Abstract
The recent Middle East respiratory syndrome coronavirus (MERS-CoV) outbreak poses a serious threat to public health. Here, we summarize recent advances in identifying human neutralizing monoclonal antibodies (mAbs) against MERS-CoV, describe their mechanisms of action, and analyze their potential for treatment of MERS-CoV infections.
Keywords: Coronavirus, MERS-CoV, mAbs, Receptor binding domain (RBD)
1. Introduction
In September 2012, a novel human coronavirus, Middle East respiratory syndrome (MERS) coronavirus (MERS-CoV), was isolated from a Saudi Arabian patient suffering with a SARS-like disease characterized by fever, cough and shortness of breath. The patient later died of respiratory and renal failure [1]. Most people diagnosed with MERS-CoV infection have developed severe acute respiratory illness. As of November 7, 2014, 909 laboratory-confirmed cases of MERS-CoV infection have been reported to the World Health Organization, including at least 331 related deaths (http://www.who.int/csr/don/07-november-2014-mers). The outbreak of MERS-CoV poses a serious threat to global public health and highlights an urgent need for the development of effective therapeutic and prophylactic agents to treat and prevent MERS-CoV infection [2], [3].
Therapeutic modalities based on monoclonal antibodies (mAbs) have shown clinical success in the treatment of many diseases [4], [5], [6], [7], [8]. The therapeutic potential of antibodies targeting coronaviruses was well recognized during the SARS outbreak [8], [9], [10], [11], [12], [13], [14]. In this review, we summarize the recent progress in identifying human neutralizing mAbs against MERS-CoV, describe their mechanisms of action, and analyze their potential for the therapy and prophylaxis of MERS. We also discuss future directions towards developing a strategy for the rapid development of antibody-based antivirals to combat emerging viruses and diseases in an outbreak setting.
2. From SARS to MERS: the threat of novel coronaviruses
Coronaviruses are a large family of viruses that typically infect the respiratory and gastrointestinal tract of mammals. Based on observation under electron microscopy, they are named for solar corona-like surface projections created by viral spike (S) glycoproteins. The first two coronaviruses infecting human, HCoV-229E and HCoV-OC43, were identified in the 1960s from the nasal cavities of patients presenting with the common cold [15], [16], [17]. They were found to cause only mild to moderate upper respiratory tract illnesses [18], [19]; as a result, coronaviruses were considered relatively harmless to humans. However, during the winter of 2002–2003, clinicians recognized a new, deadly coronavirus strain able to infect both the upper and lower respiratory tract and cause severe acute respiratory syndrome (SARS) [20], [21]. This coronavirus, subsequently named SARS-CoV, rapidly spread around the world and caused a worldwide outbreak with 8096 confirmed cases, including 774 deaths from 2002 to 2003 (http://www.who.int/csr/sars/country/table2004_04_21).
The SARS epidemic was contained in 2003, and no known cases of SARS-CoV infection have been reported since 2004. Two more human coronaviruses, NL63 and HKU1, were discovered from 2004 to 2005 [22], [23]. These coronaviruses, like HCoV-229E and HCoV-OC43, are only common cold viruses which circulate worldwide and generally cause relatively mild respiratory symptoms [22], [23], [24]. However, the threat of coronaviruses has not disappeared. In 2012, a novel coronavirus, MERS-CoV, was identified, and its human infection results in a higher mortality rate (∼36%) than that of SARS-CoV (∼10%) [1]. Similar to SARS-CoV, MERS-CoV can cause atypical pneumonia, acute respiratory distress syndrome, and, potentially, renal failure in infected individuals [25]. MERS-CoV cases have been reported in more than twenty countries, including Saudi Arabia, Qatar, Egypt, the United Arab Emirates, France, the United Kingdom, and the United States, and it has been shown to spread between people who are in close contact [2], [3].
Despite sharing several clinical and epidemiological features in common with SARS-CoV, the two coronaviruses can still be distinguished. MERS-CoV is a lineage C betacoronavirus, while SARS-CoV is a lineage B betacoronavirus [1]. MERS-CoV is phylogenetically distinct from any human coronavirus, including SARS-CoV, but it is more related to the bat coronaviruses HKU4 and HKU5 [26], [27]. Importantly, MERS-CoV uses dipeptidyl peptidase-4 (DPP4, also named CD26) as the receptor on the surface of human cells, while SARS-CoV uses angiotensin-converting enzyme 2 (ACE2) as the cellular receptor [28]. The differences in receptor binding by MERS-CoV and SARS-CoV may be a reflection of their ability and preference to infect cells. DPP4 is widely expressed on nonciliated bronchial epithelium and the epithelial cells in kidney, small intestine, liver, parotid gland, and even testis and prostate [2]. Its wide distribution may explain the diversity of clinical manifestations in MERS-CoV infections.
The emergence of novel coronavirus MERS-CoV, which comes only a decade after the appearance of the first highly pathogenic coronavirus, SARS-CoV, suggests that coronaviruses may represent a continuous and long-term threat to human health. Both SARS-CoV and MERS-CoV are believed to have originated from bats [29], [30]. Since MERS-CoV-specific antibodies and RNA fragments have been detected in camels from Saudi Arabia, Egypt, Tunisia, Nigeria, and Kenya, dromedary camels in the Middle East and Africa are considered to be an intermediate transmitter of MERS-CoV from bats to humans [31], [32], [33], [34]. Importantly, Yang et al. recently examined the cross-species transmissibility of bat coronavirus HKU4, which is genetically related to MERS-CoV, and found that HKU4 also uses DPP4 as the cellular receptor, but prefers bat DPP4 over human DPP4, while MERS-CoV prefers human DPP4 over bat DPP4 [35]. These results suggest that MERS-CoV has adapted to human DPP4 to gain entry into host cells. Since bats are the natural host for a wide range of coronaviruses, the risks posed by the cross-species transmission of these zoonotic coronaviruses for human infections must never be underestimated.
3. MAbs for treatment of viral infections
Although the emergence of highly pathogenic MERS-CoV highlights an urgent need for potent therapeutic and prophylactic agents, no approved antiviral treatments for any human coronavirus infections are currently available. Recently, tremendous efforts have been made in the search for an effective anti-MERS-CoV agent, and a number of antiviral agents have been identified. For example, some compounds with inhibitory activities in the low micromolar range on MERS-CoV replication in cell cultures have been identified from the libraries of FDA-approved drugs [36], [37]. Falzarano et al. also reported that rhesus macaques treated with a cocktail of IFN-α2b with ribavirin, a nucleoside analog, exhibited reduced MERS-CoV replication and an improved clinical outcome [38]. Interestingly, Lu et al. also found that HR2P, a synthesized peptide derived from the HR2 domain of MERS-CoV spike protein, could specifically bind to the HR1 domain of the viral spike protein and block viral fusion core formation, resulting in the inhibition of MERS-CoV replication and its spike protein-mediated cell–cell fusion [39]. HR2P is being optimized to further improve its inhibitory activity, and these HR2P analogs have the potential to be further developed as effective viral fusion inhibitors for treatment of MERS-CoV.
MAbs are enjoying significant clinical success, and they have been used for the effective treatment of a number of diseases, in particular, cancer and immune disorders [4], [5], [6]. Although Synagis (palivizumab), a humanized mAb against respiratory syncytial virus, is still the only mAb approved by the FDA for clinical use against a viral disease [40], a number of antiviral mAbs have been developed in recent years, and some are now in clinical development [41], [42], [43], [44], [45]. Recently, for example, a human mAb, m102.4 [41], made history by being the first mAb administered on a compassionate basis to humans exposed to Hendra virus based on its efficacy in vitro and in vivo [42], [43]. More recently, ZMapp, a cocktail of three mAbs, which showed promising results when administered to rhesus monkeys infected with Ebola virus [46], has been administered to several Ebola patients. Broadly protective antibodies against HIV-1, or influenza A viruses, are also being tested in clinical trials [44], [45].
During the SARS outbreak, neutralizing antibodies were detected in SARS patients, as well as animals infected with the virus [47], [48], [49]. The antibodies also protected uninfected animals from SARS-CoV infection. Specifically, passive transfer of immune serum to naïve mice prevented virus replication in the lower respiratory tract following intranasal virus challenge [50]. Thus, a vast effort has been devoted to developing mAbs that can neutralize the virus and have the potential for treatment and prevention of SARS-CoV infection [9], [10], [11], [13], [14]. The generation and mechanism of neutralization of these mAbs have been thoroughly reviewed elsewhere [8]. In general, neutralizing antibodies against SARS-CoV can be isolated from the memory B-cell repertoire of patients who have recovered from SARS-CoV infection, generated from transgenic mice with human immunoglobulin genes immunized with recombinant SARS-CoV S glycoprotein, or identified from non-immune human antibody libraries constructed from the B lymphocytes of healthy donors. The prophylactic and therapeutic efficacies of these human mAbs have been demonstrated in mice or ferret models of SARS-CoV infection [10], [13], [14]. Most of the neutralizing antibodies target the receptor binding domain (RBD) of the SARS-CoV S glycoprotein, suggesting a possible mechanism of neutralization by preventing virus attachment to its receptor. Some antibodies recognize epitopes on the S2 domain of S glycoprotein, suggesting that other mechanisms could also be involved in the inhibition of SARS-CoV infection, including steric hindrance that indirectly prevents virus attachment, or antibody Fc-mediated effector functions, for example, antibody-dependent cellular cytotoxicity (ADCC).
4. Human mAbs against MERS-CoV: development and efficacy evaluation
Since the SARS epidemic was contained in July 2003, the clinical development of the above-mentioned antibodies targeting SARS-CoV has not been pursued. Fortunately, a wealth of knowledge has been accumulated through the experience of managing the SARS outbreak, and the therapeutic potential of coronavirus-targeting antibodies has been well recognized. When the novel coronavirus MERS-CoV emerged in Saudi Arabia in 2012, this body of knowledge allowed the development of an effective response to the threat of MERS at an unprecedented pace.
First, it has been found that the RBD of MERS-CoV S1 glycoprotein is capable of inducing significant neutralizing antibody responses in mice [51], [52], [53]. Thus, neutralizing mouse mAbs could be developed to block MERS-CoV entry into human cells. For example, Du et al. generated some neutralizing mAbs by immunizing mice with recombinant MERS-CoV S1 fused to IgG1 Fc [53]. Anti-MERS-CoV mAbs were identified by screening positive clones from hybridoma cell lines and testing their inhibition of MERS-CoV pseudovirus entry mediated by S protein, as well as neutralization against live MERS-CoV infection, in DPP4-expressing Vero E6 cells. Mersmab1, the most potent anti-MERS-CoV mAb, was selected on the basis of its efficacy in blocking the entry of MERS-CoV pseudoviruses into DPP4-expressing Huh-7 cells and inhibiting the formation of MERS-CoV-induced CPE during MERS-CoV infection of permissive Vero E6 cells and Calu-3 cells. Thus, mouse mAbs, such as Mersmab1, can be humanized for development as potent therapeutic and prophylactic agents against MERS-CoV infections.
In April 2014, three studies conducted by separate laboratories around the world reported the development of fully human neutralizing mAbs against MERS-CoV [54], [55], [56]. It is noteworthy that all these mAbs target the RBD of the MERS-CoV S1 glycoprotein and that they were all identified from non-immune human antibody libraries. Specifically, among these antibodies, three highly potent mAbs (m336, m337, m338) were identified from a very large phage-displayed antibody Fab library that we recently generated by using B cells from the blood of 40 healthy donors [54]. This library was panned against recombinant MERS-CoV RBD to enrich for high affinity binders. The three identified mAbs, all derived from the VH gene 1–69, which has been the source of many other antiviral antibodies, exhibited exceptionally potent activity and neutralized pseudotyped MERS-CoV with 50% inhibitory concentration (IC50), ranging from 0.005 to 0.017 μg/ml. Notably, the most potent mAb, m336, inhibited >90% MERS-CoV pseudovirus infection (IC90) in DPP4-expressing Huh-7 cells at a concentration of 0.039 μg/ml. Similarly, m336 showed the most potent live MERS-CoV neutralizing activity in inhibiting the formation of MERS-CoV-induced CPE during live MERS-CoV infection of permissive Vero E6 cells, with an IC50 of 0.07 μg/ml. Jiang et al. also identified two potent RBD-specific neutralizing mAbs, MERS-4 and MERS-27, by using a non-immune yeast-displayed scFv library to screen against the recombinant MERS-CoV RBD [55]. The most potent mAb, MERS-4, neutralized the pseudotyped MERS-CoV infection in DPP4-expressing Huh-7 cells with an IC50 of 0.056 μg/ml and inhibited the formation of MERS-CoV-induced CPE during live MERS-CoV infection of permissive Vero E6 cells with an IC50 of 0.5 μg/ml. Tang et al. also identified neutralizing mAbs by using a non-immune phage-displayed scFv library [56]. The panning was performed by sequentially using MERS-CoV spike-containing paramagnetic proteoliposomes and MERS-CoV S glycoprotein-expressing 293T cells as antigens. A panel of 7 anti-S1 scFvs was identified and expressed in both scFv-Fc and IgG1 formats, and their neutralizing activity against pseudotyped MERS-CoV in DPP4-expressing 293T cells, as well as live MERS-CoV infection in Vero cells, was measured. The most potent antibody, 3B11, neutralized live MERS-CoV in the plaque reduction neutralization tests with an IC50 of 1.83 μg/ml and 3.50 μg/ml in the scFv-Fc and IgG format, respectively.
Although all the above-mentioned human mAbs exhibited potent neutralizing activity against MERS-CoV infection in vitro, the evaluation of their efficacy in animal models of infection is still necessary before any of them can be further developed as therapeutic or prophylactic agents. However, unlike SARS-CoV, which can effectively infect and replicate in several cell types in human and animals, including mice and rhesus macaques, MERS-CoV cannot infect small laboratory animals, e.g., mice, hamsters or ferrets, while only causing mild to moderate symptoms in rhesus macaques, thus effectively stalling further research efforts [57], [58], [59]. Variations in DPP4 among animal species are considered to determine susceptibility to MERS-CoV infection [59]. Thus, Zhao et al. developed a mouse model for MERS by using an adenovirus expressing the human DPP4 to sensitize mice for infection [60]. With prior transduction of adenoviral human DPP4-expressing vectors, mice became more susceptible to MERS-CoV infection. This method could be used for rapid evaluation of an anti-MERS vaccine and an antiviral therapy. However, whether the infected mice treated by this inhaled-adenovirus method underwent the same disease progression and immune response as typically observed in human remains unknown. A better model could be a transgenic mouse model with the human DPP4 gene integrated into the genome. Recently, Falzarano et al. found that MERS-CoV infection in marmosets closely mimics the severe pneumonia experienced by people infected with MERS-CoV [59]. Most of the animals infected with MERS-CoV developed a progressive severe pneumonia leading to euthanasia of some animals. Extensive lesions were evident, and high viral loads were detected in the lungs of all marmosets. Marmoset DPP4 has an amino acid sequence identical to that of human DPP4 in the MERS-CoV S glycoprotein binding region, resulting in the observed susceptibility to MERS-CoV infection. These results suggest that the marmoset model is an important advance in the ability to assess the efficacy of intervention strategies against MERS, in turn allowing the preclinical evaluation of neutralizing mAbs for treatment of MERS-CoV infection.
5. Human mAbs against MERS-CoV: mechanisms of virus neutralization
To realize the preventive or therapeutic potential of neutralizing mAbs in the clinical setting, it is also necessary to investigate the mechanisms by which the antibodies modulate the biological behaviors of MERS-CoV. In order to explore the mechanism of action of human mAbs m336, m337, and m338, we first defined the MERS-CoV epitopes recognized by these mAbs [54]. The binding of mAbs to fragments of the S glycoprotein was measured, and, as expected, the mAbs only bound to the S fragments containing the RBD, specifically residues 377 to 588. We further analyzed the mAbs binding to a series of alanine mutants of the RBD and found that the three mAbs had overlapping, but distinct, binding sites. For example, residues 510 and 553 were important for binding of all three mAbs. Residues 536 and 539, on the other hand, were uniquely bound by m336, which turned out to be the most potent neutralizer among the three mAbs, suggesting that these residues may be crucial for m336 interaction and, hence, important for vaccine development. We next found that the three mAbs competed with each other for binding the MERS-CoV RBD and also competed with RBD for binding to a soluble version of DPP4. Consistent with the neutralization results, m336 was slightly more potent than m337 and m338 in blocking the binding of RBD to DPP4. The IC50s of m336, m337, and m338 were 0.034, 0.044, and 0.041 μg/ml, respectively, values which were in a range similar to the neutralizing activity of the mAbs against pseudotyped MERS-CoV in DPP4-expressing Huh-7 cells (0.005–0.017 μg/ml). These results support the idea that the mAbs have overlapping epitopes and neutralize MERS-CoV by competing with the receptor binding. Using the mutagenesis data and the MERS-CoV RBD crystal structure, we generated three-dimensional molecular docking models of the mAbs interacting with the RBD [54]. Good superimposition of DPP4 with the mAbs-RBD structures confirmed that the competition with MERS-CoV for receptor binding is an important mechanism of action of the mAbs in neutralizing MERS-CoV cell entry. The extensive overlapping between the mAb epitopes and the receptor binding sites on RBD explains the exceptional neutralizing potency of these mAbs. Similarly, Jiang et al. also found that the neutralizing mAbs MERS-4 and MERS-27 could inhibit the binding of soluble RBD to DPP4-expressing Huh7 cells [55]. From the binding competition assays, Tang et al. found that 2B11 and six other mAbs could recognize at least three different epitopes on MERS-CoV RBD and that the antibodies could block DPP4 binding to MERS-CoV RBD and inhibit the attachment of pseudovirus to DPP4-expressing cells [56]. DPP4 could also block the antibodies from RBD binding. Taken together, these results suggest that the neutralizing mechanism of all the above-mentioned human mAbs, despite recognizing different epitopes on RBD, occurred through the blocking of MERS-CoV binding to its cellular receptor DPP4. Interestingly, Jiang et al. also demonstrated that the combination of MERS-4 and MERS-27, which were found to recognize different epitopes on RBD, resulted in a synergistic neutralizing effect against pseudotyped MERS-CoV [55].
6. Future directions
Emerging viruses, e.g. SARS-like or MERS-like novel coronaviruses, are highly likely to continue posing a serious threat to human health in the near future. In this context, it is prudent to develop strategies to provide a quick response. Polyclonal human immunoglobulin has been used with various degrees of success for viral diseases for more than a century, and some are still in clinical use against, for example, hepatitis A, hepatitis B, cytomegalovirus, rabies, measles and vaccinia [7]. However, a number of toxicity-related problems have arisen, including a risk for allergic reactions, pathogen transmission, and lot-to-lot variation. In addition, only a very small portion of the total antibodies in a polyclonal preparation is typically neutralizing, while the remainder is not only ineffective, but could be immunogenic, or even toxic. These limitations could be overcome by the use of human mAbs. Nevertheless, a major obstacle comes from the difficulty of generating highly potent neutralizing mAbs in a relatively short amount of time. In this review, we summarized the recent developments in human neutralizing mAbs against MERS-CoV which have been identified by different laboratories around the globe at an unprecedented pace. Improvement in antibody identification techniques, combined with the recent advances in antibody production technologies, highlights the potential of human mAbs for application toward a strategy designed to combat future emerging viruses and diseases in an outbreak setting.
Apart from the above limitations, the use of antiviral mAbs may produce escape mutants. Although the mutation rates in SARS-CoV and MERS-CoV are only moderate compared to those in other RNA viruses, the frequency of the emergence of escape mutants should not be underestimated. A promising solution may lie in the use of “mAb cocktails” in which two or more mAbs targeting different epitopes are mixed. Such mAb cocktails would not only provide more potent antiviral activity than a single mAb by additive and/or synergistic effects [61], but also prevent escape variants for many viruses, including SARS-CoV [13]. Interestingly, even though all recently developed human mAbs against MERS-CoV target the RBD of S glycoprotein [54], [55], [56], most of them recognize distinct epitopes, as discussed above. Therefore, it might be expected that the emergence of resistant viruses to these RBD-specific mAbs, if any, would exert a toll on virus fitness because the escape mutants could have lower affinity for the cellular receptor DPP4. Furthermore, Jiang et al. have demonstrated that the combination of two human mAbs, MERS-4 and MERS-27, resulted in a synergistic neutralizing effect against pseudotyped MERS-CoV [55]. Thus, it would be interesting to test if other combinations of the available mAbs would give similar, or better, synergistic effects and if so, whether these mAb cocktails could neutralize mutant viruses emerging in the future.
High production cost is a substantial obstacle for the commercial development of antiviral human mAbs, especially mAbs against emerging viruses. One strategy to reduce the overall cost is to develop exceptionally potent neutralizing mAbs, resulting in a reduction in the dose required to achieve efficacy. This could be achieved by developing human mAbs with ultra high binding affinity and then rationally designing the products to target the critical neutralization site, e.g., RBD. For example, the exceptionally potent mAb m336 can bind to the MERS-CoV RBD with picomolar affinity, and the potency for virus neutralization (IC50) is also in the picomolar range, suggesting that a low dose of mAbs could be clinically effective. Furthermore, recent progress in antibody engineering enables the generation of more effective antibody-based therapeutics, e.g., bispecific antibodies, antibody-drug conjugates, or antibodies with improved effector functions, such as antibody-dependent cell-mediated cytotoxicity (ADCC) [5]. Notably, antibody fragments with a reduced size (12–50 kDa) could achieve enhanced tissue penetration, as well as a wider range of possible targets, and, importantly, require much lower production costs, providing the potential to overcome the fundamental limitations of full-size mAbs [62]. However, compared to mAbs, antibody fragments, such as VH or Fab, display greatly reduced half-lives, and as such, they have limited clinical potential. We recently generated some novel IgG1 Fc-based antibody fragments, which have small size (14–27 kDa), good stability and solubility, and relatively long in vivo half-life [63], [64], [65], [66], [67]. When fused to VH, the resulting novel antibody constructs can still be solubly expressed in Escherichia coli with high yields [64]. Therefore, these small, long-acting antibodies have the potential to be developed as commercially attractive prophylactic and therapeutic antivirals for a wide array of indications (Fig. 1 ).
Fig. 1.
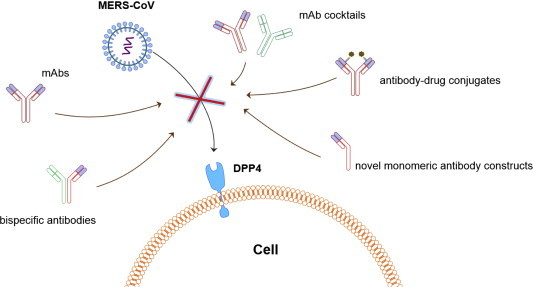
Schematic illustration of the inhibition of MERS-CoV cell entry by neutralizing mAbs, mAb cocktails, bispecific antibodies, antibody-durg conjugates, and novel monomeric antibody constructs.
7. Conclusions
The threat of emerging infectious diseases, such as SARS and MERS, emphasizes the need for a fast and versatile approach that allows us to rapidly identify effective antivirals to combat the viruses. We have reviewed here the recent success in identifying fully human neutralizing mAbs against MERS-CoV. The rapid identification of these antibodies suggests the possibility of using non-immune human antibody libraries and related methodologies for a quick response to these emerging viruses with pandemic potential. All the mAbs, despite being developed by different laboratories, target the RBD of MERS-CoV S glycoprotein and may have a similar mechanism of action, i.e., blocking the binding of MERS-CoV to its cellular receptor DPP4. Notably, some human mAbs have exhibited exceptionally potent neutralizing activity against MERS-CoV infection in vitro, whereas the evaluation of their efficacy in an effective animal infection model, like the marmoset MERS-CoV infection model, is yet to be performed. Most mAbs recognize different epitopes on MERS-CoV RBD, suggesting that mAb cocktails may exhibit more potent anti-MERS activity based on additive and/or synergistic effects, also helping to prevent the occurrence of viral escape mutants. We expect that recent advances in antibody engineering will foster the development of more effective and affordable therapeutic or prophylactic agents by improving effector functions, making antibody-drug conjugates, or generating small-sized and long-acting novel antibody constructs based on these human neutralizing mAbs.
Conflict of interest
The authors declare no competing financial interest.
Acknowledgment
This work was supported by grants from the National Natural Science Foundation of China and the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research.
References
- 1.Zaki A.M., van Boheemen S., Bestebroer T.M., Osterhaus A.D., Fouchier R.A. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N Engl J Med. 2012;367:1814–1820. doi: 10.1056/NEJMoa1211721. [DOI] [PubMed] [Google Scholar]
- 2.Lu L., Liu Q., Du L., Jiang S. Middle east respiratory syndrome coronavirus (MERS-CoV): challenges in identifying its source and controlling its spread. Microb Infect. 2013;15:625–629. doi: 10.1016/j.micinf.2013.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hotez P.J., Bottazzi M.E., Tseng C.T., Zhan B., Lustigman S., Du L. Calling for rapid development of a safe and effective MERS vaccine. Microb Infect. 2014;16:529–531. doi: 10.1016/j.micinf.2014.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dimitrov D.S. Therapeutic proteins. Methods Mol Biol. 2012;899:1–26. doi: 10.1007/978-1-61779-921-1_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carter P.J. Potent antibody therapeutics by design. Nat Rev Immunol. 2006;6:343–357. doi: 10.1038/nri1837. [DOI] [PubMed] [Google Scholar]
- 6.Schrama D., Reisfeld R.A., Becker J.C. Antibody targeted drugs as cancer therapeutics. Nat Rev Drug Discov. 2006;5:147–159. doi: 10.1038/nrd1957. [DOI] [PubMed] [Google Scholar]
- 7.Casadevall A., Dadachova E., Pirofski L.A. Passive antibody therapy for infectious diseases. Nat Rev Microbiol. 2004;2:695–703. doi: 10.1038/nrmicro974. [DOI] [PubMed] [Google Scholar]
- 8.Zhang M.Y., Choudhry V., Xiao X., Dimitrov D.S. Human monoclonal antibodies to the S glycoprotein and related proteins as potential therapeutics for SARS. Curr Opin Mol Ther. 2005;7:151–156. [PubMed] [Google Scholar]
- 9.Sui J., Li W., Murakami A., Tamin A., Matthews L.J., Wong S.K. Potent neutralization of severe acute respiratory syndrome (SARS) coronavirus by a human mAb to S1 protein that blocks receptor association. Proc Natl Acad Sci U S A. 2004;101:2536–2541. doi: 10.1073/pnas.0307140101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.ter Meulen J., Bakker A.B., van den Brink E.N., Weverling G.J., Martina B.E., Haagmans B.L. Human monoclonal antibody as prophylaxis for SARS coronavirus infection in ferrets. Lancet. 2004;363:2139–2141. doi: 10.1016/S0140-6736(04)16506-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Traggiai E., Becker S., Subbarao K., Kolesnikova L., Uematsu Y., Gismondo M.R. An efficient method to make human monoclonal antibodies from memory B cells: potent neutralization of SARS coronavirus. Nat Med. 2004;10:871–875. doi: 10.1038/nm1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yang Z.Y., Werner H.C., Kong W.P., Leung K., Traggiai E., Lanzavecchia A. Evasion of antibody neutralization in emerging severe acute respiratory syndrome coronaviruses. Proc Natl Acad Sci U S A. 2005;102:797–801. doi: 10.1073/pnas.0409065102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.ter Meulen J., van den Brink E.N., Poon L.L., Marissen W.E., Leung C.S., Cox F. Human monoclonal antibody combination against SARS coronavirus: synergy and coverage of escape mutants. PLoS Med. 2006;3:e237. doi: 10.1371/journal.pmed.0030237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhu Z., Chakraborti S., He Y., Roberts A., Sheahan T., Xiao X. Potent cross-reactive neutralization of SARS coronavirus isolates by human monoclonal antibodies. Proc Natl Acad Sci U S A. 2007;104:12123–12128. doi: 10.1073/pnas.0701000104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tyrrell D.A., Bynoe M.L. Cultivation of a novel type of common-cold virus in organ cultures. Br Med J. 1965;1:1467–1470. doi: 10.1136/bmj.1.5448.1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hamre D., Procknow J.J. A new virus isolated from the human respiratory tract. Proc Soc Exp Biol Med. 1966;121:190–193. doi: 10.3181/00379727-121-30734. [DOI] [PubMed] [Google Scholar]
- 17.McIntosh K., Dees J.H., Becker W.B., Kapikian A.Z., Chanock R.M. Recovery in tracheal organ cultures of novel viruses from patients with respiratory disease. Proc Natl Acad Sci U S A. 1967;57:933–940. doi: 10.1073/pnas.57.4.933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bradburne A.F., Bynoe M.L., Tyrrell D.A. Effects of a “new” human respiratory virus in volunteers. Br Med J. 1967;3:767–769. doi: 10.1136/bmj.3.5568.767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bradburne A.F., Somerset B.A. Coronative antibody tires in sera of healthy adults and experimentally infected volunteers. J Hyg (Lond) 1972;70:235–244. doi: 10.1017/s0022172400022294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ksiazek T.G., Erdman D., Goldsmith C.S., Zaki S.R., Peret T., Emery S. A novel coronavirus associated with severe acute respiratory syndrome. N Engl J Med. 2003;348:1953–1966. doi: 10.1056/NEJMoa030781. [DOI] [PubMed] [Google Scholar]
- 21.Hafner S. SARS attacks! Microb Infect. 2013;15:85–87. doi: 10.1016/j.micinf.2012.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.van der Hoek L., Pyrc K., Jebbink M.F., Vermeulen-Oost W., Berkhout R.J., Wolthers K.C. Identification of a new human coronavirus. Nat Med. 2004;10:368–373. doi: 10.1038/nm1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Woo P.C., Lau S.K., Chu C.M., Chan K.H., Tsoi H.W., Huang Y. Characterization and complete genome sequence of a novel coronavirus, coronavirus HKU1, from patients with pneumonia. J Virol. 2005;79:884–895. doi: 10.1128/JVI.79.2.884-895.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pyrc K., Berkhout B., van der Hoek L. The novel human coronaviruses NL63 and HKU1. J Virol. 2007;81:3051–3057. doi: 10.1128/JVI.01466-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Assiri A., Al-Tawfiq J.A., Al-Rabeeah A.A., Al-Rabiah F.A., Al-Hajjar S., Al-Barrak A. Epidemiological, demographic, and clinical characteristics of 47 cases of Middle East respiratory syndrome coronavirus disease from Saudi Arabia: a descriptive study. Lancet Infect Dis. 2013;13:752–761. doi: 10.1016/S1473-3099(13)70204-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.van Boheemen S., de Graaf M., Lauber C., Bestebroer T.M., Raj V.S., Zaki A.M. Genomic characterization of a newly discovered coronavirus associated with acute respiratory distress syndrome in humans. MBio. 2012;3 doi: 10.1128/mBio.00473-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lau S.K., Li K.S., Tsang A.K., Lam C.S., Ahmed S., Chen H. Genetic characterization of betacoronavirus lineage C viruses in bats reveals marked sequence divergence in the spike protein of pipistrellus bat coronavirus HKU5 in Japanese pipistrelle: implications for the origin of the novel Middle East respiratory syndrome coronavirus. J Virol. 2013;87:8638–8650. doi: 10.1128/JVI.01055-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Raj V.S., Mou H., Smits S.L., Dekkers D.H., Muller M.A., Dijkman R. Dipeptidyl peptidase 4 is a functional receptor for the emerging human coronavirus-EMC. Nature. 2013;495:251–254. doi: 10.1038/nature12005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Annan A., Baldwin H.J., Corman V.M., Klose S.M., Owusu M., Nkrumah E.E. Human betacoronavirus 2c EMC/2012-related viruses in bats, Ghana and Europe. Emerg Infect Dis. 2013;19:456–459. doi: 10.3201/eid1903.121503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Anthony S.J., Ojeda-Flores R., Rico-Chavez O., Navarrete-Macias I., Zambrana-Torrelio C.M., Rostal M.K. Coronaviruses in bats from Mexico. J Gen Virol. 2013;94:1028–1038. doi: 10.1099/vir.0.049759-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hemida M.G., Perera R.A., Wang P., Alhammadi M.A., Siu L.Y., Li M. MIDDLE EAST respiratory syndrome (MERS) coronavirus seroprevalence in domestic livestock in Saudi Arabia, 2010 to 2013. Euro Surveill. 2013;18:20659. doi: 10.2807/1560-7917.es2013.18.50.20659. [DOI] [PubMed] [Google Scholar]
- 32.Reusken C.B., Ababneh M., Raj V.S., Meyer B., Eljarah A., Abutarbush S. Middle East respiratory syndrome coronavirus (MERS-CoV) serology in major livestock species in an affected region in Jordan, June to September 2013. Euro Surveill. 2013;18:20662. doi: 10.2807/1560-7917.es2013.18.50.20662. [DOI] [PubMed] [Google Scholar]
- 33.Barlan A., Zhao J., Sarkar M.K., Li K., McCray P.B., Jr., Perlman S. Receptor variation and susceptibility to Middle East respiratory syndrome coronavirus infection. J Virol. 2014;88:4953–4961. doi: 10.1128/JVI.00161-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Reusken C.B., Haagmans B.L., Muller M.A., Gutierrez C., Godeke G.J., Meyer B. Middle East respiratory syndrome coronavirus neutralising serum antibodies in dromedary camels: a comparative serological study. Lancet Infect Dis. 2013;13:859–866. doi: 10.1016/S1473-3099(13)70164-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yang Y., Du L., Liu C., Wang L., Ma C., Tang J. Receptor usage and cell entry of bat coronavirus HKU4 provide insight into bat-to-human transmission of MERS coronavirus. Proc Natl Acad Sci U S A. 2014;111:12516–12521. doi: 10.1073/pnas.1405889111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.de Wilde A.H., Jochmans D., Posthuma C.C., Zevenhoven-Dobbe J.C., van Nieuwkoop S., Bestebroer T.M. Screening of an FDA-approved compound library identifies four small-molecule inhibitors of Middle East respiratory syndrome coronavirus replication in cell culture. Antimicrob Agents Chemother. 2014;58:4875–4884. doi: 10.1128/AAC.03011-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dyall J., Coleman C.M., Hart B.J., Venkataraman T., Holbrook M.R., Kindrachuk J. Repurposing of clinically developed drugs for treatment of Middle East respiratory syndrome coronavirus infection. Antimicrob Agents Chemother. 2014;58:4885–4893. doi: 10.1128/AAC.03036-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Falzarano D., de Wit E., Rasmussen A.L., Feldmann F., Okumura A., Scott D.P. Treatment with interferon-alpha2b and ribavirin improves outcome in MERS-CoV-infected rhesus macaques. Nat Med. 2013;19:1313–1317. doi: 10.1038/nm.3362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lu L., Liu Q., Zhu Y., Chan K.H., Qin L., Li Y. Structure-based discovery of Middle East respiratory syndrome coronavirus fusion inhibitor. Nat Commun. 2014;5:3067. doi: 10.1038/ncomms4067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shadman K.A., Wald E.R. A review of palivizumab and emerging therapies for respiratory syncytial virus. Expert Opin Biol Ther. 2011;11:1455–1467. doi: 10.1517/14712598.2011.608062. [DOI] [PubMed] [Google Scholar]
- 41.Zhu Z., Dimitrov A.S., Bossart K.N., Crameri G., Bishop K.A., Choudhry V. Potent neutralization of hendra and nipah viruses by human monoclonal antibodies. J Virol. 2006;80:891–899. doi: 10.1128/JVI.80.2.891-899.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bossart K.N., Zhu Z., Middleton D., Klippel J., Crameri G., Bingham J. A neutralizing human monoclonal antibody protects against lethal disease in a new ferret model of acute nipah virus infection. PLoS Pathog. 2009;5:e1000642. doi: 10.1371/journal.ppat.1000642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bossart K.N., Geisbert T.W., Feldmann H., Zhu Z., Feldmann F., Geisbert J.B. A neutralizing human monoclonal antibody protects african green monkeys from hendra virus challenge. Sci Transl Med. 2011;3:105ra3. doi: 10.1126/scitranslmed.3002901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lingwood D., McTamney P.M., Yassine H.M., Whittle J.R., Guo X., Boyington J.C. Structural and genetic basis for development of broadly neutralizing influenza antibodies. Nature. 2012;489:566–570. doi: 10.1038/nature11371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mascola J.R., Haynes B.F. HIV-1 neutralizing antibodies: understanding nature's pathways. Immunol Rev. 2013;254:225–244. doi: 10.1111/imr.12075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Qiu X., Wong G., Audet J., Bello A., Fernando L., Alimonti J.B. Reversion of advanced ebola virus disease in nonhuman primates with ZMapp. Nature. 2014;514:47–53. doi: 10.1038/nature13777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hofmann H., Hattermann K., Marzi A., Gramberg T., Geier M., Krumbiegel M. S protein of severe acute respiratory syndrome-associated coronavirus mediates entry into hepatoma cell lines and is targeted by neutralizing antibodies in infected patients. J Virol. 2004;78:6134–6142. doi: 10.1128/JVI.78.12.6134-6142.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hsueh P.R., Huang L.M., Chen P.J., Kao C.L., Yang P.C. Chronological evolution of IgM, IgA, IgG and neutralisation antibodies after infection with SARS-associated coronavirus. Clin Microbiol Infect. 2004;10:1062–1066. doi: 10.1111/j.1469-0691.2004.01009.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nie Y., Wang G., Shi X., Zhang H., Qiu Y., He Z. Neutralizing antibodies in patients with severe acute respiratory syndrome-associated coronavirus infection. J Infect Dis. 2004;190:1119–1126. doi: 10.1086/423286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Subbarao K., McAuliffe J., Vogel L., Fahle G., Fischer S., Tatti K. Prior infection and passive transfer of neutralizing antibody prevent replication of severe acute respiratory syndrome coronavirus in the respiratory tract of mice. J Virol. 2004;78:3572–3577. doi: 10.1128/JVI.78.7.3572-3577.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Du L., Kou Z., Ma C., Tao X., Wang L., Zhao G. A truncated receptor-binding domain of MERS-CoV spike protein potently inhibits MERS-CoV infection and induces strong neutralizing antibody responses: implication for developing therapeutics and vaccines. PLoS One. 2013;8:e81587. doi: 10.1371/journal.pone.0081587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mou H., Raj V.S., van Kuppeveld F.J., Rottier P.J., Haagmans B.L., Bosch B.J. The receptor binding domain of the new Middle East respiratory syndrome coronavirus maps to a 231-residue region in the spike protein that efficiently elicits neutralizing antibodies. J Virol. 2013;87:9379–9383. doi: 10.1128/JVI.01277-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Du L., Zhao G., Yang Y., Qiu H., Wang L., Kou Z. A conformation-dependent neutralizing monoclonal antibody specifically targeting receptor-binding domain in Middle East respiratory syndrome coronavirus spike protein. J Virol. 2014;88:7045–7053. doi: 10.1128/JVI.00433-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ying T., Du L., Ju T.W., Prabakaran P., Lau C.C., Lu L. Exceptionally potent neutralization of Middle East respiratory syndrome coronavirus by human monoclonal antibodies. J Virol. 2014;88:7796–7805. doi: 10.1128/JVI.00912-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jiang L., Wang N., Zuo T., Shi X., Poon K.M., Wu Y. Potent neutralization of MERS-CoV by human neutralizing monoclonal antibodies to the viral spike glycoprotein. Sci Transl Med. 2014;6:234ra59. doi: 10.1126/scitranslmed.3008140. [DOI] [PubMed] [Google Scholar]
- 56.Tang X.C., Agnihothram S.S., Jiao Y., Stanhope J., Graham R.L., Peterson E.C. Identification of human neutralizing antibodies against MERS-CoV and their role in virus adaptive evolution. Proc Natl Acad Sci U S A. 2014;111:E2018–E2026. doi: 10.1073/pnas.1402074111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.de Wit E., Prescott J., Baseler L., Bushmaker T., Thomas T., Lackemeyer M.G. The Middle East respiratory syndrome coronavirus (MERS-CoV) does not replicate in syrian hamsters. PLoS One. 2013;8:e69127. doi: 10.1371/journal.pone.0069127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Coleman C.M., Matthews K.L., Goicochea L., Frieman M.B. Wild-type and innate immune-deficient mice are not susceptible to the Middle East respiratory syndrome coronavirus. J Gen Virol. 2014;95:408–412. doi: 10.1099/vir.0.060640-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Falzarano D., de Wit E., Feldmann F., Rasmussen A.L., Okumura A., Peng X. Infection with MERS-CoV causes lethal pneumonia in the common marmoset. PLoS Pathog. 2014;10:e1004250. doi: 10.1371/journal.ppat.1004250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhao J., Li K., Wohlford-Lenane C., Agnihothram S.S., Fett C., Zhao J. Rapid generation of a mouse model for Middle East respiratory syndrome. Proc Natl Acad Sci U S A. 2014;111:4970–4975. doi: 10.1073/pnas.1323279111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bregenholt S., Jensen A., Lantto J., Hyldig S., Haurum J.S. Recombinant human polyclonal antibodies: a new class of therapeutic antibodies against viral infections. Curr Pharm Des. 2006;12:2007–2015. doi: 10.2174/138161206777442173. [DOI] [PubMed] [Google Scholar]
- 62.Nelson A.L., Reichert J.M. Development trends for therapeutic antibody fragments. Nat Biotechnol. 2009;27:331–337. doi: 10.1038/nbt0409-331. [DOI] [PubMed] [Google Scholar]
- 63.Ying T., Chen W., Gong R., Feng Y., Dimitrov D.S. Soluble monomeric IgG1 Fc. J Biol Chem. 2012;287:19399–19408. doi: 10.1074/jbc.M112.368647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ying T., Chen W., Feng Y., Wang Y., Gong R., Dimitrov D.S. Engineered soluble monomeric IgG1 CH3 domain: generation, mechanisms of function, and implications for design of biological therapeutics. J Biol Chem. 2013;288:25154–25164. doi: 10.1074/jbc.M113.484154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ying T., Ju T.W., Wang Y., Prabakaran P., Dimitrov D.S. Interactions of IgG1 CH2 and CH3 domains with FcRn. Front Immunol. 2014;5:146. doi: 10.3389/fimmu.2014.00146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ying T., Gong R., Ju T.W., Prabakaran P., Dimitrov D.S. Engineered Fc based antibody domains and fragments as novel scaffolds. Biochim Biophys Acta. 2014;1844:1977–1982. doi: 10.1016/j.bbapap.2014.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ying T., Feng Y., Wang Y., Chen W., Dimitrov D.S. Monomeric IgG1 Fc molecules displaying unique Fc receptor interactions that are exploitable to treat inflammation-mediated diseases. mAbs. 2014;6:1201–1210. doi: 10.4161/mabs.29835. [DOI] [PMC free article] [PubMed] [Google Scholar]


