Abstract
Cells employ a plethora of signaling pathways to make their life-and-death decisions. Extensive genetic, biochemical, and physiological studies have led to the accumulation of knowledge about signaling components and their interactions within signaling networks. These conventional approaches, though useful, lack the ability to control the spatial and temporal aspects of signaling processes. The recently emerged optogenetic tools open up exciting opportunities by enabling signaling regulation with superior temporal and spatial resolution, easy delivery, rapid reversibility, fewer off-target side effects, and the ability to dissect complex signaling networks. Here we review recent achievements in using light to control intracellular signaling pathways, and discuss future prospects for the field, including integration of new genetic approaches into optogenetics.
Keywords: optogenetics; intracellular signaling pathways; signal transduction; light-induced protein-protein interaction; photoactivatable proteins; phytochrome, cryptochrome, LOV; UVR8; Dronpa; oligomerization
Challenges in accessing the dynamic information of intracellular signal transduction
Cells are constantly sensing and responding to extracellular stimuli in their environment. A central question in cell biology is how intracellular signal pathways respond to external environment to make appropriate decisions and how decision-making processes go awry in disease conditions. Genomics and proteomics have been continuously expanding our knowledge base of genes and proteins that are responsible for specific cellular functions. However, much less is known about the dynamic nature of signal mechanisms, primarily due to lack of appropriate tools for accessing this dynamic information.
From an engineering point of view, intracellular signal pathways serve as circuits for processing extracellular inputs, computing net results, and executing outputs. For instance, multiple signaling pathways are activated by growth factors (inputs) in order to regulate proliferation, differentiation, migrations, and apoptosis (outputs). Intriguingly, distinct cellular outputs that are elicited by different growth factors often utilize the same set of intracellular signaling pathways [1]. It has been suggested that the output specificity is achieved by regulating intracellular signaling transduction in space and time. However, a better understanding of the spatiotemporal aspect is hindered by the technical challenges inherent in controlling specific signal cascades in space and time.
Conventional methods for studying signal transduction primarily involve pharmacological and genetic approaches. These approaches characterize cellular outputs in response to changes in certain signaling components elicit by chemical (agonist or antagonist) or genetic (gain- or loss-of-function mutations) perturbations. Such approaches proved to be extremely crucial for identifying components involved in signaling pathways. However, these approaches lack the spatial and temporal control to decode the dynamic information in intracellular signal transduction. Chemical genetic approaches have been developed to improve the flexibility of signaling control by using chemical inducers to trigger the activation of engineered proteins [2, 3]. Unfortunately, the diffusive nature of chemicals still hampers their capacity for precise spatiotemporal control.
Emerging optogenetic approaches have led to novel ways of studying signal transduction in live biological systems. Initial successes in optogenetics used light to regulate neuroelectric activities and havetransformed experimental neurobiology [4, 5, 6, 7, 8]. The field of optochemical control of cell signaling, which primarily used photo-uncaging of small molecules [9, 10, 11] or unnatural amino acids [12, 13, 14] to trigger the activation states of signaling molecules, has also seen success. However, we will focus on optical control of intracellular signaling pathways based on genetically-encoded photoactivatable proteins. In this type of optogenetic control, activities of intracellular signaling components are coupled to light-induced conformational changes of photoactivatable proteins [15, 16, 17, 18, 19]. We summarize current achievements in optogenetic control of signaling pathways, highlight advantages of precise spatiotemporal control, and explore future prospects.
Optogenetic control of cell signaling
Photoactivatable Proteins
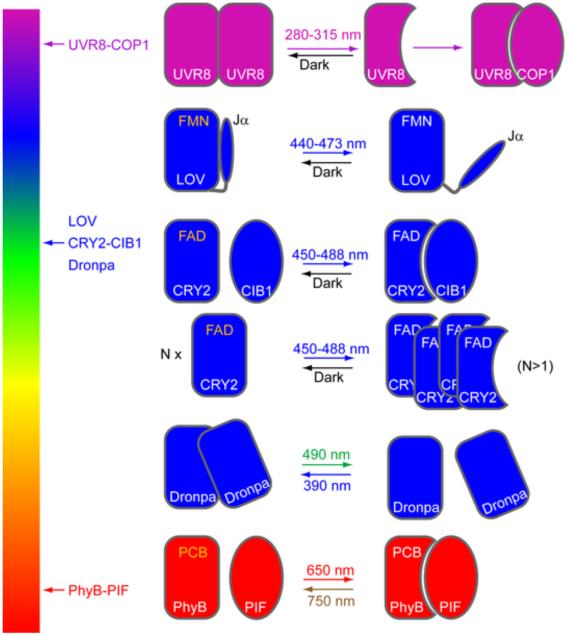
Photoactivatable proteins are core components for optogenetic control of intracellular signal transduction. Pioneering work by a number of research groups has led to the discovery of several photoactivatable proteins, such as LOV (light, oxygen, and voltage) domains [20, 21, 22], phytochrome B (PhyB) [23, 24], cryptochrome 2 (CRY2) [25], UV resistance Locus 8 (UVR8) [26, 27], and Dronpa [28] (Box 1 and Table 1). Some photoactivatable proteins, such as split GFPs [29, 30] have yet to be used in controlling live-cell signal transduction, but there has been recent success in using light-controlled protein-protein interactions to regulate intracellular signaling pathways in live cells (Table 2). The mechanisms of these photoactivatable systems are well known [19]. By absorbing energy from the photons in excitation light, photoactivatable proteins undergo conformational changes, rearrange inter or intra-protein contacts, and modulate inter- or intra-protein interactions (Figure 1).
Box 1.
Photoactivatable proteins
Photoactivatable proteins or photoreceptors are core components for optogenetic control of intracellular signal transduction:
Light-oxygen-voltage (LOV) domain
LOV is a small domain (125 amino acids) with a PAS (PER-ARNT-SIM) core that binds flavin mononucleotide (FMN ), an endogenous cofactor that is ubiquitously produced by mammalian cells [70]. No external cofactors are needed when used in mammalian cells. Blue light photoactivates LOV domain by inducing formation of a covalent adduct between FMN and the Sγ sulfur on a conserved cysteine residue, which causes the C-terminal Jα alpha helix to swing out from the LOV core domain. Hydrolysis of this cysteinyl-flavin bond in dark returns the LOV domain to the ground state [71].
Phytochrome B-PIF
Phytochromes are signaling photoreceptors with five identified members (PhyA through PhyE), which mediate many light-sensitive processes in plants, such as seed germination, seedling de-etiolation, and shade avoidance [78, 79]. PhyB responds to red and infrared light through reversible conformational change induced by photoisomerization of a covalently bound chromophore PCB. The inactive form (Pr) changes into the active form (Pfr) after being exited with 650-nm light; the active form (Pfr), which initiates biological responses, can be converted back to the inactive form (Pr) by absorbing infrared light at 750nm. A disadvantage of the PhyB-PIF protein pair is that its function requires the exogenous cofactor PCB.
Cryptochrome
Cryptochromes are involved in light-regulated cell elongation and photoperiodic flowering in Arabidopsis thaliana. Cryptochrome 2 (CRY2) binds to cryptochrome-interacting bHLH (CIB1) under blue light, and the CRY2-CIB1 complex dissociate in dark. Interestingly, CRY2 has also been found to undergo blue light-induced protein oligomerization [34], enabling applications that use a single protein to control signal transduction. A mutant of CRY2 (E490G or ‘CRY2olig’) that significantly enhances oligomerization has been identified recently by a yeast two-hybrid assay [47]. Both full-length CRY2 (612 amino acids) and its N-terminal photolyase homology region (CRY2PHR, 1-498 amino acids) can bind to either full-length CIB1 (335 amino acid) or CIBN (amino acid 1-170 of CIB1). For simplicity, we use CRY2-CIB1 to cover all combinations of CRY2 and CIB1 interactions.
UV Resistance Locus 8 (UVR8)
Unlike other photoreceptors such as LOV, phytochrome, and cryptochrome, UVR8 has no small-molecule cofactors and uses tryptophan residues as the light perception elements. In the absence of ultraviolet light, UVR8 forms a homodimer. Ultraviolet light can induce disruption of the cation-π interactions between tryptophan and arginine residues at the homodimer interface, resulting in instantaneous dissociation of the homodimer [74]. UVR8 monomer can bind to Constitutively Photomorphogenic 1 (COP1) [26]. Redimerization of UVR8 monomer takes several hours in vitro [76, 77]. The rate of redimerization appears much faster in plant, possibly mediated by Repressor of UV-B Photomorphogenesis (RUP)1 and RUP2 [75].
Dronpa
The fluorescent protein Dronpa forms a tetramer under blue light. Upon cyan light (500 nm) stimulation, the tetramer dissociates into monomers [28]. Like UVR8, Dronpa has no small-molecule cofactors and uses tryptophan for light sensing. The association/dissociation reaction is reversible.
Tables 1.
Characterization of individual light-sensitive protein pairs in optogenetic toolboxes
| Photoactivatable proteins |
Size (a.a.) | Cofactor | As./Dis. λ(nm) |
As./Dis. time |
Refs |
|---|---|---|---|---|---|
|
| |||||
| PhyB(FL)/PIF3 | 1211/524 | PCB | 650/750 | sec/sec | [64, 65] |
| PhyB(NT)/PIF3 | 621/524 | PCB | 650/750 | sec/sec | [24] |
| PhyB/PIF6 | 908/100 | PCB | 650/750 | msec/msec | [23, 66] |
| CRY2/CIB1 | 612/335 | FAD | 450/ dark | sec/6 min | [67, 68, 69] |
| CRY2/CIB1 | 498/170 | FAD | 450/ dark | sec/6 min | [25] |
| CRY2/CRY2 | 498/498 | FAD | 450/ dark | sec/6 min | [34] |
| CRY2olig | 498 | FAD | 450/ dark | sec/23 min | [47] |
|
EL222 (LOV fast
cycler) |
150 | FMN | 450/ dark | sec/sec | [70, 71] |
|
FKF1/G1 (LOV fast
cycler) |
619/1173 | FMN | 450/ dark | min/hr | [21] |
| LOVpep/ePDZ | 153/194 | FMN | 450/ dark | sec/sec | [22, 31] |
|
VVD/VVD (LOV slow
cycler) |
150/150 | FAD | 450/ dark | sec/sec-day | [72, 73] |
| Dronpa/Dronpa | 257/257 | None | 400/500 | sec/sec | [16, 28] |
| UVR8/COP1C340 | 440/340 | None | dark/290-310 | 1-4 h/sec | [26, 74] |
| UVR8/UVR8 | 440/440 | None | dark/280-315 | 2-24h/sec | [27, 75, 76, 77] |
|
|
|||||
Tables 2.
Current applications of optogenetic control of signaling pathways.
| Cellular function |
Photoactivatab le proteins |
Controlling mechanism |
Signaling proteins | Model system |
Refs |
|---|---|---|---|---|---|
|
| |||||
| MAPK | LOV | Translocation | Ste5 | Yeast | [31] |
| Ras/ERK | PhyB-PIF6 | Translocation | SOScat | Mammalian | [32] |
| Raf/ERK | CRY2-CIB1 | Translocation | Raf1 | Mammalian | [33] |
| Raf/ERK | CRY2-CRY2 | Translocation | Raf1 | Mammalian | [35] |
| Phosphatase | CRY2-CIB1 | Translocation | 5-phosphotases | Mammalian | [40] |
| PI3K | CRY2-CIB1 | Translocation | SH2 of p85α | Mammalian | [40] |
| PIP3 | CRY2-CIB1 | Translocation | SH2 of p85β | Neuron | [39] |
| Rho GTPase | PhyB-PIF6 | Translocation | Tiam, Tim, Intersectin |
Mammalian | [23] |
| Rho GTPase | LOV | Uncaging | Rac1 | Mammalian | [20] |
| Rho GTPase | FKF1-G1 | Translocation | Rac1 | Mammalian | [21] |
| Rho GTPase | LOVpep/ePDZ | Translocation | Cdc24 | Yeast | [31] |
| Rho GTPase | Dronpa | Uncaging | Intersectin | Mammalian | [28] |
| RTK | CRY2-CRY2 | Translocation | FGFR | Mammalian | [41] |
| RTK | LOV | Translocation | FGFR | Mammalian | [42] |
| Protease | Dronpa | Uncaging | NS3-4A protease | Mammalian | [28] |
| Apoptosis | LOV | Uncaging | Caspase-7 | Mammalian | [43] |
| Protein splicing | PhyB-PIF3 | Translocation | Vacuolar ATPase (VMA) intein | Yeast | [45] |
| Protein secretion |
UVR8 | Dissociation | Vesicular stomatitis virus glycoprotein (VSVG)-YFP |
Mammalian, Neuron | [27] |
| Protein inactivation |
CRY2- CIB1(MP) |
Translocation | Vav2, Tiam1, Rac1, Cdc42, RhoG, VHH antibody |
Mammalian | [46] |
| Protein inactivation |
CRY2(E490G) | Translocation | Clathrin light chain (CLC) | Mammalian | [47] |
| Actin polymerization |
CRY2(E490G) | Translocation | Nck, Verprolin-homology, Central, Acidic (VCA) domain of N-WASP |
Mammalian | [47] |
| Protein degradation |
LOV | Uncaging | degron | Mammalian | [49] |
| Protein degradation |
LOV | Uncaging | degron | Mammalian, Zebrafish |
[50] |
| DNA transcription |
PhyB-PIF3 | Translocation | Gal4-DNA-binding domain(GBD)/Gal4-activation domain(GAD) |
Yeast | [24] |
| DNA transcription |
PhyB-PIF6 | Translocation | GBD/GAD | Yeast | [52] |
| DNA transcription |
CRY2-CIB1 | Translocation | Split Cre recombinase | Mammalian | [25] |
| DNA transcription |
CRY2-CIB1 | Translocation | Split Cre recombinase | Drosophila | [57] |
| DNA transcription |
CRY2-CIB1 | Translocation | LexA/VP16 | Yeast | [52] |
| DNA transcription |
CRY2-CIB1 | Translocation | GBD/GAD | Zebrafish | [53] |
| DNA transcription |
FKF1-G1 | Translocation | GBD/VP16 | Mammalian | [21] |
| DNA transcription |
FKF1-G1 | Translocation | Zinc Finger Protein (ZFP)/VP16 | Mammalian | [55] |
| DNA transcription |
EL222 | Uncaging | Helix-turn-helix (HTH)/VP16 | T-cell, Zebrafish | [54] |
| DNA transcription |
UVR8-COP1 | Translocation | NF-κB activation domain/GBD | Mammalian | [26] |
| EndogenousD NA transcription |
CRY2-CIB1 | Translocation | Transcription activator-like effector (TALE)-DNA binding/VP64 |
Mammalian | [61] |
|
|
|||||
Figure 1.
Scheme of light-induced conformational change in various photoactivatable proteins. The left bar illustrates the color of light (wavelength) that is used to stimulate photoactivation. Various protein pairs are shown on the right with light-induced inter-molecular change (UVR8, CRY2-CIB1, CRY2 alone, and PhyB-PIF) or intra-molecular change (LOV, Dronpa). For proteins containing cofactors (FMN, FAD, PCB), the yellow color marks the ground state and the white color marks the photo-activated state. UVR8 and Dronpa do not have cofactors and primarily use their tryptophan residues for photo reception.
The mitogen-activated protein kinase (MAPK) signaling pathway
The MAPK signaling pathway plays important roles in controlling cell proliferation, differentiation, survival, and apoptosis. Light-controlled activation of the MAPK signaling pathway was first demonstrated in yeast by membrane recruitment of the scaffold protein Ste5, which was known to activate the MAPK pathway when tethered to the plasma membrane [31]. Ste5 was fused to a PDZ domain, which bound to a membrane-anchored LOV-epitope upon blue light stimulation and subsequently activated the MAPK pathway. In mammalian cells, a light-induced MAPK (Ras/Raf/MEk/ERK module) activation system was built based on the PhyB-PIF6 system [32]. PhyB was anchored to the plasma membrane, and PIF6 was fused to the catalytic segment of the protein SOS (SOScat). Red light induced PhyB-PIF6 binding and membrane recruitment of SOS, which subsequently activated the Ras/Raf/MEK/ERK signaling pathway. Light-controlled activation of the Raf/MEK/ERK pathway in mammalian cells has also been achieved by the CIB1-CRY2 system [33]. CIB1 was anchored to the plasma membrane, and CRY2PHR was fused to Raf1. Blue light stimulation recruited Raf1 to the plasma membrane, where Raf1 was activated to activate its downstream kinases. This approach used Raf1 as the controlling component to avoid potential crosstalk with other signaling pathways, which may be induced by upstream factors such as SOS. Light-induced activation of the Raf/MEK/ERK pathway stimulated significant neurite outgrowth in PC12 cells in the absence of nerve growth factors. Interestingly, neurite outgrowth did not require constant ERK activation. Intermittent on-off light control revealed a 45-min threshold for the light-off interval, which still supported maximum neurite outgrowth [33]. In addition to light-induced binding between CRY2 and CIB1, CRY2PHR has been shown to oligomerize upon blue light illumination [34]. Such a property allows light-induced aggregation of CRY2-Raf1 in the cytoplasm [35] , which was able to activate Raf1 and the downstream Raf/MEK/ERK signaling pathway. It is worth nothing that heterodimerization between Raf1-CIB1 and Raf1-CRY2 in the cytoplasm did not induce ERK activation, likely due to steric effects that block Raf/Raf interaction.
The phosphatidylinositol 3-kinases (PI3K) signaling pathway
PI3Ks phosphorylate the 3-hydroxyl group of phosphatidylinositol (PtdIns) to produce signaling lipids, such as PIP3, which activate downstream pathways such as AKT, Rac/actin, and PKC to regulate diverse biological functions including cell growth, survival, migration, and cell cycle progression [36, 37]. Light-controlled activation of the PI3K/PIP3 pathway has been achieved by a membrane recruitment assay based on PIF6-PhyB [38], in which PhyB was anchored to the plasma membrane and PIF6 was fused to the SH2 domain of a PI3K binding protein p85α. Red light induced PhyB-PIF6 binding and recruited the SH2 domain to the plasma membrane. Membrane-bound SH2 recruited PI3K to the membrane and led to consequential production of PIP3. A similar scheme for controlling PI3K activity using the CIB1-CRY2 protein pair was reported recently [39]. Local photoactivation of PI3K in the growth cone of mouse hippocampal neurons induced growth cone expansion, and increased the numbers of filopodia and lamellipodia. The CIB1-CRY2 protein pair has also been used to control the phosphoinositide metabolism [40]. Whereas the membrane recruitment of PI3K produced PIP3, the membrane recruitment of a phosphatase resulted in dephosphorylation of PIP3. Local activation of the phosphatase caused loss of membrane ruffling as well as collapse and retraction of the cell edge.
The Rho family GTPase activity
The Rho family of GTPase is a subfamily of the Ras superfamily and plays a role in regulating actin dynamics, cell mobility, and organelle development. Three Rho family members, Rac1, Cdc42, and RhoA, have been well studied. Light-controlled activation of Rho GTPases was initiated by light-controlled membrane recruitment of their guanine nucleotide exchange factors (GEFs) [23]. PhyB was anchored to the plasma membrane and PIF was fused to various GEFs including Tiam (RacGEF), intersectin (Cdc42GEF), and Tim (RhoGEF). Global recruitment of Tiam and intersectin generated lamellipodia and filopodia, whereas global membrane recruitment of Tim induced cell body contraction. Subcellular local activation of Tiam enabled a directed extrusion of lamellipodia in live 3T3 cells. The Rho GTPase activity has also been controlled in a translocation assay by the FKF1-GI [21] and LOVpep-ePDZ systems [22, 31].
In addition to the light-induced protein translocation methods, Rho family GTPase activity has also been controlled optogenetically by a LOV-based photo-uncaging system. In this case, a photoactivatable Rac1 was constructed by fusing the LOV2-Jα sequence to the amino terminus of a constitutively active Rac1 [20]. In the dark state, the LOV domain sterically blocked the effector binding site of Rac1; light-induced unwinding of the Jα helix released the steric inhibition and activated Rac1.
The receptor tyrosine kinase activity
The Ras/Raf/ERK, PI3K, and PLC signaling pathways are downstream of receptor tyrosine kinase activation. Blue light can be used to control the activity of fibroblast growth factor receptor (FGFR) and activate all three downstream signaling pathways through blue light-induced CRY2 oligomerization. When a CRY2 domain was fused between a membrane targeting sequence and the cytosolic catalytic domain of FGFR, light-induced oligomerization led to auto-activation of the engineered receptor and subsequent activation of all downstream signaling pathways [41]. In another example, a chimeric receptor was made by linking the LOV domain of Aureochrome 1 from V. frigida to the intracellular catalytic domain of murine FGFR1 (Opto-mFGFR1) [42]. Photoactivated LOV domains dimerized and activated Opto-mFGFR1, which activated downstream ERK, AKT, and PLCγ signaling pathways.
Light-induced programmed cell death
Programmed cell death, or apoptosis, is a regulated cellular suicide mechanism. Caspases, a family of cysteine proteases, are critical regulators of programmed cell death. Once activated, initial caspases cleave and activate downstream effector caspases, which in turn degrade other cellular proteins and cause apoptosis. A light-controlled caspase has been constructed by fusing a LOV domain to the apoptosis-executing domain of caspase-7. Under dark conditions, the LOV domain caged the caspase activity; blue light illumination induced conformational changes in the LOV domain and released the caspase activity, causing cell apoptosis within one hour [43]. This method can potentially be used to induce cell apoptosis of non-transfected cancer cells. At low pH (<6.0), membrane fusion formed syncytia (cells with multiple nuclei) from engineered and co-cultured cancer cells; subsequent illumination induced cellular apoptosis of syncytia and thereby the targeted cancer cells [44].
Light-controlled production, inactivation, and degradation of proteins
The ability to rapidly induce the production, inactivation, or degradation of specific proteins is imperative for dissecting complex signaling network and identifying novel therapeutic targets. A light-inducible protein production system has been constructed through protein splicing in yeast [45]. Protein splicing is a post-translational modification that can generate new proteins by removing an internal segment (intein) followed by ligation of the remaining N- and C-terminal segments (extein). In the light-inducible protein-splicing system, protein fragments were fused to 177 two halves of an intein. Maltose Binding Protein (MBP) was fused to intein-N (IN) and Flag was fused to intein-C (IC). The two epitopes MBP-IN and Flag-IC were then fused to PIF3 and PhyB, respectively. PhyB-PIF3 binding upon red light illumination reconstituted the full-length intein, which autocatalytically excised itself to produce the splice product MBP-Flag. Such an experiment implies that light can be used to control the activity of proteins at the post-translational level in live cells.
A light-induced clustering strategy has been recently developed to inactivate target proteins [46]. The system uses both light-mediated heterodimerization of CRY2 and CIB1, as well as light-mediated homodimerization of CRY2. CIB1 was fused to a multimeric protein Ca2+/calmodulin-dependent protein kinase IIα (CIB1-MP). The target protein was fused to CRY2. Photoactivated CRY2 proteins simultaneously oligomerized and bound to CIB1-MP, which induced the formation of higher-order clusters by promoting interconnection among MPs. Such clusters served as traps to inactivate target proteins including various GEFs (Vav2 and Tiam1) and GTPases (Rac1, Cdc42, and RhoG) [46]. Although CRY2 homo-oligomerization has been shown upon photoactivation, hetero-oligomerization between CRY2 and CIB1-MP is significantly enhanced. A CRY2 mutant (E490G or ‘CRY2olig’) with significantly improved photo-induced clustering effect was recently identified [47]. This mutation resulted in clustering of ~40-90% cytosolic CRY2olig in 100% cells illuminated, compared to clustering of ~6% cytosolic wild-type CRY2 in ~12% cells illuminated. Photoactivation of a fusion protein CRY2olig-CLC (clathrin light chain) resulted in enhanced transferrin uptake via endocytosis. Clustering of Nck SH3 domains induced localized actin polymerization.
In addition to light-induced protein production and inactivation, 200 light has also been used to induce protein degradation. Degrons, or destabilizing sequences, are small peptide sequences that can be recognized by specific proteasomes for protein degradation in live cells [48]. A light-controlled protein degradation system has been constructed by fusing a 23-amino-acid degron sequence to the C-terminal of LOV2. Upon blue light illumination, the normally caged degron sequence was exposed, resulting in rapid degradation of the fusion protein [49]. In another example, a smaller degron (five amino-acids) was fused to the C-terminus of the LOV domain. The LOV-degron was in turn fused to the proteins of target (YFP, mCherry, and β-actin-mCherry) for light-induced degradation [50]. In both groups’ work, photo-uncaging of LOV domains resulted in exposure of degrons to cause protein degradation.
Light-controlled protein trafficking and secretion
After being synthesized by ribosomes, proteins that are targeted for secretion are translocated into the endoplasmic reticulum (ER) to be glycosylated and correctly folded. If proteins aggregate within the ER, they will be retained within the ER until the aggregates dissociate, and this phenomenon has been used in a ligand-induced protein secretion system [51]. By fusing two or three copies of UVR8 to the C-terminal intracellular domain of vesicular stomatitis virus glycoprotein (VSVG), a light-induced protein secretion system has been engineered [27]. In the dark state, UVR8s formed oligomers and the VSVG-YFP-UVR8s were trapped in the ER. One 7-sec pulse of UV light was sufficient to dissociate the VSVG-YFP-UVR8 clusters and induce protein trafficking from the ER to the Golgi complex and to the plasma membrane via the secretory pathway.
Light-controlled DNA transcription
Light-induced protein-protein interaction has been used to recruit transcription activators to promoters and subsequently regulate transcription of specific genes. The first implementations utilized the PhyB-PIF3 system. Two chimeric proteins, PhyB fused to GAL4 DNA binding domains (PhyB-GBD) and PIF3 fused to GAL4-activation-domains (PIF3-GAD), were expressed in yeast cells. PhyB-PIF3 binding upon red light illumination brought GBD and GAD in close proximity, which activated transcription of marker genes containing promoters with a GAL4 DNA-binding site. Gene expression can be repressed by far-red light illumination, which dissociates the GBD/GAD complex [24]. Similar systems have also been constructed using the PhyB-PIF6 pair [52], CRY2-CIB1 pair [52, 53], and the LOV2 domain [21, 54, 55]. A different strategy used light-activated Cre recombinase to regulate DNA transcription [25] based on a split Cre recombinase assay [56]. CRY2 and CIB1 were fused to the N- and C-domains of Cre, which can be reconstituted by photoactivation-induced CRY2-CIB1 association. Reconstitution of split Cre recovered its recombinase activity and activated the transcription of a reporter gene (EGFP preceded by a transcriptional stop sequence flanked by loxP sites). This light-induced split Cre system has been used recently in Drosophila [57]. Different protein pairs have also been used in light-inducible systems to control mammalian cell gene expression [58].
Spatial and temporal control of intracellular signaling transduction
Light-controlled intracellular signal transduction is blessed with a unique capacity for spatial and temporal resolution. Light can be turned on or off instantaneously and can be directed to specific subcellular locations. Here we present a few examples to illustrate the powerful spatiotemporal control of light-activated intracellular signaling pathways.
Spatial control of subcellular signal activation
By activating specific signaling pathway in a subcellular region, one can understand how localized signaling activation affects cell behavior. The most striking example is localized activation of the Rho family GTPase that controls actin dynamics. Several studies have demonstrated the light control of Rac1, Cdc42, and RhoA [20, 21, 23, 28]. In one case, local photoactivation of the LOV-based photoactivatable Rac1 (PA-Rac1) allowed reversible induction of membrane ruffles and protrusions [20]. Interestingly, activating Rac1 in one spot near the cell edge also produced retraction on the opposite side of the cell. Other experiments using a protein translocation assay [21, 23] and an uncaging assay [28] also showed that localized activation of the Rho GTPase activity was sufficient to establish cell polarity. The process was associated with actin polymerization, translocation of downstream effectors, and localized PAK phosphorylation. Localized activation of the FGFR signaling at the edge of a cell also resulted in cell polarity [41]. The PI3K signaling pathway was found to be primarily responsible for such an effect, consistent with previous findings that PI3K is an upstream activator of Rac1 [59].
The spatial resolution of optogenetic control may be degraded by the diffusion of activated signaling components. Using a PhyB-PIF6 system, such degradation in resolution can be rescued by deactivating the active species in the undesired areas [23]. In such a configuration, the activation can be confined to a highly localized area. This scheme is not necessary for LOV domain where dissociation is fast, which limits the existence of activated signaling components in unilluminated area. However, when the dissociation is slow (CRY2-CIB1 and UVR8) or when the binding is not reversible (VVD), the spatial resolution of localized signaling in cells is expected to be lower than what the light can achieve.
Temporal control of intracellular signaling pathways
How fast can cells catch up with changes in their environment? This time scale determines the adaptive capacity of cells. To understand this adaptive capacity, it would be ideal to stimulate cells at various frequencies and characterize cellular responses accordingly. Light exposure can be precisely controlled in frequency and duration, thus enabling the measurement of the frequency of cell response. A PhyB-PIF6 based system was used to measure how fast ERK activity caught up with the intermittent light stimulation [32]. It was found that the Ras/ERK module acts as a wide-bandwidth, low-pass filter, transmitting signals from the time scale of 4 min to 2 h.
On longer time scales, light activation of the Raf/MEK/ERK signaling module is sufficient to induce PC12 cell differentiation in 1-3 days [33]. A series of ON/OFF light patterns was used to activate the Raf/MEK/ERK pathway in PC12 cells and revealed that the duration of the light-OFF dark interval was more crucial in determining the neurite length than the light-ON duration. When the dark interval was equal to or less than 45 min, regardless of the light-ON duration, the neurite length was the same as under continuous light illumination. On the other hand, when the dark duration was longer than 45 min, the neurite length decreased considerably. These studies demonstrate that light is a powerful tool to dissect the kinetic responses of intracellular signaling pathways.
Precautions in designing optogenetic control of intracellular signaling
Some precautions should be used, however, in designing experimental schemes with optogenetic tools. Many photoreceptors, such as full-length phytochrome and cryptochrome, undergo light-mediated oligomerization. Oligomerization may orient the signaling components into different configurations than those that arise from naturally occurring ligand-induced dimerization. Such a difference may lead to differential outputs of subsequent signalling pathways. Additionally, basal signaling activities should always be checked in both the translocation and uncaging assays. In the translocation assay, overexpression of the optogenetic protein(s) may lead to false binding; in the uncaging assay, basal activities may arise from imperfect allosteric inhibition. Screening and protein engineering are often needed for optimized performance. Negative controls, such as using dark conditions and activity-dead mutants should be performed. The excitation light may also induce phototoxicity, depending on the wavelength, exposure time, and the intensity, especially for applications that require long-term illumination. In such cases, protein pairs that can be activated by light with long wavelengths and low intensities would be preferred.
Concluding remarks and future perspectives
Recently developed genome engineering techniques enable precise editing (deletion, insertion, or mutation) of genome sequences in mammalian cells [60]. Current photoactivatable proteins can be combined with these new genome engineering tools to control the endogenous gene transcription. One such integration has been demonstrated by the engineering of light-inducible transcriptional effectors (LITEs), in which a TALE DNA-binding domain was fused to CRY2 and an effector (VP16) was fused to CIB1 [61], where . Light-induced binding between CRY2 and CIB1 activated endogenous gene (Neurog2) transcription in mammalian cells. Such a technique can be generalized for precise regulation of gene expression and epigenetic states. Optogenetics could also extend its application in synthetic biology. The power of dynamically modulating signaling activity offers guidance for the design of customized signaling circuits [62]. As the optogenetic control of neuronal activity has already spearheaded in translational research [63], the optogenetic control of intracellular signaling could potentially serve as a powerful tool in complement to current treatment procedures, such as gene therapy, in clinical settings.
Box 2.
Outstanding Questions
How can we improve the photophysical properties of photoactivatable proteins such as tunable activation wavelength, sensitivity and dynamic range in response to the stimulation light, the association and dissociation kinetics, and light-dependent oligomerization?
Can orthogonal optogenetic tools be developed for simultaneous control of multiple signaling pathways?
What is required to streamline the design and characterization of optogenetic tools for signaling control?
Will new devices for better light delivery and interfacing with biological systems be developed?
How do we implement optogenetics in therapeutic applications?
Highlights.
We explain mechanisms of light-induced conformation change of photoactivatable proteins.
We describe strategies and studies of using photoactivatable proteins to control intracellular signaling pathways.
We highlight the advantages of using light to control intracellular signaling pathways with superior spatial and temporal resolutions.
We discuss precautions to be used in designing experimental schemes of optogenetic control of cell signaling.
Figure 2.
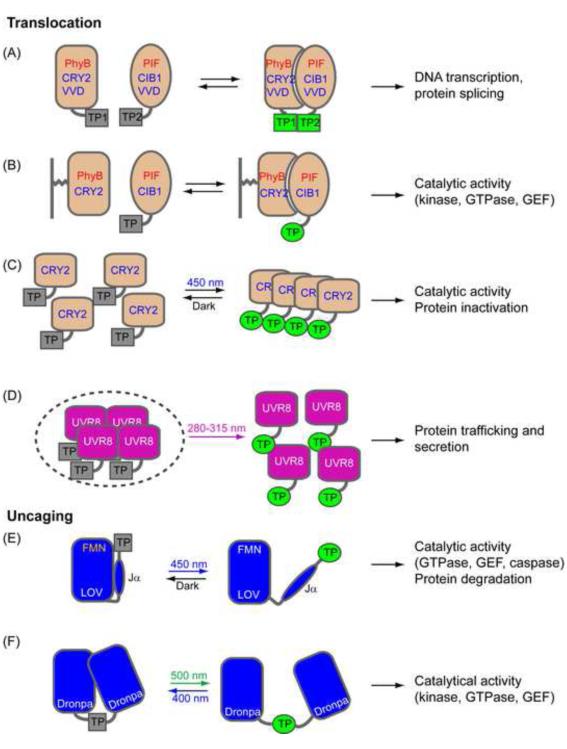
Modes of signaling control by photoactivatable proteins. The target proteins (TP) can be activated by either photo-induced protein translocation (A-D) or uncaging (E-F). (A) Binding between TP1 (e.g. DNA binding domain) and TP2 (e.g. activation domain) can lead to activation of DNA transcription. TP1 and TP2 can also be split inteins which leads to protein splicing after protein binding. (B) Light can recruit signaling proteins to certain subcellular locations (e.g. the cytoplasmic leaflet of the plasma membrane) and activate downstream signaling pathways. (C) Light-induced oligomerization of CRY2 allows increase in the local concentration of signaling protein (e.g. receptor tyrosine kinase) and subsequent activation of downstream pathways. Oligomerization can also be used to conditionally inactivate protein activities. (D) The oligomeric states of UVR8 can be used to trap or release proteins from organelles (e.g. ER). (E-F) Photo-uncaging can release the steric inhibition of signaling components and activate downstream pathways.
Acknowledgements
We thank Ling-chun Chen from Stanford University for giving comments on our manuscript. This work is supported by NIH Innovator award NS082125 and a Packard Science and Engineering Fellowship (BC) and University of Illinois at Urbana-Champaign (KZ).
Glossary Box
- Optogenetics
Optogenetics combines the power of light and genetics and uses light-mediated protein-protein interactions to control the open/closed state of channels or the activation/inactivation states of signaling components within live cells.
- Photoactivatable proteins
Also referred to as photoreceptors. These proteins undergo light-induced conformational change to initiate signal transduction.
- Photoexcitation
The process of converting photon energy to conformational changes of photoreceptors.
- Cofactor
Photosensitive small molecules bound to photoactivatable proteins. Cofactors are required for the photoactivation of photoactivatable proteins. Common cofactors include flavin (blue light sensitive) and bilin (red light sensitive) and their derivatives. Some photoactivatable proteins (such as UVR8 and Dronpa) do not have cofactors and use intrinsic amino acids such as tryptophan residues to mediate their conformational changes.
- Association/Dissociation wavelength
the wavelength of light used to stimulate the association and dissociation of photoactivatable proteins. Some photoactivatable proteins (such as CRY2 and LOV) do not have a light-driven dissociation mechanism. Instead, the complex dissociates spontaneously in dark.
- Association/Dissociation time
The average time it takes to induce association and dissociation of photoactivatable proteins.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Santos SD, Verveer PJ, Bastiaens PI. Growth factor-induced MAPK network topology shapes Erk response determining PC-12 cell fate. Nat Cell Biol. 2007;9:324–330. doi: 10.1038/ncb1543. [DOI] [PubMed] [Google Scholar]
- 2.Inoue T, Do Heo W, Grimley JS, Wandless TJ, Meyer T. An inducible translocation strategy to rapidly activate and inhibit small GTPase signaling pathways. Nature Methods. 2005;2:415–418. doi: 10.1038/nmeth763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Karginov AV, Ding F, Kota P, Dokholyan NV, Hahn KM. Engineered allosteric activation of kinases in living cells. Nat Biotechnol. 2010;28:743–747. doi: 10.1038/nbt.1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Deisseroth K. Optogenetics. Nat Methods. 2011;8:26–29. doi: 10.1038/nmeth.f.324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Airan RD, Thompson KR, Fenno LE, Bernstein H, Deisseroth K. Temporally precise in vivo control of intracellular signalling. Nature. 2009;458:1025–1029. doi: 10.1038/nature07926. [DOI] [PubMed] [Google Scholar]
- 6.Boyden ES, Zhang F, Bamberg E, Nagel G, Deisseroth K. Millisecond-timescale, genetically targeted optical control of neural activity. Nat Neurosci. 2005;8:1263–1268. doi: 10.1038/nn1525. [DOI] [PubMed] [Google Scholar]
- 7.Zhang F, Wang LP, Boyden ES, Deisseroth K. Channelrhodopsin-2 and optical control of excitable cells. Nat Methods. 2006;3:785–792. doi: 10.1038/nmeth936. [DOI] [PubMed] [Google Scholar]
- 8.Nagel G, Szellas T, Huhn W, Kateriya S, Adeishvili N, Berthold P, Bamberg E. Channelrhodopsin-2, a directly light-gated cation-selective membrane channel. Proc Natl Acad Sci U S A. 2003;100:13940–13945. doi: 10.1073/pnas.1936192100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Karginov AV, Zou Y, Shirvanyants D, Kota P, Dokholyan N.V., Young, D.D., … Deiters, A. Light Regulation of Protein Dimerization and Kinase Activity in Living Cells Using Photocaged Rapamycin and Engineered FKBP. J Am Chem Soc. 2010 doi: 10.1021/ja109630v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu QY, Deiters A. Optochemical Control of Deoxyoligonucleotide Function via a Nucleobase-Caging Approach. Accounts of Chemical Research. 2014;47:45–55. doi: 10.1021/ar400036a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shestopalov IA, Sinha S, Chen JK. Light-controlled gene silencing in zebrafish embryos. Nat Chem Biol. 2007;3:650–651. doi: 10.1038/nchembio.2007.30. [DOI] [PubMed] [Google Scholar]
- 12.Arbely E, Torres-Kolbus J, Deiters A, Chin JW. Photocontrol of tyrosine phosphorylation in mammalian cells via genetic encoding of photocaged tyrosine. J Am Chem Soc. 2012;134:11912–11915. doi: 10.1021/ja3046958. [DOI] [PubMed] [Google Scholar]
- 13.Gautier A, Deiters A, Chin JW. Light-activated kinases enable temporal dissection of signaling networks in living cells. J Am Chem Soc. 2011;133:2124–2127. doi: 10.1021/ja1109979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nguyen DP, Mahesh M, Elsasser SJ, Hancock SM, Uttamapinant C, Chin JW. Genetic Encoding of Photocaged Cysteine Allows Photoactivation of TEV Protease in Live Mammalian Cells. Journal of the American Chemical Society. 2014;136:2240–2243. doi: 10.1021/ja412191m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tucker CL. Manipulating cellular processes using optical control of protein-protein interactions. Prog Brain Res. 2012;196:95–117. doi: 10.1016/B978-0-444-59426-6.00006-9. [DOI] [PubMed] [Google Scholar]
- 16.Kim B, Lin MZ. Optobiology: optical control of biological processes via protein engineering. Biochemical Society Transactions. 2013;41:1183–1188. doi: 10.1042/BST20130150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Toettcher JE, Gong DQ, Lim WA, Weiner OD. Light Control of Plasma Membrane Recruitment Using the Phy-Pif System. Method Enzymol. 2011;497:409–423. doi: 10.1016/B978-0-12-385075-1.00017-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zoltowski BD, Gardner KH. Tripping the light fantastic: blue-light photoreceptors as examples of environmentally modulated protein-protein interactions. Biochemistry. 2011;50:4–16. doi: 10.1021/bi101665s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tischer D, Weiner OD. Illuminating cell signalling with optogenetic tools. Nat Rev Mol Cell Bio. 2014;15:551–558. doi: 10.1038/nrm3837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wu YI, Frey D, Lungu OI, Jaehrig A, Schlichting I, Kuhlman B, Hahn KM. A genetically encoded photoactivatable Rac controls the motility of living cells. Nature. 2009;461:104–108. doi: 10.1038/nature08241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yazawa M, Sadaghiani AM, Hsueh B, Dolmetsch RE. Induction of protein-protein interactions in live cells using light. Nature Biotechnology. 2009;27:941–U105. doi: 10.1038/nbt.1569. [DOI] [PubMed] [Google Scholar]
- 22.Strickland D, Yao X, Gawlak G, Rosen MK, Gardner KH, Sosnick TR. Rationally improving LOV domain-based photoswitches. Nat Methods. 2010;7:623–626. doi: 10.1038/nmeth.1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Levskaya A, Weiner OD, Lim WA, Voigt CA. Spatiotemporal control of cell signalling using a light-switchable protein interaction. Nature. 2009;461:997–1001. doi: 10.1038/nature08446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shimizu-Sato S, Huq E, Tepperman JM, Quail PH. A light-switchable gene promoter system. Nat Biotechnol. 2002;20:1041–1044. doi: 10.1038/nbt734. [DOI] [PubMed] [Google Scholar]
- 25.Kennedy MJ, Hughes RM, Peteya LA, Schwartz JW, Ehlers MD, Tucker CL. Rapid blue-light-mediated induction of protein interactions in living cells. Nat Methods. 2010;7:973–975. doi: 10.1038/nmeth.1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Crefcoeur RP, Yin RH, Ulm R, Halazonetis TD. Ultraviolet-B-mediated induction of protein-protein interactions in mammalian cells. Nature Communications. 2013:4. doi: 10.1038/ncomms2800. [DOI] [PubMed] [Google Scholar]
- 27.Chen D, Gibson ES, Kennedy MJ. A light-triggered protein secretion system. J Cell Biol. 2013;201:631–640. doi: 10.1083/jcb.201210119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhou XX, Chung HK, Lam AJ, Lin MZ. Optical control of protein activity by fluorescent protein domains. Science. 2012;338:810–814. doi: 10.1126/science.1226854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kent KP, Boxer SG. Light-activated reassembly of split green fluorescent protein. J Am Chem Soc. 2011;133:4046–4052. doi: 10.1021/ja110256c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Do K, Boxer SG. GFP Variants with Alternative beta-Strands and Their Application as Light-driven Protease Sensors: A Tale of Two Tails. J Am Chem Soc. 2013;135:10226–10229. doi: 10.1021/ja4037274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Strickland D, Lin Y, Wagner E, Hope CM, Zayner J, Antoniou C, Glotzer M. TULIPs: tunable, light-controlled interacting protein tags for cell biology. Nat Methods. 2012;9:379–384. doi: 10.1038/nmeth.1904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Toettcher JE, Weiner OD, Lim WA. Using optogenetics to interrogate the dynamic control of signal transmission by the Ras/Erk module. Cell. 2013;155:1422–1434. doi: 10.1016/j.cell.2013.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang K, Duan L, Ong Q, Lin Z, Varman P, Sung K, Cui B. Light-Mediated Kinetic Control Reveals the Temporal Effect of the Raf/MEK/ERK Pathway in PC12 Cell Neurite Outgrowth. PLoS ONE. 2014;9:e92917. doi: 10.1371/journal.pone.0092917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bugaj LJ, Choksi AT, Mesuda CK, Kane RS, Schaffer DV. Optogenetic protein clustering and signaling activation in mammalian cells. Nat Methods. 2013;10:249–252. doi: 10.1038/nmeth.2360. [DOI] [PubMed] [Google Scholar]
- 35.Wend S, Wagner HJ, Muller K, Zurbriggen MD, Weber W, Radziwill G. Optogenetic Control of Protein Kinase Activity in Mammalian Cells. ACS Synth Biol. 2013 doi: 10.1021/sb400090s. [DOI] [PubMed] [Google Scholar]
- 36.Vanhaesebroeck B, Guillermet-Guibert J, Graupera M, Bilanges B. The emerging mechanisms of isoform-specific PI3K signalling. Nat Rev Mol Cell Biol. 2010;11:329–341. doi: 10.1038/nrm2882. [DOI] [PubMed] [Google Scholar]
- 37.Cantley LC. The phosphoinositide 3-kinase pathway. Science. 2002;296:1655–1657. doi: 10.1126/science.296.5573.1655. [DOI] [PubMed] [Google Scholar]
- 38.Toettcher JE, Gong D, Lim WA, Weiner OD. Light-based feedback for controlling intracellular signaling dynamics. Nat Methods. 2011;8:837–839. doi: 10.1038/nmeth.1700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kakumoto T, Nakata T. Optogenetic control of PIP3: PIP3 is sufficient to induce the actin-based active part of growth cones and is regulated via endocytosis. PLoS One. 2013;8:e70861. doi: 10.1371/journal.pone.0070861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Idevall-Hagren O, Dickson EJ, Hille B, Toomre DK, De Camilli P. Optogenetic control of phosphoinositide metabolism. Proc Natl Acad Sci U S A. 2012;109:E2316–2323. doi: 10.1073/pnas.1211305109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kim N, Kim JM, Lee M, Kim CY, Chang KY, Heo WD. Spatiotemporal control of fibroblast growth factor receptor signals by blue light. Chem Biol. 2014;21:903–912. doi: 10.1016/j.chembiol.2014.05.013. [DOI] [PubMed] [Google Scholar]
- 42.Grusch M, Schelch K, Riedler R, Reichhart E, Differ C, Berger W, Janovjak H. Spatio-temporally precise activation of engineered receptor tyrosine kinases by light. Embo J. 2014;33:1713–1726. doi: 10.15252/embj.201387695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mills E, Chen X, Pham E, Wong S, Truong K. Engineering a photoactivated caspase-7 for rapid induction of apoptosis. ACS Synth Biol. 2012;1:75–82. doi: 10.1021/sb200008j. [DOI] [PubMed] [Google Scholar]
- 44.Nagaraj S, Mills E, Wong SS, Truong K. Programming membrane fusion and subsequent apoptosis into mammalian cells. ACS Synth Biol. 2013;2:173–179. doi: 10.1021/sb3000468. [DOI] [PubMed] [Google Scholar]
- 45.Tyszkiewicz AB, Muir TW. Activation of protein splicing with light in yeast. Nat Methods. 2008;5:303–305. doi: 10.1038/nmeth.1189. [DOI] [PubMed] [Google Scholar]
- 46.Lee S, Park H, Kyung T, Kim NY, Kim S, Kim J, Heo WD. Reversible protein inactivation by optogenetic trapping in cells. Nat Methods. 2014;11:633–636. doi: 10.1038/nmeth.2940. [DOI] [PubMed] [Google Scholar]
- 47.Taslimi A, Vrana JD, Chen D, Borinskaya S, Mayer BJ, Kennedy MJ, Tucker CL. An optimized optogenetic clustering tool for probing protein interaction and function. Nat Commun. 2014;5:4925. doi: 10.1038/ncomms5925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jungbluth M, Renicke C, Taxis C. Targeted protein depletion in Saccharomyces cerevisiae by activation of a bidirectional degron. Bmc Syst Biol. 2010:4. doi: 10.1186/1752-0509-4-176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Renicke C, Schuster D, Usherenko S, Essen LO, Taxis C. A LOV2 Domain-Based Optogenetic Tool to Control Protein Degradation and Cellular Function. Chemistry & Biology. 2013;20:619–626. doi: 10.1016/j.chembiol.2013.03.005. [DOI] [PubMed] [Google Scholar]
- 50.Bonger KM, Rakhit R, Payumo AY, Chen JK, Wandless TJ. General Method for Regulating Protein Stability with Light. Acs Chemical Biology. 2014;9:111–115. doi: 10.1021/cb400755b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rivera VM, Wang XR, Wardwell S, Courage NL, Volchuk A, Keenan T, Clackson T. Regulation of protein secretion through controlled aggregation in the endoplasmic reticulum. Science. 2000;287:826–830. doi: 10.1126/science.287.5454.826. [DOI] [PubMed] [Google Scholar]
- 52.Hughes RM, Bolger S, Tapadia H, Tucker CL. Light-mediated control of DNA transcription in yeast. Methods. 2012;58:385–391. doi: 10.1016/j.ymeth.2012.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Liu H, Gomez G, Lin S, Lin C. Optogenetic control of transcription in zebrafish. PLoS One. 2012;7:e50738. doi: 10.1371/journal.pone.0050738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Motta-Mena LB, Reade A, Mallory MJ, Glantz S, Weiner OD, Lynch KW, Gardner KH. An optogenetic gene expression system with rapid activation and deactivation kinetics. Nat Chem Biol. 2014;10:196–202. doi: 10.1038/nchembio.1430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Polstein LR, Gersbach CA. Light-Inducible Spatiotemporal Control of Gene Activation by Customizable Zinc Finger Transcription Factors. Journal of the American Chemical Society. 2012;134:16480–16483. doi: 10.1021/ja3065667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jullien N, Sampieri F, Enjalbert A, Herman JP. Regulation of Cre recombinase by ligand-induced complementation of inactive fragments. Nucleic Acids Res. 2003;31:e131. doi: 10.1093/nar/gng131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Boulina M, Samarajeewa H, Baker JD, Kim MD, Chiba A. Live imaging of multicolor-labeled cells in Drosophila. Development. 2013;140:1605–1613. doi: 10.1242/dev.088930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Muller K, Naumann S, Weber W, Zurbriggen MD. Optogenetics for gene expression in mammalian cells. Biological chemistry. 2014 doi: 10.1515/hsz-2014-0199. [DOI] [PubMed] [Google Scholar]
- 59.Raftopoulou M, Hall A. Cell migration: Rho GTPases lead the way. Developmental Biology. 2004;265:23–32. doi: 10.1016/j.ydbio.2003.06.003. [DOI] [PubMed] [Google Scholar]
- 60.Gaj T, Gersbach CA, Barbas CF., 3rd ZFN, TALEN, and CRISPR/Cas-based methods for genome engineering. Trends Biotechnol. 2013;31:397–405. doi: 10.1016/j.tibtech.2013.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Konermann S, Brigham MD, Trevino A, Hsu PD, Heidenreich M, Le C, Zhang F. Optical control of mammalian endogenous transcription and epigenetic states. Nature. 2013 doi: 10.1038/nature12466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lim WA. Designing customized cell signalling circuits. Nat Rev Mol Cell Biol. 2010;11:393–403. doi: 10.1038/nrm2904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chow BY, Boyden ES. Optogenetics and Translational Medicine. Science Translational Medicine. 2013:5. doi: 10.1126/scitranslmed.3003101. [DOI] [PubMed] [Google Scholar]
- 64.Ni M, Tepperman JM, Quail PH. Binding of phytochrome B to its nuclear signalling partner PIF3 is reversibly induced by light. Nature. 1999;400:781–784. doi: 10.1038/23500. [DOI] [PubMed] [Google Scholar]
- 65.Ni M, Tepperman JM, Quail PH. PIF3, a phytochrome-interacting factor necessary for normal photoinduced signal transduction, is a novel basic helix-loop-helix protein. Cell. 1998;95:657–667. doi: 10.1016/s0092-8674(00)81636-0. [DOI] [PubMed] [Google Scholar]
- 66.Khanna R, Huq E, Kikis EA, Al-Sady B, Lanzatella C, Quail PH. A novel molecular recognition motif necessary for targeting photoactivated phytochrome signaling to specific basic helix-loop-helix transcription factors. Plant Cell. 2004;16:3033–3044. doi: 10.1105/tpc.104.025643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Liu H, Yu X, Li K, Klejnot J, Yang H, Lisiero D, Lin C. Photoexcited CRY2 Interacts with CIB1 to Regulate Transcription and Floral Initiation in Arabidopsis. Science. 2008;322:1535–1539. doi: 10.1126/science.1163927. [DOI] [PubMed] [Google Scholar]
- 68.Brautigam CA, Smith BS, Ma Z, Palnitkar M, Tomchick DR, Machius M, Deisenhofer J. Structure of the photolyase-like domain of cryptochrome 1 from Arabidopsis thaliana. Proc Natl Acad Sci U S A. 2004;101:12142–12147. doi: 10.1073/pnas.0404851101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Yu X, Liu H, Klejnot J, Lin C. The Cryptochrome Blue Light Receptors. Arabidopsis Book. 2010;8:e0135. doi: 10.1199/tab.0135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Huala E, Oeller PW, Liscum E, Han IS, Larsen E, Briggs WR. Arabidopsis NPH1: a protein kinase with a putative redox-sensing domain. Science. 1997;278:2120–2123. doi: 10.1126/science.278.5346.2120. [DOI] [PubMed] [Google Scholar]
- 71.Harper SM, Neil LC, Gardner KH. Structural basis of a phototropin light switch. Science. 2003;301:1541–1544. doi: 10.1126/science.1086810. [DOI] [PubMed] [Google Scholar]
- 72.Nihongaki Y, Suzuki H, Kawano F, Sato M. Genetically engineered photoinducible homodimerization system with improved dimer-forming efficiency. ACS Chem Biol. 2014;9:617–621. doi: 10.1021/cb400836k. [DOI] [PubMed] [Google Scholar]
- 73.Zoltowski BD, Vaccaro B, Crane BR. Mechanism-based tuning of a LOV domain photoreceptor. Nature Chemical Biology. 2009;5:827–834. doi: 10.1038/nchembio.210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Rizzini L, Favory JJ, Cloix C, Faggionato D, O'Hara A, Kaiserli E, Ulm R. Perception of UV-B by the Arabidopsis UVR8 protein. Science. 2011;332:103–106. doi: 10.1126/science.1200660. [DOI] [PubMed] [Google Scholar]
- 75.Heijde M, Ulm R. Reversion of the Arabidopsis UV-B photoreceptor UVR8 to the homodimeric ground state. Proc Natl Acad Sci U S A. 2013;110:1113–1118. doi: 10.1073/pnas.1214237110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wu D, Hu Q, Yan Z, Chen W, Yan C, Huang X, Shi Y. Structural basis of ultraviolet-B perception by UVR8. Nature. 2012;484:214–219. doi: 10.1038/nature10931. [DOI] [PubMed] [Google Scholar]
- 77.Christie JM, Arvai AS, Baxter KJ, Heilmann M, Pratt AJ, O'Hara A, Getzoff ED. Plant UVR8 photoreceptor senses UV-B by tryptophan-mediated disruption of cross-dimer salt bridges. Science. 2012;335:1492–1496. doi: 10.1126/science.1218091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Rockwell NC, Su Y-S, Lagarias JC. Phytochrome structure and signaling mechanisms. Annual Review of Plant Biology. 2006;57:837–858. doi: 10.1146/annurev.arplant.56.032604.144208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wagner JR, Brunzelle JS, Forest KT, Vierstra RD. A light-sensing knot revealed by the structure of the chromophore-binding domain of phytochrome. Nature. 2005;438:325–331. doi: 10.1038/nature04118. [DOI] [PubMed] [Google Scholar]