function of these molecules did not seem to be HSAF-related, and we are currently investigating their roles.
DSF-like signal molecules are long-chain fatty acids and do not contain a lactone or a furan ring. These signal molecules have been isolated from several Gram negative bacteria, particularly plant pathogens in the class of gamma-proteobacteria (Deng et al. 2011). In plant pathogens such as Xanthomonas campestris and other Gram negative bacteria, DSF are involved in the regulation of several important processes, such as virulence, motility, biofilm dispersal, extracellular enzyme and production of extracellular polymeric substances, but to our knowledge DSF had not been shown to be involved in the regulation of a specific antibiotic. Lysobacter, which also belongs to gamma proteobacteria, produces several structurally very interesting antibiotics (Xie et al. 2012), but the regulation of these antibiotics is largely unknown. Our work represents the first DSF isolated from a Lysobacter species. It is also the first time to link a specific antibiotic to a specific DSF signal through the two-component regulators RpfG/C and the global regulator Clp. The induction of HSAF analogs by LeDSF3 became detectable when the concentration of exogenously added LeDSF3 was above 0.2 μM (Fig. 4). Although the detectable concentration (13 μg/L, or 53.7 nM) of LeDSF3 in the wild type is lower than the action concentration (0.2 μM) under our experimental conditions, we believe the detectable concentration does not fully reflect the actual concentration of LeDSF3 in vivo. DSFs are quorum sensing signal molecules, and their concentrations can change dramatically due to the auto-induction mechanism in the actual situations (Deng et al. 2011). Furthermore, due to the chemical nature of long-chain fatty acids, the extraction and isolation of LeDSF3 from the bacterial culture would not be very efficient. This is evident from the ~5 mg LeDSF3 isolated from 300 liters of culture of rpfC mutant (a LeDSF3 “high-yield strain”), although the total detectable LeDSF3 would have been ~21.3 mg in rpfC mutant (with a detectable concentration of 71 μg/L, or 293.4 nM, under the experimental conditions).
In summary, this is the first reported case connecting quorum sensing-related diffusible signaling factors to antibiotic production in the ubiquitous environmental Lysobacter species. Our data revealed a regulatory mechanism for HSAF production: a fatty acid-derived LeDSF3 functions as an extracellular signal, sensed and transducted by a two-component regulatory system (RfpC/RfpG), mediated by a global regulator (Clp), to control the production of antifungal natural products (HSAF analogs) in L. enzymogenes. We believe these findings will be useful in applied genetics and molecular biotechnology to improve antibiotic production in Lysobacter, a group of ubiquitous yet underexplored microorganisms.
Supplementary Material
Acknowledgments
This work was supported in part by the 973 Project (2013CB734002), the NIH (R01AI097260), and Program for Changjiang Scholars and Innovative Research Team in University (IRT13028). We thank Prof. Lindow for the generous gift of DSF reporter strain X. campestris 8523/pKLN55. Ron Cerny, Martha Morton, Kurt Wulser, and Javier Seravalli are thanked for technical assistance.
References
- Almeida RP, Killiny N, Newman KL, Chatterjee S, Ionescu M, Lindow SE. Contribution of rpfB to cell-to-cell signal synthesis, virulence, and vector transmission of Xylella fastidiosa. Mol Plant Microbe Interact. 2012;25:453–462. doi: 10.1094/MPMI-03-11-0074. [DOI] [PubMed] [Google Scholar]
- Amari DT, Marques CN, Davies DG. The putative enoyl-coenzyme A hydratase DspI is required for production of the Pseudomonas aeruginosa biofilm dispersion autoinducer cis-2-decenoic acid. J Bacteriol. 2013;195:4600–4610. doi: 10.1128/JB.00707-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barber CE, Tang JL, Feng JX, Pan MQ, Wilson TJ, Slater H, Dow JM, Williams P, Daniels MJ. A novel regulatory system required for pathogenicity of Xanthomonas campestris is mediated by a small diffusible signal molecule. Mol Microbiol. 1997;24:555–566. doi: 10.1046/j.1365-2958.1997.3721736.x. [DOI] [PubMed] [Google Scholar]
- Batchelor JG, Cushley RJ, Prestegard JH. Carbon-13 Fourier transform nuclear magnetic resonance. 8. Role of steric and electric field effects in fatty acid spectra. J Org Chem. 1974;39:1698–1705. doi: 10.1021/jo00925a023. [DOI] [PubMed] [Google Scholar]
- Bi H, Christensen QH, Feng Y, Wang H, Cronan JE. The Burkholderia cenocepacia BDSF quorum sensing fatty acid is synthesized by a bifunctional crotonase homologue having both dehydratase and thioesterase activities. Mol Microbiol. 2012;83:840–855. doi: 10.1111/j.1365-2958.2012.07968.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boon C, Deng Y, Wang LH, He Y, Xu JL, Fan Y, Pan SQ, Zhang LH. A novel DSF-like signal from Burkholderia cenocepacia interferes with Candida albicans morphological transition. ISME J. 2008;2:27–36. doi: 10.1038/ismej.2007.76. [DOI] [PubMed] [Google Scholar]
- Case RJ, Labbate M, Kjelleberg S. AHL-driven quorum-sensing circuits: their frequency and function among the Proteobacteria. ISME J. 2008;2:345–349. doi: 10.1038/ismej.2008.13. [DOI] [PubMed] [Google Scholar]
- Chatterjee S, Wistrom C, Lindow SE. A cell-cell signaling sensor is required for virulence and insect transmission of Xylella fastidiosa. Proc Natl Acad Sci USA. 2008;105:2670–2675. doi: 10.1073/pnas.0712236105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Z, He YW, Lim SC, Qamra R, Walsh MA, Zhang LH, Song H. Structural basis of the sensor-synthase interaction in autoinduction of the quorum sensing signal DSF biosynthesis. Structure. 2010;18:1199–1209. doi: 10.1016/j.str.2010.06.011. [DOI] [PubMed] [Google Scholar]
- Chin KH, Lee YC, Tu ZL, Chen CH, Tseng YH, Yang JM, Ryan RP, McCarthy Y, Dow JM, Wang AH, Chou SH. The cAMP receptor-like protein CLP is a novel c-di-GMP receptor linking cell-cell signaling to virulence gene expression in Xanthomonas campestris. J Mol Biol. 2010;396:646–662. doi: 10.1016/j.jmb.2009.11.076. [DOI] [PubMed] [Google Scholar]
- Christensen P, Cook FD. Lysobacter, a new genus of non-fruiting, gliding bacteria with a high base ratio. Internat J Syst Bacteriol. 1978;28:367–393. [Google Scholar]
- Comella N, Grossman AD. Conservation of genes and processes controlled by the quorum response in bacteria: characterization of genes controlled by the quorum-sensing transcription factor ComA in Bacillus subtilis. Mol Microbiol. 2005;57:1159–1174. doi: 10.1111/j.1365-2958.2005.04749.x. [DOI] [PubMed] [Google Scholar]
- Corre C, Song L, O’Rourke S, Chater KF, Challis GL. 2-Alkyl-4-hydroxymethylfuran-3-carboxylic acids, antibiotic production inducers discovered by Streptomyces coelicolor genome mining. Proc Natl Acad Sci USA. 2008;105:17510–17515. doi: 10.1073/pnas.0805530105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng Y, Boon C, Eberl L, Zhang LH. Differential modulation of Burkholderia cenocepacia virulence and energy metabolism by the quorum-sensing signal BDSF and its synthase. J Bacteriol. 2009;191:7270–7278. doi: 10.1128/JB.00681-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng Y, Wu J, Eberl L, Zhang LH. Structural and functional characterization of diffusible signal factor family quorum-sensing signals produced by members of the Burkholderia cepacia complex. Appl Environ Microbiol. 2010;76:4675–4683. doi: 10.1128/AEM.00480-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng Y, Wu J, Tao F, Zhang LH. Listening to a new language: DSF-based quorum sensing in Gram-negative bacteria. Chem Rev. 2011;111:160–173. doi: 10.1021/cr100354f. [DOI] [PubMed] [Google Scholar]
- Dusane DH, Matkar P, Venugopalan VP, Kumar AR, Zinjarde SS. Cross-species induction of antimicrobial compounds, biosurfactants and quorum-sensing inhibitors in tropical marine epibiotic bacteria by pathogens and biofouling microorganisms. Curr Microbiol. 2011;62:974–980. doi: 10.1007/s00284-010-9812-1. [DOI] [PubMed] [Google Scholar]
- Galloway WR, Hodgkinson JT, Bowden SD, Welch M, Spring DR. Quorum sensing in Gram-negative bacteria: small-molecule modulation of AHL and AI-2 quorum sensing pathways. Chem Rev. 2011;111:28–67. doi: 10.1021/cr100109t. [DOI] [PubMed] [Google Scholar]
- Fuqua C, Greenberg EP. Listening in on bacteria: acyl-homoserine lactone signaling. Nat Rev Mol Cell Biol. 2002;3:685–695. doi: 10.1038/nrm907. [DOI] [PubMed] [Google Scholar]
- He YW, Wang C, Zhou L, Song H, Dow JM, Zhang LH. Dual signaling functions of the hybrid sensor kinase RpfC of Xanthomonas campestris involve either phosphorelay or receiver domain-protein interaction. J Biol Chem. 2006;281:33414–33421. doi: 10.1074/jbc.M606571200. [DOI] [PubMed] [Google Scholar]
- He YW, Wu J, Cha JS, Zhang LH. Rice bacterial blight pathogen Xanthomonas oryzae pv. oryzae produces multiple DSF-family signals in regulation of virulence factor production. BMC Microbiol. 2010;10:187. doi: 10.1186/1471-2180-10-187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He YW, Wu J, Zhou L, Yang F, He YQ, Jiang BL, Bai L, Xu Y, Deng Z, Tang JL, Zhang LH. Xanthomonas campestris diffusible factor is 3-hydroxybenzoic acid and is associated with xanthomonadin biosynthesis, cell viability, antioxidant activity, and systemic invasion. Mol Plant Microbe Interact. 2011;24:948–957. doi: 10.1094/MPMI-02-11-0031. [DOI] [PubMed] [Google Scholar]
- Horinouchi S, Beppu T. A-factor as a microbial hormone that controls cellular differentiation and secondary metabolism in Streptomyces griseus. Mol Microbiol. 1994;12:859–864. doi: 10.1111/j.1365-2958.1994.tb01073.x. [DOI] [PubMed] [Google Scholar]
- Hou J, Robbel L, Marahiel MA. Identification and characterization of the lysobactin biosynthetic gene cluster reveals mechanistic insights into an unusual termination module architecture. Chem Biol. 2011;18:655–664. doi: 10.1016/j.chembiol.2011.02.012. [DOI] [PubMed] [Google Scholar]
- Huang TP, Wong ACL. Extracellular fatty acids facilitate flagella-independent translocation by Stenotrophomonas maltophilia. Res Microbiol. 2007;158:702–711. doi: 10.1016/j.resmic.2007.09.002. [DOI] [PubMed] [Google Scholar]
- Khoklov AS, Tovarova II, Borisova LN, Pliner SA, Schevchenko LA, Kornitskaya EY, Ivkina NS, Rapoport IA. A-factor responsible for the biosynthesis of streptomycin by a mutant strain of Actinomyces streptomycini. Dokl Akad Nauk SSSR. 1967;177:232–235. [PubMed] [Google Scholar]
- Klimentova J, Kosak P, Vavrova K, Holas T, Hrabalek A. Influence of terminal branching on the transdermal permeation-enhancing activity in fatty alcohols and acids. Bioorg Med Chem. 2006;14:7681–7687. doi: 10.1016/j.bmc.2006.08.013. [DOI] [PubMed] [Google Scholar]
- Kobayashi DY, Reedy RM, Palumbo JD, Zhou JM, Yuen GY. A clp gene homologue belonging to the Crp gene family globally regulates lytic enzyme production, antimicrobial activity, and biological control activity expressed by Lysobacter enzymogenes strain C3. Appl Environ Microbiol. 2005;71:261–269. doi: 10.1128/AEM.71.1.261-269.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Du L, Yuen G, Harris SD. Distinct ceramide synthases regulate polarized growth in the filamentous fungus Aspergillus nidulans. Mol Biol Cell. 2006;17:1218–1227. doi: 10.1091/mbc.E05-06-0533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Huffman J, Li Y, Du L, Shen Y. 3-Hydroxylation of the polycyclic tetramate macrolactam in the biosynthesis of antifungal HSAF from Lysobacter enzymogenes C3. Med Chem Comm. 2012;9:982–986. [Google Scholar]
- Li Y, Chen H, Ding Y, Xie Y, Wang H, Cerny RL, Shen Y, Du L. Iterative assembly of two separate polyketide chains by the same single-module bacterial polyketide synthase in the biosynthesis of HSAF. Angew Chem Int Ed. 2014;53:7524–7530. doi: 10.1002/anie.201403500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu G, Chater KF, Chandra G, Niu G, Tan H. Molecular regulation of antibiotic biosynthesis in Streptomyces. Microbiol Mol Biol Rev. 2013;77:112–143. doi: 10.1128/MMBR.00054-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lou L, Qian G, Xie Y, Hang J, Chen H, Zaleta-Rivera K, Li Y, Shen Y, Dussault PH, Liu F, Du L. Biosynthesis of HSAF, a tetramic acid-containing macrolactam from Lysobacter enzymogenes. J Am Chem Soc. 2011;133:643–645. doi: 10.1021/ja105732c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lou L, Chen H, Cerny RL, Li Y, Shen Y, Du L. Unusual activities of the thioesterase domain for the biosynthesis of the polycyclic tetramate macrolactam HSAF in Lysobacter enzymogenes C3. Biochem. 2012;51:4–6. doi: 10.1021/bi2015025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller MB, Bassler BL. Quorum sensing in bacteria. Annu Rev Microbiol. 2001;55:165–199. doi: 10.1146/annurev.micro.55.1.165. [DOI] [PubMed] [Google Scholar]
- Newman KL, Almeida RP, Purcell AH, Lindow SE. Cell-cell signaling controls Xylella fastidiosa interactions with both insects and plants. Proc Natl Acad Sci USA. 2004;101:1737–1742. doi: 10.1073/pnas.0308399100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian G, Wang Y, Liu Y, Xu F, He YW, Du L, Venturi V, Fan J, Hu B, Liu F. Lysobacter enzymogenes uses two distinct cell-cell signaling systems for differential regulation of secondary-metabolite biosynthesis and colony morphology. Appl Environ Microbiol. 2013a;79:6604–6616. doi: 10.1128/AEM.01841-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian G, Zhou Y, Zhao Y, Song Z, Wang S, Fan J, Hu B, Venturi V, Liu F. Proteomic analysis reveals novel extracellular virulence-associated proteins and functions regulated by the diffusible signal factor (DSF) in Xanthomonas oryzae pv. oryzicola. J Proteome Res. 2013b;12:3327–3341. doi: 10.1021/pr4001543. [DOI] [PubMed] [Google Scholar]
- Ryan RP, Dow JM. Communication with a growing family: diffusible signal factor (DSF) signaling in bacteria. Trends Microbiol. 2011;19:145–152. doi: 10.1016/j.tim.2010.12.003. [DOI] [PubMed] [Google Scholar]
- Sarpe VA, Kulkarni SS. Synthesis of maradolipid. J Org Chem. 2011;76:6866–6870. doi: 10.1021/jo200979n. [DOI] [PubMed] [Google Scholar]
- Schuster M, Sexton DJ, Diggle SP, Greenberg EP. Acyl-homoserine lactone quorum sensing: from evolution to application. Annu Rev Microbiol. 2013;67:43–63. doi: 10.1146/annurev-micro-092412-155635. [DOI] [PubMed] [Google Scholar]
- Simionato AVC, da Silva DS, Lambais MR, Carrilho E. Characterization of a putative Xylella fastidiosa diffusible signial factor by HRGC-EI-MS. J Mass Spectromet. 2007;42:490–496. doi: 10.1002/jms.1181. [DOI] [PubMed] [Google Scholar]
- Slater H, Alvarez-Morales A, Barber CE, Daniels MJ, Dow JM. A two-component system involving an HD-GYP domain protein links cell-cell signalling to pathogenicity gene expression in Xanthomonas campestris. Mol Microbiol. 2000;38:986–1003. doi: 10.1046/j.1365-2958.2000.02196.x. [DOI] [PubMed] [Google Scholar]
- Sullivan RF, Holtman MA, Zylstra GJ, White JF, Kobayashi DY. Taxonomic positioning of two biological control agents for plant diseases as Lysobacter enzymogenes based on phylogenetic analysis of 16S rDNA, fatty acid composition and phenotypic characteristics. J Appl Microbiol. 2003;94:1079–1086. doi: 10.1046/j.1365-2672.2003.01932.x. [DOI] [PubMed] [Google Scholar]
- Wang LH, He Y, Gao Y, Wu JE, Dong YH, He C, Wang SX, Wang LX, Xu JL, Tay L, Fang RX, Zhang LH. A bacterial cell-cell communication signal with cross-kingdom structural analogues. Mol Microbiol. 2004;51:903–912. doi: 10.1046/j.1365-2958.2003.03883.x. [DOI] [PubMed] [Google Scholar]
- Wang Y, Qian G, Li Y, Wright S, Shen Y, Liu F, Du L. Biosynthetic mechanism for sunscreens of the biocontrol agent Lysobacter enzymogenes. PLoS One. 2013a;8:e66633. doi: 10.1371/journal.pone.0066633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Qian G, Liu F, Li YZ, Shen Y, Du L. Facile method for site-specific gene integration in Lysobacter enzymogenes for yield improvement of the anti-MRSA antibiotics WAP-8294A and the antifungal antibiotic HSAF. ACS Synth Biol. 2013b;2:670–678. doi: 10.1021/sb4000806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winzer K, Hardie KR, Williams P. LuxS and autoinducer-2: their contribution to quorum sensing and metabolism in bacteria. Adv Appl Microbiol. 2003;53:291–396. doi: 10.1016/s0065-2164(03)53009-x. [DOI] [PubMed] [Google Scholar]
- Xie Y, Wright S, Shen Y, Du L. Bioactive natural products from Lysobacter. Nat Prod Rep. 2012;19:1277–1287. doi: 10.1039/c2np20064c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu F, Zaleta-Rivera K, Zhu X, Huffman J, Millet JC, Harris SD, Yuen G, Li XC, Du L. Structure and biosynthesis of heat-stable antifungal factor (HSAF), a broad-spectrum antimycotic with a novel mode of action. Antimicrob Agents Chemother. 2007;51:64–72. doi: 10.1128/AAC.00931-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuen GY, Steadman JR, Lindgren DT, Schaff D, Jochum CC. Bean rust biological control using bacterial agents. Crop Prot. 2001a;20:395–402. [Google Scholar]
- Yuen GY, Zhang Z. Control of brown patch using the bacterium Stenotrophomonas maltophilia C3 and culture fluid. Int Turfgrass Soc Res J. 2001b;9:742–747. [Google Scholar]
- Zhang W, Li Y, Qian G, Wang Y, Chen H, Li YZ, Liu F, Shen Y, Du L. Identification and characterization of the anti-methicillin-resistant Staphylococcus aureus WAP-8294A2 biosynthetic gene cluster from Lysobacter enzymogenes OH11. Antimicrob Agents Chemother. 2011;55:5581–5589. doi: 10.1128/AAC.05370-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Yuen GY. Biological control of Bipolaris sorokiniana on tall fescue by Stenotrophomonas maltophilia strain C3. Phytopath. 1999;89:817–822. doi: 10.1094/PHYTO.1999.89.9.817. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.