Summary
The spindle assembly checkpoint (SAC) plays a critical role in preventing mitotic errors by inhibiting anaphase until all kinetochores are correctly attached to spindle microtubules. In spite of the economic and medical importance of filamentous fungi, relatively little is known about the behavior of SAC proteins in these organisms. In our efforts to understand the role of γ-tubulin in cell cycle regulation, we have created functional fluorescent protein fusions of four SAC proteins in Aspergillus nidulans, the homologs of Mad2, Mps1, Bub1/BubR1 and Bub3. Time-lapse imaging reveals that SAC proteins are in distinct compartments of the cell until early mitosis when they co-localize at the spindle pole body. SAC activity is, thus, spatially regulated in A. nidulans. Likewise, Cdc20, an activator of the anaphase promoting complex/cyclosome, is excluded from interphase nuclei, but enters nuclei at mitotic onset and accumulates to a higher level in mitotic nuclei than in the surrounding nucleoplasm before leaving in anaphase/telophase. The activity of this critical cell cycle regulatory complex is likely regulated by the location of Cdc20. Finally, the γ-tubulin mutation mipAD159 causes a nuclear-specific failure of nuclear localization of Mps1 and Bub1/R1 but not of Cdc20, Bub3 or Mad2.
Keywords: Cell cycle, γ-tubulin, Aspergillus, mitosis, checkpoint, fungi
Introduction
The spindle assembly checkpoint has a well-established role in inhibiting anaphase until all kinetochores (KTs) are attached in a bipolar manner to spindle microtubules (Li and Murray, 1991; Hoyt et al., 1991; Musacchio and Hardwick, 2002; reviewed in Musacchio and Salmon, 2007). The SAC negatively regulates Cdc20, an activator of the anaphase-promoting complex/cyclosome (APC/C) (Hwang et al., 1998; Kim et al., 1998). This results in inhibition of APC/C activity, which, in turn, prevents ubiquitination and subsequent hydrolysis of the key proteins securin and cyclin B. Hydrolysis of securin is required for anaphase and hydrolysis of cyclin B is required for mitotic exit (Peters, 2006). The SAC proteins Mad2, Bub3, and BubR1/Mad3p, as well as the APC/C activator Cdc20, form a complex known as the mitotic checkpoint complex (MCC) that localizes to KTs (Hardwick et al., 2000; Sudakin et al., 2001; Musacchio and Salmon, 2007). The MCC plays a key role in keeping the APC/C inactive until the SAC is satisfied (Sudakin et al., 2001; Musacchio and Salmon, 2007). Only then does the MCC dissociate from KTs, thereby inactivating the SAC and allowing activation of the APC/C.
MCC components localize to KTs in prometaphase, but the data as to their locations in interphase are contradictory. Some studies indicate that MCCs are present in interphase (Sudakin et al., 2001) and exist independent of KTs (Fraschini et al., 2001) while others suggest, in aggregate, that they assemble in mitosis, on KTs and in a defined order (Vigneron et al., 2004; Johnson et al., 2004; Hardwick et al., 2000; Kulukian et al., 2009). There has been surprisingly little live imaging of MCC components, which would likely be informative with respect to how they come to form a complex at KTs.
Other proteins have been identified that have a role in the SAC and are not part of the MCC, including Mad1, Bub1, and Mps1. Although they are not part of the MCC, they are linked to KT localization of MCC components. Data from Xenopus laevis demonstrate that the KT localization of Mad2 requires Mad1 (Chen et al., 1998) and Mps1 (Abrieu et al., 2001). Bub1 is required for the KT localization of BubR1 and Mad2 in mammalian cells (Johnson et al., 2004) and for BubR1, Mad2, and Mad1 in X. laevis (Sharp-Baker and Chen, 2001).
We have recently found that γ-tubulin has an important role in regulating the APC/C in interphase (Nayak et al., 2010; Edgerton-Morgan and Oakley, 2012). At restrictive temperatures a recessive, cold-sensitive γ-tubulin allele, mipAD159, causes a nuclear-autonomous failure of cyclin B (encoded by the nimE gene), cyclin-dependent kinase 1 (Cdk1, encoded by the nimX gene) and the Aspergillus nidulans Cdc14 phosphatase homolog, AnCdc14, to accumulate in a subset of nuclei. We designate such nuclei as cyclin B negative (CB−). Cyclin B and Cdk1 are key cell cycle regulatory proteins and their failure to accumulate at appropriate times results in CB− nuclei being permanently removed from the cell cycle, and the percentage of CB− nuclei increases with each round of mitosis. We have found these proteins fail to accumulate due to a failure of inactivation of the APC/C complexed with the APC/C activator CdhA at the G1/S boundary (CdhA is the A. nidulans Cdh1 homolog) (Edgerton-Morgan and Oakley, 2012). This results in continuous activity of APC/CCdhA and, as a consequence, the continuous targeting of cyclin B and other substrates of APC/CCdhA for destruction.
A. nidulans is coenocytic and nuclei in the same cell normally go through mitosis in synchrony. When other nuclei in the cell undergo mitosis, the nuclear pore complexes in CB− nuclei partially disassemble as they do in normal nuclei, and the chromatin partially condenses. No mitotic spindle is formed in CB− nuclei, and there is no nuclear division, but the nuclear pore complexes (NPCs) reassemble at the end of the failed mitosis. Interestingly, CdhA disappeared from CB− nuclei in mitosis (Edgerton-Morgan and Oakley, 2012), and this raised the question of why the SAC did not inactivate the APC/C in mitosis, resetting the cell cycle. [Note that prolonged treatment with the antimicrotubule agent benomyl, which activates the SAC (Hoyt, 1997; Li and Murray, 1991), did reset nuclei such that cyclin B accumulated in formerly CB− nuclei (Nayak et al., 2010)]. We wished to answer this question in order to enhance our understanding of the functions of γ-tubulin in cell cycle regulation. We consequently created fluorescent fusion proteins of SAC components and performed time-lapse imaging of them in strains carrying mipAD159 and in strains wild-type at the γ-tubulin locus (mipA+). In addition, although filamentous fungi are hugely important medically and economically, there is relatively little data on the behavior of SAC proteins in these organisms. Mad1 and Mad2 orthologs have been imaged in live cells of A. nidulans (De Souza et al., 2009), but data on other SAC proteins in fungi is sparse. Understanding the fundamental cell biology of mitosis and the cell cycle in filamentous fungi is important if one is to control their growth or kill them when necessary.
Results
Identification of Cdc20 and SAC gene homologs in A. nidulans and construction of fluorescent protein fusions
Gene and protein nomenclature for A. nidulans has become inconsistent in recent years. Standard gene symbols for A. nidulans have the form abcD, and the proteins encoded by the genes have the form AbcD or ABCD. However, where genes and proteins have well studied homologs, other designations are often used such as AnabcD or An-abcD for the gene and AnAbcD or An-AbcD for the protein. For previously studied genes and proteins, we will use the initial published designations although they do not all follow a consistent format.
The A. nidulans Mad2 homolog has been previously identified (gene = md2A, protein = An-Mad2) (Prigozhina et al., 2004; De Souza et al., 2009) as has the Bub3 homolog (Efimov and Morris, 1998) (gene = sldB, protein = SldB; since readers may not be familiar with SldB, in the interest of clarity we will use SldBBub3 to refer to the protein) and the Mps1 homolog (gene = An-mps1, protein = An-Mps1) (De Souza et al., 2013). We ran BLASTP searches of the Aspergillus genome database (http://www.aspergillusgenome.org/) using published amino acid sequences of Cdc20 homologs from Homo sapiens, Saccharomyces cerevisiae and Schizosaccharomyces pombe, and we were easily able to identify the A. nidulans Cdc20 homolog (AN0814). We considered naming the gene cdcT using the standard gene format, but this designation does not clearly indicate orthology to cdc20 and cdcT might be confused with nimT, a mitotic regulatory phosphatase that has been studied extensively in A. nidulans (O'Connell et al., 1992; Son and Osmani, 2009). We will instead use An-cdc20 to refer to the gene and An-Cdc20 to refer to its protein product.
The situation with the Bub1/BubR1/Mad3 homolog is more complex. Many organisms have two BUB family proteins, Bub1 and BubR1/Mad3p. Phylogenetic analyses demonstrate that these proteins have arisen from at least nine separate gene duplication events followed by subfunctionalization of the duplicated genes including loss of kinase activity in one of the duplicated genes (Suijkerbuijk et al., 2012). A putative A. nidulans Bub1 homolog, SldA, the predicted gene product of the sldA gene, was previously identified by Efimov and Morris (Efimov and Morris, 1998). Our BLASTP searches using Bub1or BubR1 amino acid sequences returned only one hit in the Aspergillus genome database, sldA. Bub1 proteins have a functional C-terminal kinase domain but lack the conserved N-terminal KEN boxes that are recognized by APC/CCdh1. BubR1 homologs lack the C-terminal kinase domain in many species and, in vertebrates, bear non-conserved, degenerate kinase domains that probably lack kinase activity (Suijkerbuijk et al., 2012). A. nidulans, in common with Neurospora crassa and other species of Aspergillus, has a single Bub1/R1 gene, sldA, that retains the two N-terminal KEN boxes and all the functional domains associated with both Bub1 and BubR1. The sldA kinase domain is closely related to the Bub1 kinase domain (Suijkerbuijk et al., 2012; De Souza et al., 2013). The sldA gene, thus, likely never duplicated and retains the activities of the ancestral Bub1/R1 (De Souza et al., 2013). In the interest of clarity and familiarity we will use SldABub1/R1 to refer to the protein.
We created fluorescent protein fusions for each of these genes using approaches we have previously published (Nayak et al., 2006; Szewczyk et al., 2006). In all cases, except for N-terminal tagging of An-Cdc20 (discussed subsequently), the fluorescent protein fusion sequence was fused in frame to the 3’ end of the coding sequence of the target gene, and the fusion gene was the only copy of the gene in the genome. Each gene was under control of its normal promoter and the fusions were functional (Fig. S1). (A list of strains and their genotypes used in this study are listed in Table 1.) In addition, we created a fusion of GFP to SepK, the A. nidulans homolog of the spindle pole body (SPB) marker Nud1 (Kim et al., 2009b), a gene we have previously tagged with tdTomato (Xiong and Oakley, 2009). We also created a fusion of histone H1 (encoded by the hhoA gene) to the fluorescent protein T-Sapphire (Zapata-Hommer and Griesbeck, 2003). A strain expressing a KT marker, An-Ndc80, fused to mCherry and a strain expressing the nucleoporin (Nup), An-Nup49, fused to mCherry (De Souza et al., 2009; Nayak et al., 2010; Edgerton-Morgan and Oakley, 2012) were kindly provided by Drs. Colin De Souza and Steve Osmani (The Ohio State University).
Table 1.
Strains used in this study.
| Strain number | Genotype |
|---|---|
| FGSC4 | Glasgow wild type |
| LO1160 | pyrG89; md2A::pyrG; pabaA1; fwA1 |
| LO1390 | pyrG89?; md2A-GFP-AfpyrG; hhoA-mRFP-AfpyrG; pabaA1; yA2 |
| LO1438 | pyrG89; nimE-GFP-AfpyrG; hhoA-mRFP-AfpyrG; pyroA4; nkuA::argB?; riboB2; yA2 |
| LO1439 | pyrG89; nimE-GFP-AfpyrG; hhoA-mRFP-AfpyrG; pyroA4; nkuA::argB?; mipAD159 |
| LO1479 | wA3; pyrG89?; An-mps1-GFP-AfpyrG; hhoA-mRFP-AfpyrG; pabaA1; nkuA::argB? |
| LO1533 | pyrG89?; md2A-GFP-AfpyrG; hhoA-mRFP-AfpyrG; pabaA1; nkuA::argB?; mipAD159; yA2 |
| LO2585 | pyrG89; sldB-GFP-AfpyrG; pyroA4; hhoA-mCherry-AfpyroA; nkuA::argB?; mipAD159; fwA1 |
| LO2834 | wA3; pyrG89; hhoA-T-Sapphire-AfpyrG; sepK-GFP-AfpyrG; pabaA1?; pyroA4; An-ndc80-mCherry-AfpyroA; nkuA::argB; nirA14?; sE15?; riboB2; fwA1?; chaA1? |
| LO3105 | pyrG89; An-mps1-GFP-AfpyrG; hhoA-T-Sapphire-AfpyrG; pabaA1; pyroA4; An-ndc80-mCherry-AfpyroA; nkuA::argB?; chaA1 |
| LO3317 | pyrG89; nimE-GFP-AfpyrG; pabaA1; pyroA4; nkuA::argB?; riboB2; hhoA-mRFP-AfriboB |
| LO3406 | pyrG89; sldB-GFP-AfpyrG; pyroA4; hhoA-mCherry-AfpyroA; nkuA::argB |
| LO4223 | pyrG89?; An-mps1-GFP-AfpyrG; hhoA-mRFP-AfpyrG; pabaA1; nkuA:: argB?; mipAD159 |
| LO4580 | pyrG89; sldA-GFP-AfpyrG; pyroA4; nkuA::argB; riboB2; hhoA-mRFP-AfriboB |
| LO4676 | pyrG89; sldA-GFP-AfpyrG; nkuA::argB?; hhoA-mRFP-AfriboB; mipAD159; fwA1 |
| LO5538/5539 | pyrG89; pyroA4; AfpyroA-GFP-An-cdc20; nkuA::argB; riboB2; hhoA-mRFP-AfriboB |
| LO5690 | pyrG89; pyroA4; AfpyroA-GFP-An-cdc20; nkuA::argB?; hhoA-mRFP-AfriboB; mipAD159 |
| LO5849 | pyrG89; mipA-GFP-AfpyrG; pyroA4; sldB-GFP-AfpyroA; argB2?; nkuA::argB; riboB2; hhoA-T-Sapphire-AfriboB |
| LO5851 | pyrG89; An-mps1-GFP-AfpyrG; pyroA4; nirA14?; sE15?; nkuA::argB?; riboB2; mipA-mCherry-AfriboB; chaA1 |
| LO5908 | pyrG89; sldA-GFP-AfpyrG; pyroA4; An-ndc80-mCherry-AfpyroA; nkuA::argB?; fwA1 |
| LO6518 | pyrG89; An-nup49-mCherry-AfpyrG; pyroA4; AfpyroA-GFP-An-cdc20; nkuA::argB?; riboB2 |
| LO7554 | pyrG89; An-nup49-mCherry-AfpyrG; sldA-GFP-AfpyrG; nimX-mCherry-AfriboB; mipAD159 |
| LO8308 | pyrG89; An-mps1-GFP-AfpyrG; pyroA4; hhoA-T-Sapphire-AfpyroA; nkuA::argB?; nimX-mCherry-AfriboB; mipAD159 |
Question marks indicate alleles that were present in one of the parents of a cross but have not been tested in the progeny. All strains also carry veA1.
Localization of KTs with respect to the SPB during the cell cycle
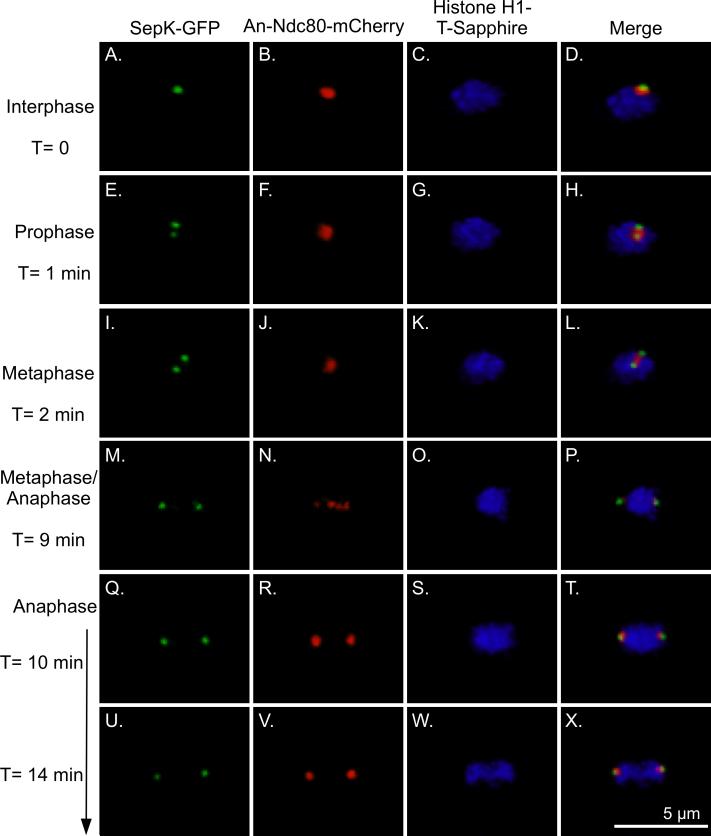
In A. nidulans, the SPB and KTs are in proximity throughout interphase (Yang et al., 2004; De Souza et al., 2009). Only during mitosis, from prometaphase through early anaphase, are the two physically separated enough to be distinguished. This period is very short in A. nidulans because mitosis lasts only about 10 min at 25°C. We created a strain (LO2834) that carried histone H1-T-Sapphire, SepK fused to GFP, and An-Ndc80 fused to mCherry, and imaged it by time-lapse confocal microscopy (Fig. 1). Since the SAC proteins are expected to localize to the SPB or KTs in mitosis, these images serve as a reference for the localization pattern of these structures during mitosis. In instances in which the SPB and KTs are too close to distinguish as separate structures we will refer to the structures as the SPB/KT complex.
Fig. 1.
Histone H1-T-Sapphire, SepK-GFP, An-Ndc80 localization in mitosis. Images are three-dimensional projections of Z-series stacks collected at 30 sec intervals at 25°C. An-Ndc80 overlaps with the Nud1 homolog SepK (an SPB marker) in interphase (A-D). In prophase and metaphase as the SPBs separate, An-Ndc80 is between the SPBs (E-L). Individual KTs are visible at metaphase/anaphase (M-P). (Since chromosomes do not form a distinct metaphase plate in A. nidulans it is difficult to distinguish metaphase from very early anaphase.) In Q-T An-Ndc80, and thus the KTs, have moved to the poles. Anaphase A, chromosome to pole movement, is thus complete but chromatids have not visibly separated. Chromosomal separation occurs as the poles move further apart (U-X).
Localization of An-Mad2
An-Mad1 and An-Mad2 have been localized previously in A. nidulans and reported to have essentially the same localization pattern (De Souza et al., 2009). Our observations with An-Mad2 (using strain LO1390) confirm the previously published data. An-Mad2 localized predominantly to the nuclear periphery in interphase nuclei (Fig. S2A-C) although it was also present in the cytoplasm. This localization is similar to the localization in H. sapiens and Drosophila melanogaster cells (Buffin et al., 2005; Campbell et al., 2001; Shah et al., 2004), S. cerevisiae cells (where there is a low concentration as well in the nucleoplasm) (Iouk et al., 2002) and S. pombe (Mayer et al., 2006) although one report indicates that it is associated with chromatin as well as the nuclear envelope in S. pombe (Ikui et al., 2002).
Upon entry into mitosis, An-Mad2 translocated from the nuclear periphery, concentrating at the SPB/KT complex (Fig. S2D-F) as in H. sapiens and D. melanogaster (Howell et al., 2000; Shah et al., 2004; Buffin et al., 2005). In animal cells, Mad2 is also present along the spindle and at spindle poles in addition to KTs (Howell et al., 2000; Howell et al., 2004; Shah et al., 2004). In S. cerevisiae and S. pombe Mad2 localizes to unattached KTs in mitosis (Ikui et al., 2002; Mayer et al., 2006; Gillett et al., 2004). However, some Mad2 remains at NPCs in S. cerevisiae throughout mitosis (Iouk et al., 2002).
The An-Mad2 signal next spread through a portion of the nucleus (Fig. S2G-L). From anaphase through telophase, An-Mad2 faintly localized to a region extending from one separating chromatin mass to the other (Fig. S2M-O), a localization that, we believe, has only been reported for A. nidulans (De Souza et al., 2009). An-Mad2 then became briefly undetectable and then returned first to the nucleoplasm (Fig. S2P-R) and eventually to the nuclear periphery in early G1.
Localization of An-Mps1
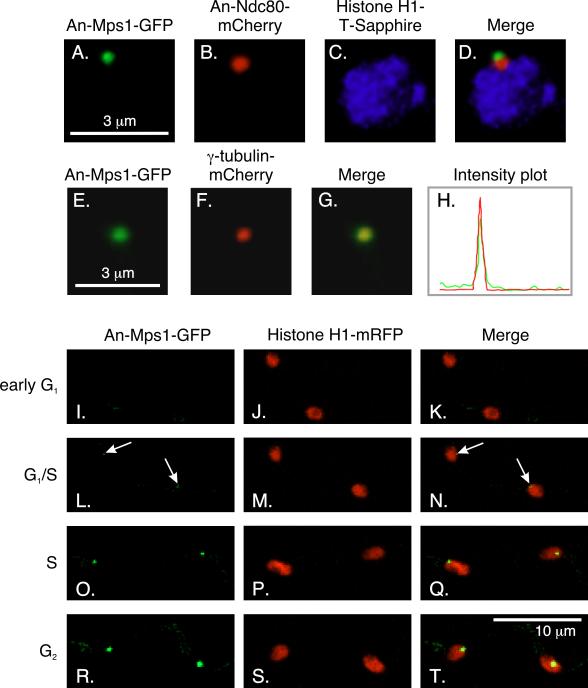
An-Mps1 is a protein kinase and unlike most SAC proteins, it is essential for viability (De Souza et al., 2013). To determine the localization of An-Mps1 through the cell cycle, we created a strain (LO1479) that expressed An-Mps1-GFP and histone H1-mRFP and imaged it using time-lapse microscopy (Fig. 2I-T). Through most of the cell cycle, An-Mps1 localized to a single dot in each nucleus, which we deduced to be the SPB since Mps1 localizes to the centrosome or SPB in other organisms (Fischer et al., 2004; Fisk and Winey, 2001; Kasbek et al., 2007; Winey et al., 1991). To verify that An-Mps1 indeed localized to the SPB, we created a strain (LO3105) that expressed An-Mps1-GFP, histone H1-T-Sapphire, and An-Ndc80-mCherry. An-Mps1 localized immediately adjacent to An-Ndc80, suggesting that it localized to the SPB, but, perhaps, not to KTs, in interphase (Fig. 2A-D). Furthermore, imaging of a strain (LO5851) that expressed An-Mps1-GFP and γ-tubulin-mCherry revealed that the two proteins co-localized in interphase, and, thus, verified that An-Mps1 localized to the SPB (Fig. 2E-H).
Fig. 2.
Localization of An-Mps1. A-D: An-Mps1 localizes adjacent to An-Ndc80 in interphase. An-Ndc80 is a KT marker that localizes adjacent to the SPB in interphase. E-H: An-Mps1 co-localizes with the SPB marker γ-tubulin in interphase. An-Mps1-GFP and γ-tubulin-mCherry co-localize perfectly. The co-localization is verified by intensity traces in the mCherry (red) and GFP (green) channels across the same region. The trace was vertical and covered a region slightly larger than the field shown. Images A-D and E-G are three-dimensional projections of through-focus Z-series image stacks. I-T: Z-stack projection images from a time-lapse data set collected at 10 min intervals at 25°C showing the localization of An-Mps1 through the cell cycle. I-K: An-Mps1 is not visible through most of G1, but it becomes faintly visible at G1/S (arrows; L-N). The An-Mps1-GFP signal becomes brighter through S (O-Q) and G2 (R-T).
We know that the cell cycle duration in A. nidulans at 25°C is 199 ± 49 min. (Edgerton-Morgan and Oakley, 2012). At 32°C G1 lasts about 15% of the cell cycle (Bergen and Morris, 1983). At 25°C, 15% of the cell cycle would correspond to 30 min. Long-term time-lapse imaging of 132 nuclei (strain LO1479) revealed that An-Mps1-GFP became detectable 28 ± 12 min (mean ± standard deviation) after mitotic exit, which would correspond to the G1/S transition (Fig. 2L-N). In comparison, CdhA remains at the SPB for 39 ± 12 min after mitotic exit (Edgerton-Morgan and Oakley, 2012), although it could be inactivated by phosphorylation before it leaves the SPB. Throughout the rest of interphase, An-Mps1 fluorescence at the SPB increased in intensity (Fig. 2R-T).
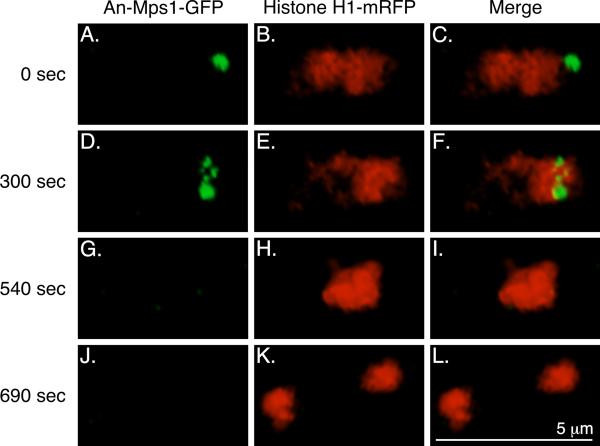
At mitotic entry (as judged by the beginning of chromosomal condensation) (Fig. 3D-F) An-Mps1 moved from the SPB to several dots associated with the chromatin. Mps1 localizes to KTs in mitotic cells of other organisms (Abrieu et al., 2001; Liu et al., 2003; Stucke et al., 2002; Winey et al., 1991), and the localization pattern we observed was consistent with it localizing to KTs in A. nidulans. The An-Mps1 signal was completely gone prior to anaphase (Fig. 3G-I).
Fig. 3.
An-Mps1-GFP localization during mitosis. A-C: In G2, An-Mps1 is at the SPB. At mitotic entry (D-F), as judged by the beginning of chromosomal condensation, An-Mps1 translocates from the SPB to the KTs. 240 sec later (G-I), still prior to anaphase, the An-Mps1 signal is completely gone. J-L: This nucleus then divides, and An-Mps1 is still not present. Images are three dimensional projections from a time-lapse data set collected at 30 sec intervals at 25°C.
Localization of SldBBub3, the A. nidulans Bub3 homolog
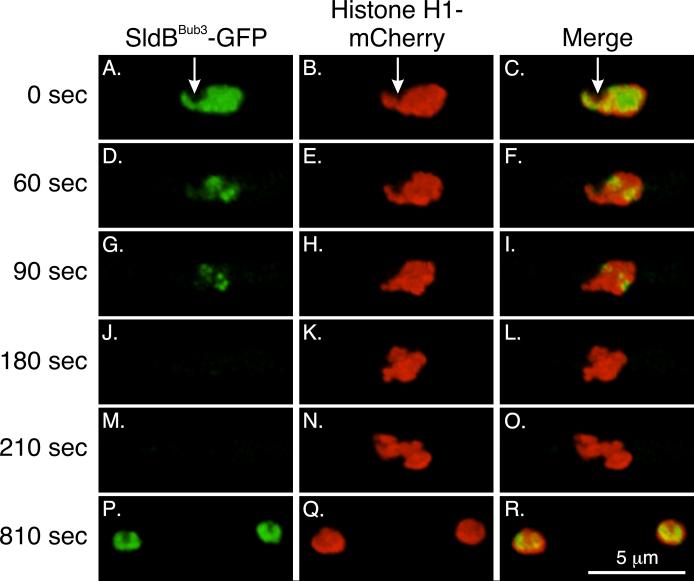
We created a strain (LO3406) that expressed SldBBub3-GFP and histone H1-mCherry to determine the localization of SldBBub3 through the cell cycle. In interphase, we found that SldBBub3 was abundant in the nucleoplasm (Fig. 4A-C), as is the case for Bub3 homologs in other organisms (Campbell and Hardwick, 2003; Kadura et al., 2005; Taylor et al., 1998), but it was excluded from the nucleolus (arrows; Fig. 4A-C). As nuclei entered mitosis, the intensity of SldBBub3 decreased in the nucleoplasm (Fig. 4D-F). This corresponds to the time that NPCs partially disassemble (De Souza et al., 2004; Osmani et al., 2006; De Souza and Osmani, 2007) and the decrease of SldBBub3 likely reflects diffusion from the nucleoplasm. However, as the nucleoplasmic signal decreased, SldBBub3 remained localized to dots in the nucleoplasm (Fig. 4G-I). By imaging strain LO5849 (SldBBub3-mCherry, γ-tubulin-GFP, Histone H1-T-sapphire), SldBBub3-mCherry was verified to localize to the position of KTs, between separating SPBs (Fig.S3). Based on their location and appearance, and the localization of Bub3 in other organisms, we deduce that the dots are KTs. SldBBub3 was only at KTs for a short period of time before the signal was lost prior to anaphase (Fig. 4J-L). It reappeared in the nucleoplasm in early G1 (Fig. 4P-R).
Fig. 4.
SldBBub3 localization in interphase and mitosis. A-C: In interphase, the Bub3 homolog SldBBub3 localizes to the nucleoplasm but is excluded from the nucleolus (arrows). D-I: Upon mitotic entry, it begins to disappear from the nucleoplasm but is left at KTs. J-L: Prior to anaphase, its signal is completely gone, and it remains absent through the rest of mitosis (M-O shows early anaphase). SldBBub3 returns to the nucleoplasm in daughter nuclei in G1. Images are three dimensional projections from a time-lapse data set collected at 30 sec intervals at 25°C.
Localization of SldABub1/R1, the A. nidulans Bub1/R1 homolog
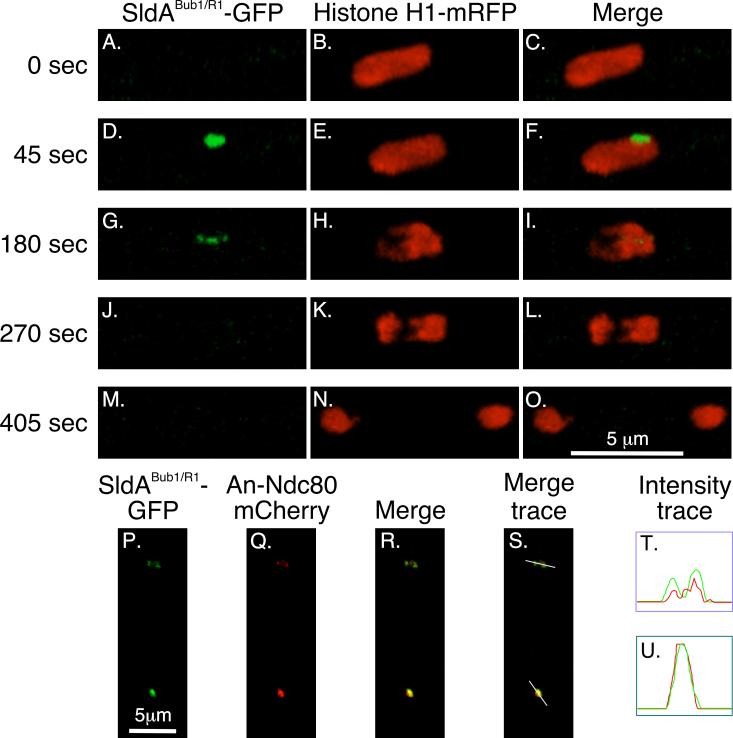
As discussed previously, sldA is the only Bub1/R1 homolog in A. nidulans, and it encodes a protein that has features of both Bub1 and BubR1. To determine the localization of SldABub1/R1 through the cell cycle, we created a strain (LO4580) that expresses SldABub1/R1-GFP and histone H1-mRFP. In interphase, SldABub1/R1 was not detectable (Fig. 5A-C). At the onset of mitosis, however, it quickly appeared at the SPB/KT complex (Fig. 5D-F). It then localized briefly to dots in the nucleoplasm that appeared to be KTs (Fig. 5G-I) before disappearing prior to anaphase (Fig. 5J-L). By creating a strain (LO5908) expressing SldABub1/R1 -GFP and An-Ndc80-mCherry and analyzing it by confocal microscopy, we determined that SldABub1/R1 -GFP colocalized with An-Ndc80-mCherry verifying that the nucleoplasmic dots are, indeed, KTs (Fig. 5P-T). This KT localization is similar to that reported for BubR1 and Bub1 in H. sapiens (Taylor et al., 2001; Johnson et al., 2004), PtK2 cells (Howell et al., 2004) and D. melanogaster (Buffin et al., 2005; Logarinho et al., 2004).
Fig. 5.
Localization of the Bub1/R1 homolog SldABub1/R1 in mitosis. A-C: In interphase, SldABub1/R1 is not detectable. D-F: At mitotic entry, SldABub1/R1 appears at the SPB/KT complex. G-I: It then goes to KTs as chromosomes further condense. J-L: Its signal is gone by anaphase and remains absent through the rest of mitosis (M-O shows late telophase/early G1). Images are three dimensional projections from a time-lapse data set collected at 45 sec intervals at 25°C. SldABub1/R1 -GFP and An-Ndc80-mCherry are shown in two nuclei in the same cell in P-S. The images are projections of a Z-stack. In the bottom nucleus the KTs are clustered at the pole and in the top nucleus the KTs are strung out between two poles that have separated (see Fig. 1). In both cases SldABub1/R1 co-localizes with the KT marker as is shown by the intensity traces (green = SldABub1/R1 -GFP, red = An-Ndc80-mCherry) for the top nucleus (T) and the bottom nucleus (U).
Localization of An-Cdc20
To observe the localization pattern of An-Cdc20 through the cell cycle, we first fused GFP to the 3’ end of the An-cdc20 coding sequence, keeping expression of the fusion gene under control of the endogenous An-cdc20 promoter. Viable transformants were obtained and correct integration of the transforming fragment was verified by diagnostic PCR. However, no GFP signal was detected in 10 different verified transformants. Since no signal was detected, we decided to try an N-terminal GFP fusion. Again, this fusion was kept under the control of the endogenous An-cdc20 promoter using a fusion PCR approach we have published previously (Wong et al., 2008). Viable transformants were obtained and verified by diagnostic PCR and Southern hybridization to have a correct integration. These transformants produced a faint, but detectable, GFP signal. The N-terminal fusion was functional, supporting normal growth over a range of temperatures (20°C-42°C; Fig. S1), and it was the only copy of the An-cdc20 gene in the genome.
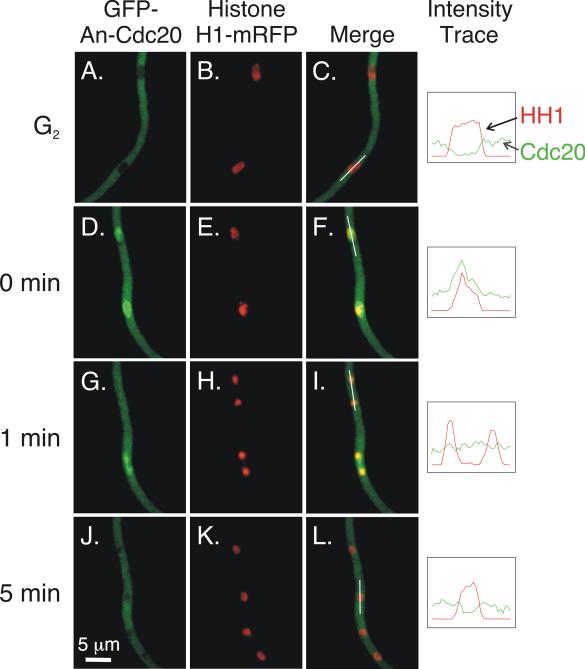
To determine the localization of An-Cdc20 through the cell cycle, we created strains LO5538 and LO5539 that express GFP-An-Cdc20 and histone H1-mRFP, as well as strain LO6518 that expresses GFP-An-Cdc20 and An-Nup49-mCherry. Time-lapse data sets revealed that, in interphase, An-Cdc20 levels in the nucleus were significantly lower than in the cytoplasm (Fig. 6A-C; Fig. S4A-C). At mitotic entry, as judged by the beginning of histone H1-mRFP condensation (and based on NPC breakdown as revealed by the dispersal of An-Nup49-mCherry), An-Cdc20 entered the nucleus, briefly concentrating in a small area that, on the basis of morphology, is likely to be the SPB or very early forming spindle (Szewczyk and Oakley, 2011) (arrows; Fig. S4D and F). It then spread through the nucleoplasm, accumulating to higher levels than in the cytoplasm (Fig. 6D-F; Fig. S4G-I). It occupied a larger volume than the condensed chromosomes but was not excluded from chromatin. In anaphase and telophase, the concentration of GFP-An-Cdc20 dropped such that its concentration in the nucleus was similar to its concentration in the cytoplasm (Fig. 6G-I, top nucleus). We noted some variation in the timing of this concentration drop. Sometimes it occurred just before anaphase and sometimes it occurred later such that there was a visible concentration between separating chromosome masses (Fig. 6G-I, bottom nucleus). After the drop, it remained at the same concentration as the surrounding cytoplasm until mitotic exit, when it was rapidly depleted from the nucleoplasm (Fig. 6J-L; Fig. S4J-L).
Fig. 6.
Localization of An-Cdc20 in interphase and mitosis. Images are Z-stack projections captured at 30°C and were taken from the same data set. Intensity traces of histone H1-mRFP and GFP-An-Cdc20 over the path of the white lines shown in C, F, I, and L are in the right-most column. The traces were created by Volocity software and were made using one optical section in the Z-series stack to minimize signal from out of focus fluorescence. A-C: Two nuclei in an interphase cell. An-Cdc20 is at relatively low levels in the cytoplasm. D-L: Another hypha in the same data set that is going through mitosis. Two nuclei are in prophase in D-F, and An-Cdc20 has become concentrated in the nucleoplasm. G-I show the same field captured one min later. In the telophase nucleus at the top, An-Cdc20 is present at approximately the same level as in the surrounding cytoplasm. A concentration of An-Cdc20 is visible in the bottom nucleus, which is in late anaphase. J-L: At the five min time point, in early G1, GFP-Cdc20 levels in the daughter nuclei have decreased.
This pattern was quite different from that reported for animal cells, where it is intranuclear in interphase and enriched at KTs and centrosomes, which are functionally similar to SPBs, in mitosis (Kallio et al., 2002; Kim et al., 2009a; Li et al., 2010; Raff et al., 2002). The only time that GFP-An-Cdc20 was possibly enriched at SPBs was in prophase when SPBs and KTs are very close together and the spindle is beginning to form. At this stage, it is difficult to determine if the localization is at the SPB, spindle, or KTs, or all three. Nevertheless, there was clearly no enrichment at SPBs at later stages of mitosis.
A γ-tubulin mutation, mipAD159, causes a nuclear-autonomous failure of localization of An-Mps1 and SldABub1/R1 at restrictive temperatures
A. nidulans is coenocytic and nuclei in each tip cell progress through the cell cycle synchronously. Cell cycle regulatory proteins, such as cyclin B, accumulate and are destroyed at essentially the same time in all nuclei within a tip cell. However, in strains carrying mipAD159, incubated at a restrictive temperature of 25°C, failure of inactivation of APC/CCdhA at the G1/S boundary leads to a failure of cyclin B accumulation in a subset of nuclei (designated CB− nuclei) and to such nuclei being taken out of the cell cycle (Nayak et al., 2010; Edgerton-Morgan and Oakley, 2012). Other nuclei in the same cell accumulate cyclin B normally (CB+ nuclei) and continue to progress through the cell cycle. When CB+ nuclei go through mitosis, CB− nuclei in the same cell go through a failed mitosis in which chromatin partially condenses and then decondenses, nuclear pores partially disassemble and reassemble, and CdhA disappears and reappears (Nayak et al., 2010; Edgerton-Morgan and Oakley, 2012). Mitotic spindles do not form in CB− nuclei (Nayak et al., 2010), and one might expect that when CB− nuclei enter mitosis the SAC would be activated, the APC/C would be inactivated, and the cell cycle would be reset. This is not the case, although prolonged mitotic blockage with the antimicrotubule agent benomyl does allow some CB− nuclei to reset to become CB+ (Nayak et al., 2010). One possible explanation is that, at restrictive temperatures, mipAD159 causes mislocalization or destruction of one or more SAC components so a functional SAC cannot be established.
To determine if mipAD159 causes a failure of accumulation or mislocalization of An-Mad2 and SldBBub3, which are associated with nuclei in interphase, we examined the localization of each protein fused to GFP in strains carrying mipAD159 and expressing histone H1 fused to either mRFP or mCherry. Strains were incubated at 25°C for 21 hours, a time by which CB− nuclei accumulate (Nayak et al., 2010; Edgerton-Morgan and Oakley, 2012). Z-series stacks of random fields were captured over a one-hour period, and tip cell nuclei were scored for the presence or absence of An-Mad2-GFP or SldBBub3-GFP. Note that we only scored tip cells in which at least one nucleus had accumulated An-Mad2-GFP or SldBBub3-GFP to make sure we were only counting cells that were actively going through the cell cycle. An-Mad2 localized to the nuclear periphery of almost all mipAD159 nuclei as only 3.6 ± 0.7% of tip cell nuclei were negative for An-Mad2 (mean ± standard deviation of three experiments; strain LO1533) compared to 0.3 ± 0.5% in a mipA+ strain (LO1390; three experiments). Likewise, SldBBub3 accumulated in almost all mipAD159 nuclei as only 0.9 ± 0.4% of tip cell nuclei failed to accumulate SldBBub3 (three experiments; strain LO2585) compared to 0.3 ± 0.5% in a mipA+ strain (LO3406; three experiments). In comparison, we previously found that under the same conditions 45.2 ± 13% of mipAD159 tip cell nuclei were CB− (strain LO1439; three experiments) compared to 1.5 ± 1.6% in two mipA+ strains (six experiments; strains LO1438 and LO3317) (Edgerton-Morgan and Oakley, 2012). Therefore, mipAD159 does not cause destruction or a gross mislocalization of An-Mad2 or SldBBub3.
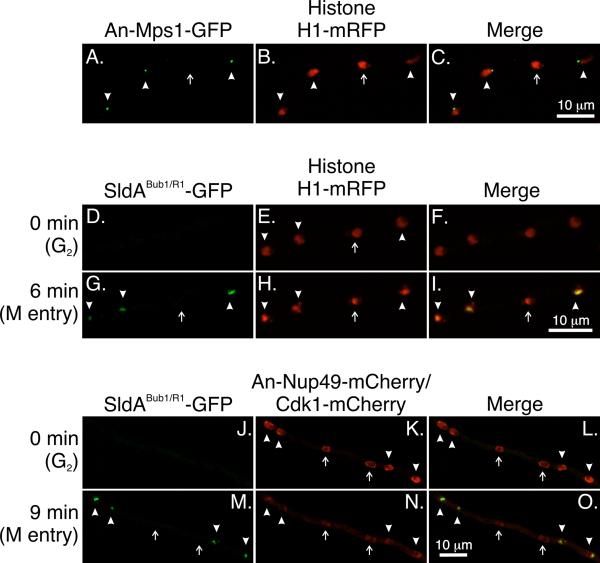
In contrast to An-Mad2 and SldBBub3, An-Mps1-GFP is undetectable in G1. We performed the same type of experiment as we did for cyclin B, An-Mad2, and SldBBub3, making sure to score only tip cells in which at least one nucleus had accumulated An-Mps1-GFP to ensure that we were not counting nuclei as negative when, in reality, they were in G1. In a mipAD159 strain, An-Mps1 failed to accumulate to the SPB in 23.1 ± 10.3% of nuclei (three experiments; strain LO4223) (Fig. 7A-C) compared to 1.9 ± 1.7% in a mipA+ strain (three experiments; strain LO1479). mipAD159, thus, causes a nuclear autonomous failure of accumulation of An-Mps1 at the SPB at restrictive temperatures. To determine if nuclei that failed to accumulate An-Mps1 (designated An-Mps1− nuclei) were also CB− we created a mipAD159 strain that expressed An-Mps1-GFP, histone H1-T-Sapphire, and Cdk1-mCherry (LO8308). Since Cdk1 forms a complex with cyclin B (Nurse, 1990), CB− nuclei also fail to accumulate Cdk1 (Nayak et al., 2010). Cdk1 has essentially the same localization pattern as cyclin B as well, localizing to the nucleoplasm and SPB in S and G2 and disappearing during mitosis (Nayak et al., 2010; Edgerton-Morgan and Oakley, 2012). In addition, in our hands, Cdk1-mCherry is more amenable to time-lapse imaging than cyclin B-mCherry (Nayak et al., 2010). LO8308 was incubated at 25°C for 21 hours and Z-series stacks of random fields were captured over a one-hour period. Tip cell nuclei were then scored for the presence or absence of An-Mps1-GFP and Cdk1-mCherry. We found that 72.9 ± 9.1% (mean ± standard deviation of three experiments) of An-Mps1− nuclei were CB−. Reciprocally, 74.1 ± 23.1% of CB− nuclei were An-Mps1−. Therefore, the majority of, but not all, mipAD159 nuclei that are An-Mps1− are CB− as well.
Fig. 7.
An-Mps1 and SldABub1/R1 fail to accumulate in a subset of nuclei at restrictive temperatures in strains carrying mipAD159. Images are maximum intensity projections from Z-series stacks captured at at 25°C. The Z-series stacks captured all of the nuclei in their entirety so the absence of fluorescence is not due to a portion of a nucleus simply being beyond the limits of the Z-series stack. A-C: Four nuclei are shown. An-Mps1 fluorescence is visible at the SPBs of three nuclei (arrowheads) but not the fourth (arrow). D-I: SldABub1/R1 does not accumulate in a subset of nuclei. Images were from a time-lapse data set with stacks captured at 3 min intervals. At T = 0 SldABub1/R1 -GFP is not detectable in any nuclei. At T = 6 min it is present in three of four nuclei (arrowheads) but not in one nucleus (arrow). J-O: SldABub1/R1 does not accumulate in Cdk1− nuclei. At T = 0 SldABub1/R1 is not present in any nuclei. All nuclei show nuclear periphery fluorescence due to An-Nup49-mCherry. Cdk1+ nuclei (arrowheads) show additional fluorescence at the SPBs and Cdk1 is present in the nucleoplasm except for the nucleolar region (visible as a small clear area). Two nuclei are Cdk1− (arrows). They do not show SPB fluorescence and the nucleoplasm is clear. At T = 9 min SldABub1/R1 has accumulated in the Cdk1+ nuclei (arrowheads) but not in the Cdk1− nuclei (arrows). At this time point, the nuclear pores are partially disassembling and the An-Nup49-mCherry fluorescence is, consequently, fainter than at T = 0. Also at this time, Cdk1 leaves Cdk1+ nuclei as previously reported (Nayak et al., 2010) so there is little difference in nuclear Cdk1 levels between Cdk1+ and Cdk1− nuclei. Most fluorescence in the nuclei is due to out of focal plane fluorescence from An-Nup49-mCherry and cytoplasmic Cdk1-mCherry.
To determine if mipAD159 caused a mislocalization, or failure of accumulation, of An-Cdc20 or SldABub1/R1 we collected time-lapse images of mipAD159 strains going through mitosis that expressed histone H1-mRFP and either GFP-An-Cdc20 (strain LO5690) or SldABub1/R1 -GFP (strain LO4676). Strains were incubated at a restrictive temperature of 25°C for 17-24 hours before imaging. We chose this range of incubation times because mipAD159 nuclei with a constitutively active APC/C have accumulated by this time (Nayak et al., 2010). An-Cdc20 accumulated in virtually all mitotic nuclei as only one nucleus failed to accumulate it (n = 71). mipAD159, thus, does not appear to affect An-Cdc20 accumulation.
With SldABub1/R1, however, a subset of mitotic mipAD159 nuclei failed to accumulate the protein (Fig. 7D-I, J-O). To determine if the nuclei in which SldABub1/R1 failed to accumulate were nuclei with a constitutively active APC/CCdhA, we created a mipAD159 strain (LO7554) expressing SldABub1/R1 -GFP, An-Nup49-mCherry, and Cdk1-mCherry. Although An-Nup49-mCherry and Cdk1-mCherry fluoresce at the same wavelength, they are distinguishable because An-Nup49-mCherry is at the nuclear periphery while Cdk1-mCherry is in the nucleoplasm and at the SPB (Fig. 7K). We scored 208 nuclei as they passed into and through mitosis, scoring whether the nuclei were Cdk1− prior to mitosis and whether SldABub1/R1 -GFP localized to the SPB/KT complex in mitosis. 131/208 nuclei (63%) were Cdk1+ and SldABub1/R1 -GFP localized to the SPB/KT complex in 129 of these. 77/208 nuclei (37%) were Cdk1− and SldABub1/R1 -GFP failed to localize to the SPB/KT complex in 74 of these. There were only two nuclei that were Cdk1+ in which SldABub1/R1 -GFP failed to localize to the SPB/KT complex, and there were three Cdk1− nuclei in which SldABub1/R1 -GFP localized to the SPB/KT complex. There is, thus, an excellent correlation between the failure of Cdk1 to accumulate in nuclei in interphase due to a constitutively active APC/CCdhA and the failure of SldABub1/R1 to accumulate at the SPB/KT complex in mitosis.
Discussion
Spatial regulation of the spindle assembly checkpoint
Given the importance of the SAC, there is surprisingly little live imaging data on SAC proteins. We report the first time-lapse in vivo localization data for Mps1, Bub3, Bub1/R1, and Cdc20 homologs in a filamentous fungus. Our results reveal the key role that localization of Cdc20 and SAC proteins plays in the regulation of mitosis in A. nidulans. We have found that the location of An-Cdc20 is regulated during the cell cycle in A. nidulans in a way that is likely to control APC/C activity. Previous localization data in A. nidulans revealed that the APC/C is located in the nucleoplasm in interphase and mitosis (Nayak et al., 2010). The low level of An-Cdc20 in the nucleoplasm prior to mitotic entry likely limits the formation of APC/CAn-Cdc20 and, thus, APC/C activity. At mitotic entry, the influx of An-Cdc20 allows increased formation of APC/CAn-Cdc20 potentiating APC/C activity. This regulatory mechanism is apparently unavailable to mammalian cells because Cdc20 is present in interphase nuclei (Kallio et al., 2002).
We have found that the localization patterns of An-Mad2, An-Mps1, SldBBub3 and An-Cdc20 are distinct from each other during interphase. An-Mad1 has been localized previously to the nuclear envelope (De Souza et al., 2009). An-Mad2 most obviously localizes to the nuclear periphery in A. nidulans, as in other organisms, although some is in the cytoplasm. An-Mps1 is at the SPB from G1/S through G2, SldBBub3 is concentrated in the nucleoplasm and An-Cdc20 is in the cytoplasm. It is interesting to note that while both Cdc20 and Mad2 localize to centrosomes in mitosis in mammalian cells (Howell et al., 2000; Kallio et al., 2002), the A. nidulans homologs do not show any clear, KT-independent, localization to the SPB. While SldABub1/R1 was difficult to detect in interphase, as there was little or no SldABub1/R1 in the nucleoplasm, it may be present in the cytoplasm but not at levels that we can detect above background levels. Cytoplasmic localization of SldABub1/R1 in interphase would be consistent with reports on BubR1 localization in H. sapiens (Burum-Auensen et al., 2007; Taylor et al., 1998) and D. melanogaster (Buffin et al., 2005).
The physical separation of these proteins likely limits or prevents the formation of a functional SAC complex until mitosis when they all localize to the SPB/KT complex. In mammalian cells there are contradictory data as to whether the MCC exists in interphase or forms only in mitosis (Sudakin et al., 2001; Fraschini et al., 2001; Vigneron et al., 2004; Johnsonet al., 2004; Hardwick et al., 2000; Kulukian et al., 2009). In A. nidulans, while An-Cdc20, An-Mad2 and SldABub1/R1 might associate in the cytoplasm in interphase, they are physically separated from SldBBub3, and possibly An-Mps1, and these proteins do not come together to form an obvious complex until after mitotic entry when they localize to the SPB/KT complex.
These data also strongly support the notion that Nups play a central role in mitotic regulation in A. nidulans (De Souza et al., 2004; De Souza and Osmani, 2007; Osmani et al., 2006; De Souza et al., 2011). A. nidulans undergoes a semi-open mitosis in which peripheral Nups disassemble from the nuclear envelope at mitotic entry (De Souza et al., 2004; De Souza and Osmani, 2007; Osmani et al., 2006). Our data, in combination with the data of De Souza et al. (De Souza et al., 2009), suggest that partial disassembly of NPCs allows (1) An-Mad1 and An-Mad2 to dissociate from the nuclear periphery and enter the nucleus, concentrating at the SPB/KT complex, (2) SldABub1/R1 and An-Cdc20 to enter the nucleus from the cytoplasm with SldABub1/R1 concentrating at the SPB/KT complex and An-Cdc20 concentrating first at the SPB/KT/forming spindle region before spreading through the nucleoplasm, and (3) SldBBub3 to leave the nucleoplasm except for the portion that remains at KTs.
γ-tubulin function is important for localization of An-Mps1 and SldABub1/R1 in mitosis
Our data also address, in part, why the SAC does not inactivate the constitutively active APC/C in CB−, mipAD159 nuclei when they enter mitosis. The proximal explanation is that many CB− nuclei (74%) lack An-Mps1 and almost all (96%) lack SldABub1/R1. Virtually all CB− nuclei, thus, lack at least one of these key SAC components, and the absence of either of them would prevent activation of the SAC and, consequently, SAC inhibition of the APC/C (Musacchio and Salmon, 2007).
Failure of accumulation of SldABub1/R1 could be due, in principle, to a failure of inactivation of APC/CCdhA since SldABub1/R1 contains two KEN boxes. We know, however, that destruction of SldABub1/R1 in the cytoplasm is not responsible for the lack of SldABub1/R1 in CB− nuclei because SldABub1/R1 accumulates normally at SPB/KTs in CB+ nuclei entering mitosis in the same cytoplasm. If the absence of SldABub1/R1 at KTs of CB− nuclei were due to constitutive APC/CCdhA activity, it would have to be destroyed after mitotic onset. However, our data reveal that in all nuclei, whether CB+, CB−, mipA+, or mipAD159, CdhA disappears from the SPB and nucleoplasm in mitosis (Edgerton-Morgan and Oakley, 2012). Therefore, APC/CCdhA is probably not active in the nucleus at this stage.
In X. laevis egg extracts, Mps1 is required for BubR1 to localize to KTs (Vigneron et al., 2004; Wong and Fang, 2006). If this is the case in A. nidulans, An-Mps1− nuclei would not be expected to accumulate SldABub1/R1 at KTs in mitosis. However, not all CB− nuclei are An-Mps1− so the absence of An-Mps1 can not fully explain why SldABub1/R1 is absent from all CB− nuclei.
An-Mps1 has a canonical destruction box (RXXLXXXXN) (Glotzer et al., 1991) that can act as a recognition sequence for either APC/CCdc20 or APC/CCdh1 (Bashir and Pagano, 2004; Pfleger and Kirschner, 2000; Tian et al., 2012; Visintin et al., 1997; Zur and Brandeis, 2002). The destruction box of human Mps1 has been demonstrated to be a target of APC/CCdc20 in mitotically arrested cells and of APC/CCdh1 in G1-arrested cells (Cui et al., 2010). These data raise the possibility that constitutively active APC/CCdhA targets An-Mps1 for destruction in CB− nuclei. This does not seem to be the case, however, for two reasons. First, our data reveal that in wild-type cells An-Mps1 becomes visible at G1/S, before APC/CCdhA is inactivated and cyclin B becomes visible (Nayak et al., 2010; Edgerton-Morgan and Oakley, 2012). Second, not all CB− nuclei are An-Mps1−. APC/CCdhA appears to be active in all CB− nuclei and An-Mps1 is not being destroyed constitutively in a substantial fraction of these nuclei (26%). The important implication of these data is that γ-tubulin has roles in localizing SldABub1/R1 and An-Mps1 that are independent of its function in inactivating APC/CCdhA. mipAD159 is a recessive allele (Jung et al., 2001) and, thus, its phenotypes are much more likely to be due to loss or reduction of function(s) rather than to interfering functions.
The failure of localization of An-Mps1 due to mipAD159 is interesting and has implications for the mitotic phenotypes of mipAD159 and the functions of γ-tubulin. It is important to remember that, at restrictive temperatures, mipAD159 causes significant defects in the regulation of mitotic progression such as chromosomal nondisjunction and extremely stretched chromatin (Prigozhina et al., 2004) that are not easily explained by the failure of inactivation of APC/CCdhA. In addition, deletion of cdhA does not rescue the cold sensitivity of mipAD159 (unpublished data from our lab). We have found that a substantial fraction of An-Mps1− nuclei are CB+ (27%). Mps1 has been reported to carry out a number of important roles in mitosis that might be defective in our Mps1− nuclei. In vertebrates, Mps1 is required for KT localization of Mad1 and Mad2, as well as two other SAC components, Plk1 and CENP-E (Abrieu et al., 2001; Liu et al., 2003; Martin-Lluesma et al., 2002; Vigneron et al., 2004; Wong and Fang, 2006). Mps1 also plays an important role in recruiting the Aurora B kinase to centromeres (van der Waal et al., 2012) and it phosphorylates Borealin, a member of the complex that regulates the Aurora B kinase (Jelluma et al., 2008). The Aurora B complex is multifunctional, but one of its key functions is correcting errors in attachment of chromosomes to the mitotic spindle (Ditchfield et al., 2003; Hauf et al., 2003; Ruchaud et al., 2007; Tanaka et al., 2002). In S. cerevisiae, MPS1, Bub1 and Sgo1 are key elements in a chromosome biorientation promoting pathway that is apparently independent of Aurora B (Storchova et al., 2011). In An-Mps1− CB+ nuclei, mitosis would likely proceed, but the SAC, and other aspects of mitotic regulation, are strongly predicted to be defective. The failure of localization of An-Mps1 could, thus, account for at least some of the defects in mitotic regulation caused by mipAD159.
The failure of localization of An-Mps1 is unlikely to account fully for the mitotic defects caused by mipAD159 and other mutant γ-tubulin alleles. The phenotypes of mipAD159 and several other alleles (all recessive conditionally-growth inhibited) have been examined in enough detail to give important clues as to the functions in which they are defective. In the original paper that reported the creation of mutant mipA alleles through alanine scanning mutagenesis, it was reported that mipAD159, mipAR338 and mipAK408 exited mitosis before mitosis was successfully completed (Jung et al., 2001). It was subsequently shown that mipAD159 causes an earlier mitotic exit than mipA+ controls even in concentrations of benomyl that prevent spindle formation (Prigozhina et al., 2004). mipAD159, mipAD123 and mipAR63 all show a high frequency of elongate spindles with chromosomes along the spindles, suggesting that chromosomal disjunction is inhibited while spindle elongation proceeds (Prigozhina et al., 2001; Prigozhina et al., 2004; Li et al., 2005), and time lapse microscopy of mipAD159 confirmed that chromosomal disjunction is often defective in this allele at restrictive temperatures (Prigozhina et al., 2004). Live imaging of mipAR63 also revealed that initiation of anaphase A (i.e. initiation of chromosomal disjunction) is inhibited at a restrictive temperature in this mutant (Li et al., 2005). All of these results are consistent with mutant γ-tubulin alleles causing failure of APC/C regulation in mitosis. However, if failure of APC/C regulation is the cause of these phenotypes, the failure must occur in two directions. Failure of chromosomal disjunction would occur because of inadequate APC/C activity that, in turn, results in the failure of securin being targeted for destruction. Premature mitotic exit, on the other hand, would reflect inappropriate APC/C activity targeting cyclin B prematurely for destruction. One speculative possibility is that mipAD159 has opposite effects on the activities of APC/CAn-Cdc20 and APC/CCdhA. The observed phenotypes are consistent with mipAD159 causing a reduction of APC/CAn-Cdc20 activity resulting in failure of chromosomal disjunction due to reduction in targeting of securin for destruction. mipAD159 has already been shown to cause a failure of inactivation of APC/CCdhA and it is certainly possible that it causes enhanced or premature APC/CCdhA activity late in mitosis which results in premature mitotic exit.
Experimental Procedures
Strains and Media
A list of strains used in this study and their genotypes is given in Table 1. YAG [5 g liter−1 yeast extract, 20 g liter−1 D-glucose, 15 g liter−1 agar, supplemented with 400 μl liter−1 of a trace element solution (Vishniac and Santer, 1957) was used as a solid complete medium. Yeast extract does not provide enough pyrimidines to supplement the pyrG89 mutation nor enough riboflavin to fully supplement the riboB2 mutation, so uridine (2.442 mg ml−1), uracil (1 mg ml−1), and riboflavin (2.5 μg ml−1) were added to YAG if needed. Liquid minimal medium (MM) for imaging consisted of 6 g liter−1 NaNO3, 0.52 g liter−1 KCl, 0.52 g liter−1 MgSO4·7H2O, 1.52 g liter−1 KH2PO4, 10 g liter−1 D-glucose, 400 μl liter−1 of a trace element solution (Vishniac and Santer, 1957) and any additional nutrients required to supplement mutations. pH was adjusted to 6.0-6.5.
Generation of linear constructs for transformation
Linear constructs for transformation of A. nidulans were generated by fusion PCR as described previously (Nayak et al., 2006; Szewczyk et al., 2006; Yang et al., 2004; Yu et al., 2004; Zarrin et al., 2005). Generation of a fusion PCR product for C-terminal tagging or deleting a gene followed the rationale and procedure given in (Szewczyk et al., 2006). N-terminally tagging An-Cdc20 was achieved by following a five-piece fusion PCR protocol similar to that in (Wong et al., 2008). In summary, three fragments were amplified from A. nidulans genomic DNA: 1) approximately 1000 bp of a region of An-cdc20's 5’ untranslated region (5’UTR), 2) approximately 500 bp of An-cdc20's promoter region, and 3) approximately 1000 bp of An-cdc20's coding sequence. GFP was amplified such that it included an ATG at its N-terminus and a flexible glycine-alanine (GA) linker at its C-terminus. The selectable marker, AfpyroA, was amplified from a plasmid and inserted between the 5’ UTR flank fragment and the promoter.
Transformation and verification of transformants
Protoplast formation, purification, and subsequent transformation were carried out following the methods of (Szewczyk et al., 2006; Oakley et al., 2012). For diagnostic PCR, genomic DNA was isolated by one of three methods, all of which follow previously published protocols (Edgerton-Morgan and Oakley, 2012; Hervás-Aguilar et al., 2007; Lee and Taylor, 1990). Primers for diagnostic PCR usually consisted of a pair of primers outside of the targeted region, as well as a pair consisting of one outside primer and one internal primer (usually inside the selectable marker), especially when outside primers would give a similar size PCR product whether the gene was correctly targeted or not. PCR was carried out using either Platinum Taq (Invitrogen) or OneTaq Hot Start Quick-Load DNA polymerase (New England Biolabs).
For Southern hybridizations, A. nidulans genomic DNA was isolated using one of two published procedures (Lee and Taylor, 1990; Oakley et al., 1987). Genomic DNA was then digested with an appropriate restriction enzyme. The hybridization was carried out using a dried gel method (Oakley et al., 1987). Radioactively labeled full-length transforming DNA fragments were used as probes and were labeled using the Prime-It II Random Primer Labeling Kit (Agilent Technologies) and purified using spin-column chromatography through Sephadex G-50 (Maniatis et al., 1982).
Microscopy
For imaging, spores were inoculated in liquid minimal medium plus appropriate nutritional supplements in eight-chamber cover glasses (Lab-Tek; Thermo Fisher Scientific). We used two systems for imaging. One was an inverted Olympus IX71 microscope equipped with Prior shutters, filter wheels, Z-axis drives, an ORCA ERAG camera (Hamamatsu Photonics), an environmental chamber to maintain stable temperatures and a mercury light source. Filter sets used were a GFP/DsRed2X2M-B dual-band Sedat filter set (Semrock) with a 459-481 nm bandpass excitation filter for GFP, a 546-566 nm excitation filter for mCherry and mRFP, a dual reflection band dichroic (457-480 nm and 542-565 nm reflection bands, 500-529 and 584-679 nm transmission bands), a 499-529 nm emission filter for GFP, and a 580-654 nm emission filter for mCherry/mRFP. Images were acquired with a 60x 1.42 NA planapochromatic objective (Olympus) using Volocity software (PerkinElmer). The second system was an UltraView VoX spinning disk confocal system (PerkinElmer) mounted on an Olympus IX71 inverted microscope. It was equipped with a controlled temperature chamber and a software-controlled piezoelectric stage for rapid Z-axis movement. Images were collected using a 60x 1.42 NA panapochromatic objective (Olympus) (in some cases with a 1.6x Optovar) and an ORCA ERAG camera (Hamamatsu Photonics). Solid state 405-, 488-, and 561-nm lasers were used for excitation. Fluorochrome-specific emission filters were used to prevent emission bleed through between fluorochromes. This system was controlled by Volocity software (PerkinElmer). Both systems were calibrated with a stage micrometer and the Ultraview/Vox optical system was determined to give correct register of the GFP and mCherry channels using fluorescent beads that fluoresce at both wavelengths. Minimum and maximum intensity cutoffs (black and white levels) for each channel were chosen in Volocity before images were exported. Where noted in the figures, three-dimensional projections of z-series stacks were made with Volocity software. No other adjustments were made to the images. Figures were prepared from exported images using CorelDraw (Corel Corporation).
Acknowledgements
We would like to thank Dr. Tania Nayak for creating mipA+ and mipAD159 strains expressing An-Mad2-GFP and histone H1-mRFP; the mipA+ and mipAD159 strains expressing cyclin B-GFP and histone H1-mRFP; and an SldBBub3-GFP, histone H1-mCherry, mipAD159 strain. We would also like to thank Dr. Oliver Griesbeck (Max-Planck-Institut für Neurobiologie) for the T-Sapphire clone, the laboratory of Dr. Stephen Osmani (Ohio State University) for An-Nup49-mCherry and An-Ndc80-mCherry strains and C. Elizabeth Oakley for advice in editing the manuscript. This work was supported by NIH grant GM031837 and by the Irving S. Johnson fund of the University of Kansas Endowment.
References
- Abrieu A, Magnaghi-Jaulin L, Kahana JA, Peter M, Castro A, Vigneron S, et al. Mps1 is a kinetochore-associated kinase essential for the vertebrate mitotic checkpoint. Cell. 2001;106:83–93. doi: 10.1016/s0092-8674(01)00410-x. [DOI] [PubMed] [Google Scholar]
- Bashir T, Pagano M. Don't skip the G1 phase: how APC/CCdh1 keeps SCFSKP2 in check. Cell Cycle. 2004;3:850–852. [PubMed] [Google Scholar]
- Bergen LG, Morris NR. Kinetics of the nuclear division cycle of Aspergillus nidulans. J Bacteriol. 1983;156:155–160. doi: 10.1128/jb.156.1.155-160.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buffin E, Lefebvre C, Huang J, Gagou ME, Karess RE. Recruitment of Mad2 to the kinetochore requires the Rod/Zw10 complex. Curr Biol. 2005;15:856–861. doi: 10.1016/j.cub.2005.03.052. [DOI] [PubMed] [Google Scholar]
- Burum-Auensen E, De Angelis PM, Schjolberg AR, Kravik KL, Aure M, Clausen OP. Subcellular localization of the spindle proteins Aurora A, Mad2, and BUBR1 assessed by immunohistochemistry. J Histochem Cytochem. 2007;55:477–486. doi: 10.1369/jhc.6A7077.2007. [DOI] [PubMed] [Google Scholar]
- Campbell L, Hardwick KG. Analysis of Bub3 spindle checkpoint function in Xenopus egg extracts. J Cell Sci. 2003;116:617–628. doi: 10.1242/jcs.00255. [DOI] [PubMed] [Google Scholar]
- Campbell MS, Chan GKT, Yen TJ. Mitotic checkpoint proteins HsMAD1 and HsMAD2 are associated with nuclear pore complexes in interphase. J Cell Sci. 2001;114:953–963. doi: 10.1242/jcs.114.5.953. [DOI] [PubMed] [Google Scholar]
- Chen R-H, Shevchenko A, Mann M, Murray AW. Spindle checkpoint protein Xmad1 recruits Xmad2 to unattached kinetochores. J Cell Biol. 1998;143:283–295. doi: 10.1083/jcb.143.2.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui Y, Cheng X, Zhang C, Zhang Y, Li S, Wang C, Guadagno TM. Degradation of the human mitotic checkpoint kinase Mps1 is cell cycle-regulated by APC-CCdc20 and APC-CCdh1 ubiquitin ligases. J Biol Chem. 2010;285:32988–32998. doi: 10.1074/jbc.M110.140905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Souza CP, Hashmi SB, Nayak T, Oakley B, Osmani SA. Mlp1 acts as a mitotic scaffold to spatially regulate spindle assembly checkpoint proteins in Aspergillus nidulans. Mol Biol Cell. 2009;20:2146–2159. doi: 10.1091/mbc.E08-08-0878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Souza CP, Hashmi SB, Osmani AH, Andrews P, Ringelberg CS, Dunlap JC, Osmani SA. Functional analysis of the Aspergillus nidulans kinome. PLoS One. 2013;8:e58008. doi: 10.1371/journal.pone.0058008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Souza CP, Hashmi SB, Yang X, Osmani SA. Regulated inactivation of the spindle assembly checkpoint without functional mitotic spindles. EMBO J. 2011;30:2648–2661. doi: 10.1038/emboj.2011.176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Souza CP, Osmani AH, Hashmi SB, Osmani SA. Partial nuclear pore complex disassembly during closed mitosis in Aspergillus nidulans. Curr Biol. 2004;14:1973–1984. doi: 10.1016/j.cub.2004.10.050. [DOI] [PubMed] [Google Scholar]
- De Souza CP, Osmani SA. Mitosis, not just open or closed. Eukaryot Cell. 2007;6:1521–1527. doi: 10.1128/EC.00178-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ditchfield C, Johnson VL, Tighe A, Ellston R, Haworth C, Johnson T, et al. Aurora B couples chromosome alignment with anaphase by targeting BubR1, Mad2, and Cenp-E to kinetochores. J Cell Biol. 2003;161:267–280. doi: 10.1083/jcb.200208091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgerton-Morgan H, Oakley BR. γ-Tubulin plays a key role in inactivating APC/CCdh1 at the G1-S boundary. J Cell Biol. 2012;198:785–791. doi: 10.1083/jcb.201203115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Efimov VP, Morris NR. A screen for dynein synthetic lethals in Aspergillus nidulans identifies spindle assembly checkpoint genes and other genes involved in mitosis. Genetics. 1998;149:101–116. doi: 10.1093/genetics/149.1.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer MG, Heeger S, Häcker U, Lehner CF. The mitotic arrest in response to hypoxia and of polar bodies during early embryogenesis requires Drosophila Mps1. Curr Biol. 2004;14:2019–2024. doi: 10.1016/j.cub.2004.11.008. [DOI] [PubMed] [Google Scholar]
- Fisk HA, Winey M. The mouse Mps1p-like kinase regulates centrosome duplication. Cell. 2001;106:95–104. doi: 10.1016/s0092-8674(01)00411-1. [DOI] [PubMed] [Google Scholar]
- Fraschini R, Beretta A, Sironi L, Musacchio A, Lucchini G, Piatti S. Bub3 interaction with Mad2, Mad3 and Cdc20 is mediated by WD40 repeats and does not require intact kinetochores. EMBO J. 2001;20:6648–6659. doi: 10.1093/emboj/20.23.6648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillett ES, Espelin CW, Sorger PK. Spindle checkpoint proteins and chromosome-microtubule attachment in budding yeast. J Cell Biol. 2004;164:535–546. doi: 10.1083/jcb.200308100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glotzer M, Murray AW, Kirschner MW. Cyclin is degraded by the ubiquitin pathway. Nature. 1991;349:132–138. doi: 10.1038/349132a0. [DOI] [PubMed] [Google Scholar]
- Hardwick KG, Johnston RC, Smith DL, Murray AW. MAD3 encodes a novel component of the spindle checkpoint which interacts with Bub3p, Cdc20p, and Mad2p. J Cell Biol. 2000;148:871–882. doi: 10.1083/jcb.148.5.871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauf S, Cole RW, LaTerra S, Zimmer C, Schnapp G, Walter R, et al. The small molecule Hesperadin reveals a role for Aurora B in correcting kinetochore-microtubule attachment and in maintaining the spindle assembly checkpoint. J Cell Biol. 2003;161:281–294. doi: 10.1083/jcb.200208092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hervás-Aguilar A, Rodríguez JM, Tilburn J, Arst HNJ, Peñalva MA. Evidence for the direct involvement of the proteasome in the proteolytic processing of the Aspergillus nidulans zinc finger transcription factor PacC. J Biol Chem. 2007;282:34735–34747. doi: 10.1074/jbc.M706723200. [DOI] [PubMed] [Google Scholar]
- Howell BJ, Hoffman DB, Fang G, Murray AW, Salmon ED. Visualization of Mad2 dynamics at kinetochores, along spindle fibers, and at spindle poles in living cells. J Cell Biol. 2000;150:1233–1249. doi: 10.1083/jcb.150.6.1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howell BJ, Moree B, Farrar EM, Stewart S, Fang G, Salmon ED. Spindle checkpoint protein dynamics at kinetochores in living cells. Curr Biol. 2004;14:953–964. doi: 10.1016/j.cub.2004.05.053. [DOI] [PubMed] [Google Scholar]
- Hoyt MA, Totis L, Roberts BT. S. cerevisiae genes required for cell cycle arrest in response to loss of microtubule function. Cell. 1991;66:507–517. doi: 10.1016/0092-8674(81)90014-3. [DOI] [PubMed] [Google Scholar]
- Hoyt MA. Eliminating all obstacles: Regulated proteolysis in the eukaryotic cell cycle. Cell. 1997;91:149–151. doi: 10.1016/s0092-8674(00)80396-7. [DOI] [PubMed] [Google Scholar]
- Hwang LH, Lau LF, Smith DL, Mistrot CA, Hardwick KG, Hwang ES, et al. Budding yeast Cdc20: a target of the spindle checkpoint. Science. 1998;279:1041–1044. doi: 10.1126/science.279.5353.1041. [DOI] [PubMed] [Google Scholar]
- Ikui AE, Furuya K, Yanagida M, Matsumoto T. Control of localization of a spindle checkpoint protein, Mad2, in fission yeast. J Cell Sci. 2002;115:1603–1610. doi: 10.1242/jcs.115.8.1603. [DOI] [PubMed] [Google Scholar]
- Iouk T, Kerscher O, Scott RJ, Basrai MA, Wozniak RW. The yeast nuclear pore complex functionally interacts with components of the spindle assembly checkpoint. J Cell Biol. 2002;159:807–819. doi: 10.1083/jcb.200205068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jelluma N, Brenkman AB, van den Broek NJF, Cruijsen CWA, van Osch MHJ, Lens SMA, et al. Mps1 phosphorylates Borealin to control Aurora B activity and chromosome alignment. Cell. 2008;132:233–246. doi: 10.1016/j.cell.2007.11.046. [DOI] [PubMed] [Google Scholar]
- Johnson VL, Scott MIF, Holt SV, Hussein D, Taylor SS. Bub1 is required for kinetochore localization of BubR1, Cenp-E, Cenp-F and Mad2, and chromosome congression. J Cell Sci. 2004;117:1577–1589. doi: 10.1242/jcs.01006. [DOI] [PubMed] [Google Scholar]
- Jung MK, Prigozhina N, Oakley CE, Nogales E, Oakley BR. Alanine-scanning mutagenesis of Aspergillus γ-tubulin yields diverse and novel phenotypes. Mol Biol Cell. 2001;12:2119–2136. doi: 10.1091/mbc.12.7.2119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadura S, He X, Vanoosthuyse V, Hardwick KG, Sazer S. The A78V mutation in the Mad3-like domain of Schizosaccharomyces pombe Bub1p perturbs nuclear accumulation and kinetochore targeting of Bub1p, Bub3p, and Mad3p and spindle assembly checkpoint function. Mol Biol Cell. 2005;16:385–395. doi: 10.1091/mbc.E04-07-0558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kallio MJ, Beardmore VA, Weinstein J, Gorbsky GJ. Rapid microtubule-independent dynamics of Cdc20 at kinetochores and centrosomes in mammalian cells. J Cell Biol. 2002;158:841–847. doi: 10.1083/jcb.200201135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasbek C, Yang CH, Yusof AM, Chapman HM, Winey M, Fisk HA. Preventing the degradation of Mps1 at centrosomes is sufficient to cause centrosome reduplication in human cells. Mol Biol Cell. 2007;18:4457–4469. doi: 10.1091/mbc.E07-03-0283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim AH, Puram SV, Bilimoria PM, Ikeuchi Y, Keough S, Wong M, et al. A centrosomal Cdc20-APC pathway controls dendrite morphogenesis in postmitotic neurons. Cell. 2009a;136:322–336. doi: 10.1016/j.cell.2008.11.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JM, Zeng CJ, Nayak T, Shao R, Huang AC, Oakley BR, Liu B. Timely septation requires SNAD-dependent spindle pole body localization of the septation initiation network components in the filamentous fungus Aspergillus nidulans. Mol Biol Cell. 2009b;20:2874–2884. doi: 10.1091/mbc.E08-12-1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SH, Lin DP, Matsumoto S, Kitazono A, Matsumoto T. Fission yeast Slp1: an effector of the Mad2-dependent spindle checkpoint. Science. 1998;279:1045–1047. doi: 10.1126/science.279.5353.1045. [DOI] [PubMed] [Google Scholar]
- Kulukian A, Han JS, Cleveland DW. Unattached kinetochores catalyze production of an anaphase inhibitor that requires a Mad2 template to prime Cdc20 for BubR1 binding. Dev Cell. 2009;16:105–117. doi: 10.1016/j.devcel.2008.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SB, Taylor JW. Isolation of DNA from fungal mycelia and single spores. In: Innis MA, Gelfand DH, Sninsky JJ, White TJ, editors. PCR Protocols: A Guide to Methods and Applications. Academic Press, Inc.; San Diego: New York: Berkeley: Boston: London: Sydney: Tokyo: Toronto: 1990. pp. 282–287. [Google Scholar]
- Li D, Morley G, Whitaker M, Huang JY. Recruitment of Cdc20 to the kinetochore requires BubR1 but not Mad2 in Drosophila melanogaster. Mol Cell Biol. 2010;30:3384–3395. doi: 10.1128/MCB.00258-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R, Murray AW. Feedback control of mitosis in budding yeast. Cell. 1991;66:519–531. doi: 10.1016/0092-8674(81)90015-5. [DOI] [PubMed] [Google Scholar]
- Li S, Oakley CE, Chen G, Han X, Oakley BR, Xiang X. Cytoplasmic dynein's mitotic spindle pole localization requires a functional anaphase-promoting complex, γ-tubulin, and NUDF/LIS1 in Aspergillus nidulans. Mol Biol Cell. 2005;16:3591–3605. doi: 10.1091/mbc.E04-12-1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S-T, Chan GKT, Hittle JC, Fujii G, Lees E, Yen TJ. Human MPS1 kinase is required for mitotic arrest induced by the loss of CENP-E from kinetochores. Mol Biol Cell. 2003;14:1638–1651. doi: 10.1091/mbc.02-05-0074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logarinho E, Bousbaa H, Dias JM, Lopes C, Amorim I, Antunes-Martins A, Sunkel CE. Different spindle checkpoint proteins monitor microtubule attachment and tension at kinetochores in Drosophila cells. J Cell Sci. 2004;117:1757–1771. doi: 10.1242/jcs.01033. [DOI] [PubMed] [Google Scholar]
- Maniatis T, Fritsch EF, Sambrook J. A laboratory manual. Cold Spring Harbor Laboratory; Cold Spring Harbor; New York: 1982. Molecular cloning. p. 545. [Google Scholar]
- Martin-Lluesma S, Stucke VM, Nigg EA. Role of Hec1 in spindle checkpoint signaling and kinetochore recruitment of Mad1/Mad2. Science. 2002;297:2267–2270. doi: 10.1126/science.1075596. [DOI] [PubMed] [Google Scholar]
- Mayer C, Filopei J, Batac J, Alford L, Paluh JL. An extended anaphase signaling pathway for Mad2p includes microtubule organizing center proteins and multiple motor-dependent transitions. Cell Cycle. 2006;5:1456–1463. doi: 10.4161/cc.5.13.2912. [DOI] [PubMed] [Google Scholar]
- Musacchio A, Hardwick KG. The spindle checkpoint: structural insights into dynamic signalling. Nat Rev Mol Cell Biol. 2002;3:731–741. doi: 10.1038/nrm929. [DOI] [PubMed] [Google Scholar]
- Musacchio A, Salmon ED. The spindle-assembly checkpoint in space and time. Nat Rev Mol Cell Biol. 2007;8:379–393. doi: 10.1038/nrm2163. [DOI] [PubMed] [Google Scholar]
- Nayak T, Edgerton-Morgan H, Horio T, Xiong Y, De Souza CP, Osmani SA, Oakley BR. γ-Tubulin regulates the anaphase-promoting complex/cyclosome during interphase. J Cell Biol. 2010;190:317–330. doi: 10.1083/jcb.201002105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nayak T, Szewczyk E, Oakley CE, Osmani A, Ukil L, Murray SL, et al. A versatile and efficient gene-targeting system for Aspergillus nidulans. Genetics. 2006;172:1557–1566. doi: 10.1534/genetics.105.052563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nurse P. Universal control mechanism regulating onset of M-phase. Nature. 1990;344:503–508. doi: 10.1038/344503a0. [DOI] [PubMed] [Google Scholar]
- O'Connell MJ, Osmani AH, Morris NR, Osmani SA. An extra copy of nimEcyclinB elevates pre-MPF levels and partially suppresses mutation of nimTcdc25 in Aspergillus nidulans. EMBO J. 1992;11:2139–2149. doi: 10.1002/j.1460-2075.1992.tb05273.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oakley CE, Weil CF, Kretz PL, Oakley BR. Cloning of the riboB locus of Aspergillus nidulans. Gene. 1987;53:293–298. doi: 10.1016/0378-1119(87)90019-9. [DOI] [PubMed] [Google Scholar]
- Oakley CE, Edgerton-Morgan H, Oakley BR. Tools for manipulation of secondary metabolism pathways: rapid promoter replacements and gene deletions in Aspergillus nidulans. Methods Mol Biol. 2012;944:143–161. doi: 10.1007/978-1-62703-122-6_10. [DOI] [PubMed] [Google Scholar]
- Osmani AH, Davies J, Liu HL, Nile A, Osmani SA. Systematic deletion and mitotic localization of the nuclear pore complex proteins of Aspergillus nidulans. Mol Biol Cell. 2006;17:4946–4961. doi: 10.1091/mbc.E06-07-0657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters JM. The anaphase promoting complex/cyclosome: a machine designed to destroy. Nat Rev Mol Cell Biol. 2006;7:644–656. doi: 10.1038/nrm1988. [DOI] [PubMed] [Google Scholar]
- Pfleger CM, Kirschner MW. The KEN box: an APC recognition signal distinct from the D box targeted by Cdh1. Genes Dev. 2000;14:655–665. [PMC free article] [PubMed] [Google Scholar]
- Prigozhina NL, Oakley CE, Lewis A, Nayak T, Osmani SA, Oakley BR. γ-Tubulin plays an essential role in the coordination of mitotic events. Mol Biol Cell. 2004;15:1374–1386. doi: 10.1091/mbc.E03-06-0405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prigozhina NL, Walker RA, Oakley CE, Oakley BR. γ-Tubulin and the C-terminal motor domain kinesin-like protein, KLPA, function in the establishment of spindle bipolarity in Aspergillus nidulans. Mol Biol Cell. 2001;12:3161–3174. doi: 10.1091/mbc.12.10.3161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raff JW, Jeffers K, Huang JY. The roles of Fzy/Cdc20 and Fzr/Cdh1 in regulating the destruction of cyclin B in space and time. J Cell Biol. 2002;157:1139–1149. doi: 10.1083/jcb.200203035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruchaud S, Carmena M, Earnshaw WC. Chromosomal passengers: conducting cell division. Nat Rev Mol Cell Biol. 2007;8:798–812. doi: 10.1038/nrm2257. [DOI] [PubMed] [Google Scholar]
- Shah JV, Botvinick E, Bonday Z, Funari F, Berns M, Cleveland DW. Dynamics of Centromere and Kinetochore Proteins: Implications for Checkpoint Signaling and Silencing. Curr Biol. 2004;14:942–952. doi: 10.1016/j.cub.2004.05.046. [DOI] [PubMed] [Google Scholar]
- Sharp-Baker H, Chen RH. Spindle checkpoint protein Bub1 is required for kinetochore localization of Mad1, Mad2, Bub3, and CENP-E, independently of its kinase activity. J Cell Biol. 2001;153:1239–1250. doi: 10.1083/jcb.153.6.1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Son S, Osmani SA. Analysis of all protein phosphatase genes in Aspergillus nidulans identifies a new mitotic regulator, fcp1. Eukaryot Cell. 2009;8:573–585. doi: 10.1128/EC.00346-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storchová Z, Becker JS, Talarek N, Kögelsberger S, Pellman D. Bub1, Sgo1, and Mps1 mediate a distinct pathway for chromosome biorientation in budding yeast. Mol Biol Cell. 2011;22:1473–1485. doi: 10.1091/mbc.E10-08-0673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stucke VM, Silljé HHW, Arnaud L, Nigg EA. Human Mps1 kinase is required for the spindle assembly checkpoint but not for centrosome duplication. EMBO J. 2002;21:1723–1732. doi: 10.1093/emboj/21.7.1723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudakin V, Chan GKT, Yen TJ. Checkpoint inhibition of the APC/C in HeLa cells is mediated by a complex of BUBR1, BUB3, CDC20, and MAD2. J Cell Biol. 2001;154:925–936. doi: 10.1083/jcb.200102093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suijkerbuijk SJE, van Dam TJP, Karagöz GE, von Castelmur E, Hubner NC, Duarte AMS, et al. The vertebrate mitotic checkpoint protein BUBR1 is an unusual pseudokinase. Dev Cell. 2012;22:1321–1329. doi: 10.1016/j.devcel.2012.03.009. [DOI] [PubMed] [Google Scholar]
- Szewczyk E, Nayak T, Oakley CE, Edgerton H, Xiong Y, Taheri-Talesh N, et al. Fusion PCR and gene targeting in Aspergillus nidulans. Nat Protoc. 2006;1:3111–3120. doi: 10.1038/nprot.2006.405. [DOI] [PubMed] [Google Scholar]
- Szewczyk E, Oakley BR. Microtubule dynamics in mitosis in Aspergillus nidulans. Fungal Genet Biol. 2011;48:998–999. doi: 10.1016/j.fgb.2011.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka TU, Rachidi N, Janke C, Pereira G, Galova M, Schiebel E, et al. Evidence that the Ipl1-Sli15 (Aurora kinase-INCENP) complex promotes chromosome bi-orientation by altering kinetochore-spindle pole connections. Cell. 2002;108:317–329. doi: 10.1016/s0092-8674(02)00633-5. [DOI] [PubMed] [Google Scholar]
- Taylor SS, Ha E, McKeon F. The human homologue of Bub3 is required for kinetochore localization of Bub1 and a Mad3/Bub1-related protein kinase. J Cell Biol. 1998;142:1–11. doi: 10.1083/jcb.142.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor SS, Hussein D, Wang Y, Elderkin S, Morrow CJ. Kinetochore localisation and phosphorylation of the mitotic checkpoint components Bub1 and BubR1 are differentially regulated by spindle events in human cells. J Cell Sci. 2001;114:4385–4395. doi: 10.1242/jcs.114.24.4385. [DOI] [PubMed] [Google Scholar]
- Tian W, Li B, Warrington R, Tomchick DR, Yu H, Luo X. Structural analysis of human Cdc20 supports multisite degron recognition by APC/C. Proc Natl Acad Sci U S A. 2012;109:18419–18424. doi: 10.1073/pnas.1213438109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Waal MS, Saurin AT, Vromans MJM, Vleugel M, Wurzenberger C, Gerlich DW, et al. Mps1 promotes rapid centromere accumulation of Aurora B. EMBO Rep. 2012;13:847–854. doi: 10.1038/embor.2012.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vigneron S, Prieto S, Bernis C, Labbé JC, Castro A, Lorca T. Kinetochore localization of spindle checkpoint proteins: who controls whom? Mol Biol Cell. 2004;15:4584–4596. doi: 10.1091/mbc.E04-01-0051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vishniac W, Santer M. The thiobacilli. Bacteriol Rev. 1957;21:195–213. doi: 10.1128/br.21.3.195-213.1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visintin R, Prinz S, Amon A. CDC20 and CDH1: a family of substrate-specific activators of APC-dependent proteolysis. Science. 1997;278:460–463. doi: 10.1126/science.278.5337.460. [DOI] [PubMed] [Google Scholar]
- Winey M, Goetsch L, Baum P, Byers B. MPS1 and MPS2: novel yeast genes defining distinct steps of spindle pole body duplication. J Cell Biol. 1991;114:745–754. doi: 10.1083/jcb.114.4.745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong KH, Todd RB, Oakley BR, Oakley CE, Hynes MJ, Davis MA. Sumoylation in Aspergillus nidulans: sumO inactivation, overexpression and live-cell imaging. Fungal Genet Biol. 2008;45:728–737. doi: 10.1016/j.fgb.2007.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong OK, Fang G. Loading of the 3F3/2 antigen onto kinetochores is dependent on the ordered assembly of the spindle checkpoint proteins. Mol Biol Cell. 2006;17:4390–4399. doi: 10.1091/mbc.E06-04-0346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong Y, Oakley BR. In vivo analysis of the functions of γ-tubulin-complex proteins. J Cell Sci. 2009;122:4218–4227. doi: 10.1242/jcs.059196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Ukil L, Osmani A, Nahm F, Davies J, De Souza CP, et al. Rapid production of gene replacement constructs and generation of a green fluorescent protein-tagged centromeric marker in Aspergillus nidulans. Eukaryot Cell. 2004;3:1359–1362. doi: 10.1128/EC.3.5.1359-1362.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J-H, Hamari Z, Han KH, Seo JA, Reyes-Domínguez Y, Scazzocchio C. Double-joint PCR: a PCR-based molecular tool for gene manipulations in filamentous fungi. Fungal Genet Biol. 2004;41:973–981. doi: 10.1016/j.fgb.2004.08.001. [DOI] [PubMed] [Google Scholar]
- Zapata-Hommer O, Griesbeck O. Efficiently folding and circularly permuted variants of the Sapphire mutant of GFP. BMC Biotechnol. 2003;3:5. doi: 10.1186/1472-6750-3-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarrin M, Leeder AC, Turner G. A rapid method for promoter exchange in Aspergillus nidulans using recombinant PCR. Fungal Genet Biol. 2005;42:1–8. doi: 10.1016/j.fgb.2004.10.002. [DOI] [PubMed] [Google Scholar]
- Zur A, Brandeis M. Timing of APC/C substrate degradation is determined by fzy/fzr specificity of destruction boxes. EMBO J. 2002;21:4500–4510. doi: 10.1093/emboj/cdf452. [DOI] [PMC free article] [PubMed] [Google Scholar]