Abstract
Chemotherapeutics fail to effectively treat tumors because they cannot reach quiescent regions far from blood vessels. Motile Salmonella are an attractive delivery system that could break this therapeutic barrier. However, little is known about the dissemination and tissue penetration of individual bacteria in tumors after intravenous administration. We hypothesized that eliminating the Trg receptor would improve accumulation in tumor quiescence. To test this hypothesis, we deleted the trg gene from nonpathogenic Salmonella. To quantify individual bacterial behavior, we measured tissue penetration in a tumor-on-a-chip device and measured colony localization in mouse tumors using immunofluorescence. In tumors in vitro and in mice, trg− Salmonella penetrated farther into tissue than control bacteria. This difference in localization was caused by the inability to sense sugars in well perfused tissue. Three distinct bacterial phenotypes were observed: proliferating, penetrating, and inactive. Large proliferating colonies, containing more than 40% of individual bacteria, only formed less than 60 µm from blood vessels. Small colonies, in comparison, were present both near (inactive) and far (penetrating) from vessels. The farthest was 361.2 µm from a vessel, demonstrating the ability to target avascular regions. In addition, colonization was most pronounced in poorly vascularized tumor regions. We show that deletion of trg amplifies Salmonella accumulation in quiescent tumor regions, and, for the first time, identify biological processes that control bacterial distribution in tumors. Understanding how Salmonella penetrate tissue, target quiescence and specifically replicate in tumors are essential steps toward creating a tightly controlled, tunable bacterial therapy.
Keywords: Salmonella, bacterial cancer therapy, tissue penetration, tumor quiescence, trg
Introduction
Salmonella bacteria have the potential to be potent anticancer agents [1, 2]. Motile bacteria have the unique ability to overcome the diffusion limitations that prevent chemotherapeutics from being effective [3]. Because Salmonella are self-propelled, they can penetrate deep into tumor tissue [4] and away from blood vessels [5]. Once there, they can be triggered to produce anticancer molecules that kill tumor cells [6–8]. Bacterial tissue penetration is controlled by chemotaxis toward molecules produced by living and dying cancer cells [4]. Similar to chemotherapeutics, systemically administered bacteria enter tumors through the vasculature. From there, they seek and replicate in preferable regions [5]. Replication is important because the disproportional increase in bacterial density in tumors is the major cause of selectivity over normal tissue [9, 10]. Salmonella have been shown to accumulate in tumors at densities as high as 1:10,000 compared to normal organs [10, 11]. To date, little is known about the behavior of individual Salmonella in tumors. Understanding how Salmonella migrate and replicate in tumors will enable creation of therapies able to eradicate tumor cells untouchable by conventional therapeutics.
Solid tumors do not respond optimally to chemotherapy for several reasons. Most chemotherapeutics do not actively target tumors [12, 13]. This lack of specificity leads to insufficient drug exposure, and gives cancer cells a chance to repopulate between treatments [14]. Large intercapillary distances, variable blood flows, and high interstitial pressures [15, 16] prevent chemotherapeutics from diffusing deep into tumors at effective concentrations [17]. This unfavorable distribution is compounded by the metabolic state of tumor cells located far from blood vessels. These cells do not progress through the cell cycle and are arrested in a quiescent state, which protects them against most anticancer agents, which target actively proliferating cells [18, 19].
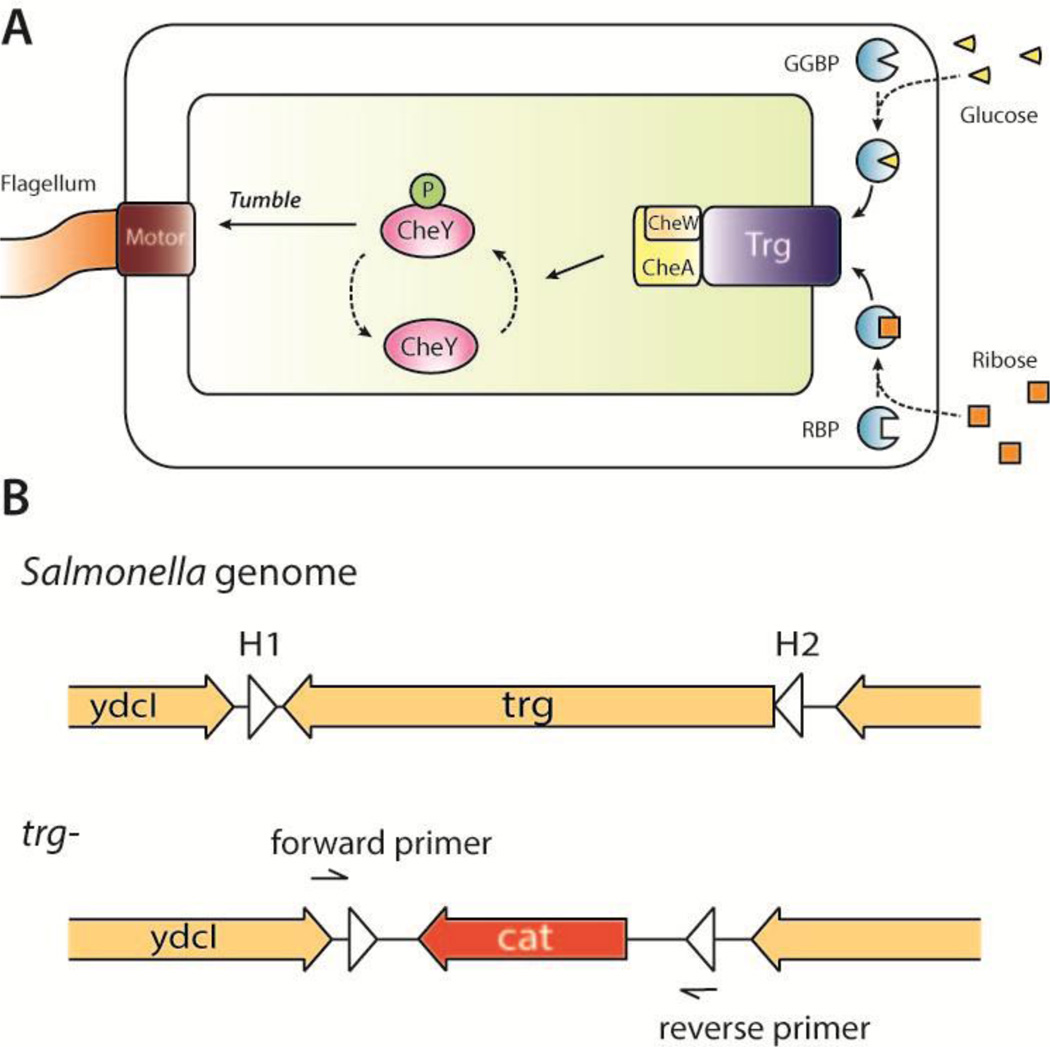
Chemotaxis machinery controls where Salmonella colonize in tumors [4, 20]. Faster swimming bacteria have increased accumulation and penetration into tumor tissue [21]. In cylindroids, Salmonella that cannot sense aspartate cannot detect tumors, and Salmonella that cannot sense serine do not penetrate tissue [20]. Salmonella mutants that cannot detect sugars penetrate tissue, but accumulate in quiescent regions between necrotic and proliferating tissue [20]. Salmonella sense sugars with the Trg receptor, which binds periplasmic binding proteins (Figure 1A) [22]. When a gradient of glucose or ribose is encountered, the sugar molecules diffuse through the outer membrane and bind to the glucose-galactose binding protein (GGBP) or the ribose binding protein (RBP). Activated GGBP or RBP bind to transmembrane complexes formed by the proteins Trg, CheW, and CheA. When activated, this complex reduces the rate of CheY phosphorylation and lowers the flagellar tumbling frequency [23], propelling bacteria towards regions with higher sugar concentrations. Salmonella without Trg would not chemotax toward sources of either glucose or ribose.
Figure 1. Role of Trg in Salmonella chemotaxis.
A) The sugar-sensing chemotaxis machinery of Salmonella includes periplasmic binding proteins (GGBP and RBP), the transmembrane chemoreceptor Trg, signal transduction components (CheW, CheA and CheY), and flagellum machinery. Extracellular glucose (triangles) and ribose (squares) bind to GGBP and RP, respectively. When complexed, these binding proteins activate Trg, which inhibits autophosphorylation of CheA kinases. Phosphorylated CheA does not transfer phosphoryl groups to CheY, which decreases tumbling frequency and results in movement up a sugar gradient. B) The trg gene was removed from the Salmonella genome by homologous recombination, and replaced by cat, which confers resistance to chloramphenicol. Recombination was mediated by homology extensions H1 and H2.
The goal of this work was to quantify and control the behavior of individual Salmonella in tumor tissue. We hypothesized that (1) Salmonella without the Trg receptor have improved accumulation in quiescent tumor regions in mice, and (2) Salmonella have distinct phenotypes that control colony localization relative to blood vessels. To test these hypotheses, the trg gene was deleted from the chromosome of a non-pathogenic Salmonella strain. The motility of trg− Salmonella was evaluated in culture and its ability to penetrate tissue was measured in a perfused tumor-on-a-chip device. The location of colonies relative to tumor blood vessels was determined in BALB/c mice with subcutaneous 4T1 tumors. Intratumoral Salmonella colonies and blood vessels were identified by immunofluorescence. Quantitative spatial analysis was used to measure colony size, distance to vessels, and local vascular density. To the best of our knowledge, this is the first time that the distribution of individual bacteria and colonies relative to tumor vasculature has been measured, and the first time that bacterial chemotaxis machinery has been manipulated to improve tumor localization. With the ability to selectively target quiescent tumor tissue, trg− Salmonella are an excellent tool to overcome the limitations of standard therapies. Understanding the principles of Salmonella intratumoral localization will enable more focused and fully controlled tumor targeting, and ultimately realize the potential of Salmonella as an anticancer agent.
Materials and Methods
Bacterial strains and cultural conditions
The attenuated Salmonella strain VNP20009 was donated by Vion Pharmaceuticals, New Haven, CT. VNP20009 (purI−, msbB−, xyl−) is a detoxified strain designed for clinical applications [9, 24]. It was necessary to create a trg deletion in the VNP20009 strain because it is non-toxic and does not induce sepsis when injected into mice [9]. The Trg-deficient strain ST832 used for in vitro cylindroid experiments [20] was derived from the virulent LT2 strain [25, 26] and would not be suitable for use in live animals. The λ-red-gene-inactivation system [27] was provided by Barry Wanner at Purdue University, West Lafayette, IN. Bacteria were grown in LB broth or on LB agar at 37°C. Exogenous DNA was introduced into Salmonella by electroporation (Bio-Rad, Hercules, CA) with 1mm cuvettes and the following settings: 1.8kv, 25µF and 200Ω.
Trg gene deletion
The trg gene was deleted from the Salmonella genome using λ-red gene inactivation [27, 28]. This system utilizes homologous recombination to replace the target gene with an antibiotic resistance gene as a screening marker. Salmonella (VNP20009) were transformed with the λ-red helper plasmid pKD46 and grown in liquid culture with 0.2% L-arabinose at 30°C to induce the expression of λ-red recombinases. The chloramphenicol resistance gene cat was amplified from plasmid pKD3 by PCR using the primer H1-P1 (ACG CGC CCG CGG CTA AAA TAG CCC GCT GGC GCG ACG CTT AGT GTA GGC TGG AGC TGC TTC) and H2-P2 (TAA CGG GCG TGT TTT ACG CAT AAA ACC TAC AAG AGA GTC GCA TAT GAA TAT CCT CCT TA). The PCR product had cat in the middle of two 40-bp DNA sequences (H1 and H2) that flank trg in the Salmonella genome (Figure 1B). This product was transformed into Salmonella by electroporation, which were plated on LB agar with 25µg/ml chloramphenicol and incubated at 37 °C to screen for isolates taking up cat in the trg locus. The trg gene deletion was confirmed by DNA sequencing (Genewiz, South Plainfield, NJ).
Salmonella motility and growth in culture
Aqueous motility was quantified by introducing a GFP-expressing plasmid into trg− and control Salmonella. Swimming velocities were measured in LB after overnight growth and resuspension. A 20 µl droplet of diluted culture was placed on a glass slide and fluorescence images were acquired every second for one minute. Fluorescent images were acquired on an inverted epifluorescent microscope (Olympus, Center Valley, PA), equipped with a Plan-APO 10X objective, a SLCPlan 40× objective, and a Peltier-cooled monochrome CCD camera (Hamamatsu, Bridgewater, NJ). All bacteria in a microscope image were tracked, X and Y coordinates were noted at each time, and average velocities were calculated. Growth rates were determined by inoculating LB cultures of trg− and control Salmonella in LB and measuring optical density at 600 nm every 30 minutes.
Salmonella penetration into tumor tissue in vitro
Microfluidic tumor-on-a-chip devices were created as described previously [21, 29]. LS174T human colorectal adenocarcinoma cells (American Tissue Type Collection ATCC, Manassas, VA) were grown at 37°C and 5% CO2 in low glucose (1 g/l) DMEM supplemented with 10% fetal bovine serum (FBS; Sigma-Aldrich). Multicellular tumor spheroids were formed by seeding 2.5×104 cells/ml on poly(2-hydroxyethyl methacrylate; Sigma-Aldrich) coated flasks for 12 days. Devices molds were fabricated by contact soft lithography [21] with silicone elastomer (Sylgard 184, Dow Corning, Midland, MI). Spheroids were introduced into 1000×300×150µm chambers and incubated for 24 hours at 37°C. Acidity was maintained by perfusion of DMEM buffered with 25 mM HEPES.
Cultures of trg− and control Salmonella were grown overnight, diluted to 105 CFU/ml in DMEM, and introduced into devices at 3 µl/min. After 1 hour, devices were flushed with bacteria-free medium, and colonization was monitored for 15 hours. Four tissues were tested for each condition. Transmitted light and green fluorescent images of device chambers were acquired at 1-hour intervals. To acquire images of entire chambers, two images (867.15 × 660.68µm each) were obtained at 10× and tiled together (IPLab, BD Bioscience, Rockville, MD). Green fluorescent images were captured using 470/40 nm excitation and 525/50 nm emission filters (Chroma, Rockingham, VT). Spatiotemporal bacterial density profiles were generated from green fluorescence images. Linear density averages for successive widths along each chamber were determined from background-subtracted images. Distances were normalized by the length of the tissue in each chamber. This procedure was automated using a customized script in MATLAB (The MathWorks Inc, Natick, MA) [21]. Penetration of trg− and control Salmonella was compared by averaging relative densities at three fractional depths: 0–0.2, 0.2–0.35, and 0.35–1, relative to the total depth of each tissue. The zero and one positions are at the front and back of the tissue, respectively.
Salmonella administration to tumor-bearing mice
Tumors were formed in mice by implanting 4T1 murine mammary carcinoma cells (American Tissue Type Collection, Manassas, VA). Prior to implantation, cells were grown in RPMI-1640 with 10% fetal bovine serum at 37°C and 5% CO2, trypsinized, and suspended in phosphate buffered saline (PBS) at 5×106 cells/ml. Ten microliters of the suspended cell solution, containing 50,000 cells, was injected subcutaneously into the right flank of 8–12-week-old, BALB/c mice. Caliper measurements were taken regularly to monitor tumor growth. Implanted tumors were grown for 3–4 weeks. When tumors reached 1000–1500 mm3, mice were injected with 2×106 CFU mid-log-phase trg− and control Salmonella. Bacteria were suspended in 100 µl PBS, and injected via the tail-vein into mice with size-matched tumors. Twelve hours after bacterial injection, mice were euthanized in a CO2 chamber and tumors were collected. Tumors were frozen in cryosectioning specimen matrix (Tissue-Tek OCT compound, VWR, Radnor, PA) on dry ice and kept in a −80°C freezer. All animal experiments were conducted in accordance with the National Institute of Health (NIH) guidelines for care and use of laboratory animals. The protocols were approved by Baystate Medical Center, Institutional Animal Care and Use Committee (IACUC).
Immunofluorescence labeling of Salmonella and endothelial cells
Tumor sections were stained to identify Salmonella colonies and tumor blood vessels. Five-micrometer, equatorial sections were cut from frozen tissue blocks. Sections were blocked with 1% bovine serum albumin (BSA) for 30 minutes, and incubated in a 1:200 dilution of polyclonal rabbit anti-Salmonella antibody (Abcam, Cambridge, MA) at 4°C overnight. The next day, sections were administered 1:250 polyclonal goat, anti-rabbit Alexa 546 (Invitrogen, Carlsbad, CA) at room temperature for 1 hour. Sections were sequentially incubated in 1:50 rat anti-mouse CD31 (BD Pharmingen, Franklin Lakes, NJ) and 1:200 donkey anti-rat Alexa 488 (Invitrogen) at room temperature for 1 hour each. Slides were counter-stained with 0.1 µg/ml DAPI (Thermo Scientific, Waltham, MA) and mounted with coverslips using mounting medium (Vectashield Hard-Set, Vector Labs, Burlingame, CA). All slides were stored at 4 °C and protected from light. Negative staining controls were performed on both bacteria-free and colonized tissues without application of primary antibodies. These controls confirmed the specificity of the antibody staining methods to Salmonella colonies.
Image acquisition and analysis of colony localization
High-resolution, immunofluorescence images of entire sections were obtained on an inverted epifluorescent microscope (Olympus), equipped with a 10× objective and an encoded, motorized stage (Ludl Electronic Products, Hawthorne, NY). To acquire composite, whole-section images, hundreds of individual frames, 867.15 × 660.68 µm in size, were acquired, scaled by a factor of 0.25, and tiled together. A 1×1 cm tissue section, for example, required 192 (12 by 16) individual images. This imaging process was controlled by automated scripts in the microscope controlling software (IPLab, BD Biosciences, Rockville, MD). Fluorescence images of Salmonella, labeled with the Alexa 546 red fluorescence dye, were acquired using a green-light excitation filter set, D546/10x-565DCXT-E590LPv2 (Chroma, Rockingham,VT). Images of blood vessels (CD31), stained with the Alexa 488 green fluorescence dye, were acquired using a blue-light excitation filter set, D470/40x-495DCXT-E515Lpv2 (Chroma). Images of the DAPI counter-stain were acquired with a UV excitation filter set, AT350/50x-400DCLP-E420LPv2 (Chroma).
The intratumoral distribution of Salmonella was quantified using built-in functions in ImageJ (National Institutes of Health, Bethesda, MD). Red and green fluorescence images of whole tissue sections were thresholded into binary images showing the locations of Salmonella and blood vessels. The sizes of Salmonella colonies were determined by measuring the number of pixels within the colony boundary. Each identified colony pixel was assumed to contain a single bacterium. Based on minced tissue plating [30], number of pixels in histological sections is equivalent to the number of bacteria in tumors. To estimate colony-to-vessel distances, Euclidean distance maps were generated around blood vessels. The value of each pixel in a Euclidean distance map indicates its distance from the nearest blood vessel. Colony-to-vessel distances were determined at the geometric centers of colonies. Tumor microenvironments were divided into three categories based on previous measurements of single-cell physiology in tumor cylindroids [31] and oxygen levels surrounding blood vessels in tumors [32]. Tumor tissue 0–60 µm from blood vessels was defined as proximal; tissue 60–300 µm from vessels was defined as quiescent; and tissue greater than 300 µm from vessels was defined as distal. Blood vessel density of tumor tissue was calculated by counting the number of blood vessels enclosed within a 50 µm radius circle.
Statistical analysis
Data are reported as means with 95% confidence intervals. Pairwise comparisons were performed using two-tailed Student's t-tests with minimal significance of 0.05.
Results
Knockout creation and localization in tissue in vitro
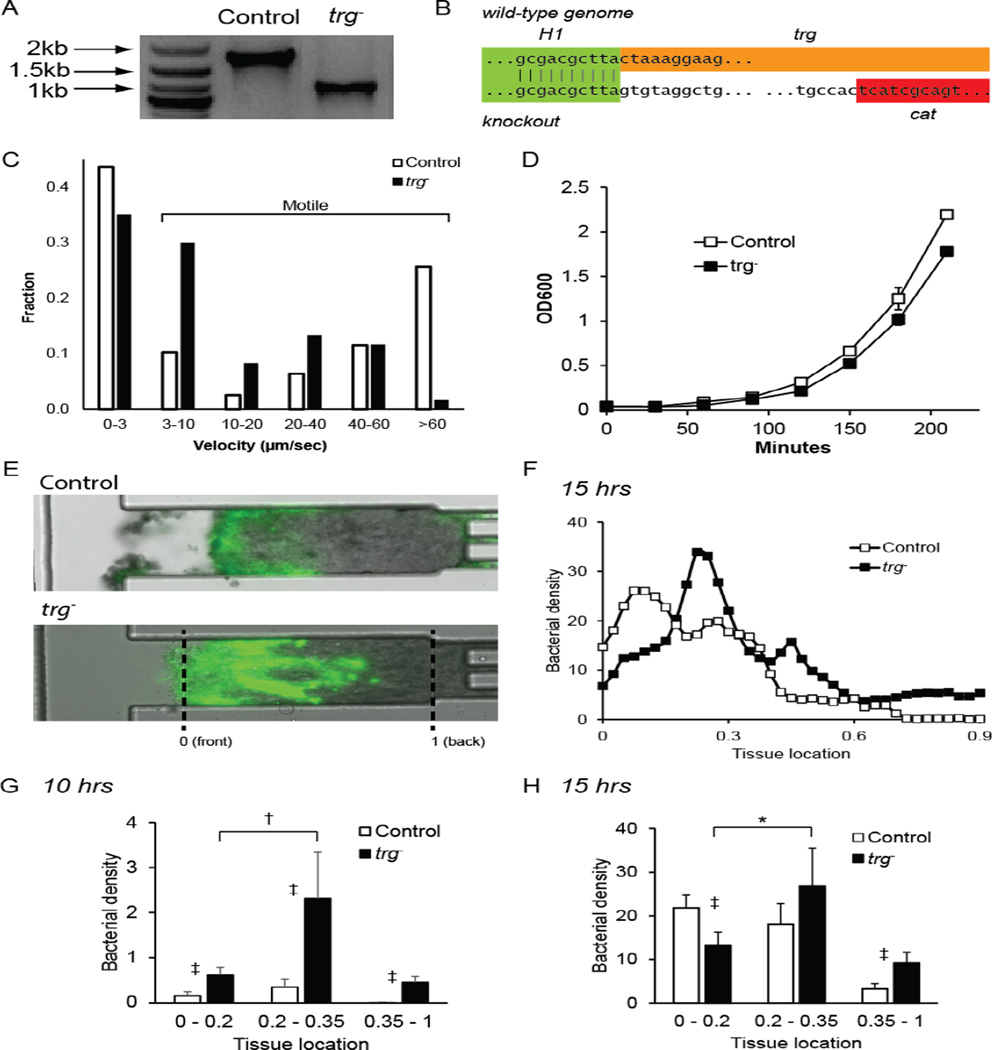
A trg− Salmonella mutant was produced with lambda-red homologous recombination (Figure 1B). Deletion of trg was confirmed by colony PCR (Figure 2A) and DNA sequencing of the chromosome (Figure 2B). The inserted genetic material, including the cat gene, is smaller than the trg by 618 bp. In culture, knocking out trg did not affect the percentage of motile bacteria (Figure 2C) or affect the growth rate (Figure 2D). Motile bacteria had velocities greater than 3 µm/sec. The doubling times for control and trg− bacteria, 31.3 ± 0.39 and 29.9 ± 1.7 minutes (Figure 2D), were not statistically different. The average velocities of control and trg− bacteria were 28.3 ± 3.6 and 13.9 ± 2.3 µm·sec−1, respectively (Figure 2C). This difference was caused by the presence of more rapidly swimming bacteria (>60 µm·sec−1) in the control population. A greater fraction of swimming trg− bacteria were in the 3–10 µm·sec−1 velocity range.
Figure 2. Design and function of trg− Salmonella.
A) Deletion of trg was confirmed by colony PCR. The length of trg between the detection primers (see Figure 1B) was 1733 bp and the insert, including cat was 1115bp. B) Comparison of genomic sequences of the trg knockout and wild-type Salmonella. The sequences match in the H1 homology region, the knockout does not contain trg, and it contains cat. C) Velocity distribution of control (n=78) and trg− Salmonella (n=60) in liquid culture. D) Growth curves of control (n=3) and trg− Salmonella (n=3). There is no difference in growth rate. E) Knockout Salmonella penetrated deeper into tissue in a tumor-on-chip-device than control bacteria. In quantitative analysis, the zero location indicates the front of the tissue and one indicates the back. F) Average bacterial density 15 hours after bacterial administration as a function of position within each tissue (n=4 chambers). Densities were scaled to the maximum value detected at 15 hours. This maximum density was set to 100. G, H) Comparison of bacterial densities at the front (0–0.2), middle (0.2–0.35) and back (0.35–1) of tumor tissue. At 10 hours (G), more trg− Salmonella were located in all three regions (‡, P<0.001) and the density in the middle was greater than the front (†, P<0.005). At 15 hours (H), more trg− were present in the back than control bacteria (‡, P<0.001), more control bacteria were located at the front (‡, P<0.001), and the density of trg− was greater in the middle than in the front (*, P<0.01).
The inability to sense glucose enabled trg− to penetrate further than control Salmonella into tumor tissue grown in a microfluidic device (Figure 2E–H). Bacterial penetration was quantified by converting fluorescence images (Figure 2E) into linear bacterial density profiles (Figure 2F), and grouping densities into three ranges: proximal (0–0.2), middle (0.2 –0.35) and distal (0.35–1.0; Figure 2G–H). The difference in swimming velocity in aqueous culture had little effect on penetration. Ten hours after administration, the accumulation of trg− was greater than control Salmonella at all locations (P<0.001). The knockout predominantly accumulated in the middle region (P<0.005; Figure 2G). By 15 hours, the overall accumulation of control bacteria was comparable to trg−, but they were in different locations (Figure 2H). Control bacteria were distributed equally between the proximal and middle regions, but trg− was predominantly in the middle compared to the proximal region (P<0.01; Figure 2H).
Salmonella distribution in tumors
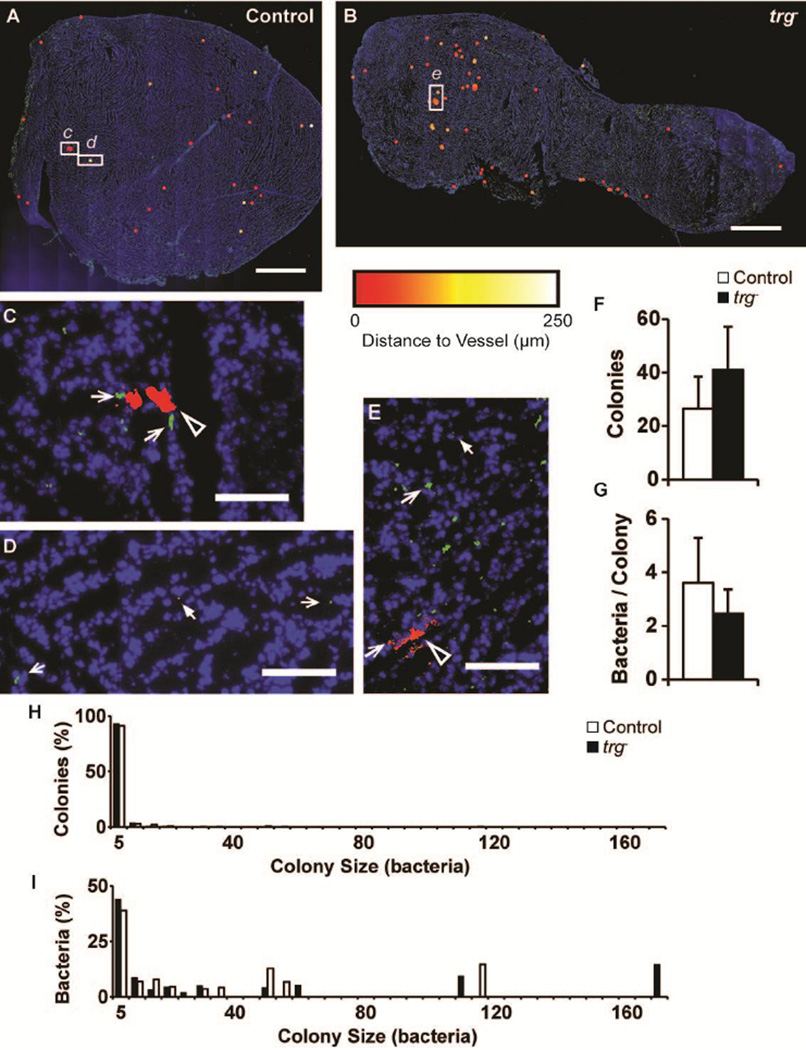
Salmonella were administered to tumor-bearing mice to determine intratumoral colonization patterns (Figure 3). Twelve hours after injection, the colony distributions of trg− (n=10) and control (n=9) Salmonella were similar. Tiled images of entire tumor sections (Figure 3A–B) were assembled from multiple, high-resolution images (Figure 3C–E) to quantify the distances between individual colonies and blood vessels. Colony color (red to white) indicates distance to nearest blood vessel (see detailed description below). In tumors, Salmonella formed both small and large colonies (Figure 3C–E). Small colonies (filled white arrows) were located near and far from blood vessels (open arrows; Figure 3D, E). Large colonies (black arrows) were only located near blood vessels (Figure 3C, E).
Figure 3. Distribution of trg− Salmonella in tumors.
A,B) Pseudocolor tumor cryosections colonized by (A) control and (B) trg− Salmonella. Salmonella colonies are colored from red to white according to their distances from blood vessels (0 to 250 µm). Blood vessels (green) were identified with anti-CD31, and tissue (blue) was counterstained with DAPI. Colonies were dilated by 50 µm to improve visibility. Scale bars are 1.5mm. C–E) High-resolution images of regions highlighted by white rectangles in A and B. Images show (C) a large control colony (black arrow) directly bordering two blood vessels (open arrows); (D) a small control colony (solid arrow) far from blood vessels; and (E) small and large trg− colonies (solid and black arrows) and the nearest vessels (open arrow). Scale bars are 100µm. F) Average number of colonies per 50mm2 for control (n=9) and trg− (n=10) Salmonella. G) Average colony size. Error bars indicate 95% confidence intervals. H) Distribution of control and trg− Salmonella colonies as a function of size (number of bacteria). The majority of colonies contained five or less bacteria. I) Frequency of individual control and trg− bacteria in colonies of different sizes. Many bacteria were located in a few large colonies.
The colony distribution of trg− and control Salmonella was sparse and uneven (Figure 3A–B). In both groups, some regions had more colonies than others, and some regions did not contain any colonies. Two regions with high vessel densities, the left edge of the control section (Figure 3A) and the central area of the trg− section (Figure 3B), both had fewer colonies than other regions. The average colonization in all mice was less than 60 colonies per 50 mm2 and there was no statistical difference between control and knockout-treated mice (Figure 3F). Similarly, there was no difference in average colony size (Figure 3D). The average colony density, 34.2 colonies per 50 mm2, was similar to previously measured densities [33].
Salmonella colony size distribution was heterogeneous
Most bacterial colonies were small, but many of the bacteria were located in large colonies (Figure 3H, I). In ten trg− and nine control Salmonella sections, 379 and 228 colonies were identified, respectively. More than 90% of the colonies formed by both strains contained less than five bacteria (Figure 3H). In these tumors, there were few large colonies. Only three colonies containing more than 60 bacteria were detected. These large colonies contained between 111 and 170 bacteria, which was a considerable fraction of the total number of colonized bacteria (Figure 3I). Many of the tumors contained only small (1–5 bacteria) or medium-sized (5–60 bacteria) colonies. Weighted by total bacterial count, small colonies contained 38.8 and 44.0% of control and trg− bacteria, respectively (Figure 3I). Comparatively, medium-sized colonies contained 46.6 and 32.2% of all bacteria, but were only 8.3 and 6.6% of total number of colonies.
Knockout trg− Salmonella preferentially localize to tumor quiescence
The number of trg− colonies in the quiescent region was greater compared to control Salmonella (Figure 4). A Euclidean mapping method was used to determine the distance of colonies to the nearest blood vessels (Figure 4A–D). The locations of blood vessels were determined in each tumor section (Figure 4A) and used to generate Euclidian distance maps (Figure 4B). The locations of bacterial colonies (Figure 4C, arrows) were transposed onto the distance maps to determine the distance of each colony from its nearest blood vessel (Figure 4D, arrows). Two-fold more trg− colonies were located in the quiescent region (60–300 µm from the nearest vessel) than control colonies (P<0.05; Figure 4E). In this region, the average trg− colony was larger than control colonies by 60% (P<0.05; Figure 4F). Only trg− colonies were located in the distal tumor region (>300µm; Figure 4E–F). There was no statistical difference in colony number or size in the proximal (0–60µm) region.
Figure 4. Colony locations relative to tumor vasculature.
A–D) Method used to quantify distance to blood vessels in tumor sections. A) Binary images were generated from immunofluorescence images that indicate the location of blood vessels. B) Euclidean distance maps were generated from the binary images, in which pixel intensities indicate the distance to the nearest vessel. C) Binary images of bacterial colonies were generated from immunofluorescence images. D) The union of colony and Euclidian maps indicate the distance of each colony to the nearest vessel. E) More trg− colonies (n=10) were located in quiescent tumor regions (60–300 µm from vessels) than control colonies (*, P<0.05; n=9). Regions were defined as proximal (0–60 µm), quiescent (60–300 µm), and distal (>300 µm). The number of control and trg− colonies were greater in proximal than quiescent regions (*, P<0.05), and quiescent than distal regions (‡, P<0.005). F) The average size of trg− colonies in quiescent regions was larger than control colonies (*, P<0.05). The size of control and trg− colonies decreased with distance from vessels (*, P<0.05). Error bars indicate 95% confidence intervals.
Colony size and density both decreased with distance from vessels (Figure 4E–F). For both control and trg− Salmonella, more colonies were located in proximal than quiescent regions (P<0.05), and more colonies were located in quiescent than distal regions (P<0.005; Figure 4E). There were 3.0 and 4.7 times more colonies in proximal than quiescent regions for trg− and control Salmonella, respectively. Similarly, colonies were larger in proximal compared to quiescent regions (P<0.05) for both strains (Figure 4F). All colonies farther than 300 µm from vessels contained single bacteria. The ratio of colony size in proximal to quiescent regions was 1.8:1 for trg− and 4.0:1 for control Salmonella.
Large colonies only formed close to blood vessels
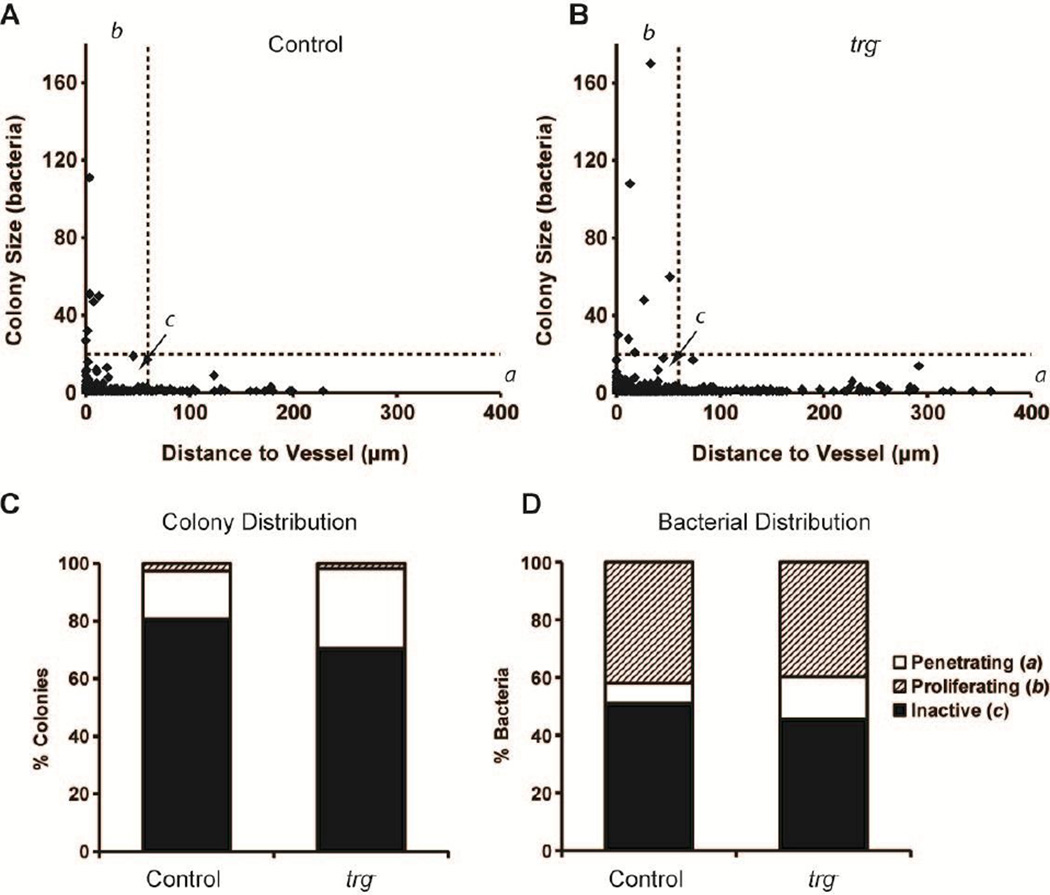
Colony size was highly dependent on location relative to blood vessels (Figure 5). Small colonies were present throughout the entire distance range, but large colonies were only located near blood vessels (Figure 5A–B). Comparison of the size and position of every bacterial colony identified three distinct behavioral groups (Figure 5A–B). Penetrating bacteria (group a) were located more than 60 µm from blood vessels; proliferating bacteria (group b) were in colonies containing more than 20 bacteria; and inactive bacteria (group c) were located less than 60 µm from vessels and in colonies with less than 20 bacteria. More than 75% of colonies (Figure 5C) and 48% of bacteria (Figure 5D) were in this inactive group.
Figure 5. Colony size was dependent on distance to vessels.
A, B) Relationship between colony size and distance for (A) control and (B) trg− Salmonella. Colonies were segregated into three groups: (a) colonies greater than 60 µm from the nearest vessel (vertical line); (b) colonies containing more than 20 bacteria (horizontal line); and (c) colonies containing less than 20 bacteria and closer than 60 µm to the nearest vessel. No colonies with more than 20 bacteria were farther than 60 µm from vessels. C) Percentage of colonies in the three categories: (a) penetrating (>60 µm), (b) proliferating (>20 bacteria), and (c) inactive (<60 µm and <20 bacteria). D) Percentage of bacteria in the three categories.
All colonies in group a (penetrating) contained less than 20 bacteria (Figure 5A–B). The greatest colony-to-blood-vessel distances for control and trg− Salmonella were 228.8 and 361.2µm, respectively. Both of these farthest-migrating colonies consisted of a single bacterium. Knockout Salmonella had more tissue-penetrating colonies (27.7 to 16.7%, Figure 5C) and more penetrating bacteria (14.6 to 7.1%; Figure 5D). Colonies containing more than 20 bacteria (group b) were all located less than 60 µm from blood vessels (dotted line in Figure 5A–B). This group contained the six largest control colonies and the seven largest trg− colonies. These colonies were 2–3% of the total number of colonies (Figure 5C), but more than 40% of the total number of bacteria (Figure 5D). The largest trg− colony contained 170 bacteria and was 32.9 µm from a vessel. The largest control colony contained 111 bacteria and was directly adjacent to two capillaries (Figures 5A and 3C).
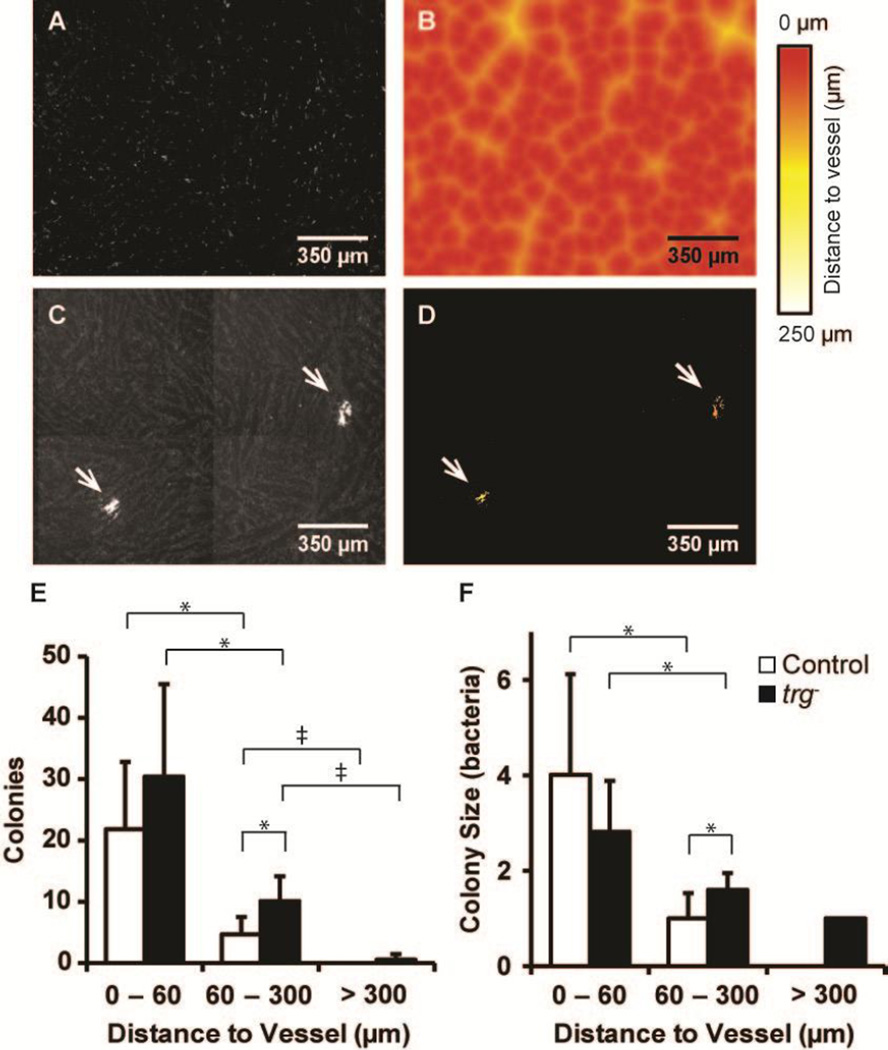
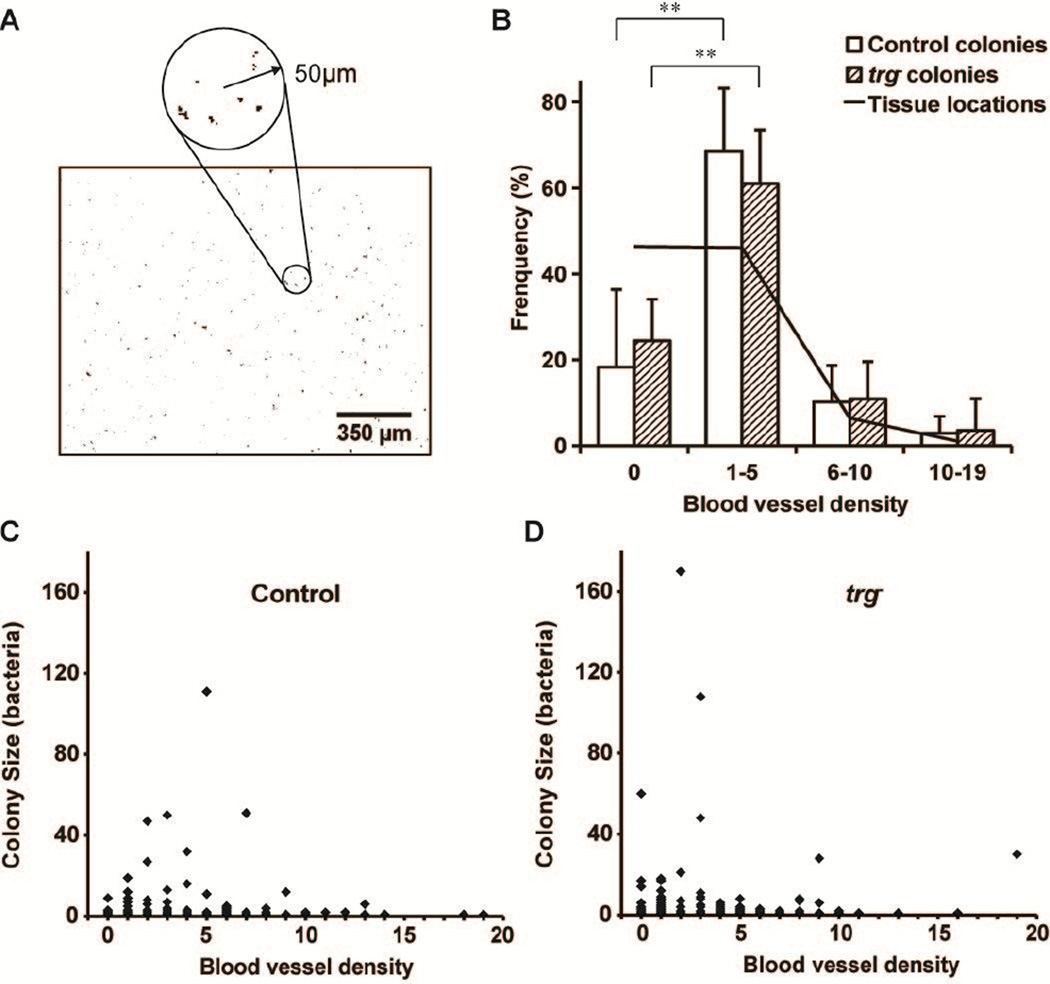
Salmonella colonized regions with low vascular density
Bacterial colonization was greatest in tumor regions with sparse vessel density and low in regions without vessels (Figure 6). Local blood vessel density was determined by measuring the number of blood vessels within 50 µm of every pixel location in a tumor section (Figure 6A). Most tumor regions (solid line in Figure 6B) did not contain vessels (density = zero) or were sparsely vascularized (one to five vessels within 50 µm). Only 8% of the tissue had moderate (6–10 vessels) or high (10–19 vessels) densities.
Figure 6. Salmonella predominantly colonized tissue with low vessel densities.
A) Local vessel densities were determined by counting the number of blood vessels in 100 µm diameter circles around every pixel in tiled binary images. B) The tissue location frequency (solid line) is the percentage of tissue pixels in the four ranges of local vessel density (0, 1–5, 6–10, and 10–19 vessels in the surrounding 50 µm circle). Most tissue was avascular (density = 0) or sparsely vascularized (1–5 vessels). Both control and trg− Salmonella preferentially colonized sparsely vascular regions. The percentage of colonies in sparse regions was greater than avascular regions (**, P<5×10−4). C, D) Colony size was moderately dependent on vessel density, for both (C) control and (D) trg− Salmonella. All large colonies were found in sparse and avascular regions.
The distribution of vessel densities for all tumor pixels (solid line in Figure 6B) served as a reference for vessel densities measured around bacterial colonies (bars in Figure 6B). If bacteria were evenly distributed throughout tissue, the percentage of bacterial colonies with a given local vessel density (bars) would have been equal to frequency of all tissue locations (solid line). The percentage of colonies located in regions with 1–5 vessels/area was greater than the tissue frequency for both strains (P<0.05), indicating a preference for sparsely vascularize tissue. Comparatively, the percentage of colonies located in regions without vessels (zero vessel density) was lower than the tissue frequency for both strains (P<0.05). There was no significant difference between the percentage of colonies in moderate and densely vascular regions and percentage of tissue with those densities. There was also no difference between control and trg− Salmonella at any density (Figure 6B).
Most large colonies, containing more than 40 bacteria, were located in regions with low vessel densities (Figure 6C,D). This relationship was unexpected because all large colonies were located near vessels (group b in Figure 5). Seven colonies with more than 40 bacteria were located in sparse regions with 1–7 vessels/area (Figure 6C,D). One colony was located in tissue with no blood vessels. By definition, colonies in regions with zero vessel density were located at least 50 µm from the nearest vessel. Combined, these results indicate that large colonies formed next to vessels that themselves were not near other vessels.
Discussion
Understanding the mechanisms that control bacterial colonization in tumors will enable the creation of more effective bacterial anticancer therapies. To that end, we created a non-pathogenic trg− knockout Salmonella strain and quantified the general behavior of bacterial colonization in tumors. The trg− knockout strain penetrated deeper than control Salmonella into tissue in a microfluidic tumor-on-a-chip device. In mice tumors, trg− Salmonella preferentially colonized quiescent tissue and formed larger colonies than controls. Overall, Salmonella colonization was sparse and unevenly distributed. Twelve hours after injection, colony size was widely distributed, with sizes ranging from single bacteria to colonies with more than 170 individuals. Many accumulated bacteria were located in a few large colonies and three different behavioral patterns were observed: penetrating, proliferating, and inactive. Large colonies, composed of proliferating bacteria, were only located close to blood vessels, and most Salmonella colonies were located in regions with low vascular density (1–5 vessels/7800um2).
Attraction of trg− Salmonella to sugars
The difference in the localization of trg− to control Salmonella was primarily caused by the inability to sense glucose. Without Trg, Salmonella were not attracted to extracellular glucose, which is at high concentrations in tumor tissue near blood vessels [34]. Colony localization of trg− Salmonella is dominated by attraction to other small molecules [20]. In cylindroids, this altered affinity caused accumulation in the annular quiescent region between viable and necrotic tissue [20]. In perfused tissue, trg− Salmonella migrated further into tissue (Figure 2) because they were not attracted to the glucose-rich region near the channel. The same effect was seen in tumors, where trg− Salmonella accumulated at higher densities in quiescent tissue compared to controls (Figure 4). The presence of trg− Salmonella in the distal regions of perfused tissue (Figure 2) and mouse tumors (Figure 4) suggests that glucose controls chemotaxis more than ribose in these environments.
Salmonella phenotypes in tumors
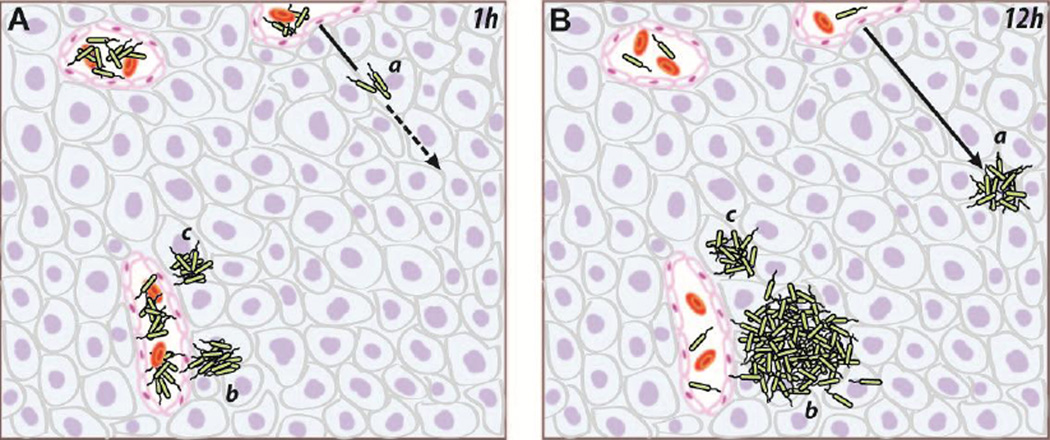
The observation of distinct accumulation patterns suggests that Salmonella have multiple behaviors when in tumors (Figure 7). Three types of Salmonella colonies were observed (Figure 5): small colonies far from vessels (group a), large colonies near vessels (group b), and small colonies near vessels (group c). Directly after injection, all bacteria entered tumors from blood vessels (Figure 7A). At that time, all colonies were small and composed of single bacteria. Over the next 12 hours, the three types of colonies formed (Figure 7B). Some bacteria penetrated into tissue, producing colonies far from vessels (group a). Other bacteria grew rapidly, forming large colonies near vessels (group b). If all large colonies formed from single bacteria, then the bacteria that seeded large colonies were only a small fraction (about 2%) of the injected population (Figure 5C). Then, because of rapid replication, those proliferating individuals produced more than 40% of the colonized bacteria (Figure 5D). The last group was inactive, neither penetrating nor growing (group c). These bacteria formed small colonies near vessels. In all 19 mice, no large colonies were found far from vessels (Figure 5). The absence of this group suggests that tumor-colonizing Salmonella either penetrate or proliferate, but not both.
Figure 7. Three behaviors of Salmonella in tumors.
The dependence of colony size on distance to vessels suggests the existence of three distinct intratumoral phenotypes. A) When bacteria were injected systemically, they entered tumors from the vasculature as individuals. B) Over the next 12 hours, three distinct phenotypes emerged. Highly motile bacteria (a) penetrated away from vasculature, but did not proliferate. Proliferating bacteria (b) replicated and formed large colonies, but did not migrate away from vasculature. Inactive bacteria (c) neither penetrated nor proliferated. The direction of migrating bacteria is indicated by a dashed line, and the solid line indicates the migration path.
There are multiple mechanisms that could explain these distinct behaviors. Most bacterial populations contain a range of phenotypes that are controlled by fluctuations in epigenetic regulators [35, 36]. The proliferation rate of individual Salmonella can vary considerably within a population, and this variation is more pronounced in small populations [37]. Changes in behavior are known to be caused by DNA methylation, when Salmonella interact with mammalian tissue [38]. In culture, Salmonella populations contain a range of motility phenotypes from non-motile to highly motile (Figure 2C). Multiple epigenetic systems regulate bacterial motility [39]. For example, the methyltransferase CheR controls adaption to varying levels of chemotactic attractors [40]. Molecular noise in these epigenetic pathways leads to behavioral changes for individual bacteria [36]. Positive feedback mechanisms stabilize these changes, creating bifurcations in gene expression and leading to phenotype switching and population bistabilities [41]. In this way, molecular noise and external influences give rise to distinct bacterial sub-populations that persist across generations [42].
Manipulating the bistable switches that control the splitting into different phenotypes could be harnessed to improve bacterial therapies. In nature, the existence of multiple phenotypes improves survival [41]. Inactive bacteria are more resistant to environmental changes, such as exposure to antibiotics. These bacteria, however, do not penetrate or proliferate (Figure 5), two functions necessary for therapy. Eliminating or reducing this population would increase therapeutic efficacy. Increasing the number of penetrating bacteria [43] would improve tissue dispersion [21]. Increasing the number of proliferating bacteria would have an exponential effect on the selectivity for tumor tissue over normal tissue. Rapid growth is the primary reason Salmonella preferentially colonize tumors over healthy organs [10]. The absence of large colonies far from vessels (Figure 5) suggests that the motile and proliferating subpopulations are mutually exclusive. Greater understanding of the compatibility of the switching between these phenotypes would enable enhanced control of Salmonella growth and distribution within tumors.
Colonization was dependent on local vessels density
The preferential accumulation of Salmonella in regions with low vessel densities identifies some mechanisms of tumor colonization. Few colonies were present in highly vascularized regions (Figure 6B). This preference is visible in the macroscopic tumor sections (Figure 3). The left edge of the control section (Figure 3A) and the central region of the trg− section (Figure 3B) both have more vessels and fewer colonies than other regions of the same sections. The absence of colonies in highly vascularized regions was unexpected because the bacteria were injected systemically via the tail vein. If colonies formed in proportion to vessel surface area, more colonies would be present in more vascularized regions. In comparison, an intravenously administered chemotherapeutic would surround all active blood vessels [44] and would have a higher concentration per volume in regions of high vessel density.
Two complementary mechanisms could explain the increased colonization of Salmonella in poorly vascularized regions. It has been previously shown that Salmonella preferentially grow in necrotic tissue [10, 33], where there is limited clearance by the immune system [45]. It has also been shown that after colonization, Salmonella destroy tumor blood vessels [46], which would cause the local vessel density to drop. The preferential localization of large colonies in tissue with few blood vessels (Figure 6C,D), suggests that bacteria were more able to replicate when supplied with nutrients from dying tissue and protected from clearance by the immune system.
The number of colonies in regions without vessels was less than the tissue-average density (Figure 6B) because bacteria were initially introduced into tumor tissue through the vasculature. Despite being lower than the tissue-average density, the number of colonies in avascular regions was significant and non-zero (18–24% of the total population; Figure 6B). All bacteria in these regions arrived by chemotaxis within the 12-hour period between injection and observation (Figure 7). In contrast, the concentration of a chemotherapeutic in these regions would be vanishingly small without vascular delivery. This difference illustrates one of the major advantages of bacterial therapy: motile bacteria target tumor tissue with low blood supply, and can overcome the transport limitations created by chaotic and dysfunctional vasculature.
The preference for low vascular regions created colonization patterns that were sparse and highly variable between mice (Figure 3). Unlike conventional therapies, micrometer-size Salmonella cannot extravasate from blood vessels through intact endothelium. They must enter tumor tissue through openings in the endothelial lining. These openings are more present in leaky intratumoral vessels [47] and are triggered by bacterial infection [48]. Their distribution might play a vital role in the scale and location of bacterial tumor invasion. Recently, we demonstrated that supplementing Salmonella injection with lipid A increases bacterial accumulation and robustness [33], by inducing intratumoral inflammation [49]. This strategy solves the problem of inadequate Salmonella tumor targeting. It also emphasizes the importance of creating passages for bacterial tumor invasion.
Conclusions
Deleting the Trg chemoreceptor from Salmonella improved its ability to target tumor quiescence, and will enable anticancer bacteria to access therapeutically-resistant regions. Quantitative analysis of Salmonella colonization revealed that individual bacteria exclusively penetrate tissue or replicate. Despite being intravenously administered, bacterial colonization favored tumor regions with low vascular density. The identification of distinct Salmonella phenotypes in tumors adds a new dimension to bacterial anticancer research. Further delineation of the genetic and epigenetic origins of these populations will enable creation of bacteria with improved tissue penetration, quiescence targeting, and tumor-specific replication. Understanding these critical mechanisms is essential to the creation of tightly controlled, tunable bacterial therapies.
Acknowledgments
The authors wish to thank Dr. Barry Wanner for supplying the λ-red gene deletion system, and National Institutes of Health for financial support (Grant No. 1R01CA120825-01A1). We would also like to thank Brooke Bentley at Pioneer Valley Life Sciences Institute, Springfield, MA, for expert technical assistance with all histological aspects of the project, Bhushan Toley for assistance with in vitro tumor tissue experiment, and Nele Van Dessel for measuring bacterial growth rate.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Forbes NS. Engineering the perfect (bacterial) cancer therapy. Nat Rev Cancer. 2010;10:785–794. doi: 10.1038/nrc2934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Leschner S, Weiss S. Salmonella-allies in the fight against cancer. J Mol Med. 2010;88:763–773. doi: 10.1007/s00109-010-0636-z. [DOI] [PubMed] [Google Scholar]
- 3.St Jean AT, Zhang M, Forbes NS. Bacterial therapies: completing the cancer treatment toolbox. Curr Opin Biotechnol. 2008;19:511–517. doi: 10.1016/j.copbio.2008.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kasinskas RW, Forbes NS. Salmonella typhimurium specifically chemotax and proliferate in heterogeneous tumor tissue in vitro. Biotechnol Bioeng. 2006;94:710–721. doi: 10.1002/bit.20883. [DOI] [PubMed] [Google Scholar]
- 5.Ganai S, Arenas RB, Sauer JP, Bentley B, Forbes NS. In tumors Salmonella migrate away from vasculature toward the transition zone and induce apoptosis. Cancer Gene Ther. 2011;18:457–466. doi: 10.1038/cgt.2011.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Swofford CA, St Jean AT, Panteli JT, Brentzel ZJ, Forbes NS. Identification of Staphylococcus aureus alpha-hemolysin as a protein drug that is secreted by anticancer bacteria and rapidly kills cancer cells. Biotechnol Bioeng. 2014;111:1233–1245. doi: 10.1002/bit.25184. [DOI] [PubMed] [Google Scholar]
- 7.St Jean AT, Swofford CA, Panteli JT, Brentzel ZJ, Forbes NS. Bacterial delivery of Staphylococcus aureus alpha-hemolysin causes regression and necrosis in murine tumors. Mol Ther. 2014;22:1266–1274. doi: 10.1038/mt.2014.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ganai S, Arenas RB, Forbes NS. Tumour-targeted delivery of TRAIL using Salmonella typhimurium enhances breast cancer survival in mice. Br J Cancer. 2009;101:1683–1691. doi: 10.1038/sj.bjc.6605403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Low KB, Ittensohn M, Le T, Platt J, Sodi S, Amoss M, Ash O, Carmichael E, Chakraborty a, Fischer J, Lin SL, Luo X, Miller SI, Zheng L, King I, Pawelek JM, Bermudes D. Lipid A mutant Salmonella with suppressed virulence and TNFalpha induction retain tumor-targeting in vivo. Nat Biotechnol. 1999;17:37–41. doi: 10.1038/5205. [DOI] [PubMed] [Google Scholar]
- 10.Forbes NS, Munn LL, Fukumura D, Jain RK. Sparse initial entrapment of systemically injected Salmonella typhimurium leads to heterogeneous accumulation within tumors. Cancer Res. 2003;63:5188–5193. [PubMed] [Google Scholar]
- 11.Bermudes D, Low B, Pawelek J. Tumor-targeted Salmonella - Highly selective delivery vectors. Cancer Gene Ther. 2000;465:57–63. doi: 10.1007/0-306-46817-4_6. [DOI] [PubMed] [Google Scholar]
- 12.Jain RK. The next frontier of molecular medicine: delivery of therapeutics. Nat Med. 1998;4:655–657. doi: 10.1038/nm0698-655. [DOI] [PubMed] [Google Scholar]
- 13.Minchinton AI, Tannock IF. Drug penetration in solid tumours. Nat Rev Cancer. 2006;6:583–592. doi: 10.1038/nrc1893. [DOI] [PubMed] [Google Scholar]
- 14.Tannock IF, Lee CM, Tunggal JK, Cowan DS, Egorin MJ. Limited penetration of anticancer drugs through tumor tissue: a potential cause of resistance of solid tumors to chemotherapy. Clin Cancer Res. 2002;8:878–884. [PubMed] [Google Scholar]
- 15.Jain RK. Determinants of tumor blood flow: a review. Cancer Res. 1988;48:2641–2658. [PubMed] [Google Scholar]
- 16.Trédan O, Galmarini CM, Patel K, Tannock IF. Drug resistance and the solid tumor microenvironment. J Natl Cancer I. 2007;99:1441–1454. doi: 10.1093/jnci/djm135. [DOI] [PubMed] [Google Scholar]
- 17.Toley BJ, Tropeano Lovatt ZG, Harrington JL, Forbes NS. Microfluidic technique to measure intratumoral transport and calculate drug efficacy shows that binding is essential for doxorubicin and release hampers Doxil. Integr Biol. 2013;5:1184–1196. doi: 10.1039/c3ib40021b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gardner SN. A mechanistic, predictive model of dose-response curves for cell cycle phase-specific and -nonspecific drugs. Cancer Res. 2000;60:1417–1425. [PubMed] [Google Scholar]
- 19.Kim JJ, Tannock IF. Repopulation of cancer cells during therapy: an important cause of treatment failure. Nat Rev Cancer. 2005;5:516–525. doi: 10.1038/nrc1650. [DOI] [PubMed] [Google Scholar]
- 20.Kasinskas RW, Forbes NS. Salmonella typhimurium lacking ribose chemoreceptors localize in tumor quiescence and induce apoptosis. Cancer Res. 2007;67:3201–3209. doi: 10.1158/0008-5472.CAN-06-2618. [DOI] [PubMed] [Google Scholar]
- 21.Toley BJ, Forbes NS. Motility is critical for effective distribution and accumulation of bacteria in tumor tissue. Integr Biol. 2012;4:165–176. doi: 10.1039/c2ib00091a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Manson MD, Armitage JP, Hoch JA, Macnab RM. Bacterial locomotion and signal transduction. J Bacteriol. 1998;180:1009–1022. doi: 10.1128/jb.180.5.1009-1022.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Alon U, Surette MG, Barkai N, Leibler S. Robustness in bacterial chemotaxis. Nature. 1999;397:168–171. doi: 10.1038/16483. [DOI] [PubMed] [Google Scholar]
- 24.Toso JF, Gill VJ, Hwu P, Marincola FM, Restifo NP, Schwartzentruber DJ, Sherry RM, Topalian SL, Yang JC, Stock F, Freezer LJ, Morton KE, Seipp C, Haworth L, Mavroukakis S, White D, MacDonald S, Mao J, Sznol M, Rosenberg Sa. Phase I study of the intravenous administration of attenuated Salmonella typhimurium to patients with metastatic melanoma. J Clin Oncol. 2002;20:142–152. doi: 10.1200/JCO.2002.20.1.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Blat Y, Eisenbach M. Tar-dependent and -independent pattern formation by Salmonella typhimurium. J Bacteriol. 1995;177:1683–1691. doi: 10.1128/jb.177.7.1683-1691.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Aswad D, Koshland DE. Isolation, characterization and complementation of Salmonella typhimurium chemotaxis mutants. J Mol Biol. 1975;97:225–235. doi: 10.1016/s0022-2836(75)80036-2. [DOI] [PubMed] [Google Scholar]
- 27.Datsenko Ka, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. P Natl Acad Sci USA. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Karlinsey JE. lambda-Red genetic engineering in Salmonella enterica serovar Typhimurium. Method Enzymol. 2007;421:199–209. doi: 10.1016/S0076-6879(06)21016-4. [DOI] [PubMed] [Google Scholar]
- 29.Walsh CL, Babin BM, Kasinskas RW, Foster JA, McGarry MJ, Forbes NS. A multipurpose microfluidic device designed to mimic microenvironment gradients and develop targeted cancer therapeutics. Lab on a chip. 2009;9:545–554. doi: 10.1039/b810571e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang M, Swofford CA, Forbes NS. Lipid A controls the robustness of intratumoral accumulation of attenuated Salmonella in mice. Int J Cancer. 2014;135:647–657. doi: 10.1002/ijc.28700. [DOI] [PubMed] [Google Scholar]
- 31.Kim BJ, Forbes NS. Single-cell analysis demonstrates how nutrient deprivation creates apoptotic and quiescent cell populations in tumor cylindroids. Biotechnol Bioeng. 2008;101:797–810. doi: 10.1002/bit.21985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kyle AH, Baker JHE, Minchinton AI. Targeting Quiescent Tumor Cells via Oxygen and IGF-I Supplementation. Cancer Res. 2012;72:801–809. doi: 10.1158/0008-5472.CAN-11-3059. [DOI] [PubMed] [Google Scholar]
- 33.Zhang M, Swofford CA, Forbes NS. Lipid A controls the robustness of intratumoral accumulation of attenuated Salmonella in mice, International journal of cancer. Journal international du cancer. 2013 doi: 10.1002/ijc.28700. [DOI] [PubMed] [Google Scholar]
- 34.Kasinskas RW, Venkatasubramanian R, Forbes NS. Rapid uptake of glucose and lactate, and not hypoxia, induces apoptosis in three-dimensional tumor tissue culture. Integr Biol. 2014;6:399–410. doi: 10.1039/c4ib00001c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Avery SV. Microbial cell individuality and the underlying sources of heterogeneity. Nat. Rev. Microbiol. 2006;4:577–587. doi: 10.1038/nrmicro1460. [DOI] [PubMed] [Google Scholar]
- 36.Korobkova E, Emonet T, Vilar JMG, Shimizu TS, Cluzel P. From molecular noise to behavioural variability in a single bacterium. Nature. 2004;428:574–578. doi: 10.1038/nature02404. [DOI] [PubMed] [Google Scholar]
- 37.Koutsoumanis KP, Lianou A. Stochasticity in Colonial Growth Dynamics of Individual Bacterial Cells. Appl. Environ. Microbiol. 2013;79:2294–2301. doi: 10.1128/AEM.03629-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Casadesus J, Low D. Epigenetic gene regulation in the bacterial world. Microbiol Mol Biol R. 2006;70:830–856. doi: 10.1128/MMBR.00016-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Avery SV. Microbial cell individuality and the underlying sources of heterogeneity. Nat. Rev. Microbiol. 2006;4:577–587. doi: 10.1038/nrmicro1460. [DOI] [PubMed] [Google Scholar]
- 40.Sourjik V, Berg HC. Receptor sensitivity in bacterial chemotaxis. P Natl Acad Sci USA. 2002;99:123–127. doi: 10.1073/pnas.011589998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Veening JW, Smits WK, Kuipers OP. Annu Rev Microbiol, Annual Reviews. Palo Alto: 2008. Bistability, Epigenetics, and Bet-Hedging in Bacteria; pp. 193–210. [DOI] [PubMed] [Google Scholar]
- 42.Snijder B, Pelkmans L. Origins of regulated cell-to-cell variability. Nat Rev Mol Cell Bio. 2011;12:119–125. doi: 10.1038/nrm3044. [DOI] [PubMed] [Google Scholar]
- 43.Derr P, Boder E, Goulian M. Changing the specificity of a bacterial chemoreceptor. J Mol Biol. 2006;355:923–932. doi: 10.1016/j.jmb.2005.11.025. [DOI] [PubMed] [Google Scholar]
- 44.Lankelma J, Dekker H, Luque FR, Luykx S, Hoekman K, van der Valk P, van Diest PJ, Pinedo HM. Doxorubicin gradients in human breast cancer. Clin Cancer Res. 1999;5:1703–1707. [PubMed] [Google Scholar]
- 45.Westphal K, Leschner S, Jablonska J, Loessner H, Weiss S. Containment of tumor-colonizing bacteria by host neutrophils. Cancer Res. 2008;68:2952–2960. doi: 10.1158/0008-5472.CAN-07-2984. [DOI] [PubMed] [Google Scholar]
- 46.Liu F, Zhang L, Hoffman RM, Zhao M. Vessel destruction by tumor-targeting Salmonella typhimurium A1-R is enhanced by high tumor vascularity. Cell Cycle. 2010;9:4518–4524. doi: 10.4161/cc.9.22.13744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.di Tomaso E, Capen D, Haskell A, Hart J, Logie JJ, Jain RK, McDonald DM, Jones R, Munn LL. Mosaic tumor vessels: cellular basis and ultrastructure of focal regions lacking endothelial cell markers. Cancer Res. 2005;65:5740–5749. doi: 10.1158/0008-5472.CAN-04-4552. [DOI] [PubMed] [Google Scholar]
- 48.Lemichez E, Lecuit M, Nassif X, Bourdoulous S. Breaking the wall: targeting of the endothelium by pathogenic bacteria. Nat. Rev. Microbiol. 2010;8:93–104. doi: 10.1038/nrmicro2269. [DOI] [PubMed] [Google Scholar]
- 49.Leschner S, Westphal K, Dietrich N, Viegas N, Jablonska J, Lyszkiewicz M, Lienenklaus S, Falk W, Gekara N, Loessner H, Weiss S. Tumor invasion of Salmonella enterica serovar Typhimurium is accompanied by strong hemorrhage promoted by TNF-alpha. PloS one. 2009;4:e6692. doi: 10.1371/journal.pone.0006692. [DOI] [PMC free article] [PubMed] [Google Scholar]